Abstract
Type IV pili (TFP) are multifunctional micrometer‐long filaments expressed at the surface of many prokaryotes. In Neisseria meningitidis, TFP are crucial for virulence. Indeed, these homopolymers of the major pilin PilE mediate interbacterial aggregation and adhesion to host cells. However, the mechanisms behind these functions remain unclear. Here, we simultaneously determined regions of PilE involved in pilus display, auto‐aggregation, and adhesion by using deep mutational scanning and started mining this extensive functional map. For auto‐aggregation, pili must reach a minimum length to allow pilus–pilus interactions through an electropositive cluster of residues centered around Lys140. For adhesion, results point to a key role for the tip of the pilus. Accordingly, purified pili interacting with host cells initially bind via their tip‐located major pilin and then along their length. Overall, these results identify functional domains of PilE and support a direct role of the major pilin in TFP‐dependent aggregation and adhesion.
Keywords: bacterial adhesion, deep mutational scanning, meningitis, pilin, type IV pilus
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Microbiology, Virology & Host Pathogen Interaction
Introduction
Prokaryotes use a great variety of surface appendages in order to interact with their environment. Type IV filaments (TFF) constitute a class of closely related proteinaceous appendages characterized by a conserved amino‐terminal signature sequence of their major constituent, the class III signal peptide (Berry & Pelicic, 2015). They comprise type IV pili (TFP), the bacterial type II secretion system (T2SS), and the archaellum (archaeal flagellum). Specifically, TFP are micrometer‐long retractile and highly dynamic filamentous appendages found at the surface of several pathogenic bacteria such as Neisseria meningitidis, Neisseria gonorrhoeae, Pseudomonas aeruginosa, and Vibrio cholerae (Berry & Pelicic, 2015). They mediate several functions including competence for transformation (Seifert et al, 1990), interbacterial aggregation (Blake et al, 1989), twitching motility (Merz et al, 2000), and adhesion to host cells (Woods et al, 1980). The multiple functions mediated by TFP place them as central components in the pathogenicity of these bacteria.
N. meningitidis (or meningococcus), used in this study as a model of TFP‐expressing organism, is a diderm bacterium found as a commensal in the human nasopharynx. Occasionally, the bacterium can breach the oropharyngeal epithelial barrier and colonize human blood vessels, eventually leading to sepsis and/or meningitis (van Deuren et al, 2000). In a humanized mouse model, this vascular colonization has been shown to be dependent on the ability of bacteria to adhere to human endothelial cells (Melican et al, 2013), resist shear stress through the reorganization of the host cell plasma membrane (Mikaty et al, 2009), and form fluid aggregates (Bonazzi et al, 2018). All these functions are dependent on the presence of TFP, thus highlighting the importance of this virulence factor in the pathogenesis of Neisseria meningitidis.
The mechanism behind the multiple functions carried by TFP remains unclear, including for adhesion and auto‐aggregation, which are central to infection. Genetic approaches have pointed to the importance of certain Pil proteins such as PilC1, PilV, and PilX. Because pilC1 mutants fail to adhere to host cells while retaining normal piliation levels and aggregation properties, this protein was for a time thought to be an adhesin (Rudel et al, 1995; Morand et al, 2004). PilV and PilX are part of a group of proteins designated as minor pilins as they all share a high degree of homology with PilE but are expressed at much lower levels. Mutants in pilX or pilV genes retain some level of piliation while loosing certain functions (Winther‐Larsen et al, 2001; Helaine et al, 2005; Mikaty et al, 2009; Imhaus & Dumenil, 2014). It has been proposed that these minor pilins could be inserted periodically in the PilE scaffold and would act as effectors, specifically mediating these functions (Helaine et al, 2007; Coureuil et al, 2010). A more recent study proposed an alternative hypothesis in which the reduced piliation levels observed in pilV and pilX mutants, ~70 and ~30% of wild‐type levels, are sufficient to explain their phenotypes (Imhaus & Dumenil, 2014). This hypothesis points back to PilE as the central mediator of TFP‐associated functions. This implies that PilE is under an unusual number of functional constraints for a 17 kDa protein. First, to assemble retractile TFP, PilE must be able to interact reversibly with several members of the piliation machinery and itself (Georgiadou et al, 2012). Second, to support pili bundling and aggregation, PilE must enable interactions between TFP in a parallel and antiparallel fashion. Third, to mediate adhesion, the major pilin needs a region that can interact with components of the host cell surface.
Using a global approach as a starting point, the present study explores the possibility that the pilin itself could directly carry out adhesive and aggregative functions. Saturating mutagenesis approaches have been successfully used in the past in N. gonorrhoeae on PorA and the N‐terminus of PilE (Chen & Seifert, 2014; Obergfell & Seifert, 2016). We decided to extend this approach by taking advantage of the deep mutational scanning method (Fowler & Fields, 2014) and applied it to the full‐length PilE. This provided functional maps of PilE for piliation, aggregation, and adhesion. Mining this extensive dataset and confirming these observations with de novo‐generated mutant now provides novel insights in the mechanisms underlying TFP‐mediated interbacterial aggregation and adhesion to human cells.
Results
Deep mutational scanning of the Neisseria meningitidis major pilin PilE
A library of point mutants in the pilE gene (sequence type pilE SB, Nassif et al, 1993) was produced using error‐prone PCR and introduced into a strain unable to undergo antigenic variation (guanine quartet mutant, Tan et al, 2015). Next‐generation sequencing of the genomic pilE locus of transformed bacteria revealed that single‐point mutations represented 35% of the mutant library and only single mutants were considered for the remainder of the analysis (Fig EV1A–C). Due to the nature of the genetic code, single nucleotide substitutions can theoretically generate four to seven amino acid substitutions. In this context, the library was close to saturation levels, containing 90% of possible amino acid variants (Fig EV1D). On average, every single amino acid of the PilE protein sequence was mutated into 6 alternative amino acids.
Figure EV1. Characterization of the libraries and selection schemes.
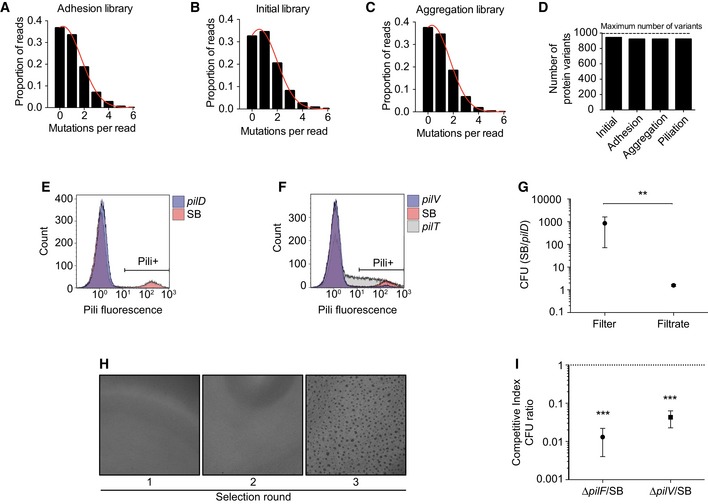
-
A–CDistribution of mutation counts per pilE read in each library assessed by NGS.
-
DNumber of single amino acid protein variants observed in each library relative to the theoretical maximum number of variants potentially obtained with single nucleotide mutations of pilE.
-
E, FPiliation selection. Flow cytometry analysis of pili expression using the 20D9 monoclonal antibody. The Pili+ gate was used to separate piliated bacteria in the wild‐type SB strain (10.5%), pilD (0.1%), pilV (3%), and pilT (28%).
-
G, HAggregation selection. (G) Ratio between the colony forming units of pilE SB and pilD after passage of a 1:1 mixture through a 5‐μm pore filters. Mean ratio ± SEM is indicated. N ≥ 3 independent experiments. Paired t‐test. P < 0.01 (**). (H) Brightfield images of aggregates after different rounds of selection.
-
IRatio between the colony‐forming units of pilE SB over pilF or pilV mutants after passage of a 1:1 mixture on HUVEC cells for 4 h of infection (MOI 100). Mean ratio ± SEM is indicated. N ≥ 3 independent experiments. Paired t‐test. P < 0.001 (***).
This initial library was then submitted to three different carefully validated selection schemes to independently assess surface piliation, auto‐aggregation, and adhesion to host cells (Figs 1A and EV1E–I). For the piliation selection, live bacteria were immunolabeled with the 20D9 anti‐pilus monoclonal antibody (Pujol et al, 1999) and sorted by FACS (Fluorescence‐Activated Cell Sorting). For the aggregation selection, bacterial suspensions from the initial mutant library were allowed to aggregate for 2 h prior to filtration through transwells with 5‐μm pores. Finally, for the adhesion selection, human umbilical vein endothelial cells (HUVEC) were infected with the initial mutant library for 4 h and washed extensively to only retain adherent bacteria. Using next‐generation sequencing, the frequency of each single‐point mutation in the three selected libraries was compared to the frequency of the same mutation in the initial library. The log2 of this ratio is indicative of whether mutations are beneficial or detrimental to the function of interest and was termed mutation score (Fig 1A). Overall, 1,147 different point mutations distributed over the whole PilE sequence were associated with three functional quantitative measurements, generating 3,441 data points.
Figure 1. Deep mutational scanning of PilE.
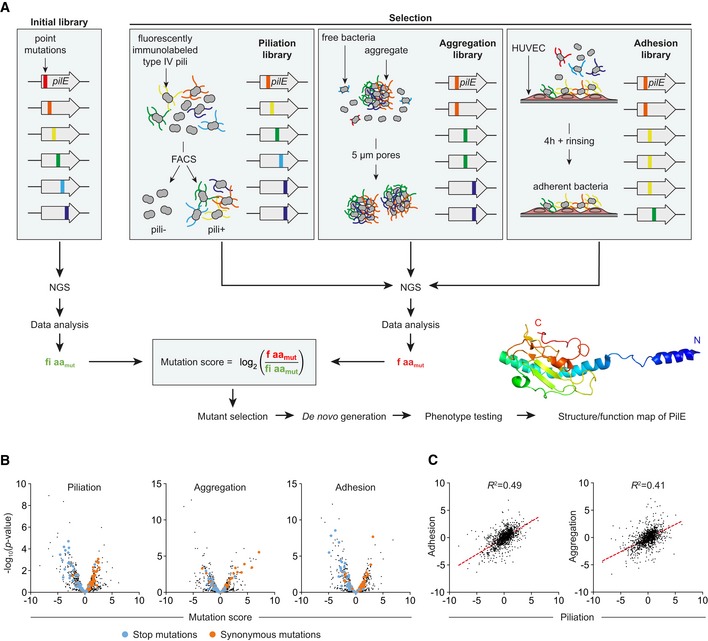
- Representation of the overall work flow used in the study with the three selection schemes of pilE mutants and subsequent NGS‐based analysis.
