Abstract
Eukaryotes arose about 1.6 billion years ago, at a time when oxygen levels were still very low on Earth, both in the atmosphere and in the ocean. According to newer geochemical data, oxygen rose to approximately its present atmospheric levels very late in evolution, perhaps as late as the origin of land plants (only about 450 million years ago). It is therefore natural that many lineages of eukaryotes harbor, and use, enzymes for oxygen-independent energy metabolism. This paper provides a concise overview of anaerobic energy metabolism in eukaryotes with a focus on anaerobic energy metabolism in mitochondria. We also address the widespread assumption that oxygen improves the overall energetic state of a cell. While it is true that ATP yield from glucose or amino acids is increased in the presence of oxygen, it is also true that the synthesis of biomass costs thirteen times more energy per cell in the presence of oxygen than in anoxic conditions. This is because in the reaction of cellular biomass with O2, the equilibrium lies very far on the side of CO2. The absence of oxygen offers energetic benefits of the same magnitude as the presence of oxygen. Anaerobic and low oxygen environments are ancient. During evolution, some eukaryotes have specialized to life in permanently oxic environments (life on land), other eukaryotes have remained specialized to low oxygen habitats. We suggest that the Km of mitochondrial cytochrome c oxidase of 0.1–10 μM for O2, which corresponds to about 0.04%–4% (avg. 0.4%) of present atmospheric O2 levels, reflects environmental O2 concentrations that existed at the time that the eukaryotes arose.
Keywords: Eukaryote anaerobes, Hydrogenosomes, Mitosomes, Euglena, Chlamydomonas, Earth history, Great oxidation event
Graphical abstract
Highlights
-
•
The first 1.5 billion years of life history took place without molecular oxygen.
-
•
The first eukaryotes appeared ca. 1.6 billion years ago, oxygen rose with land plants.
-
•
Eukaryotes arose and diversified with low oxygen, anaerobic ATP synthesis is ancient.
-
•
Anaerobic energy metabolism in mitochondria is common among eukaryotic lineages.
-
•
The Km of cytochrome c oxidase might reflect low environmental O2 at eukaryote origin.
1. Introduction
First traces of microbial life are found in rocks that are 3.95–3.8 billion years (Gy) of age [[1], [2], [3]]. At that time there was no O2 in the ocean or atmosphere, and the Earth harbored a highly reducing environment: Life on Earth originated and diversified in anoxic environments [4]. Recent studies indicate that O2 arose late in Earth history, reaching roughly present levels — 21% [v/v] in the atmosphere — only following the origin of land plants roughly 450 million years ago [[5], [6], [7]]. The reservoir of oxygen in the Earth's atmosphere stems from cyanobacterial photosynthesis [4], which started ca. 2.7–2.5 Gy and was complemented with a contribution from eukaryotic algae starting ca. 1.5 Gy [8]. The late rise in oxygen content leading to present atmospheric level is currently attributed to the emergence of land plants [5,6] stemming from the deposition of cellulose on land by the terrestrial descendants of streptophytes [9]. Carbon burial leads to oxygen accumulation [5,6]. Today land plants are estimated to comprise roughly 80% (±a factor of 1.2) of Earth's biomass [10]. The advent of terrestrial photosynthesis and biomass production accompanied by carbon burial precipitated the late rise in O2 [5,11]. Early land plants evolved from charophytic algae [12] and came onto land approximately 500 million years ago [13,14]. It was not so much that land plants generated large amounts of oxygen, rather that they removed carbon from oxidation through burial, thereby allowing O2 levels to increase.
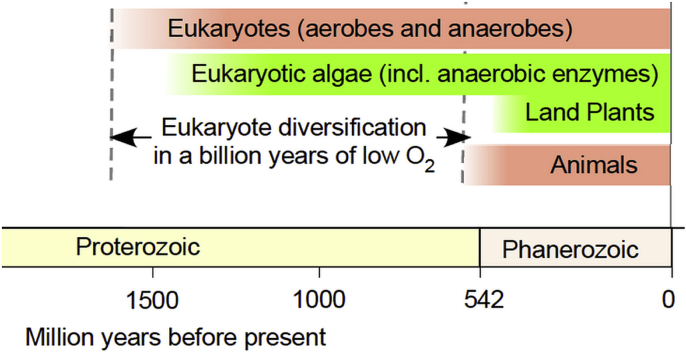
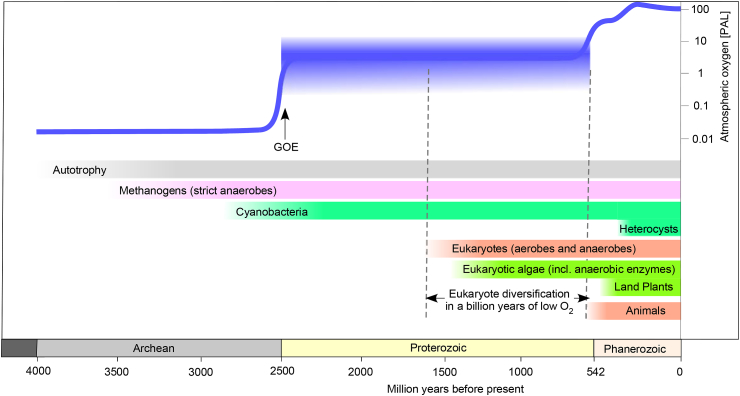
The accumulation of oxygen in the oceans was a much slower process. Oxygenation of the upper ocean to relatively persistent near-modern conditions might have only occurred as late as about 200 million years ago [15]. Many lines of evidence indicate that ocean oxygenation came to completion less than 600 My ago [6,16,17], but eukaryotes appeared by 1.6 Ga ago [8,18]. Vertebrate life on land only goes back about 350 million years (My) [19], when the Earth's atmosphere was already fully oxic. Any form of life in a permanently high-oxygen environment is a late occurrence, both in terms of evolution and in terms of Earth history. The majority of Earth history was anoxic or very low oxygen. A sketch of selected major evolutionary events with evidence in the fossil or isotope record plotted against a modern account of atmospheric oxygen is summarized in Fig. 1.
Fig. 1.
Summary of oxygen accumulation of earth history. Data for oxygen levels from Ref. [17], for the emergence of autotrophy from Ref. [1], of methanogens from Ref. [3], of cyanobacteria from Ref. [20], of animals from Ref. [24], of eukaryote anaerobes from Refs. [47,95], of eukaryote algae from Ref. [21], of algal anaerobic enzymes from Ref. [61], of land plants from Ref. [13], and for the eukaryote age from Refs. [8,30]. PAL: percent of present atmospheric level. GOE: great oxidation event. The GOE marks the appearance of continuous atmospheric O2 in the geochemical record [17,26,27,[30], [31], [32], [33]].
The appearance of the first fossil animals about 580 My ago [22] or 558–571 My ago [23,24], does not mean that the animal lineages arose at that time. Molecular data has it that metazoan radiation predated the appearance of the first fossil animals by many millions of years [25,26]. The implication is that the first animal groups, which evolved from unicellular eukaryotes (protists), arose at a time before the oceans were fully oxic. One interpretation for the sudden appearance of diverse animal groups in the Cambrian explosion is that the animals were already there, but did not leave fossil imprints until rising O2 levels permitted collagen synthesis, which is an O2-dependent process because the enzyme that hydroxylates proline in collagen, making the coiled-coil protein rigid (amenable to leaving body fossil imprints), requires O2 as a substrate [27]. Collagen provides an example of oxygen imposing its presence upon organisms that arose and existed at low oxygen levels but responded to rising O2 levels. Collagen is the most abundant protein in animals [28]. The enzymes that introduces hydroxyl residues into proline in collagen are prolyl hydroxylases [29]. In the Fe2+-dependent enzymatic mechanism, one oxygen atom of O2 becomes a hydroxyl group of hydroxyproline in collagen, the other oxygen atom cleaves 2-oxoglutarate into succinate and CO2 [29]. Proline on collagen can be hydroxylated at carbons 2, 3, or 4 and multiple hydroxylations can occur. Posttranslational hydroxylation makes the coiled-coil protein more rigid [28]. All known collagen hydroxylases are strictly O2-dependent and have a similar reaction mechanism.
2. Oxygen and energy metabolism
Understanding the oxygen requirements of the first eukaryotes can inform us about their origin and their ancestral physiological state. This requires information about eukaryote age, diversity in eukaryote energy metabolism, and the evolution of the enzymes underpinning their energy metabolism. Eukaryote age is fairly uncontroversial. Eukaryotes originated at least 1.6 Gy ago, as fossils of eukaryotic protists are found in rocks of that age [[30], [31], [32], [33], [34]] and molecular estimates for the timing of eukaryotic evolution are consistent with that view [25,26,[35], [36], [37]]. Eukaryotes can be taken to have arisen about 1.6 Gy ago, at a time when anoxic environments were more widespread than at the dawn of animal evolution, almost a billion years later.
Many protists today still inhabit environments where virtually no free O2 is available, such as anaerobic sediments [18,[38], [39], [40], [41]], and new species are still being discovered [42]. In addition, numerous species, in particular among the ciliates, harbor methanogenic archaea that live as endosymbionts within their hosts [43,44], methanogens being among the strictest anaerobes known [45]. The terms microaerophilic and anaerobic can be equally applicable for many protists. Few eukaryotes studied in detail so far are truly strict anaerobes, in that most of them regularly encounter a bit of O2 in their natural habitats. Accordingly, they have enzymes for O2 removal, including NAD(P)H oxidases (also called NAD(P)H-dependent O2 reductases or diaphorases) [46,47] and flavodiiron proteins [48]. Hence, they can readily tolerate and remove small amounts of O2. Some eukaryotic anaerobes actively make their environment anaerobic. For example, Trichomonas vaginalis liquid medium can be prepared with ambient oxygen, a dense inoculum will consume the oxygen by glycolytic glucose breakdown to pyruvate. This generates NADH that is spent to reduce the O2 to water via NADH oxidase. Once the O2 is gone the cells express their O2 sensitive enzymes which will allow pyruvate to enter the hydrogenosome, an anaerobic form of mitochondria, and provide additional ATP.
NADH oxidases are not just detoxifying enzymes, they afford redox balance. Trichomonas provides a case in point. If Trichomonas is grown with 1/1000th of today's ambient O2 levels, it grows twice as fast as when grown under strictly anaerobic conditions [49]. The reason is not that O2 helps it synthesize some required cofactor, rather the reason is that the cytosolic NADH oxidase reoxidizes NADH for the glycolytic activity faster than the H2-producing pathway of the hydrogenosome does. Redox balance is the key. ATP synthesis via glucose oxidation requires that electrons be excreted in end-products. In the presence of trace amounts of O2, Trichomonas can thus grow somewhat faster, simply because it can more readily regenerate NAD+ for glycolysis than in the complete absence of O2. The use of trace amounts of O2 alters redox balance in energy metabolism and thus leads to the excretion of different end-products [47,49]. Like many eukaryotic anaerobes, however, Trichomonas can grow indefinitely in the complete absence of O2. Some eukaryotic anaerobes such as Giardia and Entamoeba that possess NADH oxidases, but do not typically generate H2 for redox balance, excrete organic end-products like ethanol, which is more reduced than lactate or acetate, instead [47]. There are reports that the anaerobic protozoa Giardia, Entamoeba and Trichomonas can survive in culture medium exposed up to air containing up to 5% [v/v] O2, but because they possess NADH oxidases they hence can render their air-exposed culture medium anoxic as long as oxidizable substrates such as glucose or amino acids are available [50].
The oxygen-sensitivity of many eukaryote anaerobes is tightly coupled to their energy metabolism and resides in a few enzymes of pyruvate metabolism that have oxygen sensitive FeS centers: pyruvate:ferredoxin oxidoreductase (PFO or PFOR) and FeFe-hydrogenase (Fe-HYD), proteins that were first characterized in eukaryotes from the hydrogenosomes of trichomonads [51]. In trichomonad hydrogenosomes, PFO catalyzes the coenzyme A (CoA) dependent oxidative decarboxylation of pyruvate, analogous to the more familiar pyruvate dehydrogenase (PDH) reaction in mitochondria. In O2-dependent mitochondria, PDH oxidizes pyruvate to acetyl-CoA, CO2 and NADH, which is reoxidized to NAD+ by the respiratory chain, where the electrons from pyruvate are transferred to O2 as the terminal acceptor and leave the cell as the end-product, H2O. In trichomonad hydrogenosomes, PFO oxidizes pyruvate to acetyl-CoA, CO2 and reduced ferredoxin (Fd–), a FeS cluster containing protein that serves as a soluble one electron carrier. Fd– is reoxidized by Fe-HYD, an FeS cluster containing enzyme that transfers the electrons from pyruvate to protons, generating H2 gas that leaves the cell as a metabolic end-product [47,53]. In O2-respiring mitochondria, acetyl CoA is oxidized to CO2 in the Krebs cycle, the electrons enter the respiratory chain and are transferred to O2, generating transmembrane ion gradient and chemiosmotic ATP synthesis [54]. In hydrogenosomes, a two-enzyme cycle operates instead of the Krebs cycle. An acetate:succinate CoA transferase (ASCT) generates acetate as a metabolic end-product and succinyl-CoA, the substrate for succinyl-CoA synthase (SCS; also called succinate thiokinase, STK), which conserves the energy in the thioester bond as ATP via substrate level phosphorylation.
One might wonder how it can be possible that O2-sensitivity can reside in just one or a few enzymes of ATP synthesis. For amino acid or cofactor biosynthetic pathways, the situation is simple. An organism can easily circumvent an O2-sensitive enzyme for amino acid or cofactor biosynthesis by simply becoming auxotrophic for the corresponding compound (obtaining the compound from food). Indeed, the vast majority of O2-dependent reactions in metabolism are for the oxidative breakdown of compounds, rather than biosynthesis [55], and almost all eukaryote anaerobes have vitamin or amino acid auxotrophies or cannot be cultured axenically. But ATP is a different matter. Humans generate and use about one body weight of ATP per day. A microbe such as Escherichia coli weighs about 1 pg and generates about 20–60 billion ATP per cell division [56], corresponding to 16–48 body weights of ATP synthesized per cell division. If an organism cannot synthesize ATP in the presence of O2, it cannot survive, hence it makes no difference whether any of its other pathways are O2-sensitive or not. The same principle applies to the evolution of amino acid or vitamin auxotrophies: An animal with a normal diet will starve to death before it dies of a specific amino acid or vitamin deficiency. None of the eukaryotic enzymes of glycolysis or aerobic ATP synthesis in mitochondria are O2-sensitive. The O2-sensitivity of eukaryotic anaerobes resides in the FeS clusters of PFO and Fe-HYD.
There are a number of FeS centers in the electron transport chain of O2-respiring mitochondria, nine in complex I alone [57] but they are not readily inactivated by O2. However, FeS centers have a natural tendency to be oxidized by O2. That is the basis of many O2-sensing systems. In some animal lineages, a cellular oxygen-sensing system exists that employs a cytosolic homologue of the Krebs cycle enzyme aconitase, which has a 4Fe4S cluster. The cytosolic homologue is called the iron responsive element binding protein, or IRE-BP, it participates in signaling of O2 levels via O2-dependent degradation of the 4Fe4S center [58,59].
Higher plants use a similar FeS cluster degradation strategy but a different protein. The higher plant O2-sensing protein is called Gollum, it is directly derived from Fe-HYD [60]; higher plants lack active Fe-HYD, but Fe-HYD is very common in algae [61]. Bacteria use the same principle of oxidative 4Fe4S cluster decay to sense O2. Their solutions entail a handful of different FeS cluster containing proteins that have been independently recruited in different bacterial lineages to function as transcriptional regulators [62], the regulatory activity of which is modulated by O2-dependent oxidation of the FeS cluster. It is not at all surprising that plants, animals, and different groups of bacteria independently evolved FeS-dependent O2-sensing mechanisms, because high O2 levels arose late in evolution, as late as land plants origin (Fig. 1), such that evolutionarily well differentiated eukaryotic and prokaryotic lineages independently faced the challenge of physiological responses to the late appearance of high O2 levels. It should however be stressed that the O2-sensitivity of PFO and HYD, though the focal point of the anaerobic lifestyle in protists like Trichomonas, Giardia, and Entamoeba, does not stop O2-producing algae like Euglena and Chlamydomonas from using their PFO and Fe-HYD enzymes at ambient O2 levels (21% v/v) [47,61], indicating that there are differences across eukaryotic lineages regarding O2-sensitivity for PFO and Fe-HYD, founded either in inherent enzymatic properties or FeS cluster assembly and repair, that have so far not been identified at the molecular level.
3. Animal O2 sensing and the hypoxia induced factor (HIF)
The main and most important O2-sensing system in animal cells is HIF (for hypoxia-inducible factor). HIF is a protein, a transcription factor. The HIF pathway is present and operational in all animal lineages [63] tracing back to placozoa. The O2 levels at which HIF sensing elicits a response differs across lineages. Caenorhabditis elegans and other worms that live in soil live at low oxygen levels. Given a choice, Caenorhabditis prefers oxygen levels between 7 and 14% O2, below 1% Caenorhabditis accumulates HIF as a warning signal [64]. By contrast, humans require O2 levels above about 14% for good brain function [65] and above about 5%, corresponding to the O2 partial pressure at 8000 m altitude (the “death zone”) for survival [66].
