Abstract
Immunoglobulin Class switch recombination (CSR) is the gene rearrangement process by which B lymphocytes change the immunoglobulin (Ig) heavy chain constant region to permit a switch of Ig isotype from IgM to IgG, IgA, or IgE. At the DNA level, CSR occurs via generation and joining of DNA double strand breaks (DSBs) at intronic switch regions located just upstream of each of the heavy chain constant regions. Activation-induced deaminase (AID), a B cell specific enzyme, catalyzes cytosine deaminations (converting cytosines to uracils) as the initial DNA lesions that eventually lead to DSBs and CSR. Progress on AID structure integrates very well with knowledge about Ig class switch region nucleic acid structures that are supported by functional studies. It is an ideal time to review what is known about the mechanism of Ig class switch recombination and its relation to somatic hypermutation. There have been many comprehensive reviews on various aspects of the CSR reaction and regulation of AID expression and activity (Casellas et al., 2016; Chaudhuri and Alt, 2004; Chaudhuri et al., 2007; Hackney et al., 2009; Hwang et al., 2015; Matthews et al., 2014; Methot and Di Noia, 2017; Pan-Hammarstrom et al., 2007; Stavnezer et al., 2008; Yu et al., 2003). This review is focused on the relation between AID and switch region nucleic acid structures, with a particular emphasis on R-loops.
Keywords: R-loop, DNA repair, DNA recombination, immunoglobulin gene rearrangement, isotype switching, activation-induced deaminase, RNase H
1. THE KEY ELEMENTS OF IMMUNOGLOBULIN CLASS SWITCH RECOMBINATION
1.1. Overview and Biomedical Significance
Immunoglobulin (Ig) class (or isotype) is defined by the heavy (H) chain constant (C) region. Mammalian Ig loci (IGH) contain a tandem of C regions (in mouse, μ, δ, γ3, γ1, γ2b, γ2a, ε, α; and in human μ, γ3, γ1, α1, γ2, γ4, ε, α2) located downstream of the rearranged variable (V) region (variable domain exon, also known as a VDJ segment). In a humoral immune response, production of the optimal Ig class is critical for the clearance of invading pathogens as the IGH C region determines each Ig’s effector function. Switching of Ig class from IgM (μ) to IgG (γ), IgE (ε) or IgA (α) occurs as an intrachromosomal DNA deletion known as class switch recombination (CSR) (Chaudhuri and Alt, 2004; Goodman et al., 2007; Stavnezer et al., 2008; Yu and Lieber, 2003), which results in the replacement of Cμ with a downstream C region (γ, ε, or α) (Fig. 1). Each C region is preceded by a large (2-10 kb) repetitive region known as the switch (S) region that serves as the recombination target. A single CSR event involves the generation and joining of DSBs in the donor (μ) and acceptor (γ, ε or α) S regions to generate the new arrangement (Fig. 1). The IGH CSR process occurs in mature B cells in the germinal centers of the lymph nodes, spleen and Peyer’s patches of the gastrointestinal (GI) tract, as reviewed elsewhere (Chaudhuri et al., 2007; Stavnezer et al., 2008; Yu and Lieber, 2003).
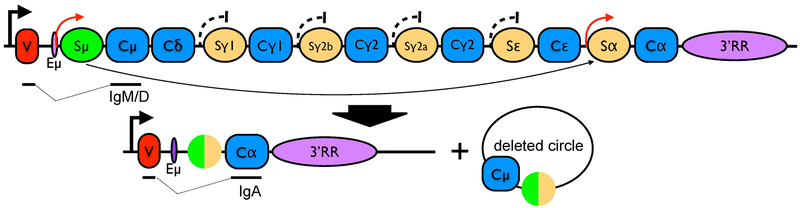
Figure 1. Diagram of Class Switch Recombination.
The exons (the constant regions have been simplified to a single rectangle) are depicted with the nearly square blue symbols. The tan circles represent class switch recombination (CSR) sequences, which are often repetitive with numerous AGCT sites and G-clusters (GGGGT or GGGCT)(Dunnick et al., 1993). A single class switch recombination event utilizes the Sμ region (often called a switch donor sequence) and one of the downstream “acceptor” switch regions, in this case Sα. The deleted region is shown as a circular DNA molecule. The VDJ exon is symbolized as a red symbol labeled with a V. The actual promoter of the gene is upstream of this V. There are sterile transcript promoters upstream of each of the switch regions, and all of these are inactive, except for the ones upstream of the Sμ and Sα switch regions (red arrow) that are, in this case, activated for recombination. A detailed diagram of the organization of the sterile transcript promoter and the Iexons is shown in Suppl. Figure 1.
Elucidating the mechanism of CSR is highly significant for human health. Without class switch recombination, humans and other mammals would only make IgM and IgD (Hackney et al., 2009; Stavnezer and Amemiya, 2004), severely limiting the effectiveness of a humoral immune response against infections. This is seen in patients who are defective for class switch recombination and develop lethal respiratory infections.
CSR is initiated by the activation-induced cytidine deaminase (AID), a B cell specific factor that also initiates somatic hypermutation (SHM) at Ig variable (V) regions, and Ig gene conversion in birds (Harris et al., 2002; Muramatsu et al., 2000; Revy et al., 2000). AID is a single-strand specific DNA enzyme (Bransteitter et al., 2003; Chaudhuri et al., 2003; Dickerson et al., 2003; Larijani and Martin, 2007; Ramiro et al., 2003; Sohail et al., 2003; Yu et al., 2004) that catalyzes DNA cytosine deamination (converting cytosine to uracil) at transcribed regions (DiNoia and Neuberger, 2002; Petersen-Mahrt et al., 2002). AID-generated uracil is recognized either as base damage that engages the base excision repair (BER) pathway or as a U:G mismatch that engages the mismatch repair (MMR) pathway (Chaudhuri and Alt, 2004; Stavnezer et al., 2008; Yu and Lieber, 2003). In most cells, the BER and MMR pathways repair these lesions with high fidelity. In contrast, repair of AID-generated uracils in activated B cells is highly mutagenic, generating mostly point mutations in the V regions during SHM and DNA double strand breaks (DSBs) in the S regions during CSR. The mechanism(s) that lead to error-prone outcomes from BER and MMR, two pathways that normally mediate high-fidelity repair, is not well-defined (Liu et al., 2008). We consider this to be among the major unsolved questions in this field. Because AID is a DNA mutator, elucidating the detailed mechanism of AID’s activity during SHM and CSR will greatly help us understand how AID occasionally targets other parts of the genome, causing genome instability and tumorigenesis (Pasqualucci et al., 2008; Pasqualucci et al., 2004).
1.2. Essential Role of Transcription
It is well established that transcription through V and S regions is absolutely required for SHM and CSR, respectively (Bransteitter et al., 2004; Canugovi et al., 2009; Chaudhuri et al., 2003; Dickerson et al., 2003; Goodman et al., 2007; Jinks-Robertson and Bhagwat, 2014; Pham et al., 2003; Sohail et al., 2003; Yu and Lieber, 2003). This is explained in part by AID’s single-strand (ss) DNA specificity, as shown originally by the Goodman lab (Bransteitter et al., 2003; Pham et al., 2003), and subsequently by others (Bransteitter et al., 2003; Chaudhuri et al., 2003; Dickerson et al., 2003; Larijani and Martin, 2007; Ramiro et al., 2003; Sohail et al., 2003; Yu et al., 2004). Transcription is thought to temporarily separate the two DNA strands to provide AID with its ssDNA substrate. However, the ssDNA specificity itself is insufficient to explain the remarkable preference of AID for its physiological targets (V and S regions) over thousands of other transcribed regions in the B cell genome. The mechanism by which CSR is targeted to V and S regions has been under great scrutiny for more than three decades but remains a major unresolved question (Dunnick et al., 1993; Goodman et al., 2007; Lebecque and Gearhart, 1990).
1.3. Essential Role of Switch Region Sequences
Mammalian S regions are highly repetitive GC-rich regions with the G-richness skewed to the nontemplate strand so as to constitute 35 to 49% of the nucleotides on that strand. Transcription through S regions induces a secondary nucleic acid structure called an R-loop that contains an RNA:DNA hybrid formed by the RNA transcript and the template DNA strand, and the displaced single-stranded non-template strand (Yu et al., 2003)(Fig. 2). The propensity to form long stable R-loops at transcribed S regions is likely due to a highly G-rich non-template strand at all mammalian switch regions. We have proposed that the G-richness evolved to generate R-loops, which provides much greater single-stranded DNA for AID to act, thereby making CSR more efficient in mammalian species relative to avians or amphibians (Yu and Lieber, 2003). Endogenous RNase H action at the R-loops can generate ssDNA on the template strand as well; creating regions we have called collapsed R-loops (Yu et al., 2003; Yu and Lieber, 2003; Zhang et al., 2014a; Zhang et al., 2014b). Other possible structures have been proposed, including G-quartets (see below).
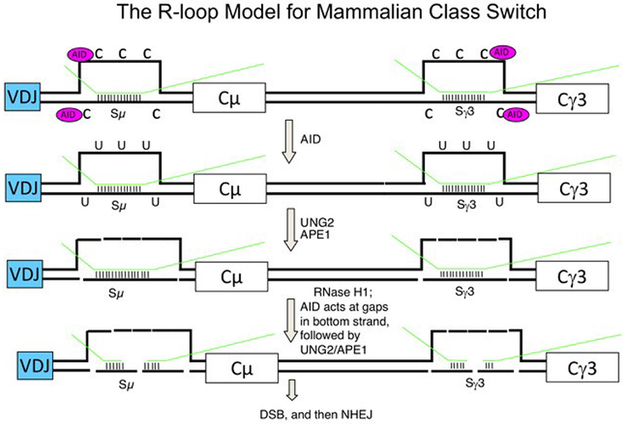
Figure 2. The R-Loop:Deaminase Model for Class Switch Recombination.
A putative CSR event to IgG3 is shown. Switch region RNA transcripts (green lines) were paired with the C-rich DNA template strand to form an R-loop structure (Yu et al., 2003; Yu and Lieber, 2003). The entire displaced G-rich DNA strand and part of the C-rich DNA at the edges of the R-loop are single-stranded, and therefore, serve as targets for AID. AID deaminates C residues located in the single-stranded region to convert them to uracil (U). UDG removes uracils in the DNA and leaves behind an abasic (apyrimidinic) site, which is cleaved by APE. The sum of nicks on both strands results in double-strand DNA breaks in the switch region, which are repaired by the NHEJ pathway to complete CSR. Coding regions (VDJ and constant region exons) are indicated by rectangles. Small dashes in the switch region indicate base pairing between the switch transcript and the C-rich DNA template.
Studies from the Alt laboratory showed that inversion of the transcriptional orientation of the S region, which is less favorable for R-loop formation, significantly reduces CSR in mouse B cells (Shinkura et al., 2003), providing evidence for the functional relevance of R-loop during CSR. R-loop formation may not be entirely dependent on skewed G/C content because transcription generates transient upstream negative superhelical tension which may permit R-loop formation at some level (Drolet et al., 2003; Masse et al., 1997). This could account for the level of single-strandedness that permits some controls to switch at detectable levels (Yeap et al., 2015; Zarrin et al., 2005).
2. CLASS SWITCH REGION FEATURES THAT ARE IMPORTANT FOR RECOMBINATION
2.1. DNA Sequence Features: WGWW Sites, G-Density, G-Clustering, and Number of Switch Repeat
AID prefers WRC sites to efficiently deaminate Cs. WGCW sites are a more narrowly defined subset of the WRC sites that are the predominant motif in mammalian class switch regions (Han et al., 2011). All of these WRC sites must be within ssDNA for AID to recognize them as substrates. Whether the ssDNA region is in the context of a DNA branch site or a more complex DNA structure is a key question.