- Volcano plots of piliation, aggregation, and adhesion mutation scores for all pilE mutations. Stop and synonymous mutations are highlighted in blue and orange, respectively.
- Adhesion and aggregation mutation scores relative to piliation. The correlation appears as a red dotted line.
Mutation scores of the stop and synonymous mutations were used to control the validity of the resulting functional mapping. Stop mutations are expected to induce a complete loss of piliation and function, while synonymous mutations should be indistinguishable from the reference strain. Indeed, the majority of stop mutations were negatively selected as shown by their distribution at the left of the volcano plots for the three functions (Fig 1B, blue dots). Conversely, synonymous mutations were distributed evenly around the 0 value (orange dots). Overall, while a large number of mutations affect piliation and associated functions, it is also worth noting that a significant number of mutations have no significant effect or a beneficial effect on pilus expression and functions. This likely reflects the robustness of the piliation system regarding sequence variations. We also observed that the mutation scores for each function were correlated to the mutation scores for piliation (Fig 1C). This is consistent with the observation that piliation is a good predictor of function (Imhaus & Dumenil, 2014) and further validates the strategy.
Mutations in the hyperconserved N‐terminal region of the pilin reveal a class of mutants with numerous but short pili that fail to aggregate
Consequently, we first sought mutations that would affect auto‐aggregation but not piliation. To tease out such mutants, we compared frequencies between the aggregation library and the piliation library (Fig 2A). Mutations altering auto‐aggregation but not piliation could be found all along the sequence (Fig 2A). Mutations in the N‐terminal part of the pilin are expected to have strong effects on the piliation level since this conserved region is buried in the core of the pilus structure (Kolappan et al, 2016), but mutants with low aggregation and normal piliation levels are unexpected. We thus first focused on the N‐terminal part of the protein and undertook a detailed analysis of the piliation of a subset of mutants in the α1N region with such properties.
Figure 2. Implication of the amino‐terminal region in aggregation.
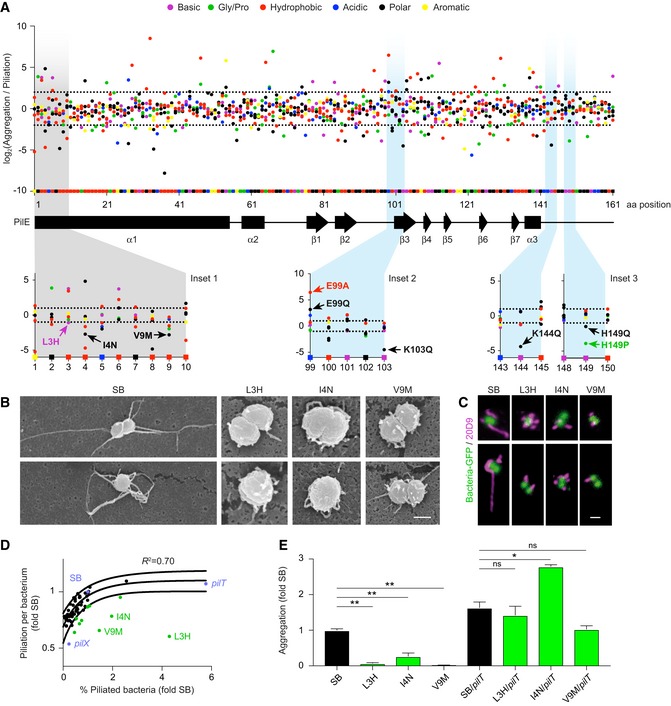
- The level of aggregation relative to piliation level is represented as the Log2 of the ratio between the frequency of each single‐point mutation in the aggregation library relative to the frequency of the same mutation in the piliation library. Each dot represents a point mutant. Mutation type is color‐coded as indicated in the legend. Squares on the x‐axis correspond to the original amino acids in the SB sequence with the protein secondary structure indicated below. Dotted lines represent a two‐fold threshold value. Bottom insets illustrate the mutations generated de novo and further analyzed in Figs 2 and 3.
- Scanning electron microscopy of single bacteria on a cellulose filter. Scale bar: 500 nm.
- Two representative immunofluorescence labeling of TFP. Scale bar: 1 μm.
- Piliation per bacterium as a function of the proportion of piliated bacteria normalized to pilE SB values as measured by flow cytometry using the 20D9 monoclonal antibody. Black dots indicate all the individual de novo‐generated PilE mutants that were characterized. Blue dots indicate mutants of the piliation machinery and green dots mutants in the α1N region. Solid lines indicate 95% confidence interval.
- Quantification of aggregation normalized to pilE SB for a subset of outliers and corresponding pilT double mutants. Mean values ± SEM are indicated for each strain. N ≥ 3 independent experiments. Paired t‐test. P < 0.05 (*), P < 0.01 (**).
Neisseria meningitidis pilE L3H, pilE I4N, and pilE V9M mutants were generated de novo and further studied (Fig 2A, inset 1). Piliation levels of these mutants were first characterized using scanning electron microscopy and immunofluorescence on single bacteria (Fig 2B and C). While the wild‐type pilE SB strain displayed a few long pili as expected, pilE L3H, pilE I4N, and pilE V9M only expressed numerous very short pili. The frequency of piliated bacteria also seemed increased compared to the wild‐type strain. These observations were then confirmed and quantified using flow cytometry (Fig 2D). Analysis of the piliation level of the pilE SB reference strain revealed that, in the conditions used here, only around 7% of the population is piliated (Fig EV2A–D). Interestingly, conducting this analysis on a large set of de novo‐generated point mutations as well as the underpiliated pilX and the hyperpiliated pilT mutants showed a good correlation between the level of piliation per bacterium assessed by flow cytometry and the percentage of piliated bacteria (Fig 2D). In contrast, the pilE L3H, pilE I4N, and pilE V9M mutants were outliers in this distribution with low piliation per bacterium but with a higher percentage of piliated bacteria, reaching an average of 30% in the case of pilE L3H (Fig 2D, in green). This phenotype did not result from a change in PilE expression levels as detected by Western blot (Fig EV2D). Detailed analysis of the piliation of three mutants in the α1N region of PilE thus reveals that they form a particular class of mutants with a larger proportion of piliated bacteria with short pili.
Figure EV2. Piliation characterization.
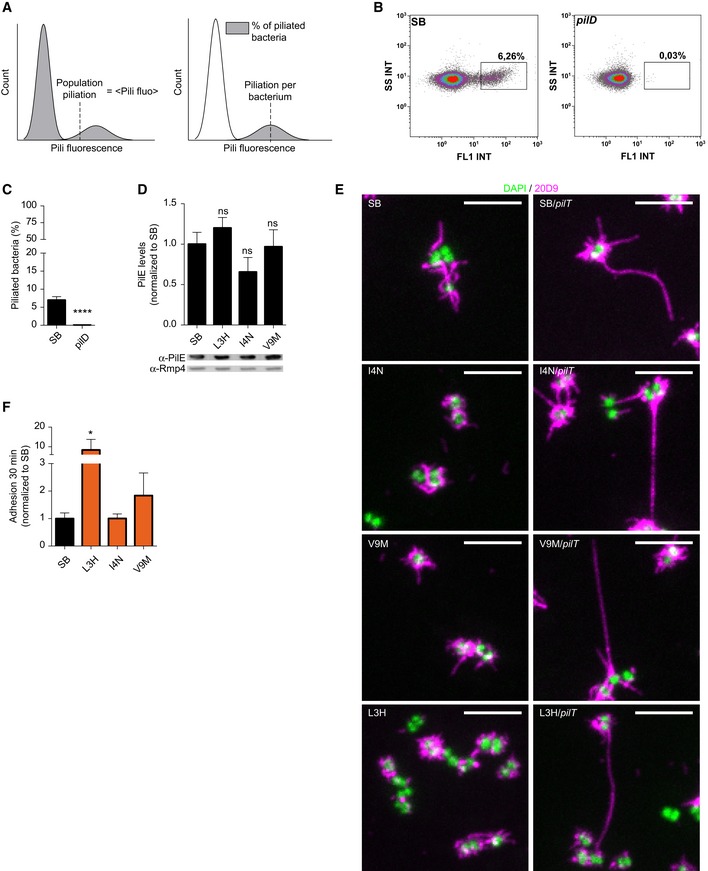
- Illustration of the three different parameters that can be extracted from analysis of piliation by flow cytometry, population piliation, piliation per bacterium, and percentage of piliated bacteria.
- Flow cytometry scatter plot (side scatter intensity as a function of pili fluorescence intensity) of pilE SB and pilD stained with the 20D9 antibody. Piliated bacteria are located in the rectangular gate.
- Percentage of piliated bacteria in the total population detected by flow cytometry in pilE SB and pilD strains. Mean ratio ± SEM is indicated. N = 7 independent experiments. Paired t‐test, P < 0.001 (***).
- Quantification of pilin expression by Western blot on whole bacterial lysates using polyclonal α‐PilE and α‐Rmp4 antibodies. PilE levels were normalized using Rmp4, and values are normalized to pilE SB values. Mean ratio ± SEM is indicated. N = 3 independent experiments. Paired t‐test.
- Representative immunofluorescence images of pilE mutants and corresponding pilT double mutants using the 20D9 monoclonal antibody; pili in magenta; Hoechst in green. Scale bar: 5 μm.
- Quantification of adhesion after infection of HUVEC for 30 min (MOI 500) by microscopy, expressed with respect to pilE SB. Mean values ± SEM are indicated for each strain. N ≥ 3 independent experiments. Paired t‐test, P < 0.05 (*).
Source data are available online for this figure.
As expected from the mutational scanning, these de novo‐generated mutants with short pili showed a near‐complete loss of ability to form aggregates (Fig 2E). Mutating the pilT retraction ATPase was sufficient to increase piliation of individual bacteria (Fig EV2E) and restore aggregation in mutants with short pili (Fig 2E). This indicates that mutations in these strains do not affect the intrinsic ability of the pili to form pili–pili interactions, and rather suggests that in these strains, pili are too short to form stable interactions. Interestingly, mutants with short but numerous pili not only had no defect in early adhesion, but even had better adhesive properties thus providing further evidence for the functionality of these pili (Fig EV2F). Together, these results support the idea that pilus length is a critical determinant of aggregation and highlight the importance of the N‐terminal region of PilE in the balance between pilus length and pilus number.