HIFs sense O2 through a mechanism involving prolyl hydroxylases (the same family of enzymes that hydroxylate collagen) [67,68]. HIFs are obligate heterodimers that consist of an oxygen regulated HIF-α subunit and a stable HIF-β subunit. HIF-α subunits heterodimerize with the constantly expressed HIF-β subunits and when this dimer accumulates it binds to hypoxia response elements (HREs), which results in increased transcription of genes for low oxygen response. However, under normal O2 levels HIF does not accumulate. That is because HIF-α is a good substrate for prolyl hydroxylases (PHDs) that hydroxylate proline residues in HIF-α in a reaction that requires O2 as a substrate. Hydroxylated HIF-α binds a protein called von Hippel-Lindau (VHL), which in turn recruits a ubiquitin protein ligase that attaches ubiquitin to the HIF-VHL complex. Ubiquitinylation is a signal that designates the complex for degradation in the proteasome (a large protein digesting complex). Thus, when O2 is around, HIF-α is degraded and the heterodimer is not formed. When O2 is lacking, the HIF dimer accumulates and when it accumulates it activates genes to elicit the physiological responses [67,68].
The main physiological response to HIF in animal cells is a shift in energy metabolism from respiratory ATP synthesis in mitochondria to glycolytic ATP synthesis by diverting glycolytic flux at pyruvate through activating the transcription of many genes [67]. Under hypoxic conditions, HIF activates genes encoding glucose transporters and glycolytic enzymes, thereby increasing the flux of glucose to pyruvate. At the same time, HIF increases transcription of pyruvate dehydrogenase kinase, which inactivates (via phosphorylation) pyruvate dehydrogenase, the mitochondrial enzyme that converts pyruvate to acetyl-CoA for the Krebs cycle. HIF also activates transcription of lactate dehydrogenase, which provides glycolytic redox balance by converting pyruvate to lactate. Furthermore, HIF activates transcription of two nuclear encoded mitochondrial proteins called BNIP3 and BNIP3L, which induce mitochondrial-selective autophagy, a process in which cells digest their own mitochondria.
The role of HIF goes beyond energy metabolism. In mammals, embryogenesis occurs at O2 concentrations of 1%–5%, rather than 21%, and HIF mediates the morphogenic role of O2 in various developmental systems [69]. There are also O2-dependent proteins that hydroxylate the methylene group of asparaginyl residues in HIF-α; the hydroxylase is called factor inhibiting HIF, or FIH, and it intervenes with the interaction between HIF-α with the co-activator p300, thereby impairing HIF transcriptional activity. The enzymatic mechanism of FIH and HIF prolyl hydroxylases is the same: one oxygen atom in O2 introduces a hydroxyl group into a methylene group of the substrate amino acid side chain while the other oxygen atom cleaves 2-oxoglutarate into succinate and CO2 [70]. There is an immense literature on HIF because cancer cells shift from mitochondrial ATP synthesis to glycolytic ATP synthesis [71] and HIF is involved in that shift [67,68]. The observation that the HIF sensing system is conserved in terms of components and function back to the metazoan common ancestor clearly indicates that the first animals arose in environments where they were confronted with low oxygen, and that the response to low oxygen has remained indispensable to the present.
4. Molecular fossils of ancient O2 levels in physiology
Though it might be tempting to try to infer ancient O2 levels at animal origin from the O2 levels that are required to induce HIF-dependent hypoxia signaling, the situation is not that simple. The reason is that the O2 affinity of prolyl hydroxylases does not by itself determine the rate of HIF protection from degradation. The reported Km values for prolyl hydroxylation of HIF vary from around 10–15 μM (the aqueous O2 concentration corresponding to roughly 5% of ambient O2 levels) to roughly 250 μM (the aqueous O2 concentration corresponding to ambient O2 levels), but the physiological hypoxia response depends on the rate at which non-hydroxylated HIF-α accumulates in the cell, which depends on the rate of degradation of the hydroxylated factor at the proteasome, which in turn depends upon the concentration of the HIF protein co-substrate as well as the prolyl hydroxylase, the Km and concentration of the co-substrate 2-oxoglutarate, and the presence of competitive 2-oxoacid inhibitors like pyruvate or oxaloacetate [72]. Nonetheless it is clear that when no O2 is available for HIF prolyl hydroxylation, the animal cell undergoes a hypoxic response [67,72].
If one was nonetheless tempted to infer environmental O2 concentrations at the time of eukaryote origin from modern enzymatic parameters, the Km for O2 of the A1 type cytochrome c oxidase, the mitochondrial terminal oxidase, would provide an estimate. The A1 type terminal oxidase that functions in mitochondria has the highest Km for O2 (lowest affinity for O2) of the terminal oxidases currently known, suggesting that it arose late in the evolution of O2 reductases that serve as bioenergetic terminal oxidases [73]. Only the alternative oxidases (AOX), which have a redox balance function for the quinone pool [74] have a higher Km for O2 among membrane associated oxygen reductases [73]. The Km of mitochondrial cytochrome c oxidase for O2 value lies in the range of 0.1–10 μM [73,75], which corresponds to about 0.04%–4% (avg. 0.4%) of present atmospheric levels, or less than 1% [v/v], which is very much in line with corresponding estimates from geochemical data [17,47]. It is well known among biochemists, from experience and observation, that enzymes tend to have a Km for their substrate that is close to the physiological concentration of the substrate [76]. In that way the enzyme will display significant activity and yet the activity will be sensitive to changes in environmental conditions, i.e. substrate concentrations. Hence the Km of mitochondrial terminal oxidase has been a kind of enigma [75], because it corresponds to an O2 concentration that is orders of magnitude lower than modern levels. Given what we now know about enzymes, anaerobes and O2 in earth history, maybe the Km of mitochondrial cytochrome c oxidase of 0.1–10 μM for O2 simply reflects environmental O2 concentrations that existed at the time that eukaryotes arose, we suggest here.
5. Living in O2 comes at a steep energetic price
In mammals, as an example of eukaryotes with O2 respiring mitochondria, the ATP yield is about 30 ATP per glucose [54]. In O2 respiring eukaryotes like yeasts, that lack complex I, the yield is lower [77]. In trichomonads the yield is 4 ATP per glucose [53]. The use of O2 as a terminal acceptor in respiratory chains is generally regarded as a substantial energetic advantage, that apparent advantage is also still discussed as an evolutionary factor in some models for mitochondrial origin [78] (but see rebuttal by Garg and Martin [79]). Curiously, the notion that oxygen (via oxidative phosphorylation in mitochondria) boosts the overall energy yield from glucose by a large factor has a caveat worth exploring. How so? The ATP yield per glucose for mammalian mitochondria is 30 [54], 7.5-fold greater that for fermentations in eukaryotic anaerobes with hydrogenosomes [47]. That would make the O2 “boost” factor 7.5, but eukaryotes that lack hydrogenosomes and have anaerobically functioning mitochondria instead, like the liver fluke, generate 5 ATP per glucose [80], reducing the ATP yield factor in the comparison of “with O2" vs. "without O2" to about 6. But a factor of 6 is still apparently a big boost, or is it? Probably not. Why?
There is an important but not widely recognized relationship between oxygen and cellular energy: The synthesis of chemical constituents of cells (amino acids, bases, lipids) from glucose and ammonium demands about 13 times more energy per cell in the presence of O2 than in the absence of O2 [81,82]. The reason is that the synthesis of the chemicals that comprise cells is thermodynamically much more favorable under anaerobic conditions than it is in the presence of oxygen [81,82]. The equilibrium in the reaction of cell mass with O2 lies very far on the side of CO2 and H2O. By contrast, in the absence of O2, the equilibrium of the reaction of H2 with CO2 lies far on the side of reduced carbon compounds [83], which is probably the main reason why the first cells on Earth were anaerobes that lived from the reaction of H2 with CO2 [84,85].
Thus, although the ATP yield from glucose is 6-fold higher in the presence of O2, the energetic cost of living in an atmosphere containing 21% oxygen is 13-fold higher than for anaerobic environments. Life on O2 turns out to be twice as expensive as for anaerobes. Fortunately, the expense is less severe if one does not have to synthesize all cell constituents from glucose and ammonium. Indeed, for many eukaryotic heterotrophs, glucose is not the main source of nutrition in nature, amino acids are [86]. But still, in the presence of O2, it costs cells more energy to assemble their components than in the absence of O2, and life in the presence of a strong oxidant comes at an energetic price [81,82] that is easy to overlook. What mitochondria did for eukaryotes, energetically, was not to boost the energy yield per glucose, but to provide internalized bioenergetic membranes that boost the energy per gene [87,88].
6. Oxygen and the first eukaryotes: two views
Currently, there are two mutually exclusive views about oxygen demands of early eukaryotes and their subsequent evolution. One view has it that the earliest eukaryotes were strict aerobes which at different occasions and by means of interdomain and intradomain lateral gene transfer (LGT) adapted to hypoxic and anoxic conditions at unspecified times during Earth history [[89], [90], [91], [92], [93], [94], [95], [96], [97]]. Proponents of the LGT view argue that eukaryotes throughout their evolutionary history were unable to survive hypoxia, and therefore had to acquire genes via lateral transfer in order to gain ecological access to anaerobic environments. The common theme behind the “aerobes first, anaerobes late” LGT theory is that the ability to survive anaerobiosis was not present in the eukaryote common ancestor, but entered the eukaryotic lineage late in evolution (after diversification of the major eukaryotic lineages) via LGT from anaerobic prokaryotes in multiple independent transfers [[90], [91], [92], [93], [94], [95], [96], [97], [98]]. A general critique of LGT theories for eukaryote anaerobe origin recently appeared [99], and a defense of the LGT theory appeared [100]. Proponents of eukaryote anaerobe origins via LGT will make their case in their contribution to this volume, we do not need to argue their case for them here, instead we will focus on our own interpretation of the evolutionary significance of eukaryotic anaerobes.
The alternative view, which we have been advocating for some time, is that the free-living ancestors of mitochondria were themselves facultative anaerobes [47,80,88,99,[101], [102], [103], [104]] with a diversity of bioenergetic enzymes suited to survival with or without O2 [47,105,106] and that the earliest eukaryotes were themselves facultative anaerobes that possessed facultatively anaerobic mitochondria [47,80,101]. This allowed them to conserve energy from their environment and to survive under a wide range of oxygen concentrations [47,101]. Under this view, the ability of eukaryotes to survive in anaerobic environments was present in the eukaryote common ancestor and that modern eukaryote anaerobes have simply persisted from the low oxygen past (Fig. 1) via vertical evolution, by virtue of not having adapted to high O2 niches.
This “anaerobes early” view directly accounts for a number of observations. First it explains why eukaryotic anaerobes from diverse lineages can survive hypoxic and anoxic conditions using overlapping subsets of a small set of only about 50 enzymes that were present in the eukaryote common ancestor [47], the core set being even smaller [80]. One might complain that 50 is not a small set of enzymes, but to put things in perspective, mitochondrial complex I alone contains 58 different subunits. Second, anaerobic eukaryote lineages are distributed all across the tree of eukaryotic life, interleaving with their aerobic relatives [47,101,102], indicating independent ecological specializations to low and high oxygen environments as the latter appeared during eukaryote evolution.
Because enzymes for aerobic and anaerobic energy metabolism were present in the eukaryote common ancestor, their vertical inheritance into the common ancestor of animals is not surprising, nor is the differential loss of unneeded enzymes in lineages that have specialized to strictly aerobic and anaerobic habitats, respectively [88,107]. Moreover, the enzymes involved in anaerobic energy metabolism are also found in some aerobes, including the algae [61]. A recent example is instructive. The genome of Naegleria gruberi, a relative of the brain-eating parasite Naegleria fowleri, was sequenced a few years back. Its genome harbored enzymes once thought to be specific to eukaryotic anaerobes, from which a metabolic map of anaerobic energy metabolism was proposed [108]. More recently it was found that Naegleria gruberi is a strict aerobe, generating ATP living from O2-dependent lipid oxidation [109]. The presence of genes for anaerobic energy metabolism in a strict aerobe is very difficult to explain as the result of selection operating on LGT events [100]. By contrast the anaerobes early view requires no special mechanisms other than normal ecophysiological specialization, and it meshes seamlessly with the evidence for late O2 accumulation in earth history (Fig. 1).
7. How do eukaryote anaerobes harness energy as ATP?
In the late 1990s it became generally accepted that hydrogenosomes are anaerobic forms of mitochondria, placing the origin of mitochondria deeper in eukaryotic history than anyone had imagined. The discoveries of very highly reduced forms of mitochondria — mitosomes [110,111] — in those eukaryotic lineages that possessed neither classical mitochondria nor hydrogenosomes, placed the origin of mitochondria as deep in eukaryotic history as any other trait that separates the eukaryotes from the prokaryotes. On top of that came the realization that eukaryotic anaerobes are distributed all across the eukaryotic tree, not just in obscure, restricted, or suspectedly primitive groups.
Today we know five different functionally characterized classes of mitochondria, all descended from one and the same endosymbiotic event [47,103]: Classical aerobic mitochondria which perform oxidative phosphorylation (OXPHOS) and use oxygen as the terminal electron acceptor (class 1), such as those found in rat liver; anaerobic mitochondria that produce ATP via OXPHOS but can use terminal electron acceptors other than oxygen, such as fumarate (class 2) as are found among several groups of invertebrates [80,112]; hydrogen-producing mitochondria possess a proton-translocating electron-transport chain but can also donate electrons of substrate oxidation via a hydrogenase to protons (class 3), which have so far only been characterized in ciliates like Nyctotherus ovalis [113]; hydrogenosomes (class 4), which occur in anaerobic groups of fungi, ciliates, trichomonads and other protists [47]; and mitosomes (class 5) which are found in some Microsporidia, Amoebozoa and Excavata [114,115]. Of the five classes of organelles of mitochondrial origin (OMOs), all but class 5 generate ATP from the breakdown of carbon compounds, but only class 1 and — dependent on environmental conditions or life cycle stage — class 2 generate ATP with the help of molecular oxygen [47]. So far, only class 1 and 2 mitochondria have been found in animal lineages. Other mitochondrial variants have been proposed from genome sequencing data [97,116], for example in Naegleria [108], but biochemical characterization revealed a functional map of mitochondrial energy metabolism for Naegleria [109] that contained none of the fermentation pathways proposed from the genome analysis.
Anaerobic forms of mitochondria and eukaryotic anaerobes occur in all of the six major groups of eukaryotes that biologists currently recognize [117], even within the animals. Anaerobic and hypoxic (oxygen-poor) environments are replete with eukaryotes [18,[38], [39], [40], [41],80,112,[118], [119], [120], [121], [122], [123]]. The following survey is adapted and updated from an earlier overview [102].
8. Anaerobic animals (opisthokonts)
The familiar map of mitochondrial ATP synthesis in eukaryotes that use oxygen as the terminal acceptor is shown in Fig. 2. Many animals exist that do not use oxygen as the terminal electron acceptor in ATP synthesis, hence they produce end-products other than water. The typical end-products are CO2, acetate, succinate, and propionate [47,120,124,125]. Among the animal lineages, the enzymatic details of anaerobic mitochondria have been studied only in a few model organisms [121]. The model systems reveal, however, minor variations on a conserved common theme called malate dismutation [121].
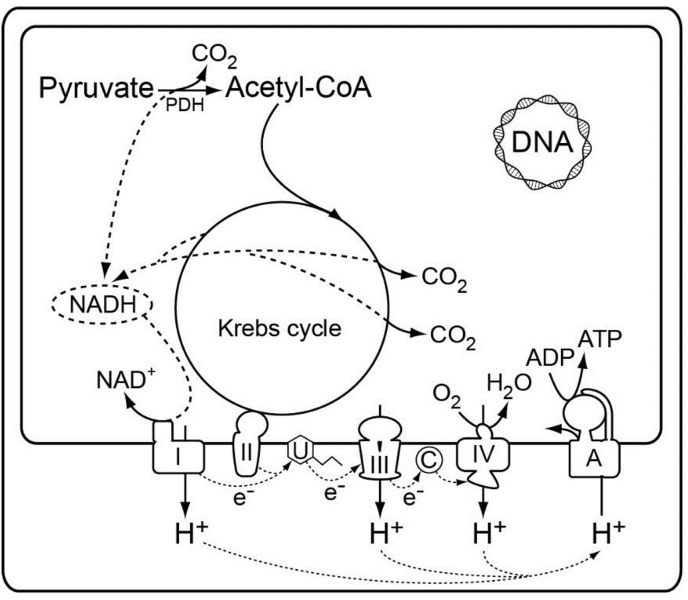
Fig. 2.
Generalized metabolic scheme of pyruvate oxidation and oxidative phosphorylation in a typical oxygen-respiring mitochondrion, for example, from rat liver. The map is redrawn after [47]. I to IV, respiratory complexes I to IV; A, ATPase; C, cytochrome c; PDH, pyruvate dehydrogenase complex; U, ubiquinone.
Among the animals, which belong to the eukaryotic supergroup called opisthokonts [117], many free-living marine invertebrates including various worms, mussels, and crustaceans, inhabit anaerobic environments or must survive under anaerobic conditions for prolonged periods of time. Their energy metabolism has been summarized in various extensive reviews [47,120,[124], [125], [126]]. The anaerobic energy metabolism of these animals very often entails the excretion of succinate as an end-product, whereby succinate is usually accompanied by propionate and acetate, and mixtures thereof are more the rule than the exception [47,80,118,120].
The anaerobic ATP-generating biochemistry of various marine invertebrates serves as an example here [118,[124], [125], [126], [127]]. Fig. 3 shows the one from the mussel Mytilus edulis. It closely parallels that characterized for several parasitic worms [80,112,[129], [130], [131]]. In succinate-producing, anaerobically functioning mitochondria (class 2), malate entering the mitochondrion is converted to fumarate, the terminal electron acceptor. Fumarate reductase (FRD) donates electrons from glucose oxidation to fumarate, yielding succinate. FRD requires a particular electron donor, rhodoquinone (RQ), that is reduced at complex I [47,80,121]. Complex I pumps protons from the matrix into the intermembrane space, allowing ATP synthesis via the mitochondrial ATPase. Succinate is either excreted as the end-product or it is participating in further reactions involving additional ATP gain through substrate level phosphorylation, whereby two additional end-products, acetate and propionate, are produced [47,80,112,124].
