Abstract

Subject Categories: Cell Cycle
The Bub1 checkpoint protein is an important regulator of spindle assembly checkpoint (SAC) signaling and chromosome segregation. While Bub1 is essential in a number of model organisms, the essentiality of Bub1 in human cell lines had been questioned by CRISPR/Cas9‐generated knockouts in RPE1 and HAP1 cells (Currie et al, 2018; Raaijmakers et al, 2018). However, we recently reported that these “Bub1 null” cell lines express residual Bub1 protein as detected by mass spectrometry (Zhang et al, 2019). Parallel work from the Jallepalli lab supported these observations and suggested that such residual Bub1 protein might be generated through nonsense‐associated alternative splicing (Rodriguez‐Rodriguez et al, 2018). Consistently, the gRNAs that had been used for generating the RPE1 and HAP1 Bub1 null cell lines target the genomic DNA encoding the N‐terminal region of the Bub1 protein, which is not required for its SAC function.
Given reports that very penetrant removal of Bub1 is required to observe an effect on the SAC, the residual Bub1 protein remaining in the RPE1 and HAP1 Bub1 knockouts might be sufficient for a functional SAC (Meraldi & Sorger, 2005). Indeed, we found that additional Bub1 depletion by RNAi in these “Bub1 null” cell lines reduced SAC strength, as monitored by the length of mitotic arrest in nocodazole. This effect was most obvious in RPE1 cells, while we observed a less dramatic effect in HAP1 cells (Zhang et al, 2019). We in parallel generated a HeLa cell line with reduced levels of Bub1 protein using CRISPR/Cas9 technology and found that depletion of Bub1 by RNAi strongly impaired the checkpoint in this cell line (Zhang et al, 2019). We could fully rescue the Bub1 RNAi phenotype in these HeLa cells with exogenous Bub1, and we detected normal levels of the mitotic checkpoint complex (MCC) at early time points. This would argue against an off‐target effect of the Bub1 RNAi oligonucleotide as cause of the observed SAC defect.
However, these recent observations failed to resolve whether it is possible to generate a full Bub1 deletion in human cell lines. Using two gRNAs to remove almost the entire Bub1 gene in HAP1 cells, Raaijmakers and Medema (2019) have now successfully achieved this. Consistent with this notion, we find here that these new cell lines are no longer sensitive to additional Bub1 depletion by RNAi under our experimental conditions (Fig 1A), in contrast to the original reported HAP1 “Bub1 null” cell line. Attempts by the Medema lab to generate a full Bub1 gene deletion in other human cell lines appear to have been unsuccessful, arguing for the essentiality of Bub1 in several model human cell lines. Our analysis of the new HAP1 cell lines with full Bub1 knockout by quantitative mass spectrometry‐based proteomics of mitotic extracts confirmed the absence of Bub1 protein (Fig 1B–D, Table EV1). This analysis also revealed changes in the abundance of a number of other proteins in the Bub1 full‐KO cell lines, including mitotic regulators. Whether these changes are needed to compensate for the complete lack of Bub1 is unclear.
Figure 1. Analysis of new full Bub1 knock‐out HAP1 cells.
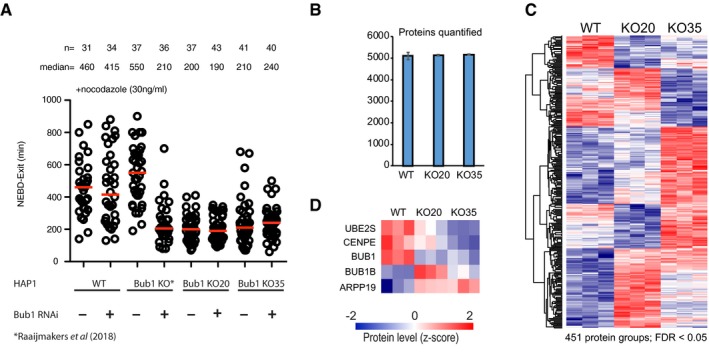
(A) Mitotic timing in nocodazole for the indicated conditions. Cells were treated either with a control RNAi oligonucleotide or a Bub1 RNAi oligonucleotide (as in Zhang et al, 2019) and followed by time‐lapse microscopy following release from a thymidine block. Cells were treated with low dose of nocodazole. Each circle represents a single cell, and cell numbers (n) and median (red bar) are indicated for each condition. (B) Number of quantified protein groups for each cell line in single‐shot LC‐MS analysis. Cell line proteomes were measured in triplicates. Data are mean values from triplicate experiments, and error bars indicate standard deviations. (C) Heatmap of all 451 ANOVA significant protein groups (FDR < 0.05, s0 = 0.1). Unsupervised hierarchical clustering was performed based on Euclidean distance for column and row‐wise clustering. Z‐scored data show relative protein level across cell lines of up‐regulated (red) and down‐regulated (blue) proteins. (D) Selection of mitotic regulatory proteins differentially expressed across cell lines.Data information: See Appendix Supplementary Methods for experimental details.
In summary, the important new result by Raaijmakers and Medema (2019) demonstrates that it is possible to generate a full Bub1 knockout at least in HAP1 cells, but also confirms our recent work that complete loss of Bub1 protein causes a SAC defect upon low‐dose nocodazole treatment (Fig 1A, Raaijmakers & Medema, 2019), consistent with checkpoint strength being graded.
Supporting information
Appendix
Table EV1
Review Process File
Acknowledgements
Work at the Novo Nordisk Foundation Center for Protein Research is supported by grant NNF14CC0001 from the Novo Nordisk Foundation.
The EMBO Journal (2019) 38: e103547
Reply to: JA Raaijmakers & RH Medema (November 2019)
References
- Currie CE, Mora‐Santos M, Smith CA, McAinsh AD, Millar JBA (2018) Bub1 is not essential for the checkpoint response to unattached kinetochores in diploid human cells. Curr Biol 28: R929–R930 [DOI] [PubMed] [Google Scholar]
- Meraldi P, Sorger PK (2005) A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J 24: 1621–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, van Heesbeen RGHP, Blomen VA, Janssen LME, van Diemen F, Brummelkamp TR, Medema RH (2018) BUB1 is essential for the viability of human cells in which the spindle assembly checkpoint is compromised. Cell Rep 22: 1424–1438 [DOI] [PubMed] [Google Scholar]
- Raaijmakers JA, Medema RH (2019) Killing a zombie: a full deletion of the BUB1 gene in HAP1 cells. EMBO J 38: e102423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez‐Rodriguez J‐A, Lewis C, McKinley KL, Sikirzhytski V, Corona J, Maciejowski J, Khodjakov A, Cheeseman IM, Jallepalli PV (2018) Distinct roles of RZZ and Bub1‐KNL1 in mitotic checkpoint signaling and kinetochore expansion. Curr Biol 28: 3422–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Kruse T, Guasch Boldú C, Garvanska DH, Coscia F, Mann M, Barisic M, Nilsson J (2019) Efficient mitotic checkpoint signaling depends on integrated activities of Bub1 and the RZZ complex. EMBO J 38: e100977 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Table EV1
Review Process File