A patch of charged amino acids surrounding K140 in the C‐terminal part of PilE is necessary for aggregation
A second region of interest regarding auto‐aggregation is the globular head domain located at the C‐terminal end of the pilin. A face of this domain is surface‐exposed and has been suggested to be involved in direct pilus–pilus interactions (Kolappan et al, 2016). Since previous studies have indicated the importance of electrostatic interactions between pili (Chamot‐Rooke et al, 2011), we focused on charged amino acids in this area of the protein (Fig 2A, insets 2 and 3). Mutations in three basic residues (K103, K144, and H149) result in a decrease in aggregation, while a mutation in one acidic residue (E99) leads to a strong increase in aggregation. Strikingly, these four amino acids delineate a cavity in the pilus structure, which is overhung by the bulky K140 (Fig 3A). We therefore focused on this patch and generated de novo several mutations around K140. Since we suspected that this region might be the epitope recognized by the 20D9 monoclonal antibody used for piliation quantification in the screening strategy, we also used an alternative antibody directed against TFP (F10 nanobody) to assess the piliation levels of these mutants (Charles‐Orszag et al, 2018). While there was an excellent agreement between the measurements made with 20D9 and F10 (R 2 = 0.94), we found two outliers in this distribution, pilE Q122E and pilE K140Q (Fig 3B), showing that these two amino acids are important constituents of the 20D9 epitope and that these two mutants present unaffected piliation levels. This observation was confirmed by immunofluorescence (Fig 3C).
Figure 3. Carboxy‐terminal region of the pilin involved in aggregation.

- Pilus structure (PDB: 5KUA) showing a cluster of charged amino acids surrounding the protruding K140 involved in aggregation. N138 is in light blue; Q122 and K140 in orange; and E99, K103, K144, and H149 in dark blue.
- Piliation per bacterium as measured by flow cytometry using the 20D9 monoclonal antibody and the F10 nanobody.
- Visualization of type IV pili by immunofluorescence either using the 20D9 antibody or the F10 nanobody as indicated (magenta). In the case of the K140Q mutant labeled with the 20D9 antibody, the background was made visible to convincingly show the absence of pilus labeling. Scale bar: 2 μm.
- Aggregation expressed as a function of piliation per bacterium measured with 20D9 antibody except for pilE Q122E and pilE K140Q for which the F10 nanobody value is reported. Each dot represents values for a PilE point mutant relative to the SB strain. Hypo‐aggregative mutants are highlighted in orange, while hyper‐aggregative mutants are in light blue. Solid lines indicate the 95% confidence interval.
- Quantification of PilX present in purified pili preparations sheared from the bacterial surface using Western blot. PilX levels were normalized using PilE as a reference and normalized to pilE SB values. Mean ratio ± SEM is indicated. N = 3 independent experiments. Paired t‐test.
We next characterized the auto‐aggregation abilities of the mutants in the K140 region. Below a certain piliation threshold, aggregation was lost (Fig 3D) as previously observed in the case of the pilX mutant (Imhaus & Dumenil, 2014). Above this threshold, aggregation increases progressively and is correlated with the piliation level. In contrast, mutations located in the K140 region formed two outlying groups: one group with lower aggregative properties (Fig 3D, indicated in orange) and a second with higher aggregation levels (Fig 3D, indicated in blue). This supports the notion that local charge and organization in this region are crucial for aggregation and further indicates a central role of the positive charge of K140 in aggregation. In addition, certain mutants with higher aggregation suggest that the presence of hydrophobic residues on the pilus surface could favor aggregation. Since PilX was described as a direct pilus‐associated pro‐aggregative factor in some studies (Helaine et al, 2005, 2007), the possibility that mutations in the K140 region might affect the association of PilX with pili was tested. Pili were purified by standard methods based on shearing in basic conditions followed by ammonium sulfate precipitation (Chamot‐Rooke et al, 2007) and the levels of associated proteins evaluated by Western blot. Mutations in the K140 area did not affect PilX copurification (Fig 3E). In addition, this mutation did not alter pilin posttranslational modifications (Table 1). Therefore, the electropositive region centered around K140 in the major pilin is essential for TFP‐mediated aggregation.
Table 1.
Characterization of non‐aggregative pili by mass spectrometry
| Pilin | PTM | M theo. | M exp. | Delta M (ppm) | Relative abundance |
|---|---|---|---|---|---|
| PilESB | Me × 1; GATDH × 1; PG × 1; Disulfide bond × 1 | 17,479.82 | 17,479.98 | −9.3 | 100 |
| Me × 1; GATDH × 1; PG × 2; Disulfide bond × 1 | 17,633.83 | 17,633.95 | −7.0 | 86 | |
| PilEK140Q | Me × 1; GATDH × 1; PG × 1; Disulfide bond × 1 | 17,479.79 | 17,479.95 | −9.2 | 100 |
| Me × 1; GATDH × 1; PG × 2; Disulfide bond × 1 | 17,633.79 | 17,633.92 | −7.6 | 38 | |
| PilEQ122E | Me × 1; × 1; PG × 1; Disulfide bond × 1 | 17,480.81 | 17,480.93 | −7.1 | 100 |
| Me × 1; GATGATDH DH × 1; PG × 2; Disulfide bond × 1 | 17,634.81 | 17,634.94 | −7.3 | 76 |
PilE proteins from the reference strain and two non‐aggregative mutants (pilE K140Q and pilE Q122E) were analyzed by liquid chromatography coupled to high‐resolution mass spectrometry. Each table summarizes the results obtained for each sample in terms of molecular weight including posttranslational modification composition. M theo is the theoretical mass; M exp, the mass measured experimentally; Me, methyl; PG, phosphoglycerol; GATDH, glyceramido‐acetamido trideoxyhexose; DeltaM, the difference between theoretical mass and experimental mass expressed in parts per million relative to theoretical mass; Relative abundance (relative intensity). The most abundant ion in the deconvolved spectrum has a relative abundance of 100%, and the relative abundances of all other peaks of the same spectrum are calculated relative to that one (definition from Proteome Deconvolution 3.0, Thermo Scientific).
Mutations specifically affecting adhesion but not aggregation are found at the tip of the pilus
We then analyzed the results of the adhesion library. Various studies have found that the ability of Neisseria meningitidis to adhere to human cells is tightly linked to their ability to form aggregates, presumably by allowing the accumulation of bacteria in three dimensions at the site of adhesion (Helaine et al, 2005). A central objective of this section was to explore whether regions of PilE specifically involved in adhesion could be identified. The ratio of the mutation score for adhesion over the mutation score for aggregation was thus determined for each mutation (Fig 4A). Averaging the adhesion/aggregation ratio over a window of five consecutive amino acids (magenta line) highlighted two regions with a severe defect in adhesion centered around amino acids Y50 and A131. Interestingly, these amino acids are located in regions of the pilin monomer found at the tip of the pilus (heat map). In particular, the conserved region surrounding Y50 is part of a EYYLN motif recognized by the SM1 monoclonal antibody and has been shown to only be exposed at the tip of the pilus when these are not under tension (Biais et al, 2010; Brissac et al, 2012).
Figure 4. Residues of the pilin localized toward the distal end of pili are involved in early adhesion.
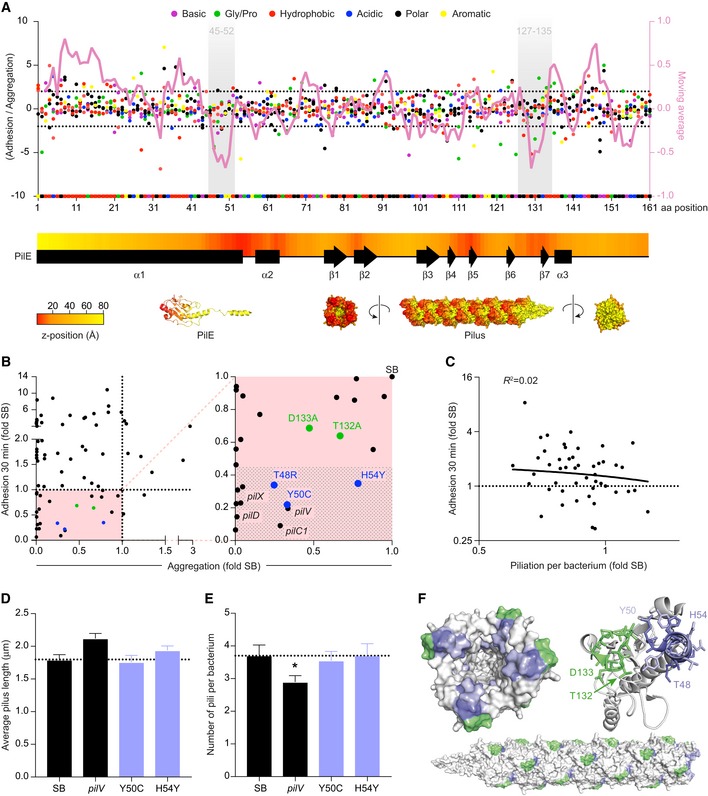
- The ratio between the adhesion and aggregation mutation scores for each amino acid in the PilE sequence is represented as dots. Mutation type is color‐coded as indicated in the legend. The superimposed magenta line represents the average ratio averaged over a moving window of 5 amino acids (right axis). Shaded areas highlight the two regions identified as important for adhesion. The yellow to red heat map along the pilin sequence indicates the axial position of the amino acid in PilE as shown on the structure of PilE (distal tip in red, proximal tip in yellow).
- Early adhesion of pilE mutants as a function of aggregation. Values were normalized to that of pilE SB. Each dot represents a pilE mutant and the pilV, pilD, pilX, and pilC1 reference mutants. Mutants in the region surrounding A131 are in green and those surrounding Y50 in purple. A blown‐up view of the zone of interest (lower left quarter) is presented on the right panel. The shaded area highlights the absence of significant adhesion.
- Early adhesion of PilE mutants as a function of piliation per bacterium normalized to that of pilE SB.
- Piliation length of a selection of mutants determined by immunofluorescence. Average pilus length ± SEM is indicated. N = 6 experiments with at least 50 bacteria analyzed in each experiment. Paired t‐test.
- Number of pili per bacteria ± SEM determined as in (E). Paired t‐test. P < 0.05 (*).
- Amino acids from the two regions identified in panel (A) are highlighted in purple and green. The SM1 epitope is colored in light purple.
To confirm these observations, mutants in these two regions were generated de novo and their ability to adhere to human endothelial cells was assessed at 30 min after initial contact. At this early time point, the contribution of aggregation is negligible and the adhesion of individual bacteria remains dependent on the presence of TFP at the bacterial surface as well as on accessory proteins such as PilC1, PilV, and PilX (Fig EV3A–F). Plotting early adhesion levels as a function of aggregation levels highlights mutations with a specific defect in adhesion (Fig 4B). Most interestingly, mutation H54Y showed a strong defect in adhesion while maintaining a near‐normal aggregation. Single amino acid mutations in PilE are sufficient to induce a loss of adhesion, thereby exhibiting more specific phenotypes than the pilV and pilC1 deletion mutants. Importantly, these phenotypes were not due to alterations in pili number and length (Figs 4D and E, and EV4A). In addition, the amount of PilV associated with purified pilus fractions was not affected by these mutations (Fig EV4B and C).