There is considerable evidence that mammalian switch regions form R-loops (Huang et al., 2007; Huang et al., 2006; Yu et al., 2003; Yu et al., 2004). The RNA transcript generated through switch regions is G-rich. It has been shown that the thermodynamic stability of a G-rich RNA paired with a C-rich DNA is much greater than that of a C-rich RNA paired with a G-rich DNA of the same sequence (Roberts and Crothers, 1992). Because R-loops contain a stable ssDNA region of extensive length, the ssDNA has been widely considered to be the recognition motif for recruiting AID during CSR. Many features of CSR can be explained by the R-loop model, especially the role of transcription. Because R-loops initiate and terminate randomly in the S region, it is also consistent with the observed random locations of CSR junctions, which suggests random locations of DSBs at S regions during CSR.
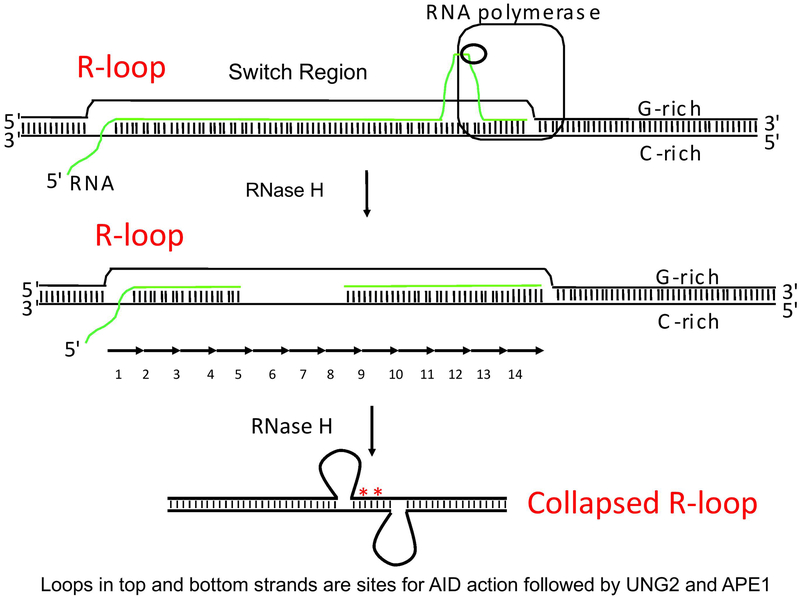
The R-loop model explains how AID accesses the template DNA strand. RNA:DNA hybrids are natural substrates for a class of enzymes called RNase H (H stands for hybrid) (Cerritelli and Crouch, 2009; Maul et al., 2017). At the time we first described long genomic R-loops, we reported that treating genomic R-loops in vitro with E.coli RNase H1 unexpectedly resulted in ss regions (50~150bp) on both DNA strands (Yu et al., 2003). We proposed the possibility that the R-loop could collapse after RNase H action with mis-alignment of the repeats, exposing ssDNA on both DNA strands (Fig. 3). We coined the hypothetical structure formed upon RNase H treatment of R-loop the “collapsed R-loop”. We suggested several advantages for how the collapsed R-loop model better explains the CSR reaction (Yu et al., 2003; Yu and Lieber, 2003).
Figure 3. Collapsed R-Loops Resulting from RNase H Action on R-loops.
AID may be unable to recognize the single-stranded character of the displaced G-rich DNA strand because that strand may be too close to the RNA:DNA helix of the R-loop. In this model, an RNase H activity is proposed to remove the RNA in the R-loop. During the degradation of the switch region RNA, the annealing of the G-rich DNA strand with the C-rich DNA strand may result in the top strand of switch region repeat number 9 annealing with the bottom strand of repeat number 6. That is, the top strand of one repeat may misalign and anneal to the bottom strand of another repeat, resulting in ssDNA loops on both the top and bottom strands in the switch region. The misaligned repeats (6 versus 9) would, therefore, have many mismatches shown by red asterisks (Yu et al., 2003; Yu and Lieber, 2003). The looped-out DNA may be the actual target of AID-mediated cytidine deamination. The rest of this process (removal of uracil by UDG, cleavage by APE and joining by NHEJ) would be similar to the R-loop:Deaminase model shown in Figure 2.
2.1.1. G-Clusters and Switch Region Strength in B Cells
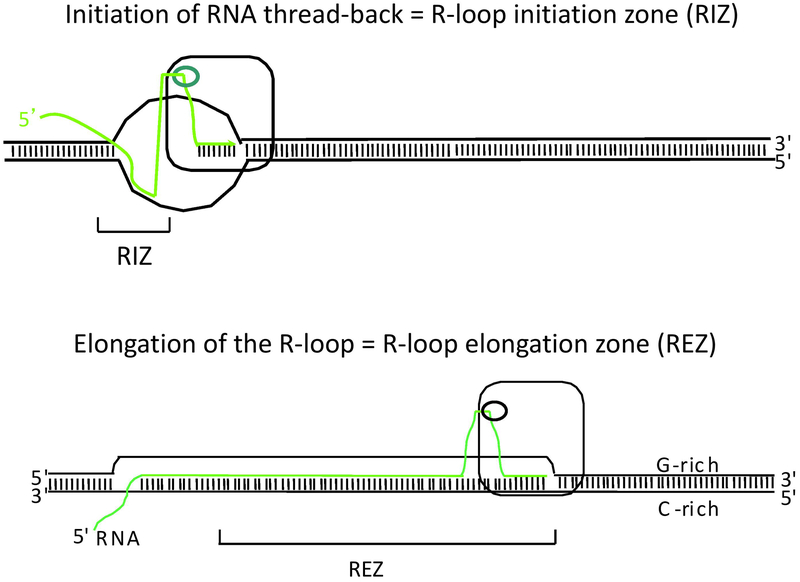
Biochemical methods using purified DNA in vitro showed that G-clusters are extremely important to initiate the thread-back of an RNA onto the DNA template in a co-transcriptional manner. This thread-back initiates the type of long R-loop formation (>30 bp) that is distinct from the smaller R-loops within or around all RNA polymerases (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010)(Figs. 4 and 5). Using the CH12 cellular CSR system, we found that G-clusters are a very important determinant of whether a switch region leads to CSR, predominantly by leading to R-loop formation. The G-clusters can be located at the leading edge of the switch region, thereby leading to R-loop formation that can allow downstream WGCW sites to be within a zone of ssDNA (and therefore be substrates for AID). In alternative arrangement, the G-clusters can be interspersed among the WGCW sites, as they are in all mammalian switch regions. We find that both of these configurations can support CSR (Zhang et al., 2014a; Zhang et al., 2014b). Murine models in which Sm repeats are deleted are consistent with an important role for G-clusters in initiating RNA:DNA hybrid and R-loop formation (Balter et al., 2012).
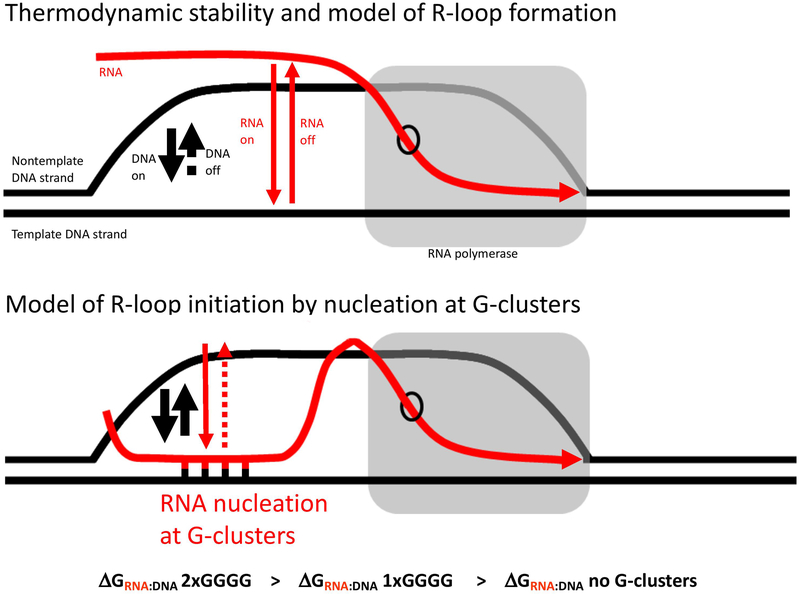
Figure 4. Competition Between the mRNA and the Nontemplate DNA Strand for Binding to the Template DNA Strand.
The G-clusters initiate R-loops in mammalian switch regions. G-clusters are important for efficient R-loop initiation within cells (Zhang et al., 2014b).
Figure 5. Initiation of Thread Back and Elongation of the R-Loop.
When the RNA base pair with the template DNA strand at G-clusters, we refer to this as the R-loop initiation zone (RIZ). Natural mammalian switch regions have many possible locations for such initiation, but for experimental cases, we often create one (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010; Zhang et al., 2014a; Zhang et al., 2014b). After the R-loop has initiated, the R-loop elongates, and we refer to this as the R-loop elongation zone or REZ.
2.1.2. Number of WGCW Sites as a Determinant of Switch Region Strength in B Cells
Variations in switch region sequences were also analyzed using the CH12 cellular system. The WGCW motif was shown to be the key DNA sequence feature required for CSR (Han et al., 2011). Building on this observation, the number and density of WGCW sites were examined, leading to the finding that the number of WGCW sites is more influential than the density of these sites, at least for total switch region lengths of less than 1 kb. Mammalian switch regions have evolved to be long relative to other DNA recombination sequences. The longer switch regions typically provide more WGCW sites and more G-clusters, but the total switch region length itself (independent of WGCW sites and G-clusters) does not appear to be critical for CSR efficiency. The distance between WGCW sites does not seem to be critical, and increased total length of the switch region is important only to the extent that it provides a greater number of WGCW sites. (Han and Yu, 2008; Hwang et al., 2015; Zhang et al., 2014a; Zhang et al., 2014b).
2.1.3. G-Density and Switch Region Strength in B Cells
Based on the much higher ratio of switched isotype (e.g., IgG in mammals or IgX in amphibians) relative to unswitched isotype (IgM) in mammals versus more distantly evolved vertebrates, we have speculated that mammalian Ig class switch recombination may be more efficient than in other species that carry out CSR. If so, some of the greater efficiency may be due to the distinctively G-rich nature of the switch sequences on the nontemplate strand (Suppl. Fig. 2). Upon transcription through switch regions, G-richness on the nontemplate DNA strand favors R-loop formation. The G-clusters have been demonstrated to be crucial for R-loop initiation in vitro and in vivo (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010; Zhang et al., 2014a). We refer to these as R-loop initiation zones (RIZ).
Downstream of the switch regions (and upstream of the first IGH coding exon), a gradual decline in G-density on the nontemplate DNA strand seems likely to account for the downstream boundary of the R-loop formation zone, which has been documented in vivo (Huang et al., 2007; Huang et al., 2006). The length of low G-density sequence through which R-loops can persist likely accounts for the wide spectrum of downstream boundaries of R-loops at mammalian switch regions. The downstream boundaries do not have a distinct point of R-loop termination at the end of the zone of high G-density (Huang et al., 2006). Rather, they are distributed up to 600 bp downstream, as the G-density gradually declines below 35%. This is consistent with a stochastic disengagement of the R-loop extension upstream of the RNA polymerase during transcription.