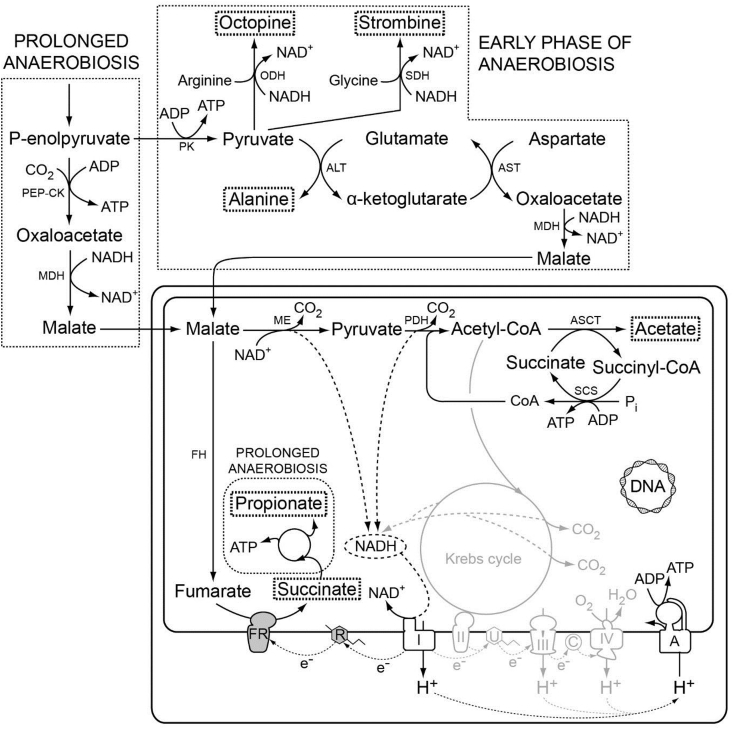
Fig. 3.
Major pathways of the facultative anaerobic energy metabolism in mitochondria of the mussel Mytilus edulis. The map is redrawn after [47]. Living attached to hard substrates, like rocks, in intertidal habitats, the bivalve has to face anaerobiosis periodically. Oxygen-independent cytosolic energy metabolism produces ATP via substrate-level phosphorylation accompanied by the formation of various end-products, including octopine, strombine, and alanine [128], which are boxed here. Under conditions of prolonged anaerobiosis, propionate is preferentially formed instead of succinate in mitochondria. Fumarate reduction is electron transfer chain coupled, and rhodoquinone serves as an electron donor to fumarate reductase. I to IV, respiratory complexes I to IV; A, ATPase; ALT, alanine aminotransferase; ASCT, acetate:succinate CoA transferase (subfamily 1B); AST, aspartate aminotransferase; C, cytochrome c; FH fumarase; FR, fumarate reductase; MDH, malate dehydrogenase; ME, malic enzyme; PDH, pyruvate dehydrogenase complex; ODH, octopine dehydrogenase; PEP-CK, phosphoenolpyruvate carboxykinase (ATP-dependent); PK, pyruvate kinase; R, rhodoquinone; SCS, succinyl-CoA synthetase; SDH, strombine dehydrogenase; U, ubiquinone.
9. Trichomonads (Excavata)
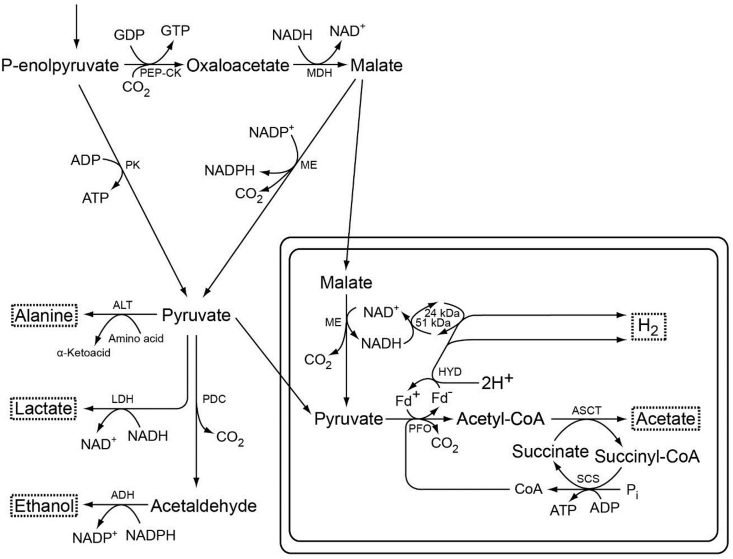
Trichomonads are anaerobic eukaryotes belonging to the large and diverse supergroup of eukaryotes called Excavata [117]. They possess hydrogenosomes (class 4), which are anaerobic forms of mitochondria that produce molecular hydrogen as an end-product of ATP synthesis as shown in Fig. 4 [44,47,[133], [134], [135]]. Hydrogenosomes were discovered in the anaerobic flagellate Tritrichomonas foetus [51] and subsequently found among ciliates [136], chytridiomycete fungi [137], and amoeboflagellates [138]. The paradigm for hydrogenosomal metabolism stems from work on the hydrogenosomes of the parabasalian flagellate Trichomonas vaginalis, the causative agent of a sexually transmitted disease in humans (trichomoniasis), and Tritrichomonas foetus, a pathogen of the bovine intestinal and reproductive tract [132,[139], [140], [141]].
Fig. 4.
Major pathways of the anaerobic, molecular hydrogen-producing, fermentative metabolism in hydrogenosomes of the flagellated protist Trichomonas vaginalis. The map is redrawn after [47]. Hydrogenosomal pyruvate breakdown involves pyruvate:ferredoxin oxidoreductase and functional 51-kDa and 24-kDa subunits of the NADH dehydrogenase module in complex I, which reoxidize NADH stemming from malate oxidation [52,139,144]. The 51-kDa and 24-kDa subunits of mitochondrial complex I function in association with [Fe]-HYD in Trichomonas [144]. Additional major end-products of cytosolic fermentations in T. vaginalis include alanine, lactate, ethanol, and glycerol [53]. End-products are boxed. ADH, alcohol dehydrogenase (NADPH-dependent); ALT, alanine aminotransferase; ASCT, acetate:succinate CoA transferase (subfamily 1C); 24 kDa/51 kDa, 24-kDa and 51-kDa subunits of the NADH dehydrogenase module of complex I; Fd, ferredoxin; HYD, hydrogenase; LDH, lactate dehydrogenase; MDH, malate dehydrogenase; ME, malic enzyme; PDC, pyruvate decarboxylase; PEP-CK, phosphoenolpyruvate carboxykinase (GTP-dependent); PFO, pyruvate:ferredoxin oxidoreductase; PK, pyruvate kinase; SCS, succinyl-CoA synthetase.
The typical end-products of the hydrogenosomal energy metabolism in trichmonads are one mol each H2, CO2, and acetate along with one mol of ATP [53]. Pyruvate and malate from glucose or glycogen degradation can be imported from the cytosol into the hydrogenosome. Malate is then converted in the organelle into pyruvate via malic enzyme [142,143]. Pyruvate is decarboxylated by pyruvate:ferredoxin oxidoreductase (PFO) in hydrogenosomes [132,143,144], generating CO2, acetyl-CoA, and reduced ferredoxin (Fd–). Fd– carries electrons to a ferredoxin-dependent [Fe]-hydrogenase [[145], [146], [147]], which donates them to protons to generate molecular hydrogen [132,144]. The CoA moiety of acetyl-CoA (from the PFO reaction) is transferred to succinate by acetate:succinate CoA transferase (ASCT), yielding acetate as an end-product, and succinyl-CoA. ATP is synthesized from ADP and Pi (inorganic phosphate) during the conversion of succinyl-CoA to succinate via substrate level phosphorylation, involving succinyl-CoA synthase (SCS), a canonical Krebs cycle enzyme [148]. The acetate-generating enzyme of hydrogenosomes, called ASCT (acetate:succinate CoA transferase), was originally described for Tritrichomonas foetus [149] and was characterized at the molecular level [150]. It is distinct from the ASCT enzyme that generates acetate as an end-product in trypanosome mitochondria [151,152].
Trichomonad hydrogenosomes contain components derived from complex I of the mitochondrial respiratory chain that help to maintain redox balance by reoxidizing NADH from the malic enzyme reaction [144]. The 51 kDa and 24 kDa proteins from mitochondrial complex I [144,153] take part in the production of H2 using NADH as electron donor [144]. The potential involvement of NADH in H2 production, as described by Hrdy et al. [144], might appear to be problematic at first sight, because the midpoint potential of NADH is not sufficiently negative to generate H2. Even though it is not known yet to occur in eukaryotes, the likely solution to this situation comes from a process discovered for prokaryotes called electron bifurcation [154,155]: Schut and Adams [156] showed that the trimeric [Fe]-hydrogenase of Thermotoga maritima operates in such a way as to accept one electron from NADH and one electron from the low-potential reduced Fd– generated by PFO per molecule of H2 produced. Despite the involvement of NADH, the overall reaction is energetically favored because of the participation of the low-potential ferredoxin, which drives the reaction forward.
Trichomonad hydrogenosomes contain two enzymes that were once thought to be specific to hydrogenosomes and specific to anaerobes: pyruvate:ferredoxin oxidoreductase (PFO) and iron-only hydrogenase ([Fe]-HYD), the latter being the enzyme that produces H2 (Fig. 4). Both enzymes occur also in the green alga Chlamydomonas reinhardtii [47,61,157,158] and in eukaryotes that were once thought to lack mitochondria altogether, such as Giardia intestinalis [47,111,159,160] or Trimastix pyriformis [161]. PFO, which has several FeS clusters, and its mechanism involves a radical intermediate [162,163], also occurs in Euglena gracilis mitochondria and in the apicomplexan Cryptosporidium parvum, but as a fusion protein that uses NADP+ as electron acceptor, rather than ferredoxin [164,165].
Trichomonas vaginalis typically experiences oxygen stress in its natural environment, for example during the transmission from one host to the other or with fluctuating vaginal oxygen levels during the menstruation cycle [[166], [167], [168]] and hence must possess mechanisms to avoid inactivation of oxygen-sensitive enzymes and to remove reactive oxygen species (ROS). T. vaginalis can readily tolerate O2 in small amounts [169]. Glutathione, a widespread antioxidant among eukaryotes, is absent in T. vaginalis, with cysteine possibly acting in its place [170]. A cytosolic NADH oxidase transfers four electrons directly to O2, yielding water [175,176]. The free energy available in this highly exergonic reaction is conserved neither as a proton gradient nor as ATP, the NADH oxidase acts as an O2 scavenger.
Individual proteins shown to be up-regulated during oxygen stress in T. vaginalis include superoxide dismutase (SOD) [[169], [170], [171]] and peroxiredoxins [169,172], ubiquitous enzymes that are thought to be central to defenses against ROS [173]. Thioredoxin reductases are also present in T. vaginalis as a component of the hydrogenosomal thioredoxin-linked antioxidant system [174].
Thus, O2 is both a toxin and a minor alternative acceptor for achieving redox balance in many anaerobic protists, and NADH oxidases of the type possessed by Trichomonas [171] are very widespread. The enzyme from Trichomonas [175,176] and that from the flagellated parasitic protist Giardia [177,178] have been characterized. Homologs of the NADH oxidase genes reported for Giardia and the pathogen Entamoeba [178] are common among eukaryotic genomes. Trichomonas possesses two diaphorases in the cytosol; the NADH-dependent enzyme yields H2O only, whereas the NADPH-dependent enzyme in addition yields H2O2 [179]. Giardia possesses a cytosolic NADH oxidase and a membrane-associated NADH peroxidase [180]. Based on their biochemical properties, the eukaryotic enzymes are similar to the prokaryotic NADH oxidases and NADH peroxidases, which produce H2O2 instead of water [181].
In addition to NADH oxidases, eukaryote anaerobes can possess flavodiiron proteins that function as O2 scavengers, as characterized for Trichomonas hydrogenosomes [48] and for Giardia [182]. They transfer electrons to O2 to produce water [48]. Thus, the parasite seems to utilize a sophisticated system to buffer oxygen stress, which includes a cytosolic and a hydrogenosomal antioxidant system in combination [172,174]. Flavodiiron proteins have close homologs encoded by several sequenced eukaryote genomes, including Entamoeba and several green algae. One might wonder why green algae, which are typically O2 producers, should possess O2-scavenging enzymes typical of anaerobes. The answer is probably one of successful generalist strategies. Some algae, such as Chlamydomonas reinhardtii, can switch from O2 production to vigorous anaerobic growth in the dark within 30 min, producing large amounts of H2 using very O2-sensitive enzymes for fermentative ATP synthesis [158,183]; accordingly, O2 detoxification is an issue for Chlamydomonas and similar algae during anaerobic growth. Since Chlamydomonas is a typical soil inhabitant [184], it can regularly encounter anaerobic conditions.
10. Fungi (opisthokonts): hydrogenosomes, denitrification, and sulfur reduction
The fungi, like the animals, belong to the opisthokont supergroup [117]. Some anaerobic fungi that live in low oxygen environments possess hydrogenosomes (class 4). The energy metabolism of representatives from the genus Neocallimastix [135,137,185] and from the genus Piromyces [186] has been studied. Typical end-products for the anaerobic fungi are acetate, lactate, hydrogen, ethanol, and formate. The production of formate is the main difference to trichomonad hydrogenosomes [187], it entails the activity of pyruvate:formate lyase (PFL). Also, anaerobic fungi produce ethanol from acetyl-CoA by a bifunctional aldehyde/alcohol dehydrogenase (ADHE; also called alcohol dehydrogenase E). The presence of PFL and ADHE distinguishes anaerobic fungi from trichomonads, but both enzymes also occur in Chlamydomonas [61,157,158], while ADHE is also found in a colorless relative of Chlamydomonas, Polytomella [157,188]. ATP is thought to be generated in fungal hydrogenosomes by the same acetate-producing SCS-ASCT route as in parabasalids, with acetate as the major end-product [186].
In Fusarium oxysporum, an ascomycete fungal pathogen of plants, an interesting anaerobic respiratory pathway of ATP synthesis occurs — denitrification, the conversion of nitrate to gaseous compounds, including nitrogen gas (N2) [[189], [190], [191], [192], [193]]. Denitrification has also been reported for benthic foraminifera [194], although the mechanisms remain unclear. The possibility that denitrification in foraminifera is likely catalyzed by endobionts, grouping in a phylogenetic analysis of 16S rRNA genes within the γ-proteobacteria [195] is debated. Recently evidence for a novel eukaryotic denitrification pathway encoded in foraminiferal genomes was reported [196]. In Fusarium, denitrification occurs under low-oxygen conditions, involves mitochondria, and entails the oxidation of reduced carbon sources, such as ethanol, to acetate with deposition of the electrons onto nitrate to generate N2O or NH3, depending upon growth conditions [[197], [198], [199], [200]]. Abe et al. [200] further showed that metabolically flexible Fusarium grows under anaerobic conditions on a variety of reduced carbon sources using elemental sulfur as the terminal electron acceptor, generating H2S as the reduced end-product in a 2:1 M ratio relative to acetate. The use of elemental sulfur as a terminal electron acceptor is unique among eukaryotes studied so far.
11. Other eukaryotes with hydrogenosomes
The ciliates belong to the major eukaryotic supergroup called ‘chromalveolates’ [117], or based on the newer phylogenetic studies to the supergroup SAR – acronym of stramenopiles, alveolates, and Rhizaria [201]. Several lineages of anaerobic ciliates possess hydrogenosomes [38,202,203]. The hydrogenosome-containing lineages are dispersed across the phylogenetic diversity of the group [103,204]. Major metabolic end-products measured for the anaerobic ciliate Trimyema compressa included acetate, lactate, ethanol, formate and hydrogen along with traces of succinate [205]. The biochemistry and the ecology of the rumen ciliates Dasytricha ruminantium and Isotricha spp., were reviewed by Williams [206].
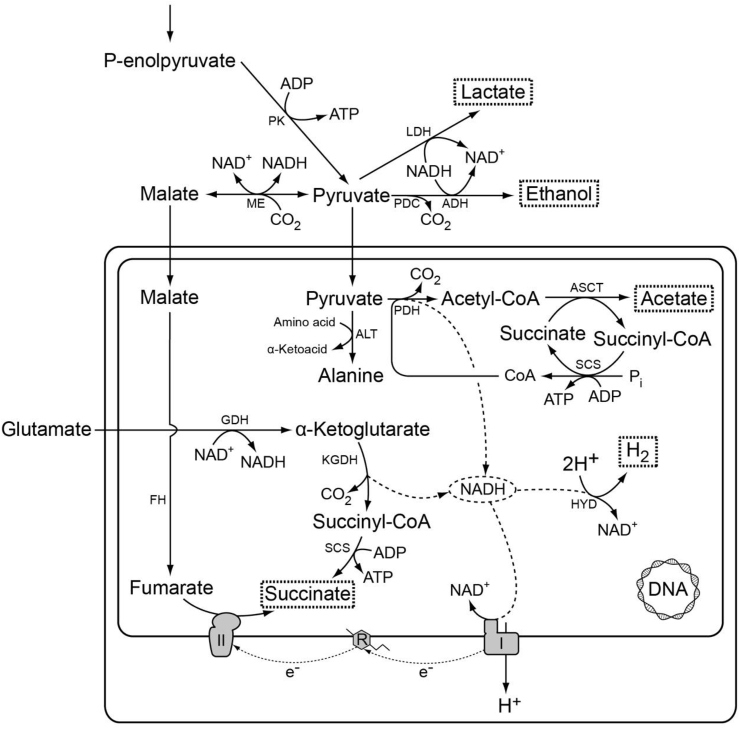
One hydrogenosome-bearing anaerobic ciliate, Nyctotherus ovalis, that inhabits the hindgut of cockroaches [207], attained attention for being the only organism known at that time whose hydrogenosomes still possess a genome [113]. According to Müller et al. [47] this protist harbors organelles of a mitochondrial origin, considered to be a link between mitochondria and hydrogenosomes, since this organelle unites the hallmark features of mitochondria and those of hydrogenosomes, a hydrogen-producing mitochondrion (class 3). On the one hand, it contains DNA coding for respiratory chain components [113,[208], [209], [210]] and an electron transport chain, and on the other hand it produces hydrogen through an iron-only hydrogenase using protons as terminal electron acceptors [113]. This finding established once and for all the evolutionary identity of mitochondria and hydrogenosomes as different manifestations of one and the same organelle.