Figure EV3. Characterization of early adhesion.
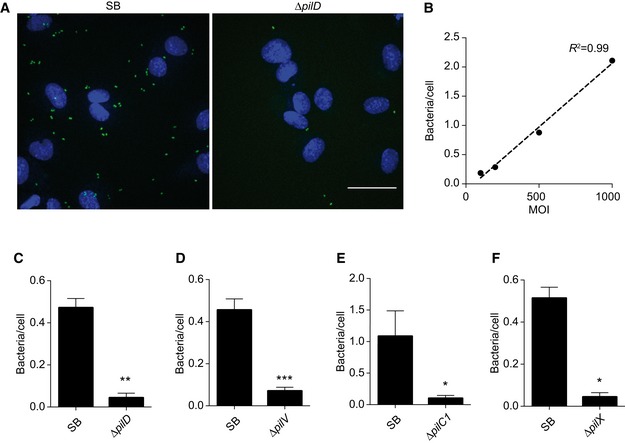
-
AZ‐projections of images captured after 30 min of infection. Adherent GFP‐expressing bacteria are in green and nuclei stained with Hoechst in blue. Scale bar: 50 μm.
-
BNumber of adherent bacteria per cell following 30 min of infection as a function of the MOI. Dotted line: linear fit of the data.
-
C–FQuantification of adhesion of the indicated mutants after infection of HUVEC cells for 30 min, expressed as number of adherent bacteria over the number of cell nuclei. Mean value ± SEM is indicated. N ≥ 3 independent experiments. Paired t‐test. P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***).
Figure EV4. Characterization of non‐adherent mutants.
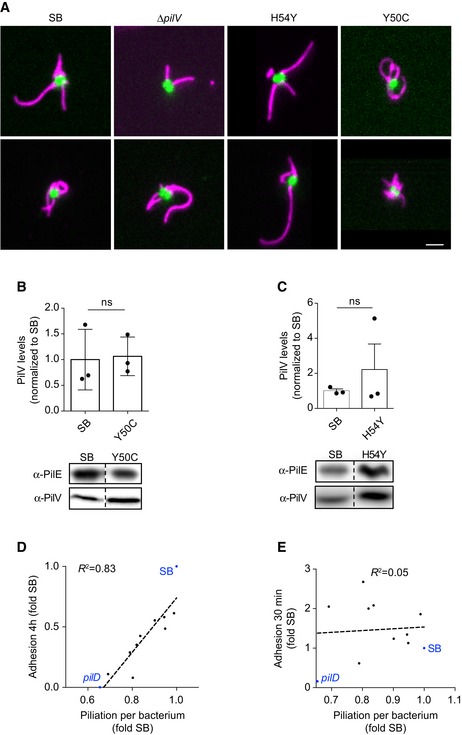
-
ATwo representative immunofluorescence images of pilE mutants using the 20D9 monoclonal antibody; pili in magenta; GFP in green. Scale bar: 1 μm.
-
B, CTop: Quantification of α‐PilV and α‐PilE blots. PilV levels were normalized using PilE as a reference and values were then normalized to pilE SB values. Mean ratio ± SEM is indicated. N = 3 independent experiments. Paired t‐test. Bottom: Representative images of Western blot membranes from purified pili preparations. Blots were probed for PilE or PilV as indicated. Dotted lines indicate that the image was cropped, but both lanes come from a single membrane.
-
D, EThe same set of mutants is presented on both graphs. Each dot represents a pilE mutant unless otherwise indicated. The dotted line corresponds to a linear fit of the data. (D) 4‐h adhesion to HUVEC cells as a function of piliation per bacterium. Values are normalized to that of pilE SB. (E) 30‐min adhesion to HUVEC cells as a function of piliation per bacterium (measured using the 20D9 antibody). Values were normalized to that of GFP‐expressing pilE SB. N ≥ 3 independent experiments.
Source data are available online for this figure.
Under the hypothesis of a pilus tip‐mediated adhesion, a prediction would be that pilus number (i.e., number of pilus tips) is more important than pilus length to promote efficient early adhesion. The piliation level per bacterium as determined by flow cytometry, which is mostly related to pilus length, would thus not correlate with early adhesion. Accordingly, we could not find any significant correlation between the piliation level per bacterium and early adhesion in the mutants characterized in this study (Fig 4C). These results are in contrast to what is observed at 4 h postinfection where aggregation, and thus pilus length, comes into play (Fig EV4D and E).
Overall, this mutational analysis points to the possibility of a tip‐mediated adhesion mechanism. At the structural level, amino acids that affect initial adhesion upon mutation form a patch at the pilus tip (Fig 4F).
TFP adhere first by their tip and then along their side
The previous findings prompted us to examine microscopically whether tip‐mediated adhesion could be visualized directly. Shear generated crude pili preparations were incubated with cells, washed, labeled with a fluorescent nanobody, and imaged by spinning‐disk confocal microscopy. With this approach, numerous pili bound to the surface of endothelial cells could be visualized. Observation of different focal planes (z1‐3) provided a three‐dimensional view showing rigid 1‐ to 2‐μm‐long pili seemingly attached from one end of the pilus and standing away from the cellular surface (Fig 5A and B). Accordingly, mechanical constraints generated by liquid flow led to the bending of pili toward cell surface in the direction of the flow (Fig 5A and B; Movie EV1). Subsequently, the ability of pili to bind along their length was evaluated with increasing flow rates. With shear stress levels below ~5 dyn cm−2, the bending of pili over the cellular surface was reversible and pili moved back to a vertical position when flow was interrupted (Fig 5B and C). In contrast, when flow was applied at a shear stress above ~5 dyn cm−2, pili remained adherent along the cellular surface after the flow was interrupted (Fig 5B and C). This result is in favor of a scenario where pili initially bind to cells by an extremity, tend to remain in a vertical position and when constrained can bind along their side.
Figure 5. Interaction of purified type IV pili with cells.
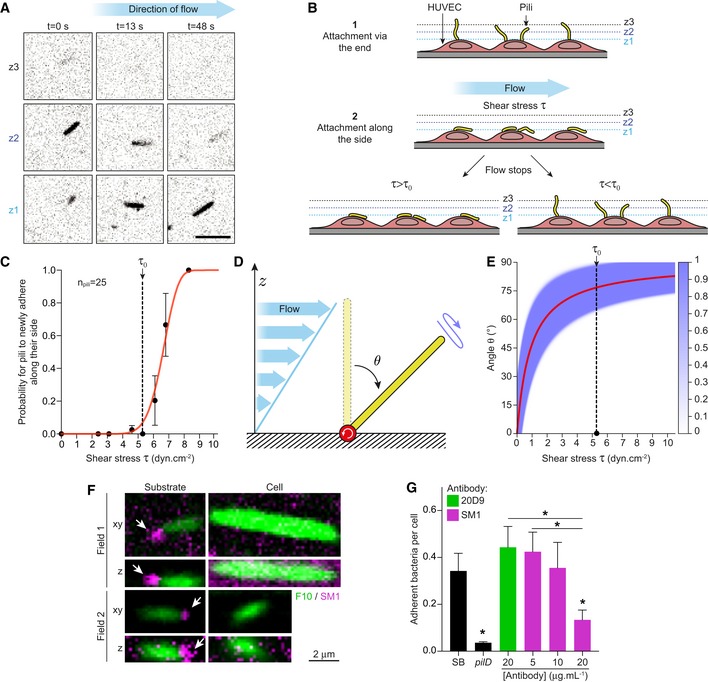
- Time‐lapse of a cell‐adhered pilus fluorescently labeled with the F10 nanobody at three different focal planes. Initiation of flow corresponds to t = 0. Scale bar: 1 μm. The video presented in Movie EV1.
- Schematic representation of the experimental setup. Changes in pili position were visualized by imaging different confocal planes (z1‐3) using spinning‐disk confocal microscopy.
- Quantification of side adhesion of pili to HUVEC as a function of the shear stress applied. Cell‐adhered pili were subjected to gradually increasing flow rates. Each increase was followed by flow arrest and the number of pili adherent along their sides determined. The probability for pili to newly adhere along their side at the i th flow was then calculated as , where N i is the number of nonadherent pili after the application of the i th flow. N = 3 independent experiments. The red curve corresponds to the fit based on the physical model described in the Appendix. Error bars correspond to mean ± SEM.
- Schematic representation of the principles of the physical model of a pilus adhering via its tip submitted to shear stress. θ is the angle relative to the vertical position that fluctuates around the equilibrium angle θ0 due to thermal fluctuations (blue arrow).
- Probability for the pilus to reach the angle θ during the 30‐s application of shear stress τ as determined by the physical model fitted to the data in panel (C). The red line is the equilibrium angle θ0 and the purple shading the thermal fluctuations.
- Spinning‐disk confocal immunofluorescence of pili stained with both SM1 (magenta) and F10 (green). The thick appearance of TFP is due to the primary and secondary reagents used to detect them as well as diffraction of light. In each field of view, a substrate‐ and a cell‐bound pilus are shown. Upper panels: z‐projection from the stacks in the xy plane; bottom panels: side view in the z plane. Scale bar: 2 μm. White arrows correspond to SM1 staining at the tip of pili.
- Quantification of adhesion after infection of HUVEC cells for 30 min, expressed as the number of adherent bacteria over the number of cell nuclei. Prior to infection, bacteria were incubated at 37°C with or without antibody or nanobody for 15 min. Mean values ± SEM are indicated for each strain. N = 5 independent experiments. Paired t‐test. P < 0.05 (*).
The occurrence of adhesion along the length of pili following application of liquid flow could be in favor of force‐dependent conformational changes that would expose additional epitopes on the pilus length (Biais et al, 2010; Lu et al, 2015). Yet, forces exerted on pili in our experimental setup were evaluated (Fig 5D and Appendix) to range between 1 and 4 pN. Such forces would probably not be sufficient to induce the previously described conformational changes that have been estimated to require forces in the tens to hundreds of pN (Beaussart et al, 2014; Lu et al, 2015). Microscopic observations of the behavior of adherent pili in flow rather suggest that their vertical position is imposed by a restoring torque. To explore whether this explanation is realistic, a physical model was elaborated based on the following assumptions: (i) A 2‐μm‐long pilus can be modeled as a rigid slender rod since their length is largely below their 5 μm persistence length (Skerker & Berg, 2001); (ii) the rigid rod is considered attached to a plane with a restoring torque bringing it back to its vertical position, and (iii) the pilus rod is under thermal fluctuations (Fig 5D). In this model, increasing shear stress results in the pilus leaning away from its vertical position toward the cell surface (Fig 5E and Appendix). Thermal fluctuations drive the pilus to explore around the equilibrium angle where shear stress and the restoring torque compensate each other, which leads to stochastic adhesion of the pilus along its length. This was sufficient to explain the experimental observations and a single parameter fit reproduced the behavior of pili under flow including the sharp change between reversion to vertical position and adhesion along the pilus length (Fig 5C, red curve), supporting the idea that the adhesion along pilus length is a secondary phenomenon following the initial tip adhesion.