We have shown that decreasing G-density at mammalian IGH switch regions impairs both R-loop formation and CSR (Zhang et al., 2014b)(Figs. 4 and 5; Suppl. Fig. 2). The persistence of R-loops through regions of low G-density (e.g., 50 bp regions of low G-density) illustrates that R-loops do not terminate acutely at a boundary transition from high to low G-density. This persistence of the R-loop beyond the end of the repetitive switch region likely reflects the propensity for the mRNA to continue threading onto the template DNA, once it has begun to do so. Downstream of the G-clusters in the RIZ, we refer to the longer region of extension of the R-loop at the R-loop elongation zone or REZ (Zhang et al., 2014a). It should be noted that majority of RNA:DNA hybrids are thermodynamically more stable than DNA:DNA duplexes of the same sequences, accounting for the persistence of the R-loop conformation for some temporary length of time.
Overall, there is very good correlation between cell-free biochemical studies of R-loop formation (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010) and in vivo cellular R-loop formation and CSR at the mammalian chromosomal level (Zhang et al., 2014a). In some studies of how G-clusters affect CSR in vivo, G-clusters are embedded in the REZ repeats. The results from those constructs are consistent with findings indicating that higher G-density in the REZ increases CSR.
We have previously noted that G-quartet formation is not necessary for R-loop formation in vitro (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010), and this continues to be supported by additional recent studies (Carrasco-Salas et al., 2019).
2.2. The Mechanism by Which the RNA Transcript Creates an R-Loop and Its Evolution
Competition between the transcript and the nontemplate strand dictates R-loop formation efficiency (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010). Conformational variation of the substrate (e.g., negative supercoiling or nicks) can affect R-loop formation by modulating the competition between the transcript and the nontemplate DNA strand for hybridization with the template DNA strand.
A single-strand DNA nick can strongly initiate R-loop formation, if it is positioned downstream of the promoter but upstream of the G-rich regions (Roy et al., 2010). The G-rich regions can help increase the level of R-loop formation, but R-loops can form even without the G-richness. Since a nick can serve as a strong R-loop initiation zone (RIZ), even in a region of sequence where the transcript does not have G-clusters, this markedly broadens the range of possible locations for R-loop formation, when AID is present to initiate the introduction of a nick.
The repetitive nature of switch regions was present earlier in evolution in avians and amphibians (Mussmann et al., 1997; Stavnezer and Amemiya, 2004; Zhu et al., 2012)(Suppl. Fig. 3). But the high G-density on the nontemplate strand is not seen until an evolutionarily later point in mammals. Amphibian repeats are A/T-rich, but also rich in AGCT sequences (one of the WGCWs). As mentioned, nicking by AID, UNG and APE1 is sufficient to generate an R-loop in any transcribed DNA, regardless of sequence; and the repetitive nature of Xenopus regions would then permit RNase H to convert that R-loop to a collapsed R-loop, likely making it a sufficient target for AID action on both strands (Hwang et al., 2015).
The duck switch regions are repetitive and G/C-rich, but not enriched for G on the nontemplate strand (Lundqvist et al., 2001). Again, AID initiated nicks would permit R-loop formation, and RNase H action on the repeats would permit formation of a collapsed R-loop, followed by AID action on both the template and nontemplate DNA strands.
2.3. Collapsed R-loop Structures
One feature of CSR sequence that is not readily explained by the simplest R-loop model is why each switch region has so many repeats (Min et al., 2005; Selsing, 2006). RNA:DNA hybrid formation in vitro only requires a few Sγ3 repeats (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010). The artificial G-rich sequence (1 kb) used by the Alt laboratory, although able to form an R-loop efficiently in vitro, only restored 7% of the CSR activity conferred by the endogenous Sγ1 sequence (Zarrin et al., 2005). In retrospect, this is likely due to a lack of repetitiveness and a paucity of WGCW sites at which AID can act.
At the beginning of our study of the in vivo R-loop, in addition to long R-loop regions (>30 bp, and often over 100 bp), we were puzzled by a low background level of bisulfite reactivity on the template (C-rich) strand at isolated C’s (Yu et al., 2003). [We later determined the bisulfite conversion of isolated C (often within stretches of 2 to 5 C’s) to be due to a B/A-duplex DNA conformation (Tsai et al., 2009).] During the work-up of this background, we considered that the background might represent stretches of shorter R-loop structures created by degraded switch transcripts. When RNase H was used to remove the RNA in these suspected short R-loops, rather than seeing ablation of the short regions of single-strandedness on the template strand, we detected long continuous single-stranded regions on the template strand in a minor but significant percentage of the DNA molecules (Yu et al., 2003). This is the case for both the in vitro and in vivo R-loops. We therefore proposed a collapsed R-loop model (Yu et al., 2003; Yu and Lieber, 2003), in which switch repeats misalign out-of-register as the RNA is being degraded. This would expose single-stranded loops on both strands, and there would be mismatches in the dsDNA between the loops. AID action at the branch sites in the collapsed R-loop (Fig. 3) might be the major deaminiation events during CSR. Additionally, the mismatches may be important in assisting the cleavage process via the MMR mechanism (Fig. 3). If the collapsed R-loop model is correct, then the R-loop would only be an intermediate structure along the path necessary to create the final AID target, namely the collapsed R-loop. The ss regions on the template strand in the collapsed R-loop can also explain where to dock the RNA-bound AID, a prerequisite that has not been addressed in the RNA-guidance model (Zheng et al., 2015). If so, the role of RNase H may be to facilitate rather than inhibit class switch recombination.
2.4. More Complex Models Related to R-loops
Elaborations of the R-loop model primarily focused on whether the G-rich strand forms a G-quartet during R-loop formation or after the R-loop has formed. In purified biochemical system with only the switch repeats and a purified prokaryotic RNA polymerase, an R-loop forms even in solutions of lithium chloride (LiCl) that lack other monovalent cations. The Li+ ion does not support G-quartet formation (Roy et al., 2008). This result indicates that R-loops do not rely on G-quartet formation.
A Li+ test cannot be done using living cells. However, using various mutant versions of the IGH mu switch region, we found secondary structure at the loxP sites that would have been very unlikely if a G-quartet were also present (Huang et al., 2007). We have taken this as one more indication for the lack of a dominant G-quartet formation by the G-rich strand in vivo.
EM and AFM methods of assessing G-quartet formation have been applied in vitro. G-quartet formation would shorten the G-rich strand and thus bend the duplex. But this is not observed in published in vitro images of R-loops (Duquette et al., 2004). More recent AFM studies further support the view that G-quartets are not essential for in vitro R-loop formation generally (Carrasco-Salas et al., 2019); but using a dot-blot method with an antibody claimed to be specific for G4 DNA, they did find some signal at switch regions. We do not regard the specificity of the G4 antibody to be sufficiently documented to permit conclusions.
It has been proposed that the RNA from the switch region forms G-quartets, binds to AID, and guides AID to the switch region (Zheng et al., 2015)(Fig. 6). An RNA debranching enzyme, DBR1, is implicated to liberate the intronic switch RNA to permit its guide role. In a more recent related paper, the Proudfoot lab has proposed that the RNA helicase, DDX1, is part of the targeting mechanism (Ribeiro de Almeida et al., 2018). They propose that DDX1 unwinds the G4 S-RNA and directs it into the S-region to favor an R-loop state. Their model proposes that RNA-bound AID comes along with the DDX1 to the switch site. It is unclear how DDX1 directs the switch RNA to the switch region. The authors conclude that the standard R-loop model (Yu et al., 2003) may operate as a parallel and independent mechanism to their proposed DDX1 pathway (Ribeiro de Almeida et al., 2018).
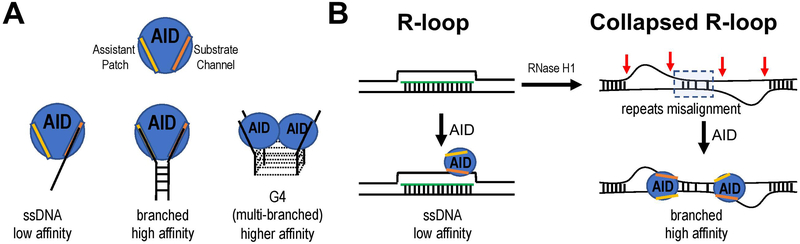
Figure 6. High affinity AID binding to collapsed R-loop.
A. Bifurcated substrate binding of AID. Adapted from a published graphic abstract (Qiao et al., 2017). B. AID binding to the displaced non-template strand in an R-loop is a low affinity binding to ssDNA. Upon RNase H treatment, S region repeats misalign to form collapsed R-loop that contains branched DNA (red arrows) for high affinity AID binding.
2.5. R-loops Outside of Ig CSR Regions and the Use of Antibodies to Detect DNA Structures
The use of antibodies to detect non-B DNA structures is vulnerable to many possible artifacts. This is because the antibody binding pocket is small relative to the larger dimensions of duplex or multi-strand DNA conformations. In addition, even minor variations of nucleic acid structure as a function of nucleotide sequence mean that an antibody binding pocket would need to be surprisingly flexible. A reliance on an antibody to document DNA structure would ideally require an X-ray crystal structure demonstrating that the antibody binds specifically to the predicted nucleic acid target. The antibodies S9.6 and BD4 have been used with the hope of identifying RNA:DNA (in R-loops) and G-quadraplexes, respectively (Biffi et al., 2013; Hu et al., 2006; Phillips et al., 2013). Both antibodies were identified using binding enrichment methods, and there is no atomic structural demonstration of the binding specificity for either. Both antibodies also bind to RNA:RNA as well (Biffi et al., 2014; Hu et al., 2006; Phillips et al., 2013).
There are many claims of R-loop formation at myriad locations in eukaryotic genomes, based on immunoprecipitation (IP) and staining methods with the S9.6 antibody. These claims have very limited functional data to support them (Balk et al., 2013; El Hage et al., 2010; Ginno et al., 2013; Ginno et al., 2012; Mischo et al., 2011; Nakama et al., 2012; Pfeiffer et al., 2013; Phillips et al., 2013; Skourti-Stathaki et al., 2014; Skourti-Stathaki et al., 2011; Stirling et al., 2012; Sun et al., 2013; Wahba et al., 2011; Yang et al., 2014). We are cautious about S9.6 as a primary reagent for identifying R-loops because we find important differences in the IP of the IGH switch regions, depending on whether the harvested genomic nucleic acid is treated with RNase A prior to IP (Zhang et al., 2015).
For IGH switch regions, we are fortunate to have functional assays for actual recombination. In addition, chemical probing and hybridization methods have been applied to define boundaries for the R-loops at Ig switch regions (Chaudhuri et al., 2007; Daniels and Lieber, 1995a; Davis et al., 1980; Dunnick et al., 1993; Reaban and Griffin, 1990; Reaban et al., 1994; Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010; Shinkura et al., 2003; Yu et al., 2003; Yu and Lieber, 2003; Yu et al., 2005; Yu et al., 2006). Application of these additional methods for claims of R-loops outside of Ig switch regions have been quite limited. For these non-Ig regions, the boundaries of S9.6-detected R-loop regions should be established using high resolution bisulfite sequencing. Then functional studies must be done to establish whether such R-loop regions are merely transient steps during transcription.
In addition to our study, there have been several reports of the limited specificity of the S9.6 antibody and of its technical complications (Hu et al., 2006; Konig et al., 2017; Phillips et al., 2013; Vanoosthuyse, 2018). Until further validation of the S9.6 antibody is available, we caution against its use as a primary readout for genomic R-loops.
If these other locations are confirmed with high resolution structural and functional studies, then the rules for R-loop formation defined here will help understand the broader contribution of R-loops to physiologic and pathologic events in the nucleus.