Nyctotherus was shown to consume glucose as a substrate and to convert it to the major metabolic end-products acetate, succinate, lactate, and ethanol, which are excreted [113]. Acetate and succinate are probably produced within the hydrogen-producing mitochondrion (see Fig. 5), in which succinate production occurs via a part of the Krebs cycle (malate-fumarate-succinate), used in a reductive direction [208]. Acetate is stemming from pyruvate, which is converted to acetyl-CoA by pyruvate dehydrogenase (PDH) and subsequently to acetate by an acetate:succinate CoA transferase (ASCT). The ASCT is belonging to subfamily 1A, with sequence similarity to the ASCT from Trypanosoma brucei [212]. ASCT transfers the CoA moiety of acetyl-CoA to succinate, yielding acetate and succinyl-CoA, which is subsequently converted by succinyl-CoA synthetase (SCS) with a concomitant ATP-production. The oxidative decarboxylation of pyruvate to acetyl-CoA by PDH results in the reduction of NAD+ to NADH. The oxidation of NADH in Nyctotherus is thought to occur in part by a truncated electron transport chain in which complex I passes the electrons from NADH through rhodoquinone to complex II, which then uses fumarate as an electron acceptor to produce succinate. Another part of NADH is possibly reoxidized by the [Fe]-hydrogenase, involving the 51- and 24-kDa subunits of complex I that are C-terminally fused to the [Fe]-HYD catalytic subunit [156,213], thereby releasing molecular hydrogen. The hydrogen-producing mitochondrion of Nyctotherus appears to generate a proton gradient [113], possibly involving complex I of the electron transport chain. This proton gradient is probably not used for ATP synthesis, as a gene encoding an ATP synthase seems to be lacking [208]. The proposed metabolism of Nyctotherus, for which the metabolic end-products have been characterized (Fig. 5), provided a good model, in terms of gene presence or absence, for the inferred organellar metabolism of the phylogenetically distant anaerobic stramenopile Blastocystis [208], for which the metabolic end-products have not been characterized. Electron microscopy revealed the presence of methanogenic bacteria in close association with these organelles in Nyctotherus, which demonstrates the in vivo production of hydrogen by the organelles [214]. The presence of the [Fe]-hydrogenase gene in the genome of Nyctotherus further indicates hydrogen production [213].
Fig. 5.
Tentative map of major pathways of energy metabolism in hydrogen-producing mitochondria of the anaerobic ciliate Nyctotherus ovalis. The map is redrawn after [47]. The incomplete Krebs cycle is likely used in the reductive direction [211]. A proton gradient is generated, probably by a functional respiratory complex I, which passes the electrons from the NADH pool through rhodoquinone to complex II, acting as fumarate reductase synthesizing succinate [208]. Redox balance is also achieved with the help of hydrogenase, releasing molecular hydrogen [113]. ATP can be synthesized by substrate-level phosphorylation, producing acetate. I, respiratory complex I; II, fumarate reductase/succinate dehydrogenase; ADH, alcohol dehydrogenase (NADH-dependent); ALT, alanine aminotransferase; ASCT, acetate:succinate CoA transferase subfamily 1A; FH fumarase; GDH, glutamate dehydrogenase; HYD, hydrogenase; KGDH, alpha-ketoglutarate dehydrogenase; LDH, lactate dehydrogenase; ME, malic enzyme; R, rhodoquinone; PDC, pyruvate decarboxylase; PDH, pyruvate dehydrogenase; PK, pyruvate kinase; SCS, succinyl-CoA synthetase.
The free-living anaerobic amoeboflagellate Psalteriomonas lanterna [138] belongs to a group of eukaryotes called ‘heteroloboseans’, which are, like the trichomonads, members of the supergroup Excavata [117]. The presence of methanogenic endosymbionts within P. lanterna and within the sister group P. vulgaris [138,215] serves as a positive biochemical bioassay that their organelles are producing hydrogen [43]. De Graaf et al. [216] identified several hydrogenosomal key-genes encoding proteins like PFO and a hydrogenase, but also mitochondrial genes, like the mitochondrial complex I subunit (51 kDa). Furthermore, a ferredoxin was identified in Psalteriomonas lanterna, which is similar to the hydrogenosomal ferredoxin of Trichomonas vaginalis [217]. Comparisons of the organellar ultrastructure and hydrogenosomes suggest that both organelles are very similar [216]. Thus, the Psalteriomonas organelles are classified as hydrogenosomes (class 4). Two additional heteroloboseans living in anoxic environments were suggested to possess hydrogenosomes: Monopylocystis visvesvarai and Sawyeria marylandensis [218]. In addition to those mentioned so far, several other eukaryotic groups are suspected to possess hydrogenosomes, too [90].
12. Groups with mitosomes
Mitosomes (class 5) are the most highly reduced forms of mitochondria known, which do not produce ATP. They were discovered independently by Tovar et al. [110] and Mai et al. [219] in the human intestinal parasite Entamoeba histolytica. Mai et al. [219] called the organelle a “crypton”, but the name “mitosome” suggested by Tovar et al. [110] has stuck. Mitosomes are smaller than mitochondria or hydrogenosomes and have been subsequently found among Microsporidia [220], in Giardia [111], with the list of organisms having previously overlooked forms of mitochondria growing rapidly [90,133,161,221,222].
Because carbon flux and energy metabolism in Entamoeba are known to be cytosolic processes [140], the role of mitosomes in the core energy metabolism, if any, can be peripheral at best. Enzymes of sulfate activation were localized to the mitosomes of Entamoeba [221,223], providing a perspective on mitosomal functions. Proteomic studies of Giardia mitosomes revealed the presence of all key components of the FeS cluster assembly machinery, including the cysteine desulfurase, IscS, the scaffold proteins IscU, Nfu, and IscA, and monothiol glutaredoxin [224,225]. Mitosomes also contain [2Fe-2S]-ferredoxin, which might provide the reducing equivalents required for the formation of FeS clusters, and a complete set of chaperones that are involved in the transfer of preassembled FeS clusters into apoproteins. Hence, mitosomes of some lineages have retained components of FeS cluster assembly [111,114] and others have retained components of sulfate activation [223]. This suggests that FeS protein assembly is the key function of mitosomes in Microsporidia [114,226]. Other possible functions might have to do with FeS cluster maturation and hydroperoxide detoxification via the rubrerythrin system [227]. The Entamoeba genome encodes several [Fe]-hydrogenase homologs [228], the presence of mRNA for a short-form [Fe]-hydrogenase in Entamoeba was shown by reverse transcription (RT)-PCR [229], and the activity of recombinant Entamoeba hydrogenase expressed in Escherichia coli was reported, but endogenous hydrogenase enzymatic activity in Entamoeba has not yet been described.
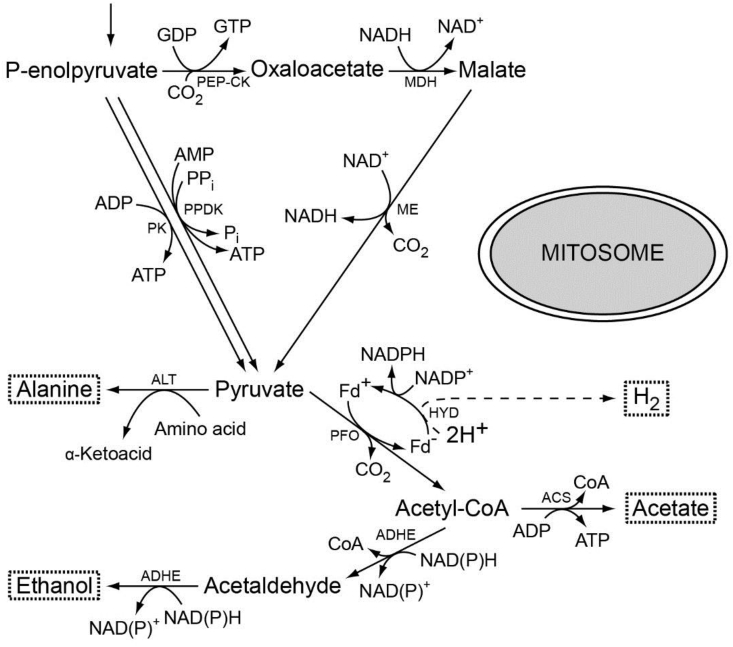
Although hydrogenosomes and mitosomes are both mitochondrion-derived organelles [103,115,133,134], there is one important difference between them — mitosomes do not produce ATP. In two biochemically well-studied taxa that possess mitosomes, Entamoeba [230,231] and Giardia, the enzymes of energy metabolism are localized to the cytosol as shown in Fig. 6 [120]. The main end-products are acetate and ethanol [139], although Giardia can also produce hydrogen under highly anoxic conditions [159]. The enzymes involved in end-product formation are PFO [232], ADHE [233], and acetyl-CoA synthase (ADP-forming) (ACS-ADP) [234,235]. AHDE allows electrons from glucose oxidation to be excreted as ethanol while ACS-ADP yields ATP through substrate-level-phosphorylation. In Entamoeba, ethanol can also be produced via two additional alcohol dehydrogenases, ADH1 and ADH3 [[236], [237], [238]].
Fig. 6.
Major pathways of the energy metabolism in the parasite Giardia intestinalis. The map is redrawn after [47]. Giardia mitosomes are not directly involved in energy metabolism but are involved in FeS cluster biogenesis [111]. The typically hydrogenosomal (and sometimes mitochondrial) enzymes PFO and [Fe]-HYD have been recompartmentalized to the cytosol during evolution. Molecular hydrogen is produced under strictly anoxic conditions [159], as indicated by the dashed line. ACS, acetyl-CoA synthetase (ADP-forming); ADHE, alcohol dehydrogenase E; ALT, alanine aminotransferase; Fd, ferredoxin; HYD, hydrogenase; MDH, malate dehydrogenase; ME, malic enzyme; PEP-CK, phosphoenolpyruvate carboxykinase (GTP-dependent); PFO, pyruvate:ferredoxin oxidoreductase; PPDK, pyruvate:orthophosphate dikinase; PK, pyruvate kinase.
13. Eukaryotes lacking typical mitochondria
The last eukaryote common ancestor possessed mitochondria and was a complex cell, but whether mitochondria or complexity came first in eukaryotic evolution is still discussed. In autogenous models (complexity first), the origin of phagocytosis poses the limiting step at eukaryote origin, with mitochondria coming late as an undigested growth substrate. In symbiosis-based models (mitochondria first), the host was an archaeon, and the origin of mitochondria was the limiting step at eukaryote origin, with mitochondria providing bacterial genes, ATP synthesis on internalized bioenergetic membranes, and mitochondrion-derived vesicles as the seed of the eukaryote endomembrane system [86]. A highly reduced eukaryote, Monocercomonoides sp., which apparently lacks mitochondria altogether [239,240], has recently been described. However, this amoeba branches within a eukaryotic group that possesses mitochondria, such that the lack of mitochondria in Monocercomonoides is a secondarily derived trait. Clearly, the ancestral eukaryote was a very complex organism with the full set of traits that distinguish eukaryotes from prokaryotes: mitochondria, an endomembrane system, a nucleus, meiosis, mitosis, a cell cycle, and the like [86,104]. Reductive evolution is very common in both prokaryotes [241] and eukaryotes [242]. Phagocytosis-first theories predicted that eukaryotes lacking mitochondria should be primitively amitochondriate, evidence of which was never more lacking than now.
A good bit of progress has been made in understanding the role of mitochondria in eukaryote evolution in recent years. First, all eukaryote lineages are now known either to have or to have had a mitochondrion in their past [243]. Second, the host that acquired the mitochondrion stems from a lineage that branches within the archaebacteria (or archaea), not as their sister [[244], [245], [246]]. Third, the presence of internalized bioenergetic membranes was the key attribute provided by mitochondrial endosymbiosis, which afforded eukaryotes many orders of magnitude more energy per gene than is available to prokaryotes [87]. Thus, while it has now been evident for some time that the common ancestor of eukaryotes possessed a mitochondrion, it is now clear why that was so: The lack of true intermediates in the prokaryote-to-eukaryote transition has a bioenergetic cause [87].
14. Two facultative anaerobes that produce oxygen
The green alga Chlamydomonas belongs to the supergroup of eukaryotes bearing primary plastids called Archaeplastida [117]. It provides an example of a facultative anaerobe among eukaryotes. Chlamydomonas reinhardtii can switch from aerobic metabolism to anaerobic growth in the dark within 30 min, generating acetate, CO2, formate, ethanol, and hydrogen as major end-products and traces of lactate and glycerol [158]. Hydrogenases are playing a central role in the H2 development [247,248]. Grown aerobically, Chlamydomonas reinhardtii respires oxygen with a normal manifestation of oxidative decarboxylation via pyruvate dehydrogenase and oxidative phosphorylation in mitochondria [249]. But when grown anaerobically, it rapidly expresses PFO [157,158], PFL [157], [Fe]-HYD [250], ADHE (bifunctional alcohol dehydrogenase E) [188], acetate kinase (ACK), and phosphotransacetylase (PTA). Chlamydomonas thus expresses many of the enzymes that are to a large extent the same as those of other anaerobic protists.
Initial metabolite both for the aerobic and anaerobic metabolism is pyruvate stemming from starch breakdown [47,61]. During anaerobiosis pyruvate is metabolized to lactate or ethanol in the cytosol and the chloroplast or it is decarboxylated into acetyl-CoA by pyruvate formate lyase (PFL), targeted to the chloroplast and the mitochondrion [157]. The catalytic mechanism of PFL involves a glycyl radical. This radical is generated by a PFL-activating enzyme (PFL-AE), which is present in Chlamydomonas and other eukaryotes that possess PFL [93,251]. The deactivating enzyme for PFL is ADHE, which is also present in Chlamydomonas and generally in eukaryotes that possess PFL [157]. In the chloroplast, the decarboxylation of pyruvate to acetyl-CoA can also be catalyzed by the oxygen-sensitive enzyme pyruvate:ferredoxin oxidoreductase (PFO) under the reduction of ferredoxin [252]. The reduced ferredoxin releases its electrons to a plastidal iron-only hydrogenase, which donates them to protons generating molecular hydrogen as the terminal electron acceptor [61]. The presence of PFO in Chlamydomonas reinhardtii was first obtained from the genome and only recently investigated [252]. While PFO in anaerobic eukaryotes is located in the cytosol or the hydrogenosomes and its alternative form PNO (pyruvate:NADP+ oxidoreductase) in the mitochondrion [47], PFO in Chlamydomonas reinhardtii, however, is located in the chloroplast [252]. Phylogenetic analysis revealed that eukaryotic PFO is a biochemical relict stemming from a facultatively anaerobic, eubacterial ancestor of mitochondria [164] and that the nuclear gene for mitochondrial, hydrogenosomal, and cytosolic PFO originated from one single eubacterial acquisition, i.e. the hydrogenosomal PFO in Trichomonas vaginalis [143].
Another facultative anaerobe is Euglena gracilis, that belongs to the supergroup Excavata [117]. Euglenids are a broad and diverse group containing many typical flagellate inhabitants of shallow freshwater environments, but relatives can also be found in anoxic marine [39,44] and anoxic freshwater [253] ecosystems, whereby some members possess organelles ultrastructurally similar to hydrogenosomes [44,253]. Only one member of the euglenids has been extensively studied from a biochemical standpoint: Euglena gracilis [254,255]. The mitochondrion has been described in detail recently [256]. Euglena has secondary plastids and can produce oxygen. Grown aerobically, Euglena gracilis expresses pyruvate dehydrogenase (PDH) in mitochondria [257] and respires O2 using a slightly modified Krebs cycle that is also found among some α-proteobacteria [258]. The shunt involves the replacement of α-ketoglutarate dehydrogenase by α-ketoglutarate decarboxylase and succinate semialdehyde dehydrogenase [254].