In principle, the purified pili used in this experiment could be interacting with cells through their distal tip or through their hydrophobic proximal base. To distinguish between these two situations, we used the SM1 monoclonal antibody, which specifically labels the distal tip of the pilus. Indeed, pili adhering on a glass surface only showed labeling by the SM1 antibody at one extremity of the fiber: the distal tip (Fig 5F, arrows). In contrast, on the same fields of view, pili adhered to cells did not display any labeling by the SM1 antibody, suggesting that TFP binding to human endothelial cells masks their distal tip.
Finally, we reasoned that addition of the SM1 antibody to bacteria prior to adhesion, should block the tip of the pilus, and prevent adhesion to host cells. Bacteria were incubated with the SM1 antibody for 15 min prior to adhesion. A dose‐dependent inhibition of early adhesion could be observed reaching statistical significance at 20 μg ml−1 antibody (Fig 5G). Incubation of bacteria with the same amount of the 20D9 monoclonal antibody, which binds along the side of the pili, had no effect on early adhesion, confirming that the effect of the SM1 antibody is not due to lateral binding along the pilus. These data demonstrate that adhesion to human cells is initiated at the tip of the pilus. Importantly, because the SM1 antibody was raised against a sequence present only in the pilin protein, these results strongly support the idea that PilE itself promotes initial adhesion at the tip.
Discussion
Studying TFP functions presents specific challenges, in particular due to their multiple interrelated functions and their highly dynamic nature. In this study, we simultaneously explored three TFP functions in an original and integrated fashion.
A first outcome of this study is the identification of regions of the major pilin involved in bacterial auto‐aggregation (Fig 6A–C), a key property that promotes an efficient vascular colonization (Bonazzi et al, 2018). Because of the tight link between piliation levels and aggregation, the introduction of a cytometry‐based technique to quantify piliation was critical to this investigation. This approach first led to an unexpected observation related to the amino‐terminal α1N region of PilE. Mutations in this region lead to a particular class of mutants with numerous but short pili that are deficient for aggregation (Fig 6B). These observations suggest that: (i) a minimum pilus length is necessary to enable aggregation and (ii) the α1N region is involved in the balance between pilus length and number. The high conservation of this region throughout diverse type IV filaments will provide means to test this hypothesis in various systems. Accordingly, similar mutations in the α1N region of the major pilin of P. aeruginosa and N. gonorrhoeae have also been shown to lead to defects in microcolony formation (Chiang et al, 1995; Park et al, 2001). Several mechanisms could account for this new role of the amino‐terminus of PilE. This region has been proposed to be involved in regulation of self‐expression of the major pilin PilA in P. aeruginosa (Kilmury & Burrows, 2016), but it is not the mechanism at play here. We would favor a hypothesis where mutations in the N‐terminal part of the protein affect interaction with the piliation machinery. Indeed, in N. meningitidis, PilE has been shown to interact with several components of the machinery (PilG, PilN, and PilO) at least partially through its N‐terminal domain (Georgiadou et al, 2012). These interactions of the major pilin with the machinery are probably critical for the initiation of pilus assembly, which would then determine the dynamics of extension and retraction.
Figure 6. Regions of the major pilin involved in the multiple functions of TFP.
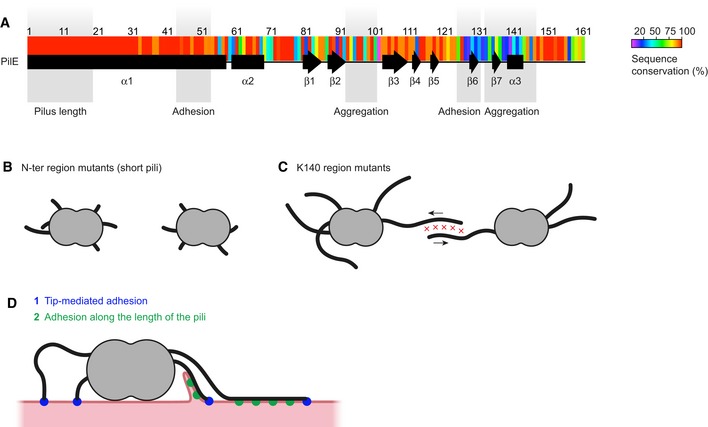
- Sequence conservation determined from the BIGSdb database of Neisseria meningitidis sequences is represented as a heat map (Jolley & Maiden, 2010). Regions of the protein involved in different functions as defined by this study are indicated as shaded areas.
- A category of mutants in the α1N amino‐terminal region of the pilin express numerous but short pili and fail to auto‐aggregate.
- Pilus–pilus interactions initiating bacterial auto‐aggregation fail to occur in mutations affecting the area around K140. Arrows indicate pilus retraction.
- When in contact with cells, pili can interact through their tips (1) and then along their sides (2). Adhesion of the plasma membrane along the side of the pili mediates one‐dimensional wetting and membrane protrusion formation (Charles‐Orszag et al, 2018).
A second region of PilE required for aggregation is located on the pilus surface around K140. The importance of K140 for pilus bundling and bacterial adhesion had been suggested based on sequence comparison of hyper‐ and hypo‐adherent variants (Marceau et al, 1995) and more recently by structural data (Kolappan et al, 2016) where it appears as a hook‐like structure protruding from the pilus, perhaps allowing the interaction with another pilus. In addition, the limited sequence conservation of these residues among N. meningitidis strains (Fig 6A) could suggest that different structural determinants in this region could participate in aggregation in different strains. The determinants of physiological or pathological protein aggregation have been studied intensively for instance in the context of amyloid fiber formation (De Baets et al, 2014). These studies reveal the presence of aggregation‐prone regions composed of 5–15 amino acids that tend to auto‐aggregate. Similar properties could explain how the major pilin can exhibit a high level of sequence diversity while retaining aggregative properties. More structural and biochemical work on these complex multimeric structures is required to further decipher the molecular mechanism of pilus–pilus interactions.
This study provides several arguments in favor of an adhesion process initiated at the tip of TFP (Fig 6D). First, global mapping of adhesion obtained through deep mutational scanning points to the tip as a key element in early adhesion. Second, early adhesion of pilin mutants in regions exposed at the pilus tip generated de novo shows decreased adhesion to human cells. Third, initial adhesion of bacterial cells is inhibited by the SM1 monoclonal antibody, which preferentially binds the tip of the pilus. This monoclonal antibody also binds along the pilus length when pili are under tension, but this is unlikely to have an impact as another monoclonal antibody (20D9), which binds along the pilus length does not inhibit adhesion. Finally, direct microscopic visualization of the adhesion of purified pili to human cells shows that they bind cells via their distal tip. Interestingly, a tip‐mediated mechanism of binding has previously been reported in P. aeruginosa by imaging fluorescently labeled pili of live bacteria adhering to quartz (Skerker & Berg, 2001; Tala et al, 2019).
This also suggests that the pilin itself would be located at the pilus tip and would interact with host cells. The ability of the SM1 antibody directed against a major pilin epitope to inhibit initial bacterial adhesion is in favor of this scenario. The exact nature of the pilus tip is currently unclear, but this result is in contradiction with studies indicating that other proteins such as minor pilins associate with pili and could potentially form the pilus tip. The existence of a structurally distinct pilus tip is well characterized for other pilus type such as the chaperone‐usher pilus present in uropathogenic E. coli (Hospenthal & Waksman, 2019). Electron microscopy images of type I pili show a thin tip fibrillum attached to the pilus itself. This tip contains the adhesin. In type IV pili, however, no identifiable structure has ever been visualized at the tip, thus providing little information. In the case of the Neisseria spp., the PilC1 protein was initially placed at the pilus tip although later studies were in favor of a location in the outer membrane (Rudel et al, 1995; Morand et al, 2004). Nevertheless, in Pseudomonas aeruginosa, PilY1, the PilC1 ortholog copurifies with sheared pili (Nguyen et al, 2015). Structural studies in the closely related type II secretion system also suggested that a minor pilin complex composed of GspIJK could fit at the pseudopilus tip (Korotkov & Hol, 2008). Transposed in the piliation system, this study suggested that the orthologous proteins PilI, J, and K could be located at the pilus tip. This idea is strengthened by evidence for pilus association of these minor pilins in P. aeruginosa (Giltner et al, 2010). A third group of proteins could also be present at the pilus tip, the non‐core minor pilin PilV, PilX, and ComP. These proteins co‐purify with pili and play a role in adhesion, aggregation, and competence (Nassif et al, 1994; Wolfgang et al, 1999; Winther‐Larsen et al, 2001; Helaine et al, 2005; Mikaty et al, 2009). Overall, evidence of tip location of these proteins is indirect, but if such proteins fully covered the tip of TFP, we should not observe inhibition of adhesion by the SM1 antibody as it specifically binds PilE. Given the multimeric nature of the pilus structure, one cannot exclude, however, the presence of proteins in combination with PilE at the pilus tip. Nevertheless, the inhibitory effect of the SM1 antibody we observe opens new therapeutic opportunities. In the case of non‐typeable Haemophilus influenzae infection, antibodies directed against the major type IV pilin triggered biofilm dispersion and immunization against the protein offered protection in a mouse model of otitis media (Novotny et al, 2015).
Observation of purified pili incubated with host cells under flow also revealed a second type of adhesion that occurs along the length of the pilus (Fig 6D). Purified pili initially interact through their tips and in the absence of flow strikingly stand away from the cell surface. This behavior likely reflects the flexibility of the particular interface formed by the pilus tip and its receptors at the cell surface as well as the relative rigidity of the pilus. Consistently, the 5 μm persistence length measured for P. aeruginosa TFP is longer than the sheared pili used here that are typically 2 μm in length (Skerker & Berg, 2001; Tala et al, 2019). It takes a liquid flow with a shear stress of a few dyn cm−2 to bend the pilus to adhere to the cellular surface, thus allowing a second form of adhesion. The molecular basis for this second adhesion remains unclear. One hypothesis could be that flow‐induced shear stress could stretch pili and uncover new cell binding sites along the length of the pilus fiber, but calculations suggest that this is unlikely to occur (Beaussart et al, 2014; Lu et al, 2015). Binding along the pilus fiber on the cellular surface evidenced here supports the recently described one‐dimensional wetting process that promotes plasma membrane remodeling (Fig 6D) and depends on adhesion along the pilus nanofiber (Charles‐Orszag et al, 2018). The specific pilin amino acids involved in this second interaction remain to be identified.