2.6. Related Factors Affecting Switch Region Nucleic Acid Structure
Eukaryotic cells have an RNA surveillance system, the RNA exosome that functions in both the nucleus and cytoplasm to process and degrade certain RNAs. (Note the intracellular RNA exosome should not be confused with the extracellular exosomes, which are lipid bilayer vesicles.) It has been reported that the RNA exosome promotes AID deamination on both DNA strands of transcribed dsDNA, and that depletion of the RNA exosome reduces CSR efficiency (Basu et al., 2011). These data suggest that the RNA exosome may function during CSR to remove S region RNA. RNA exosome also degrades myriads of cellular RNAs, including enhancer RNAs and long non-coding RNAs that are known to impact CSR (Pefanis et al., 2014; Pefanis et al., 2015). Thus, the RNA exosome seems to have multiple roles in regulating CSR.
Whether the two mammalian RNase H enzymes contribute to CSR has not been formally addressed, despite attempts with overexpression of the RNase H1 (which would conceivably improve collapsing of R-loops, and therefore, not discriminate for or against the R-loop model)(Cerritelli and Crouch, 2009). Another factor that has been implicated in resolving R-loops is an RNA helicase named Senataxi. Whether Senataxin has any role in CSR is unknown.
3. TRANSCRIPTION AND THE PROMOTERS UPSTREAM OF SWITCH REGIONS
3.1. The Requirement of Transcription
Sterile transcript promoters exist upstream of each of the switch regions (Lutzker and Alt, 1988; Manis et al., 2002). The sterile transcript promoters for the acceptor switch regions are only active when the B cells receive appropriate extracellular cytokine signals (Suppl. Fig. 1). Removal of the transcriptional control elements leads to the loss of the ability to direct recombination to a particular isotype (Jung et al., 1993; Zhang et al., 1995), while introduction of heterologous enhancer/promoters leads to recombination independent of the normal regulation (Bottaro et al., 1994; Xu et al., 1993).
The temporal linkage of sterile transcript promoter activation to class switching indicated that transcription is critical for class switch recombination targeting. There are several conceivable roles of transcription. First, the transcriptional machinery could be part of the class switch DNA recombination enzymatic complex. Second, transcription of switch DNA could alter the DNA structure in a way that is necessary for CSR. DNA structural alterations could involve changes in superhelical tension in the internucleosomal regions or changes in the conformation from a B-DNA state. Third, transcription could modulate the accessibility of the switch region DNA to the enzymes that carry out the reaction (Wang et al., 2009; Yancopoulos et al., 1986).
Exogenous integrated and extrachromosomal switch sequence substrates recapitulate many of the features of switch recombination in the genome (Daniels and Lieber, 1995b; Lepse et al., 1994; Leung and Maizels, 1992, 1994; Ott et al., 1987; Ott and Marcu, 1989). For the B lineage cell lines that score positive, control substrates with non-switch sequences instead of switch sequences recombine at levels that are significantly lower. The switch region recombination process on extrachromosomal substrates is transcription- and switch region orientation-dependent. Asymmetric mutations are seen within a few hundred nucleotides of exogenous substrates (Li et al., 1996; Stavnezer et al., 1999), just as are seen in vivo (Dunnick et al., 1993).
One study reported that the direction of transcription through the switch regions does not affect switch recombination (Kinoshita et al., 1998). The authors noted that their conclusions assume no complication by transcription entering their integrated substrate from the adjacent genomic DNA, which is difficult to validate. This controversy was addressed by a very elegant study from the Alt laboratory using a mouse model in which the entire Sγ1 region is inverted (Shinkura et al., 2003). The Sγ1 inversion markedly impaired class switch to IgG1, indicating that transcription orientation is important for CSR.
It is possible that the only role of transcription is to generate a DNA secondary structure in mammalian switch regions. Amphibian and avian switch regions may work via a somatic hypermutation role of transcription (see Evolution within Section 2), namely to cause a sufficient degree of RNA pol II-mediated distortions in the DNA duplex to permit AID action or to generate nicks that favor short lengths of transient R-loop formation (Roy et al., 2010; Zhang et al., 2014a; Zhang et al., 2014b). As mentioned, the transient negative superhelicity following passage of an RNA polymerase also may provide sufficient ssDNA for action of AID (Drolet et al., 2003; Sinden, 1994).
3.2. Distance of the Promoter from the Switch Repeats
Increasing distance between switch regions and their respective promoters reduces CSR efficiency (Zhang et al., 2014b). This agrees with the biochemical studies on R-loop formation (Roy et al., 2010). Given this, how can R-loop formation occur when the promoter for the switch regions is 200 to 1500 bp upstream of the switch regions (REZ)? One possible explanation is that a low level of AID-initiated nick formation (nicks achieved by uracil glycosylase (UNG) and AP endonuclease (APE) action on AID deamination sites) can lead to R-loop formation, which would lead to more nicks and more R-loop formation. One can think of this as rounds of nick and R-loop amplification progressing from the promoter toward the IGH switch repeat region. This conceivably explains the existence of R-loops seen in vivo at considerable distances downstream from the promoter and upstream of the G-rich class switch repeat regions (Huang et al., 2006; Roy and Lieber, 2009; Roy et al., 2008). When such nick/R-loop cycles reach the G-rich class switch repeats, more stable, rather than transient, R-loops could form.
How R-loops interfere with RNA polymerase II progression has not been formally tested. We do note that the total amount of transcripts produced in vitro using purified phage RNA polymerases through switch regions on purified DNA templates is lower than that from size-matched random sequences (Daniels and Lieber, 1995a). Perhaps RNA polymerase moves more slowly through GC-rich sequences. It is known that RNA polymerase II accumulates entering switch regions (Rajagopal et al., 2009; Wang et al., 2009). It has been speculated that an R-loop acts as a road block for the elongating RNA polymerases, but it is also possible that such accumulation of RNA polymerase contributes to the stability of R-loops once formed.
3.3. Chromatin Structure and the Role of the 3’ Regulatory Region (3’RR)
The chromatin opening of the entire IGH locus has been reviewed elsewhere (Vaidyanathan et al., 2014). There is no specific aspect of chromatin structure that has been revealed that is specific to CSR (or SHM) beyond the typical chromatin activation of the IGH locus itself.
The ∼28-kb regulatory region (3’RR), which is located at the most distal 3’ region of the Ig H chain locus, has multiple regulatory functions that control IGH expression, class-switch recombination (CSR), and somatic hypermutation. The 3’RR could function primarily via looping to activate the transcription of any of the downstream sterile transcript promoters for CSR (Guglielmi et al., 2003). The 3’RR contains four DNase I hypersensitive (HS) sites. Each individual HS (~1 kb) is a weak enhancer and dispensable for CSR (Pinaud et al., 2011). Synergistically, these four HS sites form a strong enhancer and a locus control region that confers position-independent B lineage-specific expression to associated transgenes. Although deletion of the entire 3’RR in mice severely inhibits GLT and CSR to all downstream IGH C regions (Dunnick et al., 2009; Dunnick et al., 2005; Vincent-Fabert et al., 2010), deletion of 3’RR in the CH12F3 cell line only modestly reduces CSR to IgA (Kim et al., 2016). It should be noted that there is already a basal level of germline transcription through the Sα region without cytokine stimulation. The fact that CSR to IgA in the CH12F3 cell line can occur without 3’RR indicates that 3’RR does not contain AID targeting motif per se for CSR. It is likely that the IGH locus in the CH12F3 cell line is already properly looped, bypassing the requirement for 3’RR.
4. AID PROTEIN STRUCTURE AND WHAT IT TELLS US ABOUT AID TARGETS
AID was discovered by the Honjo laboratory in a cDNA subtraction screening using a B cell line (called CH12F3) that is capable of cytokine dependent CSR in vitro (Muramatsu et al., 1999). AID shares homology to the Apobec family of enzymes, which are all polynucleotide cytosine deaminases that act on RNA or DNA. The AID knockout mouse has no detectable class switch recombination or somatic hypermutation (SHM) (Muramatsu et al., 2000). Humans with mutations in AID also show defective class switching or somatic hypermutation, and they have hyper-IgM syndrome because of the block in progression to production of the downstream heavy chain isotypes (Revy et al., 2000). AID appears to be the only lymphoid specific factor for CSR and SHM, as its ectopic expression in fibroblasts confers CSR and SHM activities to the artificial substrates integrated into these cells (Okazaki et al., 2002).
Mutations in the C-terminus of AID specifically affect CSR (Ta et al., 2003), and the reasons are unclear but could relate to AID oligomerization (Qiao et al., 2017). What is more interesting is that this C-terminus truncation confers a dominant negative trait (Al Ismail et al., 2017; Imai et al., 2005; Zahn et al., 2014), which is difficult to explain at present. The C-terminus of AID contains a nuclear exporting signal (NES), which is responsible for shuttling AID out of the nucleus. The reliance of CSR on the C-terminus of AID is somewhat counterintuitive as AID supposes to exert its function in the nucleus, suggesting complicated regulation of AID activity via active nuclear/cytoplasm trafficking. C-terminal truncated AID is generally unstable (Geisberger et al., 2009), yet it confers higher mutagenesis activity in bacterial antibiotic resistant gene revertant assays (Barreto et al., 2003), suggesting a higher specific enzyme activity. This is one of the most interesting mutations for AID as it decouples CSR from SHM or gene conversion (in birds). The general consensus is that C-terminal truncated AID is still capable of deaminating cytosines and eliciting DSBs in S regions, but somehow affect the subsequent DNA repair processes required for CSR. There are a few interactions between DNA repair factors and the C-terminus of AID that have been reported (Ucher et al., 2014; Zahn et al., 2014), but none has been adequate to explain the CSR-specific defect imposed by AID C-terminal truncation, though structural predictions anticipated some interesting aspects (King et al., 2015).
Two papers now provide atomic structures relevant to AID. An initial structure of AID was obtained containing 15 point mutations and three deletions to improve solubility (Pham et al., 2017), and this structure explained the sequence preference of AID for WRC.
The Wu lab has recently produced monomeric recombinant AID (Qiao et al., 2017). This strategy involved several mutations that are compatible with CSR when introduced into the full-length AID gene. In vitro, this monomeric AID demonstrates superior deaminase activity (~1000 fold) over WT AID (typically in an aggregated form) on ssDNA (Qiao et al., 2017). Their structural studies provided compelling evidence that AID has two DNA binding grooves (Fig. 6). This dual binding mode results in a much higher affinity (and deamination activity) for structured DNA substrates (Qiao et al., 2017) containing at least two stretches of ssDNA (e.g. Y-shaped substrates, as found in collapsed R-loops, or branched-out ssDNA adjacent to a G-quartet).
The structure from Hao Wu’s lab is entirely AID except for two aa point mutations (H130A/R131E, that do not affect AID’s in vivo function), three additional mutations (aa 42, 141, and 145), along with a 17 aa C-terminus truncation that are required to generate a form of AID that can be crystallized (Qiao et al., 2017). The Wu study identifies the catalytic portion of the active site, and the atomic structure contains dCMP in a co-crystal. The dCMP would presumably reside in a length where ssDNA would normally reside. Importantly, the Wu lab study identifies a second portion of the active site, which they call the assistant patch. The assistant patch was predicted to be an additional groove for binding ssDNA. The discovery of the assistant patch nicely explains our prior observation that a certain length (>24 nts) of ssDNA is required for efficient AID deamination (Yu et al., 2004), suggesting that the folding back of ssDNA into the assistant patch is necessary for optimal AID activity on ssDNA substrates used in biochemical studies. Interestingly, a recent study identified several arginine residues within the assistant patch that play a critical role of “licensing” AID for its activity (as opposed to only chromatin binding)(Methot et al., 2018). Some of these mutations (e.g. R174) were found in human patients with hyper-IgM syndrome attesting to a critical role of the assistant patch in AID function in vivo. This means that AID prefers to bind at branched DNA sites (shown by the Wu laboratory in biochemical studies), as would be found at the edges of R-loops. Additional AID binding sides would be present internal to the R-loop if RNase H degraded portions of the RNA. In addition, focal or complete RNase H degradation of the RNA could lead to a collapsed R-loop with multiple branched AID binding sites with the S region. Finally, some have proposed the G-rich strand could adopt a G-quartet structure, which could provide additional AID binding sites at the edges of the G-quartet (Qiao et al., 2017).