When oxygen is absent, E. gracilis uses acetyl-CoA as the terminal electron acceptor of glucose oxidation and produces an unusual end-product: wax esters [254,[259], [260], [261], [262], [263], [264]]. Some E. gracilis strains accumulate wax esters at levels up to 40 μg per 106 Euglena cells [259] or up to 65% of their dry weight [262]. Acid mine drainage biofilms mainly constituted by Euglena mutabilis contain large amounts of wax esters [265]. The wax esters are not excreted but accumulate in the cytosol instead [262,263]. Under anaerobic conditions PDH protein levels decrease in Euglena [257] and PFO is expressed, but as a fusion protein [164,165], and a trans-2-enoyl-CoA reductase (NADPH-dependent) circumvents the reversal of an O2-dependent step in β-oxidation [266,267]. Similar to the situation in anaerobic mitochondria of metazoa (Fig. 3), Euglena's wax ester fermentation involves mitochondrial fumarate reduction, and thus utilizes RQ [257] for the synthesis of propionyl-CoA [268] via the same route that the mitochondria of anaerobic animals use to excrete propionate. Fatty acids are synthesized from acetyl-CoA condensation with an acyl-CoA (starting with acetyl-CoA or propionyl-CoA), a reduction of the resulting 3-oxoacid to 3-hydroxy acid, dehydration thereof, and a reduction of the resulting trans-enoyl-CoA to the elongated acyl-CoA. In contrast to malonyl-CoA-dependent fatty acid synthesis, the acetyl-CoA-dependent Euglena route allows net fermentative ATP synthesis from glucose, because acetyl-CoA is condensed without prior ATP-dependent carboxylation to malonyl-CoA [259,261,268]. The step catalyzed by trans-2-enoyl-CoA reductase (NADPH-dependent) circumvents the reversal of an O2-dependent step in β-oxidation [266]. A portion of the fatty acids is reduced to alcohols, esterified with another fatty acid, and deposited into the cytosol as wax (wax ester fermentation). Under aerobic conditions, the wax esters can be converted back to acetyl-CoA, which can be oxidized to CO2 in the mitochondria or used to form paramylon (β-1,3-glucan) reserves [61,269,270]. This metabolism probably involves β-oxidation, the glyoxylate cycle and conversion of acetyl-CoA, which either enters the glyoxylate cycle to be converted to succinate which can be used for gluconeogenesis, or the acetyl-CoA enters the modified Krebs cycle to be oxidized to CO2 [266,271].
The presence and the use of typical components of the anaerobic energy metabolism in two distantly related eukaryotes, Chlamydomonas (archaeplastida supergroup) and Euglena (excavate supergroup), which both not only consume but even produce oxygen in the light, indicates that there is no evolutionary divide between aerobic and anaerobic eukaryotes. The divide is primarily one of ecological preference, not of evolutionary potential.
15. Conclusion
The mitochondria of diverse invertebrate lineages can respire oxygen at presently available levels and can perform malate dismutation under anaerobic conditions [47,80], this clearly suggests that the first animals could do the same and thus possessed facultatively anaerobic mitochondria with fumarate reductase and RQ. A dozen or so genes for anaerobic fermentations also entered the eukaryotic lineage at mitochondrial origin, such that aerobic respiration, anaerobic respiration and hydrogen-producing fermentations, in addition to heterotrophy in general, entered the eukaryotic lineage at mitochondrial origin, as a single inheritance from the facultatively anaerobic metabolism of the mitochondrial endosymbiont, followed by ecological specialization and differential loss in independent mitochondrion bearing eukaryotic lineages [88,107]. While aerobic eukaryotes lost the ability to survive under anoxic conditions, anaerobic eukaryotes kept the anaerobic metabolism for specialization to anaerobic niches. Some eukaryotes possess genes for enzymes pivotal for anaerobic fermentations even though they produce O2 during photosynthesis (algae) while others possess genes for anaerobic energy metabolism even though they strictly require O2 for survival (Naegleria). Anaerobic energy metabolism in eukaryotes carries the rather unmistakable imprint of a single acquisition via endosymbiosis in that it was acquired once, it represents a very narrow sample of preexisting prokaryotic metabolic diversity, and like RQ, it traces physiologically to a particular group: facultatively anaerobic α-proteobacteria. It is often stated or assumed that O2 improves the energetics of the cell and that O2 thus might have impacted the origin of mitochondria, but life in O2 is thermodynamically thirteen times more expensive than life without O2, because O2 tends to oxidize things, including the chemical substance of cells.
Conflicts of interest
The authors have declared that no competing interests exist.
Acknowledgement
WFM thanks the European Research Council (666053) and the Volkswagen Foundation (93 046) for funding. MM thanks the Slovak Grant Agency (VEGA No. 1/0798/16). The funders had no role in the preparation of the article.
Contributor Information
Verena Zimorski, Email: zimorski@hhu.de.
Marek Mentel, Email: marek.mentel@uniba.sk.
Aloysius G.M. Tielens, Email: a.tielens@erasmusmc.nl, A.G.M.Tielens@uu.nl.
William F. Martin, Email: bill@hhu.de.
References
- 1.Tashiro T., Ishida A., Hori M., Igisu M. Early trace of life from 3.95 Ga sedimentary rocks in Labrador, Canada. Nature. 2017;549:516–518. doi: 10.1038/nature24019. [DOI] [PubMed] [Google Scholar]
- 2.Arndt N., Nisbet E. Processes on the young Earth and the habitats of early life. Annu. Rev. Earth Planet Sci. 2012;40:521–549. [Google Scholar]
- 3.Ueno Y., Yamada K., Yoshida N., Maruyama S. Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature. 2006;440:516–519. doi: 10.1038/nature04584. [DOI] [PubMed] [Google Scholar]
- 4.Fischer W.W., Hemp J., Valentine J.S. How did life survive Earth's great oxygenation? Curr. Opin. Chem. Biol. 2016;31:166–178. doi: 10.1016/j.cbpa.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Lenton T.M., Dahl T.W., Daines S.J., Mills B.J.W. Earliest land plants created modern levels of atmospheric oxygen. Proc. Natl. Acad. Sci. U.S.A. 2016;113:9704–9709. doi: 10.1073/pnas.1604787113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stolper D.A., Keller B. A record of deep-ocean dissolved O2 from the oxidation state of iron in submarine basalts. Nature. 2018;553:323–327. doi: 10.1038/nature25009. [DOI] [PubMed] [Google Scholar]
- 7.Krause A.J., Mills B.J.W., Zhang S., Planavsky N.J., Lenton T.M., Poulton S.W. Stepwise oxygenation of the Paleozoic atmosphere. Nat. Commun. 2018;9:4081. doi: 10.1038/s41467-018-06383-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Javaux E.J., Lepot K. The Paleoproterozoic fossil record: implications for the evolution of the biosphere during Earth's middle-age. Earth Sci. Rev. 2018;176:68–86. [Google Scholar]
- 9.Martin W.F., Allen J.F. An algal greening of land. Cell. 2018;174:256–258. doi: 10.1016/j.cell.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 10.Bar-On Y.M., Phillips R., Milo R. The biomass distribution on Earth. Proc. Natl. Acad. Sci. U.S.A. 2018;115:6506–6511. doi: 10.1073/pnas.1711842115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daines S.J., Mills B.J.W., Lenton T.M. Atmospheric oxygen regulation at low Proterozoic levels by incomplete oxidative weathering of sedimentary organic carbon. Nat. Commun. 2017;8:14379. doi: 10.1038/ncomms14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishiyama T., Hidetoshi S., de Vries J., Buschmann H. The Chara genome: secondary complexity and implications for plant terrestrialization. Cell. 2018;174:448–464. doi: 10.1016/j.cell.2018.06.033. [DOI] [PubMed] [Google Scholar]
- 13.Morris J.L., Puttick M.N., Clark J.W., Edwards D. The timescale of early land plant evolution. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E2274–E2283. doi: 10.1073/pnas.1719588115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reski R. Enabling the water-to-land transition. Native Plants. 2018;4:67–68. doi: 10.1038/s41477-018-0101-5. [DOI] [PubMed] [Google Scholar]
- 15.Lu W., Ridgwell A., Thomas E., Hardisty D.S. Late inception of a resiliently oxygenated upper ocean. Science. 2018;361:174–177. doi: 10.1126/science.aar5372. [DOI] [PubMed] [Google Scholar]
- 16.Sahoo S.K., Planavsky N.J., Kendall B., Wang X. Ocean oxygenation in the wake of the Marinoan glaciation. Nature. 2012;489:546–549. doi: 10.1038/nature11445. [DOI] [PubMed] [Google Scholar]
- 17.Lyons T.W., Reinhard C.T., Planavsky N.J. The rise of oxygen in Earth's early ocean and atmosphere. Nature. 2014;506:307–315. doi: 10.1038/nature13068. [DOI] [PubMed] [Google Scholar]
- 18.Porter S.M., Agic H., Riedman L.A. Anoxic ecosystems and early eukaryotes. Emerg Top Life Sci. 2018;2:299–309. doi: 10.1042/ETLS20170162. [DOI] [PubMed] [Google Scholar]
- 19.Long J.A., Gordon M.S. The greatest step in vertebrate history: a paleobiological review of the fish-tetrapod transition. Physiol. Biochem. Zool. 2004;77:700–719. doi: 10.1086/425183. [DOI] [PubMed] [Google Scholar]
- 20.Summons R.E., Jahnke L.L., Hope J.M., Logan G.A. 2-Methylopanoids as biomarkers for cyanobacterial oxygenic photosynthesis. Nature. 1999;400:554–557. doi: 10.1038/23005. [DOI] [PubMed] [Google Scholar]
- 21.Butterfield N.J. Early evolution of eukaryote. Palaeontology. 2015;58:5–17. [Google Scholar]
- 22.Budd G.E. The earliest fossil record of the animals and its significance. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363:1425–1434. doi: 10.1098/rstb.2007.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maloof A.C., Rose C.V., Beach R., Samuels B.M. Possible animal-body fossils in pre-Marinoan limestones from South Australia. Nat. Geosci. 2010;3:653–659. [Google Scholar]
- 24.Hoyal Cuthill J.F., Han J. Cambrian petalonamid Stromatoveris phylogenetically links Ediacaran biota to later animals. Palaeontology. 2018;61:813–823. [Google Scholar]
- 25.Parfrey L.W., Lahr D.J., Knoll A.H., Katz L.A. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Betts H.C., Puttick M.N., Clark J.W., Williams T.A. Integrated genomic and fossil evidence illuminates life's early evolution and eukaryote origin. Nat Ecol Evol. 2018;2:1556–1562. doi: 10.1038/s41559-018-0644-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Towe K.M. Oxygen collagen priority and early metazoan fossil record. Proc. Natl. Acad. Sci. U.S.A. 1970;65:781–788. doi: 10.1073/pnas.65.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shoulders M.D., Raines R.T. Collagen structure and stability. Annu. Rev. Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myllyharju J. Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biol. 2003;22:15–24. doi: 10.1016/s0945-053x(03)00006-4. [DOI] [PubMed] [Google Scholar]
- 30.Javaux E.J., Knoll A.H., Walter M.R. Morphological and ecological complexity in early eukaryotic ecosystems. Nature. 2001;412:66–69. doi: 10.1038/35083562. [DOI] [PubMed] [Google Scholar]
- 31.Anbar A.D., Knoll A.H. Proterozoic ocean chemistry and evolution: a bioinorganic bridge? Science. 2002;297:1137–1142. doi: 10.1126/science.1069651. [DOI] [PubMed] [Google Scholar]
- 32.Knoll A.H., Javaux E.J., Hewitt D., Cohen P. Eukaryotic organisms in Proterozoic oceans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006;361:1023–1038. doi: 10.1098/rstb.2006.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knoll A.H. Paleobiological perspectives on early eukaryotic evolution. Cold Spring Harb Perspect Biol. 2014;6:a016121. doi: 10.1101/cshperspect.a016121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Butterfield N.J. The neoproterozoic. Curr. Biol. 2015;25:R859–R863. doi: 10.1016/j.cub.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 35.Douzery E.J., Snell E.A., Bapteste E., Delsuc F. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc. Natl. Acad. Sci. U.S.A. 2004;101:15386–15391. doi: 10.1073/pnas.0403984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chernikova D., Motamedi S., Csürös M., Koonin E.V. A late origin of the extant eukaryotic diversity: divergence time estimates using rare genomic changes. Biol. Direct. 2011;6:26. doi: 10.1186/1745-6150-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antcliffe J.B., Callow R.H.T., Brasier M.D. Giving the early fossil record of sponges a squeeze. Biol. Rev. 2014;4:972–1004. doi: 10.1111/brv.12090. [DOI] [PubMed] [Google Scholar]
- 38.Fenchel T., Finlay B.J., editors. Ecology and Evolution in Anoxic Worlds. Oxford University Press; Oxford, UK: 1995. [Google Scholar]
- 39.Bernhard J.M., Buck K.R., Farmer M.A., Bowser S.S. The Santa Barbara Basin is a symbiosis oasis. Nature. 2000;403:77–80. doi: 10.1038/47476. [DOI] [PubMed] [Google Scholar]
- 40.Stoeck T., Behnke A., Christen R., Amaral-Zettler L. Massively parallel tag sequencing reveals the complexity of anaerobic marine protistan communities. BMC Biol. 2009;7:72. doi: 10.1186/1741-7007-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beinart R.A., Beaudoin D.J., Bernhard L.M., Edgcomb V.P. Insights into the metabolic functioning of a multipartner ciliate symbiosis from oxygen-depleted sediments. Mol. Ecol. 2018;27:1794–1807. doi: 10.1111/mec.14465. [DOI] [PubMed] [Google Scholar]
- 42.Hanousková P., Táborsky P., Cepicka I. Dactylomonas gen. nov., a novel lineage of heterolobosean flagellates with unique ultrastructure, closely related to the amoeba Selenaion koniopes Park, De Jockheere, Simpson, 2012. J. Eukaryot. Microbiol. 2019;66:120–139. doi: 10.1111/jeu.12637. [DOI] [PubMed] [Google Scholar]
- 43.Embley T.M., Finlay B.J. The use of small subunit rRNA sequences to unravel the relationships between anaerobic ciliates and their methanogen endosymbionts. Microbiol. 1994;140:225–235. doi: 10.1099/13500872-140-2-225. [DOI] [PubMed] [Google Scholar]
- 44.Edgcomb V.P., Leadbetter E.R., Bourland W., Beaudoin D. Structured multiple endosymbiosis of bacteria and archaea in a ciliate from marine sulfidic sediments: a survival mechanism in low oxygen, sulfidic sediments? Front. Microbiol. 2011;2:1–16. doi: 10.3389/fmicb.2011.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welte C., Deppenmeier U. Bionergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim. Biophys. Acta. 2014;1837:1130–1147. doi: 10.1016/j.bbabio.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Müller M. Biochemical cytology of trichomonad flagellates. I. Subcellular localization of hydrolases, dehydrogenases, and catalase in Tritrichomonas foetus. J. Cell Biol. 1973;57:453–474. doi: 10.1083/jcb.57.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Müller M., Mentel M., van Hellemond J.J., Henze K. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 2012;76:444–495. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smutná T., Goncales V.L., Saraiva L.M., Tachezy J. Flavodiiron protein from Trichomonas vaginalis hydrogenosomes: the terminal oxygen reductase. Eukaryot. Cell. 2009;8:47–55. doi: 10.1128/EC.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paget T.A., Lloyd D. Trichomonas vaginalis requires traces of oxygen and high concentrations of carbon dioxide for optimal growth. Mol. Biochem. Parasitol. 1990;41:65–72. doi: 10.1016/0166-6851(90)90097-6. [DOI] [PubMed] [Google Scholar]
- 50.Upcroft P., Upcroft J.A. Drug targets and mechanisms of resistance in the anaerobic protozoa. Clin. Microbiol. Rev. 2001;114:150–164. doi: 10.1128/CMR.14.1.150-164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindmark D.G., Müller M. Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism. J. Biol. Chem. 1973;25:7724–7728. [PubMed] [Google Scholar]
- 52.Carlton J.M., Hirt R.P., Silva J.C., Delcher A.L. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steinbüchel A., Müller M. Anaerobic pyruvate metabolism of Tritrichomonas foetus and Trichomonas vaginalis hydrogenosomes. Mol. Biochem. Parasitol. 1986;20:57–65. doi: 10.1016/0166-6851(86)90142-8. [DOI] [PubMed] [Google Scholar]
- 54.Rich P.R., Maréchal A. The mitochondrial respiratory chain. Essays Biochem. 2010;47:1–23. doi: 10.1042/bse0470001. [DOI] [PubMed] [Google Scholar]