The approach used here provides a framework to explore the numerous multifunctional type four filaments found in bacteria and archaea. In the particular context of Neisseria meningitidis infection, this approach led to the identification of the specific structural domains of the major pilin involved in a sequence of events leading to infection: tip‐mediated adhesion, adhesion along the length of the pili followed by bacterial proliferation associated with auto‐aggregation mediated by pilus–pilus interactions.
Materials and Methods
Antibodies and chemicals
The following antibodies were used for Western blots and immunofluorescence: (i) polyclonal serum anti‐PilE (Morand et al, 2004), anti‐PilV (Mikaty et al, 2009), and anti‐Rmp4 (Morand et al, 2001); (ii) mouse monoclonal antibody anti‐PilE, clone 20D9 (Pujol et al, 1999) and clone SM1 (Virji et al, 1983); (iii) Camelidae nanobody anti‐PilE clone F10 (Charles‐Orszag et al, 2018). The following goat secondary antibodies were used for immunofluorescence, Western blot, and flow cytometry: anti‐mouse or anti‐rabbit IgG (H+L) coupled to horseradish peroxidase (Jackson Immuno‐Research Laboratories) and anti‐rabbit or anti‐mouse IgG (H+L) coupled to Alexa Fluor 488, 568, or 647 (Life Technologies) and mouse anti‐His tag (Biolegend). 4′,6‐diamidino‐2‐phenylindole (DAPI) was purchased from Life Technologies and Hoechst 33342 from Invitrogen. Trypsin‐EDTA (0.05%) was purchased from Gibco.
Bacterial growth conditions and mutagenesis
Neisseria meningitidis
8013/clone 12 (2C43) strain expressing the SB pilin variant was used in this study (Nassif et al, 1993). In particular, a strain containing a deletion of the guanine quartet upstream of the pilE gene (Tan et al, 2015) was used to minimize antigenic variation (see below for the construction of this strain). Bacteria were grown on Gonococcus Medium Base (GCB, Difco) agar plates supplemented with Kellogg's supplements (Kellogg et al, 1968) and, when required, 100 μg ml−1 kanamycin, 2 μg ml−1 erythromycin, 50 μg ml−1 spectinomycin, or 5 μg ml−1 chloramphenicol, at 37°C in moist atmosphere containing 5% CO2. Before adhesion and aggregation assays, bacteria were grown with shaking at 37°C and 5% CO2. Bacteria were inoculated at an OD600 nm of 0.05 in Human Endothelial‐SFM (Gibco) supplemented with 10% heat‐inactivated FBS and grown for 2 h‐2 h 30. Escherichia coli XL‐1 Blue was grown at 37°C on liquid or solid Luria‐Bertani medium (Difco) containing 20 μg ml−1 chloramphenicol, 50 μg ml−1 spectinomycin, 30 μg ml−1 kanamycin, or 200 μg ml−1 erythromycin when necessary. The guanine quartet mutant was generated as described by Tan and colleagues (Tan et al, 2015) using fusion PCR with primers PK‐30, 31, 32, and 33 and a chloramphenicol resistance cassette (see Dataset EV1). The insertion of the chloramphenicol resistance cassette was verified by PCR.
PilE point mutations
In order to increase transformation efficiency in N. meningitidis, we modified the previous vector (TOPO‐SB‐Kan (Imhaus & Dumenil, 2014)) used for PilE mutagenesis as follows: We amplified genomic DNA from the strain transformed with TOPO‐SB‐Kan by PCR using the primer pair PK‐10/21. This fragment was then cloned in TOPO‐pCR2.1 (Invitrogen). This plasmid was named TOPO‐SB2‐Kan and used for pilE mutagenesis. To generate point mutations in pilE, we used a single‐primer one‐step mutagenesis process (Huang & Zhang, 2017). Briefly, a mutagenic primer (listed in Dataset EV1) was used to amplify the plasmid TOPO‐SB2‐Kan by PCR using Phusion polymerase. The PCR product was digested with FastDigest DpnI at 37°C for 8 h and then inactivated at 80°C for 5 min. This product was transformed in XL1 Blue cells, which were then selected on kanamycin‐containing LB plates. Plasmids from single colonies were sequenced and transformed in N. meningitidis as described above. For primers and strains used in this study see Dataset EV1.
Mutant library generation and analysis
Random mutagenesis was performed using the GeneMorph II Random Mutagenesis Kit. Following manufacturer's instructions, the mutagenic PCR was run with Mutazyme II polymerase using the modified TOPO‐SB2‐Kan described earlier as a template and primers PK‐13 and PK‐14. Mutagenic PCR products were purified and used as megaprimers to PCR amplify TOPO‐SB‐Kan with Phusion polymerase. The PCR product was digested with FastDigest DpnI at 37°C for 8 h and then inactivated at 80°C for 5 min. This product was transformed in XL1 Blue cells and then selected on Kanamycin‐containing LB plates. Mixed plasmids from all the transformants were purified and transformed in N. meningitidis. Transformants were selected on plates containing kanamycin, collected the next day, and kept frozen until subjected to selection.
Piliation selection
Bacteria from the library were treated as described in the flow cytometry section. Bacterial suspensions were then sorted with MoFlo® Astrios for 1 h and collected in warm Human Endothelial‐SFM + 10%FBS. The sorted population was then plated on kanamycin‐containing GCB plates.
Adhesion selection
Bacteria from the library were used to run a 2‐h adhesion assay following the protocol described below at the exception that all of the final cell suspension was plated on kanamycin GCB plates.
Aggregation selection
Bacteria from the library were allowed to aggregate at an OD600 nm of 0.6 for 2 h in Millicell Cell Culture Inserts for 24‐well plates with a 5 μm pore size. Medium was flown‐through several times. The membrane was then vortexed in medium to collect the bacteria, which were then plated. This process was repeated two times to significantly enrich the library in aggregation‐positive bacteria. Genomic DNA was then extracted from the different libraries. DNA from each library was amplified by two separate PCR (20 cycles) using two reverse pairs of barcoded primers (primers PK‐34 to PK‐47) to obtain a more homogeneous coverage upon sequencing. The PCR products were purified, and concentration was normalized using Fragment analyzer (AATI) prior to sequencing. Paired‐end sequencing (2 × 300 bp) was performed using an Illumina Miseq sequencer at the Pasteur genomics platform. Resulting Fastq files were then assembled, filtered, and analyzed for single nucleotide variations using CLC Genomics workbench. Reads are available in BioSample under accession numbers SAMN08971257 to SAMN08971272. The read counts for single amino acid variation were further analyzed using the EdgeR‐dedicated script in the SARTools package (Robinson et al, 2010; Varet et al, 2016). Resulting P‐values and log values of the ratio were used for downstream analysis.
Each selection was done at least in triplicates on three separate days. The whole selection process was done twice, and the two biological replicates were sequenced twice. This means that we ran a total of 4 sequencing‐runs (two biological duplicates and two sequencing duplicates). Exhaustive data (mutation scores and associated P‐values) are presented in Dataset EV2.
Flow cytometry
Bacteria were resuspended directly from the GCB plates in human endothelial medium + 10% FBS to an OD600 nm of 0.1 and incubated for 30 min at 37°C. Bacterial suspensions were vigorously vortexed, and 1 μl of an antibody mix of 20D9 and anti‐mouse IgG (H+L) coupled to Alexa Fluor 488 or 647 each at 100 μg ml−1 or 1 μl of an antibody mix of F10‐I at 30 μg ml−1 and mouse anti‐His tag at 100 μg ml−1 and anti‐mouse IgG (H+L) coupled to Alexa Fluor 488 or 647 at 100 μg ml−1 was added to 10 μl of the bacterial suspension and incubated at 37°C for 15 min. 500 μl of warm medium was added to the tubes, samples were briefly vortexed, and samples were analyzed using a Gallios cytometer (Beckman Coulter). A total of 50,000 events were recorded. Signal was gated in order to exclude cell doublets or bigger aggregates, only removing a small proportion of bacteria.
Aggregation assays
GFP‐expressing or Hoechst‐labeled bacteria precultured for 2 h up to an OD600 nm of approximately 0.2 were pelleted (14,200 ×g for 1 min), washed, and concentrated at an OD600 nm of 0.6 in Human Endothelial‐SFM + 10% FBS. 500 μl of this suspension was dispensed in a well of a 96‐well μ‐plate with square wells (Ibidi). Plates were incubated for 30 min at 37°C in moist atmosphere containing 5% CO2, and three fluorescence images per well were captured using a 4× objective. The surface covered by bacterial aggregates was quantified in each field of view using a homemade macro in Fiji (Schindelin et al, 2012). Co‐aggregation assays were performed similarly to aggregation assays with the exception that bacteria were concentrated to an OD600 nm of 0.3 and that 250 μl of one bacterial suspension was mixed with 250 μl of another bacterial suspension. Bacterial localization in co‐aggregates was determined using the Radial Profile Plot ImageJ plugin on aggregates of similar size.
Adhesion assays
Human umbilical vein endothelial cells (HUVEC, PromoCell) were used between passages 1 and 8 and grown in Human Endothelial‐SFM (Gibco) supplemented with 10% heat‐inactivated FBS (PAA Laboratories) and 40 μg ml−1 of endothelial cell growth supplement (Sigma‐Aldrich) and passed every 2–3 days. bEnd.3 cells were grown in DMEM (Gibco) supplemented with 10% heat‐inactivated FBS. For 2–4 h adhesion, 105 cells were seeded in Costar® 24‐well TC‐Treated plates the day before infection. On the next day, cells were rinsed once and infected with midlog‐phase bacteria (multiplicity of infection (MOI) of 100) at 37°C/5% CO2. Inoculum was plated and quantified by CFU count. After 30 min of infection, cells were rinsed tree times and incubated for 90 min. Again, cells were rinsed three times. Infection was stopped at this stage for 2‐h adhesion but pursued for 2 more h for the 4‐h adhesion assay. Cells were rinsed three times and incubated in trypsin‐EDTA (0.05%) until detached. Trypsin was inactivated by adding culture medium. The infected cells were vortexed and plated at appropriate dilutions on GCB plates. Ratio of CFU after infection over CFU from the inoculum was compared between strains.
For early adhesion assays, 3.5 × 104 cells were seeded in Cellstar® 96‐well plates (Greiner) the day before infection. On the next day, cells were rinsed once. Hoechst was added to the medium at a final concentration of 1 μg ml−1, and cells were infected with midlog‐phase GFP‐bacteria (MOI 500) for 30 min at 37°C and 5% CO2. Wells were rinsed three times and then fixed with 4% formaldehyde for 30 min at room temperature. Wells were rinsed three times with PBS and then imaged with a 40× objective. Each infection was run in duplicate, and 16 images were taken per well. A homemade macro in Fiji was used to count the number of bacteria and nuclei per field of view. These values were summed for each well, and a ratio of the total number of bacteria over the total number of nuclei was used to evaluate the number of bacteria per cell.