The Wu paper does not yet reveal binding to a branched DNA structure because the crystal structures were only achieved using dCMP in the catalytic site (Qiao et al., 2017). The only informative studies with polymeric DNA used Y-structures (formally called pseudo-Y structures because the arms are single-stranded) and G4-quadraplex structures. Based upon solution binding methods (SPR, PAGE and gel shifts) and upon gel filtration elution, the authors propose that AID binds branched DNA as a monomer, and G4-quadraplex DNA potentially as a dimer. Electron microscopy (EM) raises the possibility of larger numbers (oligomers) of AID molecules, but this is difficult to distinguish from nonspecific aggregation under conditions for EM (Qiao et al., 2017). The aggregation of more than 15 AID molecules in gel filtration studies are also difficult to assess as being physiologic or artifactual.
The beautiful atomic structure from the Wu laboratory was achieved using AID lacking the C-terminal tail (CTT) because the CTT causes the AID to aggregate under crystallization conditions (Qiao et al., 2017). But the CTT is needed for CSR (Barreto et al., 2003; Kadungure et al., 2015; Ta et al., 2003; Zahn et al., 2014). In addition to lacking the CTT, the crystallization form (AID.crystal) has 5 aa point mutations that eliminate cooperativity. The in vivo contributions of these changes will be interesting to understand.
5. PROCESSING OF LESIONS AFTER AID ACTION DURING CSR AND THE CONTRAST WITH SHM
Generation of DSBs at switch regions may occur by either of two pathways. The first pathway is the simplest to envision. Because AGCT sites are palindromic, it is possible that the two nearby C’s on opposite strands within this palindrome can both be converted to U. Then action by UNG2 and then APE1 would generate a DSB.
In a second possibility, nicks on opposite strands (at WRCW sites) might be 20 or more bp apart, and thus not generate a DSB immediately (Han et al., 2011; Wuerffel et al., 1992). Given the transcription of these regions from the sterile transcript promoters, encounter of the RNA polymerase with the AID/UNG/APE lesions is more likely because these are not simple nicks (5’ P and 3’OH). Rather these are ‘complex’ nicks that have a 3’OH but a 5’ phosphodeoxyribosyl group (abasic; also known as dRP sites). Such lesions are not repaired rapidly by simple ligation because they require local nucleolytic resection. The greater longevity of such lesions would increase the probability that upstream RNA polymerases would collide with them. The stalled RNA polymerase could recruit Exo I, which is a 5’ exonuclease. Exo I nuclease extension would extend the positions of strand discontinuity and cause a DSB when the discontinuity on the two opposing strands meet each other.
For the second pathway, it is worth pointing out that if BER or MMR mechanisms arrive at an AID/UNG/APE nick before Exo I, then repair back to the original sequence may occur. Transcription may be essential to lessen this occurrence.
From a mechanistic standpoint, it is interesting to consider how SHM lesions in the same cell are handled. Elegant studies from the Alt lab have demonstrated that SHM sites in V regions harboring switch repeats can result in abundant deletions/insertions, consistent with earlier work showing that nearly 6% of SHM events in humans result in small deletions (Goossens et al., 1998). This makes one wonder how SHM lesions are processed for mutation.
For SHM, AID lesions are likely converted to nicks via UNG2 and APE1, as in CSR. Then either of three pathways may be followed. Replication over unrepaired uracils generates C to T (and G to A) transitions (Phase 1A). In wild type mice, there are more nucleotide transitions (~65%) than transversions (~35%) at G/C residues (Rada et al., 2002). Second, replication over abasic sites (upon uracil removal) generates either transition or transversion mutations depending on the translesion (TLS) DNA polymerases that are involved. In this regard, Rev1 clearly plays a role in TLS in SHM (Jansen et al., 2006; Weill and Reynaud, 2008) either via its own catalytic activity or by its capacity to serve as a scaffold protein for other TLS Pols (Jansen et al., 2006; Weill and Reynaud, 2008). Third, strand incision at abasic sites by APE1, or MMR via MutL, is extended by pol eta to generated mutations at A/T residues (Phase II).
Ig CSR requires strand breaks on both strands of the duplex. In contrast, SHM involves mostly single strand events (Faili et al., 2002), though a few percent of V regions have deletions or insertions in association with SHM (Wilson et al., 1998).
5.1. Joining of DNA Ends After DSB Formation.
Once the DSBs form in the switch regions, they can be joined by NHEJ. It is unclear whether a tightly regulated synaptic process brings the two DSBs from two different switch regions together, as some studies have proposed (Wuerffel et al., 2007). There is evidence that multiple DSBs can form within the same switch region, especially for Sμ. When these breaks are joined, it generates a small chromosomal deletion often termed intra-switch region deletion (ISD). Obviously ISDs are non-productive products for CSR. Elegant studies have demonstrated that even DSBs formed by I-SceI within engineered switch regions can be joined distally at significant levels (Gostissa et al., 2014). Thus, it appears unlikely that ISD versus productive CSR (i.e. joining distal DSBs from different switch regions) is tightly regulated (Gostissa et al., 2014). However, some DSB repair factors (e.g. 53bp1, H2AX) appear to differentially affect distal joining (CSR) versus ISD.
Regarding the actual proteins required for NHEJ of switch region DNA ends, DNA-PKcs affects switching to some of the downstream switch regions more than others (Bjorkman et al., 2015; Bosma et al., 2002; Manis et al., 2002; Woodbine et al., 2014). The essential component is a DNA ligase. Han et al have shown that DNA ligase IV is responsible for the large majority of joining events (Han and Yu, 2008). In the absence of DNA ligase IV, either DNA ligase I or ligase III can carry out joining (Masani et al., 2016), but it appears that this is much less efficient (Han and Yu, 2008). This indicates that NHEJ is the primary end joining pathway but that alternative end joining (a-EJ) may be a less efficient mechanism for joining in the rare situation where DNA ligase IV is absent(Pannunzio et al., 2018).
6. COMPARISONS of CSR and SHM
IGH CSR evolved over 100 million years after AID was already present in B cells for the purpose of mutating (and gene converting) variable domains during somatic hypermutation. Cytosine deaminase homologues are even present in ancient species such as the lampreys, before the RAG-mediated V(D)J recombination, catalyzing gene conversion to diversify the variable lymphocyte receptors (VLRs). This means that the switch regions themselves evolved much later in B cells. Therefore, the features of switch regions may have evolved to make them an increasingly optimal substrate for AID action. Are these features more optimal than the features that direct AID to Ig light chain and heavy chain variable domains for somatic hypermutation? For SHM, transcription might inefficiently provide ssDNA to permit the relatively slow process of SHM to occur over weeks of time during an immune response. But CSR occurs typically within a few days in mammals. We have proposed that AID may act more rapidly in CSR in mammals due to the high G-clustering and G-density that is the distinctive feature of all mammalian IGH switch regions (Yu et al., 2003; Yu and Lieber, 2003) and not present in organisms that evolved CSR earlier, such as avians and amphibians (Hackney et al., 2009; Zhu et al., 2012).
For SHM, cytidine sites (preferably ones within WGCW) must be within regions of ssDNA, at least transiently, to be a substrate for AID (Storb, 2014). Transcription through random DNA (containing few or no G-clusters) appears to be able to induce such transient ssDNA states, though inefficiently. Once AID causes a U:G mismatch at a WGCW site, this is converted to a nick via UNG and APE (DiNoia and Neuberger, 2007; Masani et al., 2013; Zanotti and Gearhart, 2016). In biochemical systems, R-loops can form when a nick is present in the non-template strand of the DNA, even when there are no G-clusters or the G-density is low (Roy et al., 2010). This may be the basis for how AID can act during SHM, despite a lack of G-clusters.
The ability of a nick to serve as a strong R-loop initiation zone (RIZ) could be of importance for SHM. In that process, there are no obvious RIZ or REZ sequence motifs (Liu et al., 2008). However, AID is thought to initiate (with UNG and APE) low levels of nicking in regions of somatic hypermutation. Such initial nicks could lead to short regions of R-loop formation (possibly those seen in (Ronai et al., 2007)), even though these regions lack G-richness. The single-strandedness in those short R-loops would then permit AID to much more efficiently initiate many more nicks and lead to amplification of rounds of R-loop formation and AID-initiated nicking, thereby facilitating somatic hypermutation.
7. PATHOLOGIC INVOLVEMENT OF IG SWITCH REGIONS IN CHROMOSOMAL TRANSLOCATIONS
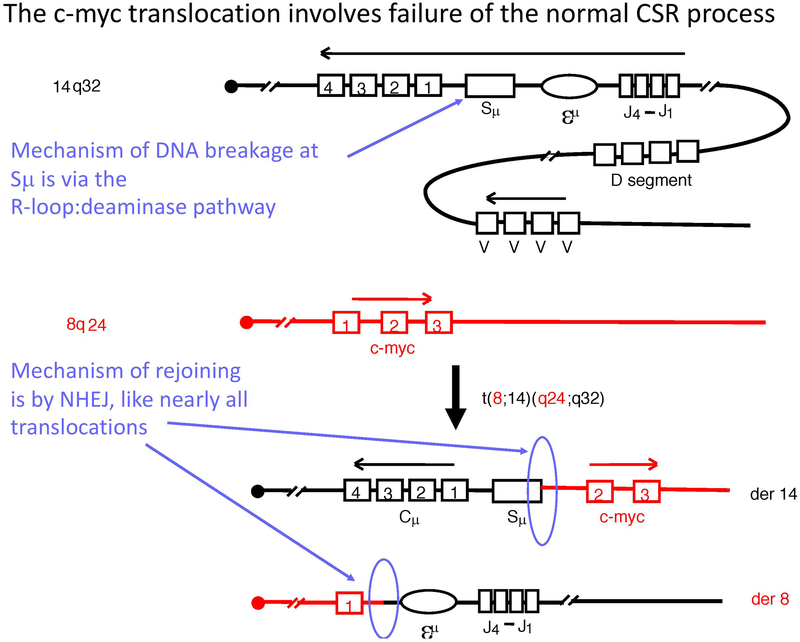
CSR is not only important for a competent immune system, but when it occurs erroneously, it can lead to oncogenic chromosomal translocations that promote B cell malignancies (Chaudhuri and Alt, 2004; Stavnezer et al., 2008; Yu and Lieber, 2003). For example, the C-MYC gene translocates to the Ig switch regions in all sporadic Burkitt’s lymphomas (small, noncleaved cell lymphomas); in ~20% of diffuse, large cell non-Hodgkins lymphomas; in a substantial fraction of HIV-associated non-Hodgkins lymphomas; and in a large fraction of multiple myelomas (Lieber, 2016)(Fig. 7).
Figure 7. Diagram of the Human c-MYC Translocation.
The diagram illustrates the translocation of the c-myc gene on human chromosome 8 with the IgH switch regions on chromosome 15. Breaks at both locations are AID-mediated. In addition to evidence for R-loops at the Ig CSR regions, there is also evidence for R-loops at c-myc.
In addition to C-MYC being a partner with Ig CSR regions, the BCL6 gene locus can also be a partner. The BCL6 translocation occurs primarily in diffuse large B cell lymphoma. The MYC translocation occurs primarily in Burkitt lymphoma, diffuse large B cell lymphomas, and in some multiple myelomas (Kuppers, 2005).