- 55.Sousa F.L., Nelson-Sathi S., Martin W.F. One step beyond a ribosome: the ancient anaerobic core. Biochim. Biophys. Acta. 2016;1857:1027–1038. doi: 10.1016/j.bbabio.2016.04.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stouthamer A.H. A theoretical study on the amount of ATP required for synthesis of microbial cell material. Antonie Leeuwenhoek. 1973;39:545–565. doi: 10.1007/BF02578899. [DOI] [PubMed] [Google Scholar]
- 57.Hinchliffe P., Sazanov L.A. Organization of iron-sulfur cluster in respiratory complex I. Science. 2005;309:771–774. doi: 10.1126/science.1113988. [DOI] [PubMed] [Google Scholar]
- 58.Klausner R.D., Rouault T.A., Harford J.B. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell. 1993;72:19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- 59.Gruer M.J., Artymiuk P.J., Guest J.R. The aconitase family: three structural variations on a common theme. Trends Biochem. Sci. 1997;22:3–6. doi: 10.1016/s0968-0004(96)10069-4. [DOI] [PubMed] [Google Scholar]
- 60.Cavazza C., Martin L., Mondy S., Gaillard J. The possible role of an [FeFe]-hydrogenase-like protein in the plant responses to changing atmospheric oxygen levels. J. Inorg. Biochem. 2008;102:1359–1365. doi: 10.1016/j.jinorgbio.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 61.Atteia A., van Lis R., Tielens A.G.M., Martin W.F. Anaerobic energy metabolism in unicellular photosynthetic eukaryotes. Biochim. Biophys. Acta. 2013;1827:210–223. doi: 10.1016/j.bbabio.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 62.Barth C., Weiss M.C., Roettger M., Martin W.F., Unden G. Origin and phylogenetic relationships of [4Fe-4S]-containing O2-sensors of bacteria. Environ. Microbiol. 2018;20:4567–4586. doi: 10.1111/1462-2920.14411. [DOI] [PubMed] [Google Scholar]
- 63.Rytkönen K.T., Storz J.F. Evolutionary origins of oxygen sensing in animals. EMBO Rep. 2011;12:3–4. doi: 10.1038/embor.2010.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Branicky R.S., Schafer W.R. Oxygen homeostasis: how the worm adapts to variable oxygen levels. Curr. Biol. 2008;18:R559–R560. doi: 10.1016/j.cub.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 65.Niedermeier M., Weisleitner A., Lamm C., Ledochowski L. Is decision making in hypoxia affected by pre-acclimatisation? A randomized controlled trial. Physiol. Behav. 2017;173:236–242. doi: 10.1016/j.physbeh.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 66.Proffitt F. Physiology. Science in the 'death zone. Science. 2005;308:1541–1542. doi: 10.1126/science.308.5728.1541. [DOI] [PubMed] [Google Scholar]
- 67.Semenza G.L. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Samanta D., Semenza G.L. Metabolic adaptation of cancer and immune cells mediated by hypoxia-inducible factors. Biochim. Biophys. Acta Rev. Canc. 2018;1870:15–22. doi: 10.1016/j.bbcan.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 69.Dunwoodie S.L. The role of hypoxia in development of the mammalian embryo. Dev. Cell. 2009;17:755–773. doi: 10.1016/j.devcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 70.Elkins J.M., Hewitson K.S., McNeill L.A., Seibel J.F. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1α. J. Biol. Chem. 2003;278:1802–1806. doi: 10.1074/jbc.C200644200. [DOI] [PubMed] [Google Scholar]
- 71.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 72.Fandrey J., Gorr T.A., Gassmann M. Regulating cellular oxygen sensing by hydroxylation. Cardiovasc. Res. 2006;71:642–651. doi: 10.1016/j.cardiores.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 73.Degli Esposti M., Mentel M., Martin W.F., Sousa F.L. Oxygen reductases in alphaproteobacterial genomes: physiological evolution from low to high oxygen environments. Front. Microbiol. 2019 doi: 10.3389/fmicb.2019.00499. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Atteia A., van Lis R., van Hellemond J., Tielens L. Identification of prokaryotic homologues indicates an endosymbiotic origin for the alternative oxidases of mitochondria (AOX) and chloroplasts (PTOX) Gene. 2004;330:143–148. doi: 10.1016/j.gene.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 75.Krab K., Kempe H., Wikström M. Explaining the enigmatic K(M) for oxygen in cytochrome c oxidase: a kinetic model. Biochim. Biophys. Acta. 2011;1807:348–358. doi: 10.1016/j.bbabio.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 76.Berg J.M., Tymoczko J.L., Gatto G.J.R., Stryer L. W.F. Freeman & Company; New York: 2015. Biochemistry. [Google Scholar]
- 77.Guerrero-Castillo S., Araiza-Olivera D., Cabrera-Orefice A., Espinasa-Jaramillo J. Physiological uncoupling of mitochondrial oxidative phosphorylation. Studies in different yeast species. J. Bioenerg. Biomembr. 2011;43:323–331. doi: 10.1007/s10863-011-9356-5. [DOI] [PubMed] [Google Scholar]
- 78.Zachar I., Szilágyi A., Számadó S., Szathmáry E. Farming the mitochondrial ancestor as a model of endosymbiotic establishment by natural selection. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E1504–E1510. doi: 10.1073/pnas.1718707115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garg S.G., Martin W.F. Asking endosymbionts to do an enzyme's job. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E4543–E4544. doi: 10.1073/pnas.1804397115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tielens A.G.M., Rotte C., van Hellemond J.J., Martin W. Mitochondria as we don't know them. Trends Biochem. Sci. 2002;27:564–572. doi: 10.1016/s0968-0004(02)02193-x. [DOI] [PubMed] [Google Scholar]
- 81.McCollom T.M., Amend J.P. A thermodynamic assessment of energy requirements for biomass synthesis by chemolithoautotrophic microorganisms in oxic and anoxic environments. Geology. 2005;3:135–144. [Google Scholar]
- 82.Lever M.A., Rogers K.L., Lloyd K.G., Overmann J. Life under extreme energy limitation: a synthesis of laboratory- and field-based investigations. FEMS Microbiol. Rev. 2015;39:688–728. doi: 10.1093/femsre/fuv020. [DOI] [PubMed] [Google Scholar]
- 83.Thauer R.K., Jungermann K., Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weiss M.C., Sousa F.L., Mrnjavac N., Neukirchen S. The physiology and habitat of the last universal common ancestor. Nat Microbiol. 2016;1:16116. doi: 10.1038/nmicrobiol.2016.116. [DOI] [PubMed] [Google Scholar]
- 85.Weiss M.C., Preiner M., Xavier J.C., Zimorski V., Martin W.F. The last common ancestor between ancient Earth chemistry and the onset of genetics. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martin W.F., Tielens A.G.M., Mentel M., Garg S.G. The physiology of phagocytosis in the context of mitochondrial origin. Microbiol. Mol. Biol. Rev. 2017;81:e00008–e00017. doi: 10.1128/MMBR.00008-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lane N., Martin W. The energetics of genome complexity. Nature. 2010;467:929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- 88.Martin W.F. Symbiogenesis, gradualism and mitochondrial energy in eukaryote evolution. Period. Biol. 2017;119:141–158. [Google Scholar]
- 89.Andersson S.G., Kurland C.G. Origins of mitochondria and hydrogenosomes. Curr. Opin. Microbiol. 1999;2:535–541. doi: 10.1016/s1369-5274(99)00013-2. [DOI] [PubMed] [Google Scholar]
- 90.Barbera M.J., Ruiz-Trillo I., Leigh J., Hug L.A. The diversity of mitochondrion-related organelles amongst eukaryotic microbes. In: Martin W.F., Müller M., editors. Origin of Mitochondria and Hydrogenosomes. Springer-Verlag; Heidelberg, Germany: 2007. pp. 239–276. [Google Scholar]
- 91.Hug L.A., Stechmann A., Roger A.J. Phylogenetic distributions and histories of proteins involved in anaerobic pyruvate metabolism in eukaryotes. Mol. Biol. Evol. 2010;27:311–324. doi: 10.1093/molbev/msp237. [DOI] [PubMed] [Google Scholar]
- 92.Hampl V., Stairs C.W., Roger A.J. The tangled past of eukaryotic enzymes involved in anaerobic metabolism. Mobile Genet. Elem. 2011;1:71–74. doi: 10.4161/mge.1.1.15588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stairs C.W., Roger A.J., Hampl V. Eukaryotic pyruvate formate lyase and its activating enzyme were acquired laterally from a firmicute. Mol. Biol. Evol. 2011;28:2087–2099. doi: 10.1093/molbev/msr032. [DOI] [PubMed] [Google Scholar]
- 94.Stairs C.W., Eme L., Brown M.W., Mutsaers C. A SUF Fe-S cluster biogenesis system in the mitochondrion-related organelles of the anaerobic protist Pygsuia. Curr. Biol. 2014;24:1176–1186. doi: 10.1016/j.cub.2014.04.033. [DOI] [PubMed] [Google Scholar]
- 95.Stairs C.W., Leger M.M., Roger A.J. Diversity and origins of anaerobic metabolism in mitochondria and related organelles. Philos Trans Soc Lond B Biol Sci. 2015;370:20140326. doi: 10.1098/rstb.2014.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leger M.M., Gawryluk R.M., Gray M.W., Roger A.J. Evidence for a hydrogenosomal-type anaerobic ATP generation pathway in Acanthamoeba castellanii. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Leger M.M., Eme L., Hug L.A., Roger A.J. Novel hydrogenosomes in the microaerophilic jakobid Stygiella incarcerata. Mol. Biol. Evol. 2016;33:2318–2336. doi: 10.1093/molbev/msw103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bekker A., Holland H.D., Wang P.L., Rumble D., 3rd Dating the rise of atmospheric oxygen. Nature. 2004;427:117–120. doi: 10.1038/nature02260. [DOI] [PubMed] [Google Scholar]
- 99.Martin W.F. Too much eukaryote LGT. Bioessays. 2017;39 doi: 10.1002/bies.201700115. [DOI] [PubMed] [Google Scholar]
- 100.Leger M.M., Eme L., Stairs C.W., Roger A.J. Demystifying eukaryote lateral gene transfer (Response to Martin 2017 DOI: 10.1002/bies.201700115) Bioessays. 2018;40 doi: 10.1002/bies.201700242. [DOI] [PubMed] [Google Scholar]
- 101.Martin W., Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 102.Mentel M., Martin W. Energy metabolism among eukaryotic anaerobes in light of Proterozoic ocean chemistry. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363:2717–2729. doi: 10.1098/rstb.2008.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zimorski V., Ku C., Martin W.F., Gould S.B. Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 2014;22:38–48. doi: 10.1016/j.mib.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 104.Martin W., Garg S., Zimorski V. Endosymbiotic theories for eukaryote origin. Phil. Trans. Roy. Soc. Lond. B. 2015;370:20140330. doi: 10.1098/rstb.2014.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Degli Esposti M. Bioenergetic evolution in proteobacteria and mitochondria. Genome Biol Evol. 2014;6:3238–3251. doi: 10.1093/gbe/evu257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Degli Esposti M., Chouaia B., Comandatore F., Crotti E. Evolution of mitochondria reconstructed from the energy metabolism of living bacteria. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ku C., Nelson-Sathi S., Roettger M., Sousa F.L. Endosymbiotic origin and differential loss of eukaryotic genes. Nature. 2015;524:427–432. doi: 10.1038/nature14963. [DOI] [PubMed] [Google Scholar]
- 108.Fritz-Laylin L.K., Prochnik S.E., Ginger M.L., Dacks J.B. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 109.Bexkens M.L., Zimorski V., Sarink M.J., Wienk H. Lipids are the preferred s ubstrate of the protist Naegleria gruberi, relative of a human brain pathogen. Cell Rep. 2018;25:537–543. doi: 10.1016/j.celrep.2018.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tovar J., Fischer A., Clark C.G. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol. Microbiol. 1999;32:1013–1021. doi: 10.1046/j.1365-2958.1999.01414.x. [DOI] [PubMed] [Google Scholar]
- 111.Tovar J., León-Avila G., Sánchez L.B., Sutak R. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature. 2003;426:172–176. doi: 10.1038/nature01945. [DOI] [PubMed] [Google Scholar]
- 112.van Hellemond J.J., van der Klei A., van Weelden S.W.H., Tielens A.G.M. Biochemical and evolutionary aspects of anaerobically functioning mitochondria. Phil. Trans. Roy. Soc. Lond. B. 2003;358:205–213. doi: 10.1098/rstb.2002.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boxma B., de Graaf R.M., van der Staay G.W., van Alen T.A. An anaerobic mitochondrion that produces hydrogen. Nature. 2005;434:74–79. doi: 10.1038/nature03343. [DOI] [PubMed] [Google Scholar]
- 114.Goldberg A.V., Molik S., Tsaousis A.D., Neumann K. Localization and [ functionality of microsporidian iron-sulphur cluster assembly proteins. Nature. 2008;452:624–628. doi: 10.1038/nature06606. [DOI] [PubMed] [Google Scholar]
- 115.van der Giezen M. Hydrogenosomes and mitosomes: conservation and evolution of functions. J. Eukaryot. Microbiol. 2009;56:221–231. doi: 10.1111/j.1550-7408.2009.00407.x. [DOI] [PubMed] [Google Scholar]
- 116.Gawryluk RMR, Kamikawa R, Stairs CW, Silberman JD, et al. The earliest stages of mitochondrial adaptation to low oxygen revealed in a novel rhizarian. Curr. Biol. 26: 2729–2738. DOI: 10.1016/j.cub.2016.08.025. [DOI] [PubMed]
- 117.Adl S.M., Simpson A.G., Farmer M.A., Andersen R.A. The new higher level c lassification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 118.Livingstone D.R. Invertebrate and vertebrate pathways of anaerobic metabolism: evolutionary considerations. J Geol Soc London. 1983;140:27–37. [Google Scholar]
- 119.Bryant C., editor. Metazoan Life without Oxygen. Chapman and Hall; London, UK: 1991. [Google Scholar]
- 120.Grieshaber M.K., Hardewig I., Kreutzer U., Pörtner H.O. Physiological and metabolic responses to hypoxia in invertebrates. Rev. Physiol. Biochem. Pharmacol. 1994;125:43–147. doi: 10.1007/BFb0030909. 10013/epic.11652. [DOI] [PubMed] [Google Scholar]
- 121.Tielens A.G.M., van Hellemond J.J. The electron transport chain in anaerobically functioning eukaryotes. Biochim. Biophys. Acta. 1998;1365:71–78. doi: 10.1016/s0005-2728(98)00045-0. [DOI] [PubMed] [Google Scholar]
- 122.Levin L.A. Oxygen minimum zone benthos: adaptation and community response to hypoxia. Oceanogr. Mar. Biol. 2003;41:1–45. [Google Scholar]
- 123.Mentel M., Tielens A.G.M., Martin W.F. Animals, anoxic environments, and reasons to go deep. BMC Biol. 2016;14:44. doi: 10.1186/s12915-016-0266-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.de Zwaan A. Molluscs. In: Bryant C., editor. Metazoan Life without Oxygen. Chapman and Hall; London, UK: 1991. pp. 186–217. [Google Scholar]
- 125.Schöttler U., Bennet E.M. Annelids. In: Bryant C., editor. Metazoan Life without Oxygen. Chapman and Hall; London, UK: 1991. pp. 165–185. [Google Scholar]
- 126.Zebe E. Arthropods. In: Bryant C., editor. Metazoan Life without Oxygen. Chapman and Hall; London, UK: 1991. pp. 218–237. [Google Scholar]
- 127.Harcet M., Perina D., Plese B. Opin dehydrogenases in marine invertebrates. Biochem. Genet. 2013;51:666–676. doi: 10.1007/s10528-013-9596-7. [DOI] [PubMed] [Google Scholar]
- 128.Al-Subiai S.N., Jha A.N., Moody A.J. Contamination of bivalve haemolymph samples by adductor muscle components: implications for biomarker studies. Ecotoxicology. 2009;18:334–342. doi: 10.1007/s10646-008-0287-9. [DOI] [PubMed] [Google Scholar]
- 129.Tielens A.G.M. Energy generation in parasitic helminths. Parasitol. Today. 1994;10:346–352. doi: 10.1016/0169-4758(94)90245-3. [DOI] [PubMed] [Google Scholar]
- 130.Bringaud F., Ebikeme C., Boshart M. Acetate and succinate production in amoebae, helminths, diplomonads and trypanosomatids: common and diverse metabolic strategies used by parasitic lower eukaryotes. Parasitology. 2010;137:1315–1331. doi: 10.1017/S0031182009991843. [DOI] [PubMed] [Google Scholar]
- 131.Harder A. The biochemistry of Haemonchus contortus and other parasitic nematodes. Adv. Parasitol. 2016;93:69–94. doi: 10.1016/bs.apar.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 132.Müller M. The hydrogenosome. J. Gen. Microbiol. 1993;139:2879–2889. doi: 10.1099/00221287-139-12-2879. [DOI] [PubMed] [Google Scholar]
- 133.Tachezy J., editor. Hydrogenosomes and Mitosomes: Mitochondria of Anaerobic Eukaryotes. Springer-Verlag; Heidelberg, Germany: 2008. [Google Scholar]
- 134.Shiflett A.M., Johnson P.J. Mitochondrion-related organelles in eukaryotic protists. Annu. Rev. Microbiol. 2010;64:409–429. doi: 10.1146/annurev.micro.62.081307.162826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Marvin-Sikkema F.D., Pedro Gomes T.M., Grivet J.P., Gottschal J.C. Characterization of hydrogenosomes and their role in glucose metabolism of Neocallimastix sp. L2. Arch. Microbiol. 1993;160:388–396. doi: 10.1007/BF00252226. [DOI] [PubMed] [Google Scholar]