Scanning electron microscopy of type IV pili
Hydrophilic mixed cellulose esters filters, 13 mm in diameter containing 0.025 μm pores (MF‐Millipore, VSWp01300), were placed in 24‐well plate, layered with 500 μl of a bacterial suspension at an OD600 nm of 1 in Human Endothelial‐SFM + 10% FBS, and incubated for 30 min at 37°C and 5% CO2 in a humidified incubator. 500 μl of pre‐warmed 8% PFA in 0.1 M HEPES pH 7.4 was then added to the well and incubated at room temperature for 45 min. After three washing steps in HEPES, type IV pili were blocked for 20 min in HEPES‐0.2% gelatin (HEPES‐G) and incubated with 2 μg ml−1 20D9 antibody followed by 10 μg ml−1 goat anti‐mouse antibody coupled to Alexa Fluor 568 in HEPES‐G, each for 1 h at room temperature. Samples were then post‐fixed overnight at 4°C with 2.5% EM‐grade glutaraldehyde in HEPES, washed in HEPES, post‐fixed in 1% OsO4 in HEPES for 1 h, washed in distilled water, dehydrated in graded series of ethanol (25, 50, 75, 90, and 100%), critical point dried in liquid CO2 in a Leica EM CPD300, mounted on aluminum stubs and sputter‐coated with 15‐nm gold/palladium in a Gatan PEC 682 gun ionic evaporator. Imaging was performed in an Auriga scanning electron microscope (Carl Zeiss) operated at 7 kV with an in‐lens secondary electrons detector.
Crude pili preparation
Crude pili preparations were obtained by suspending bacteria to an OD600 nm of 10 in cold PBS. Bacteria were vortexed for 1 min at maximum speed and then incubated on ice for 2 min. These steps were repeated two more times. To separate pili from debris and bacteria, the suspension was then centrifuged for 5 min at 9,000 ×g. This was repeated twice.
Pili under flow
40,000 HUVEC were seeded in Ibidi channels (ibiTreat μ‐Slide VI 0.4) 1 day before conducting the experiment. On the next day, crude pili of the reference strain were prepared. Cells were incubated with 5 μl of these preparations for at least 1 h. Channels were washed gently three times with cell culture medium. Pili were then labeled by incubating cells with 20D9 at a final concentration of 2.5 μg ml−1 and goat anti‐mouse secondary antibody at a final concentration of 5 μg ml−1 for at least 15 min at 37°C and 5% CO2.
Channels were then perfused using a FlowEz Fluigent pressure‐controlled pump. Pressure was computer‐controlled using the company‐built automation software. Every 30 s, pressure was alternatively turned off or increased by steps of 30 mbar (starting from 55 mbar) for a total duration of 9 min. Flow was then stopped. Cells were imaged every 5 s for a total duration of 10 min using a 40× oil immersion objective. Pilus inversion could easily be monitored by visual inspection of the resulting videos, allowing to report threshold pressure values yielding to pilus immobilization.
Flow pressure was converted to shear stress by first estimating the flow using Fluigent's conversion tool. Flow rate was then translated into shear stress using the following equation: τ = η ∙ 176.1 ∙ Φ based on Ibidi's documentation, where τ is the shear stress (dyn cm−2), η is the dynamical viscosity of the medium (dyns cm−2), and Φ is the flow rate (ml min−1).
Pili purification
Pili used for Western blots and mass spectrometry were prepared as described previously (Gault et al, 2015). Briefly, bacteria from 10 to 12 petri dishes were harvested in 5 mL of 150 mM ethanolamine at pH 10.5. Pili were sheared by vortexing for 1 min. Bacteria were centrifuged at 4,000 ×g for 30 min at 4°C, and the resulting supernatant further centrifuged at 15,000 ×g for 30 min at room temperature. The supernatant was removed, pili precipitated from the suspension by the addition of 10% (vol/vol) ammonium sulfate saturated in 150 mM ethanolamine pH 10.5 and allowed to stand for 1 h. The precipitate was pelleted by centrifugation at 4,000 ×g for 1 h at 20°C. Pellets were washed twice with PBS and suspended in 100 μl distilled water.
Mass spectrometry analysis
Samples were analyzed in LC‐MS using a Dionex UltiMate 3000 RSLC Nano System coupled to an Orbitrap Fusion Lumos mass spectrometer fitted with a nano‐electrospray ionization source (Thermo Scientific). 5 μl of protein sample was loaded at a flow rate of 5 μl min−1 onto an in‐house packed C4 (5 μm, Reprosil) trap column (0.150 mm i.d. × 35 mm) and separated at a flow rate of 0.5 μl min−1 using a C4 (5 μm, Reprosil) column (0.075 mm i.d. × 340 mm). The following gradient was used: 2.0% B from 0 to 7 min; 35% B at 8 min.; 60% B at 20 min.; 99% B from 30 to 34 min.; and 2.0% B from 34.1 to 50 min. Solvent A consisted of 98% H2O, 2% ACN, and 0.1% FA, and solvent B consisted of 20% H2O, 80% ACN, and 0.1% FA.
MS scans were acquired at 120,000 resolving power (at m/z 400) with a scan range set to 550–1,750 m/z, five microscans (μscans) per MS scan, an automatic gain control (AGC) target value of 5 × 105, and maximum injection time of 50 ms. All MS scans were charge‐state deconvoluted and deisotoped with XTRACT in Proteome Deconvolution 3.0 (Thermo Scientific) (70% fit factor, minimum S/N = 2, remainder threshold = 10%, minimum of three detected charge states, and a charge range of + 5 to + 30). Relative abundance was calculated using an extracted ion chromatogram (XIC) for each protein.
Immunofluorescence
For immunofluorescence, samples were fixed with PBS containing 4% formaldehyde for 30 min at room temperature. After rinsing, samples were blocked in PBS containing 0.2% skin fish gelatin (PBSG) for 30 min. Samples were then incubated with appropriate combinations of the following: 20D9 diluted in PBSG at 2.5 μg ml−1 and/or SM1 diluted at 2 μg ml−1 or F10‐I diluted at 4 μg ml−1 for 1 h and subsequently incubated with the secondary antibody anti‐mouse‐Alexa Fluor 491 or 568 diluted in PBSG at 10 μg ml−1 and/or DyLight 650 (Invitrogen) conjugated mouse anti‐His tag for 1 h at room temperature or overnight at 4°C.
Quantification of piliation by microscopy
Bacteria were precultured for 2 h in endothelial cell culture medium. Bacterial suspensions were normalized to an OD600 nm of 0.2 and dispensed in Ibidi channels (ibiTreat μ‐Slide VI 0.4) and incubated for 30 min at 37°C and 5% CO2. Bacteria were then rinsed four times with PBS to recover adherent bacteria only. Samples were then stained with 20D9 and DAPI as described in the immunofluorescence section. Over 100 images were captured for each sample using a 100× oil immersion objective. Quantification of piliation on individual bacteria was performed automatically using a custom‐built Fiji macro (available upon request) taking advantage of the Ridge Detection plugin. Downstream data analysis was only performed on piliated bacteria.
SDS–PAGE and Western blot
Protein samples preparation, SDS–PAGE separation, transfer, and immunoblotting were performed using standard biochemistry techniques (Harlow & Lane, 1976 a Laboratory Manual). Proteins were separated by SDS–PAGE in Tris‐glycine gels containing 12–15% polyacrylamide and transferred onto PVDF membranes (Thermo Scientific) using semi‐dry electrotransfer (Bio‐Rad). Membranes were blocked with PBS + 0.1% Tween‐20 + 5% milk for 30 min and incubated for 1 h with polyclonal antibodies (anti‐PilE at 1/5,000; anti‐PilV at 1/1,500) diluted in blocking solution. Membranes were then incubated with horseradish peroxidase (HRP)‐coupled anti‐rabbit antibody (1/10,000) in blocking solution. Peroxidase activity was detected by enhanced chemiluminescence (ECL‐plus, Pierce) and recorded with a G:BOX Chemi multi‐purpose imaging system from Syngene. Protein quantities were analyzed using the Fiji software.
Statistical analysis
GraphPad was used to plot graphs and run appropriate statistical tests. Paired t‐test was used in Figs EV1G and I, and EV2C and EV3C–F, and EV4B and C, and one‐way ANOVA was used in Figs 2E and 3E, and 4D and E, and 5G, and EV2D and F Statistical significance was defined by P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***).
Author contributions
PK and GD designed research. PK, AC‐O SG, and A‐FI performed experiments. MD and JC‐R provided the mass spectrometry analysis. DN provided the physical model of pilus adhesion under flow. PK and GD wrote the manuscript with help for the figures from AC‐O.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Movie EV1
Source Data for Expanded View
Review Process File
Acknowledgements
This work was supported by the Integrative Biology of Emerging Infectious Diseases (IBEID) laboratory of excellence; the VIP European Research Council starting grant (GD); Paris Descartes University PhD fellowship, the FRM PhD fellowship (FDT20170437205), and Programme Bettencourt (Ecole Doctorale Frontières du Vivant, FdV) funding. NGS was performed at the Genomics Platform, member of “France Génomique” consortium (ANR10‐INBS‐09‐08). We acknowledge the Ultrapole platform and cytometry platform of the Center for Innovation and Technological Research (CRT) at Institut Pasteur for support in conducting this study. We would like to thank Mumtaz Virji and Darryl J Hill for providing the SM1 antibody. We are also extremely grateful to Daria Bonazzi, Dorian Obino, and Olivera Francetic for critical reading of the manuscript and to all members of the Dumenil laboratory for helpful discussions.