For such translocation to occur, the recombination process at the IGH class switch locus (IGH-S) must fail. This failure may occur during the process in which the DSBs at Sμ and a downstream S region are brought into proximity of joining. Such a step is called synapsis. There is limited current information about this step, though progress is forthcoming (F. W. Alt, personal comm.). A failure of the DNA ends at Sμ to be joined to the ends of a DSB at a downstream switch region (for example, Sγ) allows the two Sμ ends to be joined to two ends of a DSB on another chromosome, such as at MYC or BCL6 (Lu et al., 2013) (Fig. 7). Thus, the IGH locus DSBs in these events are usually created during IGH CSR.
Studies in mice have shown that the MYC break is dependent on AID (Robbiani et al., 2008). For human translocations, the MYC and BCL6 translocations occur at DNA sequence motifs for which the AID enzyme has a known predilection (Lu et al., 2013), as discussed above. Thus, the breaks at BCL6 and MYC are AID-mediated events, and the breaks at the IGH-S regions are also AID-mediated events (as part of the IGH CSR process). Biochemical and immunoprecipitation approaches have both suggested that R-loops form at the c-MYC locus (Duquette et al., 2005; Yang et al., 2014).
8. FUTURE DIRECTIONS
Among the many mechanistic questions about Ig CSR, the following may be central.
how fundamentally different are CSR and SHM mechanisms? Is the primary difference the R-loop formation, which is so remarkably distinctive for mammalian switch regions?
The CSR and SHM mechanism(s) that lead to error-prone outcomes from BER and MMR, two pathways that normally mediate high-fidelity repair, must be further elucidated.
how are two switch regions brought into synapsis? (papers from Fred Alt may be discussed above, if the papers are out).
for proposed R-loops outside of the Ig switch regions, why are there not more translocations between switch regions and these sites? Perhaps these non-Ig R-loops are extremely shorter-lived.
For both CSR, whether the ssDNA region for AID action is in the context of a DNA branch site or a more complex DNA structure is a key question.
Indirect contributions of other factors prior to the NHEJ components will be interesting to elucidate, including effects of 53BP1, Rif1, and Shieldin (Setiaputra and Durocher, 2019).
Supplementary Material
Suppl. Figure 1. Sterile Transcription Units for Each Acceptor Switch Region. Cytokine-regulated transcription activates S region for CSR. The sterile transcript is spliced to join the I exon and the constant region exons.
Suppl. Figure 2. Switch Region G-Richness. During evolution, mammalian switch regions became increasingly G-rich. The R-loop model is the only proposed explanation for this remarkable G-rich profile.
Suppl. Figure 3 (A-C). Switch Region Sequence and Parameters Driving R-loop Formation. CSR efficiency is in direct proportion to the total number of WGCW sites and the amount of ssDNA. The amount of ssDNA is proportional to the transcription firing rate, RIZ strength and REZ strength. RIZ strength is a function of the total G-cluster number and the G nucleotide number within each G-cluster. REZ strength is dependent on overall G-density, which determines the length of the R-loop. Naturally-occurring switch regions have interspersed G-clusters, WGCW sites, and a high G-density. Hence, for naturally-occurring R-loops in cells, the ssDNA length (R-loop length) varies depending at which G-cluster the R-loop initiates (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010).
Acknowledgements.
Work in the authors labs is supported by NIH. The authors apologize to any relevant work that is not cited.
Footnotes
Disclosures. The authors have no financial or academic conflicts of interest.
REFERENCES
- Al Ismail A, Husain A, Kobayashi M, Honjo T, and Begum NA (2017). Depletion of recombination-specific cofactors by the C-terminal mutant of the activation-induced cytidine deaminase causes the dominant negative effect on class switch recombination. Int Immunol 29, 525–537. [DOI] [PubMed] [Google Scholar]
- Balk B, Maicher A, Dees M, Klermund J, Luke-Glaser S, Bender K, and Luke B (2013). Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat Struct Mol Biol 20, 1199–1205. [DOI] [PubMed] [Google Scholar]
- Balter BB, Ciccone DN, Oettinger MA, and Selsing E (2012). Mice lacking Smu tandem repeats maintain RNA polymerase patterns but exhibit histone modification pattern shifts linked to class switch site locations. Mol Immunol 52, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto V, Reina-San-Martin B, Ramiro AR, McBride KM, and Nussenzweig MC (2003). C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol Cell 12, 501–508. [DOI] [PubMed] [Google Scholar]
- Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, Eccleston J, Zhang T, Myers D, Wasserman CR, Wesemann DR, et al. (2011). The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell 144, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G, Di Antonio M, Tannahill D, and Balasubramanian S (2014). Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nature chemistry 6, 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G, Tannahill D, McCafferty J, and Balasubramanian S (2013). Quantitative visualization of DNA G-quadruplex structures in human cells. Nature chemistry 5, 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman A, Du L, Felgentreff K, Rosner C, Pankaj Kamdar R, Kokaraki G, Matsumoto Y, Davies EG, van der Burg M, Notarangelo LD, et al. (2015). DNA-PKcs Is Involved in Ig Class Switch Recombination in Human B Cells. J Immunol 195, 5608–5615. [DOI] [PubMed] [Google Scholar]
- Bosma GC, Kim J, Ulrich T, Fath DM, Cotticelli MG, Ruetsch NR, Radic MZ, and Bosma MJ (2002). DNA-dependent protein kinase activity is not required for immunoglobulin class switching. J Exp Med 196, 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottaro A, Lansford R, Xu L, Zhang J, Rothman P, and Alt FW (1994). S region transcription per se promotes basal IgE class switch recombination but additional factors regulate the efficiency of the process. EMBO J 13, 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Calabrese P, and Goodman MF (2004). Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J Biol Chem 279, 51612–51621. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, and Goodman MF (2003). Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci 100, 4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canugovi C, Samaranayake M, and Bhagwat AS (2009). Transcriptional pausing and stalling causes multiple clustered mutations by human activation-induced deaminase. FASEB J 23, 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco-Salas Y, Malapert A, Sulthana S, Molcrette B, Chazot-Franguiadakis L, Bernard P, Chedin F, Faivre-Moskalenko C, and Vanoosthuyse V (2019). The extruded non-template strand determines the architecture of R-loops. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casellas R, Basu U, Yewdell WT, Chaudhuri J, Robbiani DF, and Di Noia JM (2016). Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity. Nat Rev Immunol 16, 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli SM, and Crouch RJ (2009). Ribonuclease H: the enzymes in eukaryotes. The FEBS journal 276, 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, and Alt FW (2004). Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol 4, 541–552. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, Vuong B, Wang J, Phan RT, Datta A, et al. (2007). Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol 94, 157–214. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaus E, and Alt FW (2003). Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422, 726–730. [DOI] [PubMed] [Google Scholar]
- Daniels GA, and Lieber MR (1995a). RNA:DNA complex formation upon transcription of immunoglobulin switch regions: implications for the mechanism and regulation of class switch recombination. Nucl Acids Res 23, 5006–5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels GA, and Lieber MR (1995b). Strand-specificity in the transcriptional targeting of recombination at immunoglobulin class switch sequences. Proc Natl Acad Sci USA 92, 5625–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M, Kim SK, and Hood LE (1980). DNA Sequences Mediating Class Switching in a-Immunoglobulins. Science 209, 1360–1365. [DOI] [PubMed] [Google Scholar]
- Dickerson SK, Market E, Besmer E, and Papavasiliou FN (2003). AID mediates hypermutation by deaminating single-stranded DNA. J Exp Med 197, 1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNoia J, and Neuberger MS (2002). Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature 419, 43–48. [DOI] [PubMed] [Google Scholar]
- DiNoia JM, and Neuberger MS (2007). Molecular mechanisms of antibody somatic hypermutation. Ann Rev Biochem 76, 1–22. [DOI] [PubMed] [Google Scholar]
- Drolet M, Broccoli S, Rallu F, Hraiky C, Fortin C, Masse E, and Baaklini I (2003). The problem of hypernegative supercoiling and R-loop formation in transcription. Front Biosci 8, d210–221. [DOI] [PubMed] [Google Scholar]
- Dunnick WA, Collins JT, Shi J, Westfield G, Fontaine C, Hakimpour P, and Papavasiliou FN (2009). Switch recombination and somatic hypermutation are controlled by the heavy chain 3’ enhancer region. J Exp Med 206, 2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick WA, Hertz GZ, Scappino L, and Gritzmacher C (1993). DNA sequence at immunoglobulin switch region recombination sites. Nucl Acid Res 21, 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick WA, Shi J, Graves KA, and Collins JT (2005). The 3’ end of the heavy chain constant region locus enhances germline transcription and switch recombination of the four gamma genes. J Exp Med 201, 1459–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette ML, Handa P, Vincent JA, Taylor AF, and Maizels N (2004). Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev 18, 1618–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duquette ML, Pham P, Goodman MF, and Maizels N (2005). AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene 24, 5791–5798. [DOI] [PubMed] [Google Scholar]
- El Hage A, French SL, Beyer AL, and Tollervey D (2010). Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 24, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faili A, Aoufouchi S, Gueranger Q, Zober C, Leon A, Bertocci B, Weill JC, and Reynaud CA (2002). AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat Immunol 3, 815–821. [DOI] [PubMed] [Google Scholar]
- Geisberger R, Rada C, and Neuberger MS (2009). The stability of AID and its function in class-switching are critically sensitive to the identity of its nuclear-export sequence. Proc Natl Acad Sci U S A 106, 6736–6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lim YW, Lott PL, Korf I, and Chedin F (2013). GC skew at the 5’ and 3’ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res 23, 1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginno PA, Lott PL, Christensen HC, Korf I, and Chedin F (2012). R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 45, 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman MF, Scharff MD, and Romesberg FE (2007). AID-initiated purposeful mutations in immunoglobulin genes. Adv Immunol 94, 127–155. [DOI] [PubMed] [Google Scholar]
- Goossens T, Klein U, and Kuppers R (1998). Frequent occurrence of deletions and duplications during somatic hypermutation: implications for oncogene translocations and heavy chain disease. Proc Natl Acad Sci U S A 95, 2463–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostissa M, Schwer B, Chang A, Dong J, Meyers RM, Marecki GT, Choi VW, Chiarle R, Zarrin AA, and Alt FW (2014). IgH class switching exploits a general property of two DNA breaks to be joined in cis over long chromosomal distances. Proc Natl Acad Sci U S A 111, 2644–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunseich C, Wang IX, Watts JA, Burdick JT, Guber RD, Zhu Z, Bruzel A, Lanman T, Chen K, Schindler AB, et al. (2018). Senataxin Mutation Reveals How R-Loops Promote Transcription by Blocking DNA Methylation at Gene Promoters. Mol Cell 69, 426–437 e427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi L, Le Bert M, Truffinet V, Cogne M, and Denizot Y (2003). Insulators to improve expression of a 3(‘)IgH LCR-driven reporter gene in transgenic mouse models. Biochem Biophys Res Commun 307, 466–471. [DOI] [PubMed] [Google Scholar]