- 136.Yarlett N., Lloyd D., Williams A.G. Respiration of the rumen ciliate Dasytricha ruminantium Schuberg. Biochem. J. 1982;206:259–266. doi: 10.1042/bj2060259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yarlett N., Orpin C.G., Munn E.A., Yarlett N.C. Hydrogenosomes in the rumen fungus Neocallimastix patriciarum. Biochem. J. 1986;236:729–739. doi: 10.1042/bj2360729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Broers C.A., Stumm C.K., Vogels G.D., Brugerolle G. Psalteriomonas lanterna gen. nov., sp. nov., a free-living amoeboflagellate isolated from freshwater anaerobic sediments. Eur. J. Protistol. 1990;25:369–380. doi: 10.1016/S0932-4739(11)80130-6. [DOI] [PubMed] [Google Scholar]
- 139.Müller M. Energy metabolism of protozoa without mitochondria. Annu. Rev. Microbiol. 1988;42:465–488. doi: 10.1146/annurev.mi.42.100188.002341. [DOI] [PubMed] [Google Scholar]
- 140.Müller M. Energy metabolism. Part I: anaerobic protozoa. In: Marr J., editor. Molecular Medical Parasitology. Academic Press; London, UK: 2003. pp. 125–139. [Google Scholar]
- 141.Müller M. The road to hydrogenosomes. In: Martin W.F., Müller M., editors. Origin of Mitochondria and Hydrogenosomes. Springer-Verlag; Heidelberg, Germany: 2007. pp. 1–12. [Google Scholar]
- 142.Drmota T., Proost P., van Ranst M., Weyda F. Iron-ascorbate cleavable malic enzyme from hydrogenosomes of Trichomonas vaginalis: purification and characterization. Mol. Biochem. Parasitol. 1996;83:221–234. doi: 10.1016/s0166-6851(96)02777-6. [DOI] [PubMed] [Google Scholar]
- 143.Hrdy I., Müller M. Primary structure of the hydrogenosomal malic enzyme of Trichomonas vaginalis and its relationship to homologous enzymes. J. Eukaryot. Microbiol. 1995;42:593–603. doi: 10.1111/j.1550-7408.1995.tb05913.x. [DOI] [PubMed] [Google Scholar]
- 144.Hrdy I., Hirt R.P., Dolezal P., Bardonova L. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- 145.Bui E.T., Johnson P.J. Identification and characterization of [Fe]-hydrogenases in the hydrogenosome of Trichomonas vaginalis. Mol. Biochem. Parasitol. 1996;76:305–310. doi: 10.1016/0166-6851(96)02567-4. [DOI] [PubMed] [Google Scholar]
- 146.Payne M.J., Chapman A., Cammack R. Evidence for an [Fe]-type hydrogenase in the parasitic protozoan Trichomonas vaginalis. FEBS Lett. 1993;317:101–104. doi: 10.1016/0014-5793(93)81500-y. [DOI] [PubMed] [Google Scholar]
- 147.Zwart K.B., Goosen N.K., van Schijndel M.W., Broers C.A.M. Cytochemical localization of hydrogenase activity in the anaerobic protozoa Trichomonas vaginalis, Plagiopyla nasuta and Trimyema compressum. J. Gen. Microbiol. 1988;134:2165–2170. [Google Scholar]
- 148.Dacks J.B., Dyal P.L., Embley T.M., van der Giezen M. Hydrogenosomal succinyl-CoA synthetase from the rumen-dwelling fungus Neocallimastix patriciarum; an energy-producing enzyme of mitochondrial origin. Gene. 2006;373:75–82. doi: 10.1016/j.gene.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 149.Lindmark D.G. Acetate production in Tritrichomonas foetus. In: van den Bossche H., editor. Biochemistry of Parasites and Host-Parasite Relationships. North-Holland; Amsterdam, The Netherlands: 1976. pp. 16–21. [Google Scholar]
- 150.van Grinsven K.W., Rosnowsky S., van Weelden S.W., Pütz S. Acetate:succinate CoA-transferase in the hydrogenosomes of Trichomonas vaginalis: identification and characterization. J. Biol. Chem. 2008;283:1411–1418. doi: 10.1074/jbc.M702528200. [DOI] [PubMed] [Google Scholar]
- 151.van Hellemond J.J., Opperdoes F.R., Tielens A.G. Trypanosomatidae produce acetate via a mitochondrial acetate:succinate CoA transferase. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3036–3041. doi: 10.1073/pnas.95.6.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.van Weelden S.W., van Hellemond J.J., Opperdoes F.R., Tielens A.G. New functions for parts of the Krebs cycle in procyclic Trypanosoma brucei, a cycle not operating as a cycle. J. Biol. Chem. 2005;280:12451–12460. doi: 10.1074/jbc.M412447200. [DOI] [PubMed] [Google Scholar]
- 153.Dyall S.D., Yan W., Delgadillo-Correa M.G., Lunceford A. Non-mitochondrial complex I proteins in a hydrogenosomal oxidoreductase complex. Nature. 2004;431:1103–1107. doi: 10.1038/nature02990. [DOI] [PubMed] [Google Scholar]
- 154.Li F., Hinderberger J., Seedorf H., Zhang J. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J. Bacteriol. 2008;190:843–850. doi: 10.1128/JB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Buckel W., Thauer R.K. Flavin-based electron bifurcation, ferredoxin, flavodoxin, and anaerobic respiration with protons (Ech) or NAD+ (Rnf) as electron acceptors: a historical review. Front. Microbiol. 2018;9:401. doi: 10.3389/fmicb.2018.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schut G.J., Adams M.W.W. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J. Bacteriol. 2009;191:4451–44657. doi: 10.1128/JB.01582-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Atteia A., van Lis R., Gelius-Dietrich G., Adrait A. Pyruvate:formate lyase and a novel route of eukaryotic ATP-synthesis in anaerobic Chlamydomonas mitochondria. J. Biol. Chem. 2006;281:9909–9918. doi: 10.1074/jbc.M507862200. [DOI] [PubMed] [Google Scholar]
- 158.Mus F., Dubini A., Seibert M., Posewitz M.C. Anaerobic acclimation in Chlamydomonas reinhardtii – anoxic gene expression, hydrogenase induction, and metabolic pathways. J. Biol. Chem. 2007;282:25475–25486. doi: 10.1074/jbc.M701415200. [DOI] [PubMed] [Google Scholar]
- 159.Lloyd D., Ralphs J.R., Harris J.C. Giardia intestinalis, a eukaryote without hydrogenosomes, produces hydrogen. Microbiology. 2002;148:727–733. doi: 10.1099/00221287-148-3-727. [DOI] [PubMed] [Google Scholar]
- 160.Embley T.M., van der Giezen M., Horner D.S., Dyal P.L. Hydrogenosomes, mitochondria and early eukaryotic evolution. IUBMB Life. 2003;55:387–395. doi: 10.1080/15216540310001592834. [DOI] [PubMed] [Google Scholar]
- 161.Hampl V., Silberman J.D., Stechmann A., Diaz-Trivino S. Genetic evidence for a mitochondriate ancestry in the ‘amitochondriate’ flagellate Trimastix pyriformis. PLoS One. 2008;3 doi: 10.1371/journal.pone.0001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Docampo R., Moreno S.N., Mason R.P. Free radical intermediates in the reaction of pyruvate:ferredoxin oxidoreductase in Tritrichomonas foetus hydrogenosomes. J. Biol. Chem. 1987;262:12417–12420. [PubMed] [Google Scholar]
- 163.Ragsdale S.W. Pyruvate ferredoxin oxidoreductase and its radical intermediate. Chem. Rev. 2003;103:2333–2346. doi: 10.1021/cr020423e. [DOI] [PubMed] [Google Scholar]
- 164.Rotte C., Stejskal F., Zhu G., Keithly J.S. Pyruvate:NADP+ oxidoreductase from the mitochondrion of Euglena gracilis and from the apicomplexan Cryptosporidium parvum: a biochemical relic linking pyruvate metabolism in mitochondriate and amitochondriate protists. Mol. Biol. Evol. 2001;18:710–720. doi: 10.1093/oxfordjournals.molbev.a003853. [DOI] [PubMed] [Google Scholar]
- 165.Nakazawa M., Takenaka S., Ueda M., Inui H. Pyruvate: NADP+ oxidoreductase is stabilized by its cofactor, thiamin pyrophosphate, in mitochondria of Euglena gracilis. Arch. Biochem. Biophys. 2003;411:183–188. doi: 10.1016/s0003-9861(02)00749-x. [DOI] [PubMed] [Google Scholar]
- 166.Wagner G., Levin R. Oxygen tension of the vaginal surface during sexual stimulation in the human. Fertil. Steril. 1978;30:50–53. doi: 10.1016/s0015-0282(16)43395-9. [DOI] [PubMed] [Google Scholar]
- 167.Ellis J.E., Cole D., Lloyd D. Influence of oxygen on the fermentative metabolism of metronidazole-sensitive and resistant strains of Trichomonas vaginalis. Mol. Biochem. Parasitol. 1992;56:79–88. doi: 10.1016/0166-6851(92)90156-e. [DOI] [PubMed] [Google Scholar]
- 168.Hill D.R., Brunner M.E., Schmitz D.C., Davis C.C. In vivo assessment of human vaginal oxygen and carbon dioxide levels during and post menses. J. Appl. Physiol. 2005;99:1582–1591. doi: 10.1152/japplphysiol.01422.2004. [DOI] [PubMed] [Google Scholar]
- 169.Gould S.B., Woehle C., Kusdian G., Landan G. Deep sequencing of Trichomonas vaginalis during the early infection of vaginal epithelial cells and amoeboid transition. Int. J. Parasitol. 2013;43:707–719. doi: 10.1016/j.ijpara.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 170.Ellis J.E., Yarlett N., Cole D., Humphreys M.J. Antioxidant defences in the microaerophilic protozoan Trichomonas vaginalis: comparison of metronidazole-resistant and sensitive strains. Microbiol. 1994;140:2489–2494. doi: 10.1099/13500872-140-9-2489. [DOI] [PubMed] [Google Scholar]
- 171.Rasoloson D., Tomková E., Cammack R., Kulda J. Metronidazole- resistant strains of Trichomonas vaginalis display increased susceptibility to oxygen. Parasitology. 2001;123:45–56. doi: 10.1017/s0031182001008022. [DOI] [PubMed] [Google Scholar]
- 172.Coombs G.H., Westrop G.D., Suchan P., Puzova G. The amitochondriate e eukaryote Trichomonas vaginalis contains a divergent thioredoxin-linked peroxiredoxin antioxidant system. J. Biol. Chem. 2004;279:5249–5256. doi: 10.1074/jbc.M304359200. [DOI] [PubMed] [Google Scholar]
- 173.Gretes M.C., Poole L.B., Karplus P.A. Peroxiredoxins in parasites. Antioxidants Redox Signal. 2012;17:608–633. doi: 10.1089/ars.2011.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mentel M., Zimorski V., Haferkamp P., Martin W. Protein import into hydrogenosomes of Trichomonas vaginalis involves both N-terminal and internal targeting signals: a case study of thioredoxin reductases. Eukaryot. Cell. 2008;7:1750–1757. doi: 10.1128/EC.00206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Linstead D.J., Bradley S. The purification and properties of two soluble reduced nicotinamide:acceptor oxidoreductases from Trichomonas vaginalis. Mol. Biochem. Parasitol. 1988;27:125–134. doi: 10.1016/0166-6851(88)90032-1. [DOI] [PubMed] [Google Scholar]
- 176.Tanabe M. Trichomonas vaginalis: NADH oxidase activity. Exp. Parasitol. 1979;48:135–143. doi: 10.1016/0014-4894(79)90063-8. [DOI] [PubMed] [Google Scholar]
- 177.Castillo-Villanueva A., Méndez S.T., Torres-Arroyo A., Reyes-Vivas H. Cloning, expression and characterization of recombinant, NADH oxidase from Giardia lamblia. Protein J. 2016;35:24–33. doi: 10.1007/s10930-015-9643-9. [DOI] [PubMed] [Google Scholar]
- 178.Nixon J.E. Evidence for lateral transfer of genes encoding ferredoxins, nitroreductases, NADH oxidase, and alcohol dehydrogenase 3 from anaerobic prokaryotes to Giardia lamblia and Entamoeba histolytica. Eukaryot. Cell. 2002;1:181–190. doi: 10.1128/EC.1.2.181-190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chapman A., Linstead D.J., Lloyd D. Hydrogen peroxide is a product of oxygen consumption by Trichomonas vaginalis. J. Biosci. 1999;24:339–344. [Google Scholar]
- 180.Brown D.M., Upcroft J.A., Edwards M.R., Upcroft P. Anaerobic bacterial metabolism in the ancient eukaryote Giardia duodenalis. Int. J. Parasitol. 1998;28:149–164. doi: 10.1016/s0020-7519(97)00172-0. [DOI] [PubMed] [Google Scholar]
- 181.Gibson C.M., Mallett T.C., Claiborne A., Caparon M.G. Contribution of NADH oxidases to aerobic metabolism of Streptococcus pyogenes. J. Bacteriol. 2000;182:448–455. doi: 10.1128/jb.182.2.448-455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Di Matteo A., Scandurra F.M., Testa F., Forte E. The O2-scavenging flavodiiron protein in the human parasite Giardia intestinalis. J. Biol. Chem. 2008;283:4061–4068. doi: 10.1074/jbc.M705605200. [DOI] [PubMed] [Google Scholar]
- 183.Meuser J.E., Ananyev G., Wittig L.E., Kosourov S. Phenotypic diversity of hydrogen production in chlorophycean algae reflects distinct anaerobic metabolisms. J. Biotechnol. 2009;142:21–30. doi: 10.1016/j.jbiotec.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 184.Merchant S.S., Allen M.D., Kropat J., Moseley J.L. Between a rock and a hard place: trace element nutrition in Chlamydomonas. Biochim. Biophys. Acta. 2006;1763:578–594. doi: 10.1016/j.bbamcr.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 185.Makiuchi T., Nozaki T. Highly divergent mitochondrion-related organelles in anaerobic parasitic protozoa. Biochimie. 2014;100:3–17. doi: 10.1016/j.biochi.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 186.Boxma B., Voncken F., Jannink S., van Alen T. The anaerobic chytridiomycete fungus Piromyces sp. E2 produces ethanol via pyruvate:formate lyase and an alcohol dehydrogenase. E. Mol Microbiol. 2004;51:1389–1399. doi: 10.1046/j.1365-2958.2003.03912.x. [DOI] [PubMed] [Google Scholar]
- 187.Mountfort D.O., Orpin C.G., Dekker M. 1994. Anaerobic Fungi: Biology, Ecology and Function. (New York, NY, USA: Inc. New York) [Google Scholar]
- 188.Atteia A., van Lis R., Mendoza-Hernández G., Henze K. Bifunctional aldehyde/alcohol dehydrogenase (ADHE) in chlorophyte algal mitochondria. Plant Mol. Biol. 2003;53:175–188. doi: 10.1023/B:PLAN.0000009274.19340.36. [DOI] [PubMed] [Google Scholar]
- 189.Kobayashi M., Matsuo Y., Takimoto A., Suzuki S. Denitrification, a novel type of respiratory metabolism in fungal mitochondrion. J. Biol. Chem. 1996;271:16263–16267. doi: 10.1074/jbc.271.27.16263. [DOI] [PubMed] [Google Scholar]
- 190.Tsuruta S., Takaya N., Zhang L., Shoun H. Denitrification by yeasts and occurrence of cytochrome P450nor in Trichosporon cutaneum. FEMS Microbiol. Lett. 1998;168:105–110. doi: 10.1111/j.1574-6968.1998.tb13262.x. [DOI] [PubMed] [Google Scholar]
- 191.Morozkina E.V., Kurakov A.V. Dissimilatory nitrate reduction in fungi under conditions of hypoxia and anoxia: a review. Appl. Biochem. Microbiol. 2007;43:544–549. [PubMed] [Google Scholar]
- 192.Takaya N. Response to hypoxia, reduction of electron acceptors, and subsequent survival by filamentous fungi. Biosci. Biotechnol. Biochem. 2009;73:1–8. doi: 10.1271/bbb.80487. [DOI] [PubMed] [Google Scholar]
- 193.Zhou Z., Takaya N., Shoun H. Multi-energy metabolic mechanisms of the fungus Fusarium oxysporum in low oxygen environments. Biosci. Biotechnol. Biochem. 2010;74:2431–2437. doi: 10.1271/bbb.100482. [DOI] [PubMed] [Google Scholar]
- 194.Risgaard-Petersen N., Langezaal A.M., Ingvardsen S., Schmid M.S. Evidence for complete denitrification in a benthic foraminifer. Nature. 2006;443:93–96. doi: 10.1038/nature05070. [DOI] [PubMed] [Google Scholar]
- 195.Bernhard J.M., Edgcomb V.P., Casciotti K.L., McIlvin M.R. Denitrification likely catalyzed by endobionts in an allogromiid foraminifer. ISME J. 2012;6:951–960. doi: 10.1038/ismej.2011.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Woehle C., Roy A.-S., Glock N., Wein T. A novel eukaryotic denitrification pathway in foraminifera. Curr. Biol. 2018;28:2536–2543. doi: 10.1016/j.cub.2018.06.027. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhou Z., Takaya N., Sakairi M.A., Shoun H. Oxygen requirement for denitrification by the fungus Fusarium oxysporum. Arch. Microbiol. 2001;175:19–25. doi: 10.1007/s002030000231. [DOI] [PubMed] [Google Scholar]
- 198.Zhou Z., Takaya N., Nakamura A., Yamaguchi M. Ammonia fermentation, a novel anoxic metabolism of nitrate by fungi. J. Biol. Chem. 2002;277:1892–1896. doi: 10.1074/jbc.M109096200. [DOI] [PubMed] [Google Scholar]
- 199.Takaya N., Kuwazak S., Adachi Y., Suzuki S. Hybrid respiration in the denitrifying mitochondria of Fusarium oxysporum. J. Biochem. 2003;133:461–465. doi: 10.1093/jb/mvg060. [DOI] [PubMed] [Google Scholar]
- 200.Abe T., Hoshino T., Nakamura A., Takaya N. Anaerobic elemental sulfur reduction by fungus Fusarium oxysporum. Biosci. Biotechnol. Biochem. 2007;71:2402–2407. doi: 10.1271/bbb.70083. [DOI] [PubMed] [Google Scholar]
- 201.Burki F. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb Perspect Biol. 2014;6:a016147. doi: 10.1101/cshperspect.a016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.van Bruggen J.J., Stumm C.K., Vogels G.D. Symbiosis of methanogenic bacteria and sapropelic protozoa. Arch. Microbiol. 1983;136:89–95. [Google Scholar]
- 203.Ellis J.E., McIntyre P.S., Saleh M., Williams A.G. Influence of CO2 and low concentrations of O2 on fermentative metabolism of the rumen ciliate Dasytricha ruminantium. J. Gen. Microbiol. 1991;137:1409–1417. doi: 10.1099/00221287-137-6-1409. [DOI] [PubMed] [Google Scholar]