The EMBO Journal (2019) 38: e102145
References
- Beaussart A, Baker AE, Kuchma SL, El‐Kirat‐Chatel S, O'Toole GA, Dufrene YF (2014) Nanoscale adhesion forces of Pseudomonas aeruginosa type IV Pili. ACS Nano 8: 10723–10733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JL, Pelicic V (2015) Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS Microbiol Rev 39: 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biais N, Higashi DL, Brujic J, So M, Sheetz MP (2010) Force‐dependent polymorphism in type IV pili reveals hidden epitopes. Proc Natl Acad Sci USA 107: 11358–11363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake MS, MacDonald CM, Klugman KP (1989) Colony morphology of piliated Neisseria meningitidis . J Exp Med 170: 1727–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonazzi D, Lo Schiavo V, Machata S, Djafer‐Cherif I, Nivoit P, Manriquez V, Tanimoto H, Husson J, Henry N, Chate H et al (2018) Intermittent pili‐mediated forces fluidize Neisseria meningitidis aggregates promoting vascular colonization. Cell 174: 143–155 e16 [DOI] [PubMed] [Google Scholar]
- Brissac T, Mikaty G, Dumenil G, Coureuil M, Nassif X (2012) The meningococcal minor pilin PilX is responsible for type IV pilus conformational changes associated with signaling to endothelial cells. Infect Immun 80: 3297–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamot‐Rooke J, Rousseau B, Lanternier F, Mikaty G, Mairey E, Malosse C, Bouchoux G, Pelicic V, Camoin L, Nassif X et al (2007) Alternative Neisseria spp. type IV pilin glycosylation with a glyceramido acetamido trideoxyhexose residue. Proc Natl Acad Sci USA 104: 14783–14788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamot‐Rooke J, Mikaty G, Malosse C, Soyer M, Dumont A, Gault J, Imhaus AF, Martin P, Trellet M, Clary G et al (2011) Posttranslational modification of pili upon cell contact triggers N. meningitidis dissemination. Science 331: 778–782 [DOI] [PubMed] [Google Scholar]
- Charles‐Orszag A, Tsai FC, Bonazzi D, Manriquez V, Sachse M, Mallet A, Salles A, Melican K, Staneva R, Bertin A et al (2018) Adhesion to nanofibers drives cell membrane remodeling through one‐dimensional wetting. Nat Commun 9: 4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Seifert HS (2014) Saturating mutagenesis of an essential gene: a majority of the Neisseria gonorrhoeae major outer membrane porin (PorB) is mutable. J Bacteriol 196: 540–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang SL, Taylor RK, Koomey M, Mekalanos JJ (1995) Single amino acid substitutions in the N‐terminus of Vibrio cholerae TcpA affect colonization, autoagglutination, and serum resistance. Mol Microbiol 17: 1133–1142 [DOI] [PubMed] [Google Scholar]
- Coureuil M, Lecuyer H, Scott MG, Boularan C, Enslen H, Soyer M, Mikaty G, Bourdoulous S, Nassif X, Marullo S (2010) Meningococcus Hijacks a beta2‐adrenoceptor/beta‐Arrestin pathway to cross brain microvasculature endothelium. Cell 143: 1149–1160 [DOI] [PubMed] [Google Scholar]
- De Baets G, Schymkowitz J, Rousseau F (2014) Predicting aggregation‐prone sequences in proteins. Essays Biochem 56: 41–52 [DOI] [PubMed] [Google Scholar]
- van Deuren M, Brandtzaeg P, van der Meer JW (2000) Update on meningococcal disease with emphasis on pathogenesis and clinical management. Clin Microbiol Rev 13: 144–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Fields S (2014) Deep mutational scanning: a new style of protein science. Nat Methods 00: 801–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gault J, Ferber M, Machata S, Imhaus AF, Malosse C, Charles‐Orszag A, Millien C, Bouvier G, Bardiaux B, Pehau‐Arnaudet G et al (2015) Neisseria meningitidis type IV pili composed of sequence invariable pilins are masked by multisite glycosylation. PLoS Pathog 11: e1005162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadou M, Castagnini M, Karimova G, Ladant D, Pelicic V (2012) Large‐scale study of the interactions between proteins involved in type IV pilus biology in Neisseria meningitidis: characterization of a subcomplex involved in pilus assembly. Mol Microbiol 84: 857–873 [DOI] [PubMed] [Google Scholar]
- Giltner CL, Habash M, Burrows LL (2010) Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J Mol Biol 398: 444–461 [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D (1976) Adrenergic stimulants (or sympathomimetic medicines). Rev Infirm 26: 75–78 [PubMed] [Google Scholar]
- Helaine S, Carbonnelle E, Prouvensier L, Beretti JL, Nassif X, Pelicic V (2005) PilX, a pilus‐associated protein essential for bacterial aggregation, is a key to pilus‐facilitated attachment of Neisseria meningitidis to human cells. Mol Microbiol 55: 65–77 [DOI] [PubMed] [Google Scholar]
- Helaine S, Dyer DH, Nassif X, Pelicic V, Forest KT (2007) 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili. Proc Natl Acad Sci USA 104: 15888–15893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hospenthal MK, Waksman G (2019) The remarkable biomechanical properties of the type 1 chaperone‐usher pilus: a structural and molecular perspective. Microbiol Spectr 7: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Zhang L (2017) An in vitro single‐primer site‐directed mutagenesis method for use in biotechnology. New York, NY: Humana Press, 375–383 [DOI] [PubMed] [Google Scholar]
- Imhaus AF, Dumenil G (2014) The number of Neisseria meningitidis type IV pili determines host cell interaction. EMBO J 33: 1767–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley KA, Maiden MC (2010) BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11: 595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DS Jr, Cohen IR, Norins LC, Schroeter AL, Reising G (1968) Neisseria gonorrhoeae. II. Colonial variation and pathogenicity during 35 months in vitro . J Bacteriol 96: 596–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmury SL, Burrows LL (2016) Type IV pilins regulate their own expression via direct intramembrane interactions with the sensor kinase PilS. Proc Natl Acad Sci USA 113: 6017–6022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolappan S, Coureuil M, Yu X, Nassif X, Egelman EH, Craig L (2016) Structure of the Neisseria meningitidis type IV pilus. Nat Commun 7: 13015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov KV, Hol WG (2008) Structure of the GspK‐GspI‐GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system. Nat Struct Mol Biol 15: 462–468 [DOI] [PubMed] [Google Scholar]
- Lu S, Giuliani M, Harvey H, Burrows LL, Wickham RA, Dutcher JR (2015) Nanoscale pulling of type IV pili reveals their flexibility and adhesion to surfaces over extended lengths of the pili. Biophys J 108: 2865–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau M, Beretti JL, Nassif X (1995) High adhesiveness of encapsulated Neisseria meningitidis to epithelial cells is associated with the formation of bundles of pili. Mol Microbiol 17: 855–863 [DOI] [PubMed] [Google Scholar]
- Melican K, Michea Veloso P, Martin T, Bruneval P, Dumenil G (2013) Adhesion of Neisseria meningitidis to dermal vessels leads to local vascular damage and purpura in a humanized mouse model. PLoS Pathog 9: e1003139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz AJ, So M, Sheetz MP (2000) Pilus retraction powers bacterial twitching motility. Nature 407: 98–102 [DOI] [PubMed] [Google Scholar]
- Mikaty G, Soyer M, Mairey E, Henry N, Dyer D, Forest KT, Morand P, Guadagnini S, Prevost MC, Nassif X et al (2009) Extracellular bacterial pathogen induces host cell surface reorganization to resist shear stress. PLoS Pathog 5: e1000314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morand PC, Tattevin P, Eugene E, Beretti JL, Nassif X (2001) The adhesive property of the type IV pilus‐associated component PilC1 of pathogenic Neisseria is supported by the conformational structure of the N‐terminal part of the molecule. Mol Microbiol 40: 846–856 [DOI] [PubMed] [Google Scholar]
- Morand PC, Bille E, Morelle S, Eugene E, Beretti JL, Wolfgang M, Meyer TF, Koomey M, Nassif X (2004) Type IV pilus retraction in pathogenic Neisseria is regulated by the PilC proteins. EMBO J 23: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif X, Lowy J, Stenberg P, O'Gaora P, Ganji A, So M (1993) Antigenic variation of pilin regulates adhesion of Neisseria meningitidis to human epithelial cells. Mol Microbiol 8: 719–725 [DOI] [PubMed] [Google Scholar]
- Nassif X, Beretti JL, Lowy J, Stenberg P, O'Gaora P, Pfeifer J, Normark S, So M (1994) Roles of pilin and PilC in adhesion of Neisseria meningitidis to human epithelial and endothelial cells. Proc Natl Acad Sci USA 91: 3769–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Y, Harvey H, Sugiman‐Marangos S, Bell SD, Buensuceso RN, Junop MS, Burrows LL (2015) Structural and functional studies of the Pseudomonas aeruginosa minor pilin, PilE. J Biol Chem 290: 26856–26865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny LA, Jurcisek JA, Ward MO Jr, Jordan ZB, Goodman SD, Bakaletz LO (2015) Antibodies against the majority subunit of type IV Pili disperse nontypeable Haemophilus influenzae biofilms in a LuxS‐dependent manner and confer therapeutic resolution of experimental otitis media. Mol Microbiol 96: 276–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obergfell KP, Seifert HS (2016) The pilin N‐terminal domain maintains neisseria gonorrhoeae transformation competence during pilus phase variation. PLoS Genet 12: e1006069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HS, Wolfgang M, van Putten JP, Dorward D, Hayes SF, Koomey M (2001) Structural alterations in a type IV pilus subunit protein result in concurrent defects in multicellular behaviour and adherence to host tissue. Mol Microbiol 42: 293–307 [DOI] [PubMed] [Google Scholar]
- Pujol C, Eugene E, Marceau M, Nassif X (1999) The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus‐mediated adhesion. Proc Natl Acad Sci USA 96: 4017–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel T, Scheurerpflug I, Meyer TF (1995) Neisseria PilC protein identified as type‐4 pilus tip‐located adhesin. Nature 373: 357–359 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert HS, Ajioka RS, Paruchuri D, Heffron F, So M (1990) Shuttle mutagenesis of Neisseria gonorrhoeae: pilin null mutations lower DNA transformation competence. J Bacteriol 172: 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Berg HC (2001) Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci USA 98: 6901–6904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tala L, Fineberg A, Kukura P, Persat A (2019) Pseudomonas aeruginosa orchestrates twitching motility by sequential control of type IV pili movements. Nat Microbiol 4: 774–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan FY, Wormann ME, Loh E, Tang CM, Exley RM (2015) Characterization of a novel antisense RNA in the major pilin locus of Neisseria meningitidis influencing antigenic variation. J Bacteriol 197: 1757–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varet H, Brillet‐Gueguen L, Coppee JY, Dillies MA (2016) SARTools: a DESeq2‐ and EdgeR‐Based R pipeline for comprehensive differential analysis of RNA‐Seq data. PLoS ONE 11: e0157022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virji M, Heckels JE, Watt PJ (1983) Monoclonal antibodies to gonococcal pili: studies on antigenic determinants on pili from variants of strain P9. J Gen Microbiol 129: 1965–1973 [DOI] [PubMed] [Google Scholar]
- Winther‐Larsen HC, Hegge FT, Wolfgang M, Hayes SF, van Putten JP, Koomey M (2001) Neisseria gonorrhoeae PilV, a type IV pilus‐associated protein essential to human epithelial cell adherence. Proc Natl Acad Sci USA 98: 15276–15281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M, van Putten JP, Hayes SF, Koomey M (1999) The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol Microbiol 31: 1345–1357 [DOI] [PubMed] [Google Scholar]
- Woods DE, Straus DC, Johanson WG Jr, Berry VK, Bass JA (1980) Role of pili in adherence of Pseudomonas aeruginosa to mammalian buccal epithelial cells. Infect Immun 29: 1146–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Movie EV1
Source Data for Expanded View
Review Process File