- Hackney JA, Misaghi S, Senger K, Garris C, Sun Y, Lorenzo MN, and Zarrin AA (2009). DNA targets of AID evolutionary link between antibody somatic hypermutation and class switch recombination. Adv Immunol 101, 163–189. [DOI] [PubMed] [Google Scholar]
- Han L, Masani S, and Yu K (2011). Overlapping activation-induced cytidine deaminase hotspot motifs in Ig class-switch recombination. Proc Natl Acad Sci U S A 108, 11584–11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, and Yu K (2008). Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV--deficient B cells. J Exp Med 205, 2745–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Sale JE, Petersen-Mahrt SK, and Neuberger MS (2002). AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr Biol 12, 435–438. [DOI] [PubMed] [Google Scholar]
- Hu Z, Zhang A, Storz G, Gottesman S, and Leppla SH (2006). An antibody-based microarray assay for small RNA detection. Nucleic Acids Res 34, e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F-T, Yu K, Balter BB, Selsing E, Oruc Z, Khamlichi AA, Hsieh C-L, and Lieber MR (2007). Sequence-dependence of chromosomal R-loops at the immunoglobulin heavy chain Smu class switch region. Mol Cell Biol 27, 5921–5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F-T, Yu K, Hsieh C-L, and Lieber MR (2006). The downstream boundary of chromosomal R-loops at murine switch regions: implications for the mechanism of class switch recombination. Proc Natl Acad Sci 103, 5030–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang JK, Alt FW, and Yeap LS (2015). Related Mechanisms of Antibody Somatic Hypermutation and Class Switch Recombination. Microbiology spectrum 3, MDNA3-0037-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Zhu Y, Revy P, Morio T, Mizutani S, Fischer A, Nonoyama S, and Durandy A (2005). Analysis of class switch recombination and somatic hypermutation in patients affected with autosomal dominant hyper-IgM syndrome type 2. Clin Immunol 115, 277–285. [DOI] [PubMed] [Google Scholar]
- Jansen JG, Langerak P, Tsaalbi-Shtylik A, van den Berk P, Jacobs H, and de Wind N (2006). Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J Exp Med 203, 319–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S, and Bhagwat AS (2014). Transcription-associated mutagenesis. Annu Rev Genet 48, 341–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Rajewsky K, and Radbruch A (1993). Shutdown of Class Switch Recombination by Deletion of a Switch Region Control Element. Science 259, 984–987. [DOI] [PubMed] [Google Scholar]
- Kadungure T, Ucher AJ, Linehan EK, Schrader CE, and Stavnezer J (2015). Individual substitution mutations in the AID C terminus that ablate IgH class switch recombination. PLoS One 10, e0134397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Han L, Santiago GE, Verdun RE, and Yu K (2016). Class-Switch Recombination in the Absence of the IgH 3’ Regulatory Region. J Immunol 197, 2930–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JJ, Manuel CA, Barrett CV, Raber S, Lucas H, Sutter P, and Larijani M (2015). Catalytic pocket inaccessibility of activation-induced cytidine deaminase is a safeguard against excessive mutagenic activity. Structure 23, 615–627. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Tashiro J, Tomita S, Lee CG, and Honjo T (1998). Target specificity of immunoglobulin class switch recombination is not determined by nucleotide sequences of S regions. Immunity 9, 849–858. [DOI] [PubMed] [Google Scholar]
- Konig F, Schubert T, and Langst G (2017). The monoclonal S9.6 antibody exhibits highly variable binding affinities towards different R-loop sequences. PLoS One 12, e0178875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppers R (2005). Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer 5, 251–262. [DOI] [PubMed] [Google Scholar]
- Larijani M, and Martin A (2007). Single-stranded DNA structure and positional context of the target cytidine determine the enzymatic efficiency of AID. Mol Cell Biol 27, 8038–8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebecque SG, and Gearhart P (1990). Boundaries of somatic mutation in rearranged immunoglobulin genes: 5’ Boundary is near the promoter, and 3’ boundary is about 1 kb from V(D)J gene. J Exp Med 172, 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepse CL, Kumar R, and Ganea D (1994). Extrachromosomal eukaryotic DNA substrates for switch recombination: analysis of isotype and cell specificity. DNA Cell Biol 13, 1151–1161. [DOI] [PubMed] [Google Scholar]
- Leung H, and Maizels N (1992). Transcriptional Regulatory Elements Stimulate Recombination in Extrachromosomal Substrates Carrying Immunoglobulin Switch-Region Sequences. Proc Natl Acad Sci 89, 4154–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung H, and Maizels N (1994). Regulation and Targeting of Recombination in Extrachromosomal Substrates Carrying Immunoglobulin Switch Region Sequences. Mol Cell Biol 14, 1450–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Daniels GA, and Lieber MR (1996). Asymmetric mutation around the recombination break point of immunoglobulin class switch sequences on extrachromosomal substrates. Nucl Acids Res 24, 2104–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR (2016). Mechanisms of human lymphoid chromosomal translocations. Nat Rev Cancer 16, 387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, and Schatz DG (2008). Two levels of protection for the B cell genome during somatic hypermutation. Nature 451, 841–845. [DOI] [PubMed] [Google Scholar]
- Lu Z, Tsai AG, Akasaka T, Ohno H, Jiang Y, Melnick AM, Greisman HA, and Lieber MR (2013). BCL6 breaks occur at different AID sequence motifs in Ig-BCL6 and non-Ig-BCL6 rearrangements. Blood 121, 4551–4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundqvist ML, Middleton DL, Hazard S, and Warr GW (2001). The immunoglobulin heavy chain locus of the duck. Genomic organization and expression of D, J, and C region genes. J Biol Chem 276, 46729–46736. [DOI] [PubMed] [Google Scholar]
- Lutzker S, and Alt F (1988). Structure and Expression of Germ Line Immunoglobulin g2b Transcripts. Mol Cell Biol 8, 1849–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis JP, Dudley D, Kaylor L, and Alt FW (2002). IgH class switch recombination to IgG1 in DNA-PKcs-deficient B cells. Immunity 16, 607–617. [DOI] [PubMed] [Google Scholar]
- Masani S, Han L, Meek K, and Yu K (2016). Redundant function of DNA ligase 1 and 3 in alternative end-joining during immunoglobulin class switch recombination. Proc Natl Acad Sci U S A 113, 1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masani S, Han L, and Yu K (2013). Apurinic/apyrimidinic endonuclease 1 is the essential nuclease during immunoglobulin class switch recombination. Mol Cell Biol 33, 1468–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Phoenix P, and Drolet M (1997). DNA topoisomerases regulate R-loop formation during transcription of the rrnB operon in E. coli. J Biol Chem 272, 12816–12823. [DOI] [PubMed] [Google Scholar]
- Matthews AJ, Zheng S, DiMenna LJ, and Chaudhuri J (2014). Regulation of immunoglobulin class-switch recombination: choreography of noncoding transcription, targeted DNA deamination, and long-range DNA repair. Adv Immunol 122, 1–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul RW, Chon H, Sakhuja K, Cerritelli SM, Gugliotti LA, Gearhart PJ, and Crouch RJ (2017). R-Loop Depletion by Over-expressed RNase H1 in Mouse B Cells Increases Activation-Induced Deaminase Access to the Transcribed Strand without Altering Frequency of Isotype Switching. J Mol Biol 429, 3255–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot SP, and Di Noia JM (2017). Molecular Mechanisms of Somatic Hypermutation and Class Switch Recombination. Adv Immunol 133, 37–87. [DOI] [PubMed] [Google Scholar]
- Methot SP, Litzler LC, Subramani PG, Eranki AK, Fifield H, Patenaude AM, Gilmore JC, Santiago GE, Bagci H, Cote JF, et al. (2018). A licensing step links AID to transcription elongation for mutagenesis in B cells. Nature communications 9, 1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Rothlein LR, Schrader CE, Stavnezer J, and Selsing E (2005). Shifts in targeting of class switch recombination sites in mice that lack mu switch region tandem repeats or Msh2. J Exp Med 201, 1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, and Proudfoot NJ (2011). Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 41, 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, and Honjo T (2000). Class switch recombination and somatic hypermutation require activation-induced cytidine deaminase (AID), a member of the RNA editing cytidine deaminase family. Cell 102, 541–544. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand V, Anant S, Sugai M, Kinoshita K, Davidson N, and Honjo T (1999). Specific Expression of Activation-Induced Cytidine Deaminase (AID), a Novel Member of the RNA-Editing Deaminase Family in Germinal Center B Cells. J Biol Chem 274, 18470–18476. [DOI] [PubMed] [Google Scholar]
- Mussmann R, Courtet M, Schwager J, and Pasquier LD (1997). Microsites for immunoglobulin switch recombination breakpoints from Xenopus to mammals. Eur J Immunol 27, 2610–2619. [DOI] [PubMed] [Google Scholar]
- Nakama M, Kawakami K, Kajitani T, Urano T, and Murakami Y (2012). DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes to cells : devoted to molecular & cellular mechanisms 17, 218–233. [DOI] [PubMed] [Google Scholar]
- Okazaki IM, Kinoshita K, Muramatsu M, Yoshikawa K, and Honjo T (2002). The AID enzyme induces class switch recombination in fibroblasts. Nature 416, 340–345. [DOI] [PubMed] [Google Scholar]
- Ott DE, Alt FW, and Marcu KB (1987). Immunoglobulin Heavy Chain Switch Region Recombination within a Retroviral Vector in Murine pre-B Cells. EMBO 6, 577–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott DE, and Marcu KB (1989). Molecular Requirements for Immunoglobulin Heavy Chain Constant Region Gene Switch-Recombination Revealed with Switch -Substrate Retrovirues. Int Immunol 1, 582–591. [DOI] [PubMed] [Google Scholar]
- Pan-Hammarstrom Q, Zhao Y, and Hammarstrom L (2007). Class switch recombination: a comparison between mouse and human. Adv Immunol 93, 1–61. [DOI] [PubMed] [Google Scholar]
- Pannunzio NR, Watanabe G, and Lieber MR (2018). Nonhomologous DNA End Joining for Repair of DNA Double-Strand Breaks. J Biol Chem 293, 10512–10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HC 3rd, Nussenzweig MC, and Dalla-Favera R (2008). AID is required for germinal center-derived lymphomagenesis. Nat Genet 40, 108–112. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Guglielmino R, Houldsworth J, Mohr J, Aoufouchi S, Polakiewicz R, Chaganti RS, and Dalla-Favera R (2004). Expression of the AID protein in normal and neoplastic B cells. Blood 104, 3318–3325. [DOI] [PubMed] [Google Scholar]
- Pefanis E, Wang J, Rothschild G, Lim J, Chao J, Rabadan R, Economides AN, and Basu U (2014). Noncoding RNA transcription targets AID to divergently transcribed loci in murine B cells. Nature 514, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, et al. (2015). RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161, 774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, and Neuberger MS (2002). AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418, 99–103. [DOI] [PubMed] [Google Scholar]
- Pfeiffer V, Crittin J, Grolimund L, and Lingner J (2013). The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J 32, 2861–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham P, Afif SA, Shimoda M, Maeda K, Sakaguchi N, Pedersen LC, and Goodman MF (2017). Activation-induced deoxycytidine deaminase: Structural basis for favoring WRC hot motif specificities unique among APOBEC family members. DNA Repair (Amst) 54, 8–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham P, Bransteitter R, Petruska J, and Goodman MF (2003). Processive AID-catalyzed cytosine deamination on single-stranded DNA stimulates somatic hypermutation. Nature 424, 103–107. [DOI] [PubMed] [Google Scholar]
- Phillips DD, Garboczi DN, Singh K, Hu Z, Leppla SH, and Leysath CE (2013). The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J Mol Recognit 26, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinaud E, Marquet M, Fiancette R, Peron S, Vincent-Fabert C, Denizot Y, and Cogne M (2011). The IgH locus 3’ regulatory region: pulling the strings from behind. Adv Immunol 110, 27–70. [DOI] [PubMed] [Google Scholar]
- Qiao Q, Wang L, Meng FL, Hwang JK, Alt FW, and Wu H (2017). AID Recognizes Structured DNA for Class Switch Recombination. Mol Cell 67, 361–373 e364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, and Neuberger MS (2002). Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol 12, 1748–1755. [DOI] [PubMed] [Google Scholar]