- 204.Embley T.M., Finlay B.J., Dyal P.L., Hirt R.P. Multiple origins of anaerobic ciliates with hydrogenosomes within the radiation of aerobic ciliates. Proc. Biol. Sci. 1995;262:87–93. doi: 10.1098/rspb.1995.0180. [DOI] [PubMed] [Google Scholar]
- 205.Goosen N.K., van der Drift C., Stumm C.K., Vogels G.D. End products of metabolism in the anaerobic ciliate Trimyema compressum. FEMS Microbiol. Lett. 1990;69:171–175. [Google Scholar]
- 206.Williams A.G. Rumen holotrich ciliate protozoa. Microbiol. Rev. 1986;50:25–49. doi: 10.1128/mr.50.1.25-49.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.van Hoek A.H., van Alen T.A., Sprakel V.S., Hackstein J.H. Evolution of anaerobic ciliates from the gastrointestinal tract: phylogenetic analysis of the ribosomal repeat from Nyctotherus ovalis and its relatives. Mol. Biol. Evol. 1998;15:1195–1206. doi: 10.1093/oxfordjournals.molbev.a026027. [DOI] [PubMed] [Google Scholar]
- 208.de Graaf R.M., Ricard G., van Alen T.A., Duarte I. The organellar genome and metabolic potential of the hydrogen-producing mitochondrion of Nyctotherus ovalis. Mol. Biol. Evol. 2011;28:2379–2391. doi: 10.1093/molbev/msr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Akhmanova A., Voncken F., van Alen T., van Hoek A. A hydrogenosome with a genome. Nature. 1998;396:527–528. doi: 10.1038/25023. [DOI] [PubMed] [Google Scholar]
- 210.van Hoek A.H., Akhmanova A.S., Huynen M.A., Hackstein J.H. A mitochondrial ancestry of the hydrogenosomes of Nyctotherus ovalis. Mol. Biol. Evol. 2000;17:202–206. doi: 10.1093/oxfordjournals.molbev.a026234. [DOI] [PubMed] [Google Scholar]
- 211.Hackstein J.H.P., Akhmanova A., Boxma B., Harhangi H.R. Hydrogenosomes: eukaryotic adaptations to anaerobic environments. Trends Microbiol. 1999;7:441–447. doi: 10.1016/s0966-842x(99)01613-3. [DOI] [PubMed] [Google Scholar]
- 212.Tielens A.G.M., van Grinsven K., Henze K., van Hellemond J.J. Acetate formation in the energy metabolism of parasitic helminths and protists. Int. J. Parasitol. 2010;40:387–397. doi: 10.1016/j.ijpara.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 213.Boxma B., Ricard G., van Hoeck A.H., Severing E. The [FeFe] hydrogenase of Nyctotherus ovalis has a chimeric origin. BMC Evol. Biol. 2007;7:230. doi: 10.1186/1471-2148-7-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Gijzen H.J., Broers C.A., Barughare M., Stumm C.K. Methanogenic bacteria as endosymbionts of the ciliate Nyctotherus ovalis in the cockroach hindgut. Appl. Environ. Microbiol. 1991;57:1630–1634. doi: 10.1128/aem.57.6.1630-1634.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Broers C.A., Meijers H.H., Symens J.C., Stumm C.K., Vogels G.D., Brugerolle G. Symbiotic association of Psalteriomonas vulgaris n. spec. with Methanobacterium formicicum. Eur. J. Protistol. 1993;29:98–105. doi: 10.1016/S0932-4739(11)80302-0. [DOI] [PubMed] [Google Scholar]
- 216.de Graaf R.M., Duarte I., van Alen T.A., Kuiper J.W. The hydrogenosomes of Psalteriomonas lanterna. BMC Evol. Biol. 2009;9:287. doi: 10.1186/1471-2148-9-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Brul S., Veltman R.H., Lombardo M.C., Vogels G.D. Molecular cloning of hydrogenosomal ferredoxin cDNA from the anaerobic amoeboflagellate Psalteriomonas lanterna. Biochim. Biophys. Acta. 1994;1183:544–546. doi: 10.1016/0005-2728(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 218.O'Kelly C.J., Silberman J.D., Amaral Zettler L.A., Nerad T.A. Monopylocystis visvesvarai n. gen., n. sp. and Sawyeria marylandensis n. gen., n. sp.: two new amitochondrial heterolobosean amoebae from anoxic environments. Protist. 2003;154:281–290. doi: 10.1078/143446103322166563. [DOI] [PubMed] [Google Scholar]
- 219.Mai Z.M., Ghosh S., Frisardi M., Rosenthal B. Hsp60 is targeted to a cryptic mitochondrion-derived organelle ("crypton") in the microaerophilic protozoan parasite Entamoeba histolytica. Mol. Cell Biol. 1999;19:2198–2205. doi: 10.1128/mcb.19.3.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Williams B.A., Hirt R.P., Lucocq J.M., Embley T.M. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature. 2002;418:865–869. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- 221.van der Giezen M., Tovar J., Clark C.G. Mitochondrion-derived organelles in protists and fungi. Int. Rev. Cytol. 2005;244:175–225. doi: 10.1016/S0074-7696(05)44005-X. [DOI] [PubMed] [Google Scholar]
- 222.van der Giezen M., Tovar J. Degenerate mitochondria. EMBO Rep. 2005;6:525–530. doi: 10.1038/sj.embor.7400440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Mi-ichi F., Yousuf M.A., Nakada-Tsukui K., Nozaki T. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc Nat Acad. Sci USA. 2009;106:21731–21736. doi: 10.1073/pnas.0907106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Rada P., Smid O., Sutak R., Dolezal P. The monothiol single-domain glutaredoxin is conserved in the highly reduced mitochondria of Giardia intestinalis. Eukaryot. Cell. 2009;8:1584–1591. doi: 10.1128/EC.00181-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Jedelsky P.L., Dolezal P., Rada P., Pyrih J. The minimal proteome in the reduced mitochondrion of the parasitic protist Giardia intestinalis. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tachezy J., Dolezal P. Iron–sulfur proteins and iron–sulfur cluster assembly in organisms with hydrogenosomes and mitosomes. In: Martin W.F., Müller M., editors. Origin of Mitochondria and Hydrogenosomes. Springer-Verlag; Heidelberg, Germany: 2007. pp. 105–134. [Google Scholar]
- 227.Maralikova B., Ali V., Nakada-Tsukui K., Nozaki T. Bacterial-type oxygen detoxification and iron-sulfur cluster assembly in amoebal relict mitochondria. Cell Microbiol. 2010;12:331–342. doi: 10.1111/j.1462-5822.2009.01397.x. [DOI] [PubMed] [Google Scholar]
- 228.Loftus B., Anderson I., Davies R., Alsmark U.C. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 229.Nixon J.E., Wang A., Field J., Morrison H.G. Iron-dependent hydrogenases of Entamoeba histolytica and Giardia lamblia: activity of the recombinant entamoebic enzyme and evidence for lateral gene transfer. Biol. Bull. 2003;204:1–9. doi: 10.2307/1543490. [DOI] [PubMed] [Google Scholar]
- 230.Reeves R.E., Warren L.G., Susskind B., Lo H.S. An energy-conserving pyruvate-to-acetate pathway in Entamoeba histolytica. Pyruvate synthase and a new acetate thiokinase. J. Biol. Chem. 1977;252:726–731. [PubMed] [Google Scholar]
- 231.Reeves R.E. Metabolism of Entamoeba histolytica schaudin, 1903. Adv. Parasitol. 1984;23:105–142. doi: 10.1016/s0065-308x(08)60286-9. [DOI] [PubMed] [Google Scholar]
- 232.Townson S.M., Upcroft J.A., Upcroft P. Characterization and purification of pyruvate:ferredoxin oxidoreductase from Giardia duodenalis. Mol. Biochem. Parasitol. 1996;79:183–193. doi: 10.1016/0166-6851(96)02661-8. [DOI] [PubMed] [Google Scholar]
- 233.Sanchez L.B. Aldehyde dehydrogenase (CoA-acetylating) and the mechanism of ethanol formation in the amitochondriate protist, Giardia lamblia. Arch. Biochem. Biophys. 1998;354:57–64. doi: 10.1006/abbi.1998.0664. [DOI] [PubMed] [Google Scholar]
- 234.Sanchez L.B., Müller M. Purification and characterization of the acetate forming enzyme, acetyl-CoA synthetase (ADP-forming) from the amitochondriate protist, Giardia lamblia. FEBS Lett. 1996;378:240–244. doi: 10.1016/0014-5793(95)01463-2. [DOI] [PubMed] [Google Scholar]
- 235.Sanchez L.B., Galperin M.Y., Müller M. Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of acyl-CoA synthetases (nucleoside diphosphate-forming) J. Biol. Chem. 2000;275:5794–5803. doi: 10.1074/jbc.275.8.5794. [DOI] [PubMed] [Google Scholar]
- 236.Kumar A., Shen P.S., Descoteaux S., Pohl J. Cloning and expression of an NADP+-dependent alcohol dehydrogenase gene of Entamoeba histolytica. Proc. Natl. Acad. Sci. U.S.A. 1992;89:10188–10192. doi: 10.1073/pnas.89.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Rodríguez M.A., Báez-Camargo M., Delgadillo D.M., Orozco E. Cloning and expression of an Entamoeba histolytica NAPD+-dependent alcohol dehydrogenase gene. Biochim. Biophys. Acta. 1996;1306:23–26. doi: 10.1016/0167-4781(96)00014-0. [DOI] [PubMed] [Google Scholar]
- 238.Bruchhaus I., Tannich E. Purification and molecular characterization of the NAD+-dependent acetaldehyde/alcohol dehydrogenase from Entamoeba histolytica. Biochem. J. 1994;303:743–748. doi: 10.1042/bj3030743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Karnkowska A., Hampl V. The curious case of vanishing mitochondria. Microb Cell. 2016;3:491–494. doi: 10.15698/mic2016.10.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Karnkowska A., Vacek V., Zubácová Z., Treitli S.C. A eukaryote without a mitochondrial organelle. Curr. Biol. 2016;26:1274. doi: 10.1016/j.cub.2016.03.053. [DOI] [PubMed] [Google Scholar]
- 241.Dagan T., Roettger M., Bryant D., Martin W. Genome networks root the tree of life between prokaryotic domains. Genome Biol Evol. 2010;2:379–392. doi: 10.1093/gbe/evq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Dagley M.J., Dolezal P., Likic V.A., Smid O. The protein import channel in the outer mitosomal membrane of Giardia intestinalis. Mol. Biol. Evol. 2009;26:1941–1947. doi: 10.1093/molbev/msp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.JO McInerney, O'Connell M., Pisani D. The hybrid nature of the eukaryota and a consilient view of life on Earth. Nat. Rev. Microbiol. 2014;12:449–455. doi: 10.1038/nrmicro3271. [DOI] [PubMed] [Google Scholar]
- 244.Williams T.A., Foster P.G., Cox C.J., Embley T.M. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504:231–236. doi: 10.1038/nature12779. [DOI] [PubMed] [Google Scholar]
- 245.Williams T.A., Embley M. Archaeal “dark matter” and the origin of eukaryotes. Genome Biol Evol. 2014;6:474–481. doi: 10.1093/gbe/evu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Guy L., Saw J.H., Ettema T.J.G. The archaeal legacy of eukaryotes: a phylogenomic perspective. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a016022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Horner D.S., Heil B., Happe T., Embley T.M. Iron hydrogenases—ancient enzymes in modern eukaryotes. Trends Biochem. Sci. 2002;27:148–153. doi: 10.1016/s0968-0004(01)02053-9. [DOI] [PubMed] [Google Scholar]
- 248.Vignais P.M., Billoud B. Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 2007;107:4206–4272. doi: 10.1021/cr050196r. [DOI] [PubMed] [Google Scholar]
- 249.Cardol P., Gonzalez-Halphen D., Reyes-Prieto A., Baurain D. The mitochondrial oxidative phosphorylation proteome of Chlamydomonas reinhardtii deduced from the genome sequencing project. Plant Physiol. 2005;137:447–459. doi: 10.1104/pp.104.054148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Happe T., Naber J.D. Isolation, characterization and N-terminal amino-acid-sequence of hydrogenase from the green-alga Chlamydomonas reinhardtii. Eur. J. Biochem. 1993;214:475–481. doi: 10.1111/j.1432-1033.1993.tb17944.x. [DOI] [PubMed] [Google Scholar]
- 251.Gelius-Dietrich G., Henze K. Pyruvate formate lyase (PFL) and PFL activating enzyme in the chytrid fungus Neocallimastix frontalis: a free-radical enzyme system conserved across divergent eukaryotic lineages. J. Eukaryot. Microbiol. 2004;51:456–463. doi: 10.1111/j.1550-7408.2004.tb00394.x. [DOI] [PubMed] [Google Scholar]
- 252.van Lis R., Baffert C., Couté Y., Nitschke W. Chlamydomonas reinhardtii chloroplasts contain a homodimeric pyruvate:ferredoxin oxidoreductase that functions with FDX1. Plant Physiol. 2013;161:57–71. doi: 10.1104/pp.112.208181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Simpson A.G.B., van den Hoff J., Bernard C., Burton H.R. The ultrastructure and systematic position of the Euglenozoon Postgaardi mariagerensis, Fenchel et al. Arch. Protistenkd. 1997;147:213–225. [Google Scholar]
- 254.Buetow D.E. The mitochondrion. In: Buetow D.E., editor. vol. 4. Academic Press; San Diego, CA, USA: 1989. pp. 247–314. (The Biology of Euglena). [Google Scholar]
- 255.Schwartzbach S.D., Shigeoka S., editors. vol. 979. Springer; Berlin, Germany: 2017. Euglena: biochemistry, cell and molecular biology. (Advances in Experimental Medicine and Biology). [Google Scholar]
- 256.Zimorski V., Rauch C., van Hellemond J.J., Tielens A.G.M. The mitochondrion of Euglena gracilis. In: Schwartzbach S.D., Shigeoka S., editors. Euglena: Biochemistry, Cell and Molecular Biology. Advances in Experimental Medicine and Biology 979. Springer-Verlag; Berlin, Germany: 2017. pp. 19–37. [DOI] [PubMed] [Google Scholar]
- 257.Hoffmeister M., van der Klei A., Rotte C., van Grinsven K.W. Euglena gracilis rhodoquinone:ubiquinone ratio and mitochondrial proteome differ under aerobic and anaerobic conditions. J. Biol. Chem. 2004;279:22422–22429. doi: 10.1074/jbc.M400913200. [DOI] [PubMed] [Google Scholar]
- 258.Green L.S., Li Y., Emerich D.W., Bergersen F.J. Catabolism of α-ketoglutarate by a sucA mutant of Bradyrhizobium japonicum: evidence for an alternative tricarboxylic acid cycle. J. Bacteriol. 2000;182:2838–2844. doi: 10.1128/jb.182.10.2838-2844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Inui H., Miyatake K., Nakano Y., Kitaoka S. Wax ester fermentation in Euglena gracilis. FEBS Lett. 1982;150:89–93. doi: 10.1002/1873-3468.13276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Inui H., Miyatake K., Nakano Y., Kitaoka S. Production and composition of wax esters by fermentation of Euglena gracilis. Agric. Biol. Chem. 1983;47:2669–2671. [Google Scholar]
- 261.Inui H., Miyatake K., Nakano Y., Kitaoka S. Fatty acid synthesis in mitochondria of Euglena gracilis. Eur. J. Biochem. 1984;142:121–126. doi: 10.1111/j.1432-1033.1984.tb08258.x. [DOI] [PubMed] [Google Scholar]
- 262.Tucci S., Vacula R., Krajcovic J., Proksch P. Variability of wax ester fermentation in natural and bleached Euglena gracilis strains in response to oxygen and the elongase inhibitor flufenacet. J. Eukaryot. Microbiol. 2010;57:63–69. doi: 10.1111/j.1550-7408.2009.00452.x. [DOI] [PubMed] [Google Scholar]
- 263.Ogawa T., Kimura A., Sakuyama H., Tamoi M. Identification and characterization of cytosolic fructose-1,6-bisphosphatase in Euglena gracilis. Biosci. Biotechnol. Biochem. 2015;79:1957–1964. doi: 10.1080/09168451.2015.1069694. [DOI] [PubMed] [Google Scholar]
- 264.Tucci S., Proksch P., Martin W. Fatty acid biosynthesis in mitochondria of Euglena gracilis. In: Benning C., Ohlrogge J., editors. Current Advances in the Biochemistry and Cell Biology of Plant Lipid: Proceedings of the 17th International Symposium of Plant Lipids. Aardvark Global Publishing Company, LLC Salt Lake City UT; 2007. pp. 133–136. [Google Scholar]
- 265.Dasgupta S., Fang J., Brake S.S., Hasiotis S.T. Biosynthesis of sterols and wax esters by Euglena of acid mine drainage biofilms: implications for eukaryotic evolution and the early Earth. Chem. Geol. 2012;306:139–145. [Google Scholar]
- 266.Hoffmeister M., Piotrowski M., Nowitzki U., Martin W. Mitochondrial trans-2-enoyl-CoA reductase of wax ester fermentation from Euglena gracilis defines a new family of enzymes involved in lipid synthesis. J. Biol. Chem. 2005;280:4329–4338. doi: 10.1074/jbc.M411010200. [DOI] [PubMed] [Google Scholar]
- 267.Tucci S., Martin W. A novel prokaryotic trans-2-enoyl-CoA reductase from the spirochete Treponema denticola. FEBS Lett. 2007;581:1561–1566. doi: 10.1016/j.febslet.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 268.Schneider T., Betz A. Waxmonoester fermentation in Euglena gracilis T. Factors favouring the synthesis of odd-numbered fatty acids and alcohols. Planta. 1985;166:67–73. doi: 10.1007/BF00397387. [DOI] [PubMed] [Google Scholar]
- 269.Inui H., Miyatake K., Nakano Y., Kitaoka S. Synthesis of reserved polysaccharide from wax esters accumulated as the result of anaerobic energy generation in Euglena gracilis returned from anaerobic to aerobic conditions. Int. J. Biochem. 1992;24:799–803. [Google Scholar]
- 270.Ono K., Miyatake K., Inui H., Kitaoka S. Wax ester production by anaerobic Euglena gracilis. J. Mar. Biotechnol. 1995;2:157–161. [Google Scholar]
- 271.Ono K., Kondo M., Osafune T., Miyatake K. Presence of glyoxylate cycle enzymes in the mitochondria of Euglena gracilis. J. Eukaryot. Microbiol. 2003;50:92–96. doi: 10.1111/j.1550-7408.2003.tb00239.x. [DOI] [PubMed] [Google Scholar]