- Rajagopal D, Maul RW, Ghosh A, Chakraborty T, Khamlichi AA, Sen R, and Gearhart PJ (2009). Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J Exp Med 206, 1237–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Stavropoulos P, Jankovic M, and Nussenzweig MC (2003). Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat Immunol 4, 452–456. [DOI] [PubMed] [Google Scholar]
- Reaban ME, and Griffin JA (1990). Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature 348, 342–344. [DOI] [PubMed] [Google Scholar]
- Reaban ME, Lebowitz J, and Griffin JA (1994). Transcription induces the formation of a stable RNA.DNA hybrid in the immunoglobulin alpha switch region. J Biol Chem 269, 21850–21857. [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Lagelouse R, Gennery A, et al. (2000). Acitvation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell 102, 565–575. [DOI] [PubMed] [Google Scholar]
- Ribeiro de Almeida C, Dhir S, Dhir A, Moghaddam AE, Sattentau Q, Meinhart A, and Proudfoot NJ (2018). RNA Helicase DDX1 Converts RNA G-Quadruplex Structures into R-Loops to Promote IgH Class Switch Recombination. Mol Cell 70, 650–662 e658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, et al. (2008). AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell 135, 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RW, and Crothers DM (1992). Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science 258, 1463–1466. [DOI] [PubMed] [Google Scholar]
- Ronai D, Iglesias-Ussel MD, Fan M, Li Z, Martin A, and Scharff MD (2007). Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J Exp Med 204, 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, and Lieber MR (2009). G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol 29, 3124–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Yu K, and Lieber MR (2008). Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol 28, 50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D, Zhang Z, Lu Z, Hsieh CL, and Lieber MR (2010). Competition Between the RNA Transcript and the Nontemplate DNA Strand During R-Loop Formation In Vitro: A Nick Can Serve as a Strong R-loop Initiation Site. Mol Cell Biol 30, 146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selsing E (2006). Ig class switching: targeting the recombinational mechanism. Curr Opin Immunol 18, 249–254. [DOI] [PubMed] [Google Scholar]
- Setiaputra D, and Durocher D (2019). Shieldin - the protector of DNA ends. EMBO Rep 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkura R, Tian M, Khuong C, Chua K, Pinaud E, and Alt FW (2003). The influence of transcriptional orientation on endogenous switch region function. Nature Immunol 4, 435–441. [DOI] [PubMed] [Google Scholar]
- Sinden RR (1994). DNA Structure and Function (San Diego: Academic Press; ). [Google Scholar]
- Skourti-Stathaki K, Kamieniarz-Gdula K, and Proudfoot NJ (2014). R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, and Gromak N (2011). Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 42, 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail A, Klapacz J, Samaranayake M, Ullah A, and Bhagwat AS (2003). Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucl Acids Res 31, 2990–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, and Amemiya CT (2004). Evolution of isotype switching. Semin Immunol 16, 257–275. [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Bradley SP, Rousseau N, Pearson T, Shanmugam A, Waite DJ, Rogers PR, and Kenter AL (1999). Switch recombination in a transfected plasmid occurs preferentially in a B cell line that undergoes switch recombination of its chromosomal Ig heavy chain genes. J Immunol 163, 2028–2040. [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, and Schrader CE (2008). Mechanism and regulation of class switch recombination. Annu Rev Immunol 26, 261–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Chan YA, Minaker SW, Aristizabal MJ, Barrett I, Sipahimalani P, Kobor MS, and Hieter P (2012). R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev 26, 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storb U (2014). Why does somatic hypermutation by AID require transcription of its target genes? Adv Immunol 122, 253–277. [DOI] [PubMed] [Google Scholar]
- Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, and Dean C (2013). R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340, 619–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta VT, Nagaoka H, Catalan N, Durandy A, Fischer A, Imai K, Nonoyama S, Tashiro J, Ikegawa M, Ito S, et al. (2003). AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol 4, 843–848. [DOI] [PubMed] [Google Scholar]
- Tsai AG, Engelhart AE, Hatmal MM, Houston SI, Hud NV, Haworth IS, and Lieber MR (2009). Conformational variants of duplex DNA correlated with cytosine-rich chromosomal fragile sites. J Biol Chem 284, 7157–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ucher AJ, Ranjit S, Kadungure T, Linehan EK, Khair L, Xie E, Limauro J, Rauch KS, Schrader CE, and Stavnezer J (2014). Mismatch repair proteins and AID activity are required for the dominant negative function of C-terminally deleted AID in class switching. J Immunol 193, 1440–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan B, Yen WF, Pucella JN, and Chaudhuri J (2014). AIDing Chromatin and Transcription-Coupled Orchestration of Immunoglobulin Class-Switch Recombination. Front Immunol 5, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V (2018). Strengths and Weaknesses of the Current Strategies to Map and Characterize R-Loops. Non-coding RNA 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent-Fabert C, Fiancette R, Pinaud E, Truffinet V, Cogne N, Cogne M, and Denizot Y (2010). Genomic deletion of the whole IgH 3’ regulatory region (hs3a, hs1,2, hs3b, and hs4) dramatically affects class switch recombination and Ig secretion to all isotypes. Blood 116, 1895–1898. [DOI] [PubMed] [Google Scholar]
- Wahba L, Amon JD, Koshland D, and Vuica-Ross M (2011). RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell 44, 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wuerffel R, Feldman S, Khamlichi AA, and Kenter AL (2009). S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J Exp Med 206, 1817–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill JC, and Reynaud CA (2008). DNA polymerases in adaptive immunity. Nat Rev Immunol 8, 302–312. [DOI] [PubMed] [Google Scholar]
- Wilson PC, de Bouteiller O, Liu YJ, Potter K, Banchereau J, Capra JD, and Pascual V (1998). Somatic hypermutation introduces insertions and deletions into immunoglobulin V genes. J Exp Med 187, 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbine L, Gennery AR, and Jeggo PA (2014). The clinical impact of deficiency in DNA non-homologous end-joining. DNA Repair (Amst) 16, 84–96. [DOI] [PubMed] [Google Scholar]
- Wuerffel R, Jamieson CE, Morgan L, Merkulov GV, Sen R, and Kenter AL (1992). Switch Recombination Breakpoints are Strickly Correlated with DNA Recognition Motifs for Immunoglobulin Sg3 DNA-binding Proteins. J Exp Med 176, 339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, Alt FW, Cogne M, Pinaud E, and Kenter AL (2007). S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity 27, 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Gorham B, Li SC, Bottaro A, Alt FW, and Rothman P (1993). Replacement of germ-line e promoter by gene targeting alters control of immunoglobulin heavy chain class switching. Proc Natl Acad Sci USA 90, 3705–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos GD, DePinho RA, Zimmerman KA, Lutzker SG, Rosenberg N, and Alt FW (1986). Secondary genomic rearrangement events in pre-B cells: VhDJh replacement by a LINE-1 sequence and directed class switching. EMBO J 5, 3259–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, McBride KM, Hensley S, Lu Y, Chedin F, and Bedford MT (2014). Arginine Methylation Facilitates the Recruitment of TOP3B to Chromatin to Prevent R Loop Accumulation. Mol Cell 53, 484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeap LS, Hwang JK, Du Z, Meyers RM, Meng FL, Jakubauskaite A, Liu M, Mani V, Neuberg D, Kepler TB, et al. (2015). Sequence-Intrinsic Mechanisms that Target AID Mutational Outcomes on Antibody Genes. Cell 163, 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh C-L, Wilson TE, and Lieber MR (2003). R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nature Immunol 4, 442–451. [DOI] [PubMed] [Google Scholar]
- Yu K, Huang FT, and Lieber MR (2004). DNA substrate length and surrounding sequence affect the activation induced deaminase activity at cytidine. J Biol Chem 279, 6496–6500. [DOI] [PubMed] [Google Scholar]
- Yu K, and Lieber MR (2003). Nucleic acid structures and enzymes in the immunoglobulin class switch recombination mechanism. DNA Repair 2, 1163–1174. [DOI] [PubMed] [Google Scholar]
- Yu K, Roy D, Bayramyan M, Haworth IS, and Lieber MR (2005). Fine-structure analysis of activation-induced deaminase accessbility to class switch region R-loops. Mol Cell Biol 25, 1730–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Roy D, Huang FT, and Lieber MR (2006). Detection and structural analysis of R-loops. Methods Enzymol 409, 316–329. [DOI] [PubMed] [Google Scholar]
- Zahn A, Eranki AK, Patenaude AM, Methot SP, Fifield H, Cortizas EM, Foster P, Imai K, Durandy A, Larijani M, et al. (2014). Activation induced deaminase C-terminal domain links DNA breaks to end protection and repair during class switch recombination. Proc Natl Acad Sci U S A 111, E988–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti KJ, and Gearhart PJ (2016). Antibody diversification caused by disrupted mismatch repair and promiscuous DNA polymerases. DNA Repair (Amst) 38, 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrin AA, Tian M, Wang J, Borjeson T, and Alt FW (2005). Influence of switch region length on immunoglobulin class switch recombination. Proc Natl Acad Sci U S A 102, 2466–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Alt FW, and Honjo T (1995). Regulation of class switch recombination of the immunoglobulin heavy chain genes In Immunoglobulin genes, Honjo T, and Alt FW, eds. (London: Academic Press Ltd.). [Google Scholar]
- Zhang ZZ, Pannunzio NR, Han L, Hsieh CL, Yu K, and Lieber MR (2014a). The Strength of an Ig Switch Region Is Determined by Its Ability to Drive R Loop Formation and Its Number of WGCW Sites. Cell Rep 8, 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZZ, Pannunzio NR, Hsieh C-L, Yu K, and Lieber MR (2015). Complexities Due to Single-Stranded RNA During Antibody Detection of Genomic RNA:DNA Hybrids. BMC research notes 8, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZZ, Pannunzio NR, Hsieh CL, Yu K, and Lieber MR (2014b). The role of G-density in switch region repeats for immunoglobulin class switch recombination. Nucleic Acids Res 42, 13186–13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Vuong BQ, Vaidyanathan B, Lin JY, Huang FT, and Chaudhuri J (2015). Non-coding RNA Generated Following Lariat Debranching Mediates Targeting of AID to DNA. Cell 161, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Lee V, Finn A, Senger K, Zarrin AA, Du Pasquier L, and Hsu E (2012). Origin of immunoglobulin isotype switching. Curr Biol 22, 872–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Figure 1. Sterile Transcription Units for Each Acceptor Switch Region. Cytokine-regulated transcription activates S region for CSR. The sterile transcript is spliced to join the I exon and the constant region exons.
Suppl. Figure 2. Switch Region G-Richness. During evolution, mammalian switch regions became increasingly G-rich. The R-loop model is the only proposed explanation for this remarkable G-rich profile.
Suppl. Figure 3 (A-C). Switch Region Sequence and Parameters Driving R-loop Formation. CSR efficiency is in direct proportion to the total number of WGCW sites and the amount of ssDNA. The amount of ssDNA is proportional to the transcription firing rate, RIZ strength and REZ strength. RIZ strength is a function of the total G-cluster number and the G nucleotide number within each G-cluster. REZ strength is dependent on overall G-density, which determines the length of the R-loop. Naturally-occurring switch regions have interspersed G-clusters, WGCW sites, and a high G-density. Hence, for naturally-occurring R-loops in cells, the ssDNA length (R-loop length) varies depending at which G-cluster the R-loop initiates (Roy and Lieber, 2009; Roy et al., 2008; Roy et al., 2010).