Abstract
Oncogenic protein tyrosine phosphatases (PTPs) are overexpressed in numerous human cancers but they have been challenging pharmacological targets. The emblematic oncogenic PTP4A tyrosine phosphatase family regulates many fundamental malignant processes. 7-Imino-2-phenylthieno[3,2-c]pyridine-4,6(5H,7H)-dione (JMS-053) is a novel, potent, and selective PTP4A inhibitor but its mechanism of action has not been fully elucidated, nor has the chemotype been fully investigated. Because tyrosine phosphatases are notoriously susceptible to oxidation, we interrogated JMS-053 and three newly synthesized analogs with specific attention on the role of oxidation. JMS-053 and its three analogs were potent in vitro PTP4A3 inhibitors, but 7-imino-5-methyl-2-phenylthieno[3,2-c]pyridine-4,6(5H,7H)-dione (NRT-870-59) appeared unique among the thienopyridinediones with respect to its inhibitory specificity for PTP4A3 versus both a PTP4A3 A111S mutant and an oncogenic dual specificity tyrosine phosphatase, CDC25B. Like JMS-053, NRT-870-59 was a reversible PTP4A3 inhibitor. All of the thienopyridinediones retained cytotoxicity against human ovarian and breast cancer cells grown as pathologically relevant three-dimensional spheroids. Inhibition of cancer cell colony formation by NRT-870-59, like JMS-053, required PTP4A3 expression. JMS-053 failed to generate significant detectable reactive oxygen species in vitro or in cancer cells. Mass spectrometry results indicated no disulfide bond formation or oxidation of the catalytic Cys104 after in vitro incubation of PTP4A3 with JMS-053 or NRT-870-59. Gene expression profiling of cancer cells exposed to JMS-053 phenocopied many of the changes seen with the loss of PTP4A3 and did not indicate oxidative stress. These data demonstrate that PTP4A phosphatases can be selectively targeted with small molecules that lack prominent reactive oxygen species generation and encourage further studies of this chemotype.
SIGNIFICANCE STATEMENT
Protein tyrosine phosphatases are emerging as important contributors to human cancers. We report on a new class of reversible protein phosphatase small molecule inhibitors that are cytotoxic to human ovarian and breast cancer cells, do not generate significant reactive oxygen species in vitro and in cells, and could be valuable lead molecules for future studies of PTP4A phosphatases.
Introduction
The PTP4A (also known as phosphatase of regenerating liver or PRL) phosphatases are a unique subfamily of protein tyrosine phosphatases (PTPs) comprising three highly homologous members (PTP4A1, PTP4A2, and PTP4A3) with ∼80% amino acid sequence identity. PTP4A3, and to a lesser extent PTP4A1 and PTP4A2, are overexpressed in many types of cancer, promote tumor invasion and dissemination, and contribute to poor patient prognosis (Daouti et al., 2008; den Hollander et al., 2016). The PTP4A family members have been categorized among the most oncogenic of all phosphatases, and consequently they represent attractive cancer therapeutic targets (Yu and Zhang, 2017). However, like almost all of the other PTPs, the PTP4A family members lack readily available potent and selective small molecule inhibitors and are even thought by some investigators to be undruggable (Lazo and Sharlow, 2016; Stanford and Bottini, 2017). This notion has been sustained because PTPs are intracellular and the phosphorylated substrates are highly charged, making it difficult for mimics of the substrates to reach the phosphatase. There is also considerable amino acid conservation in the active site of many PTPs. Nonetheless, we previously synthesized 7-imino-2-phenylthieno[3,2-c]pyridine-4,6(5H,7H)-dione (JMS-053) and discovered its potent and reversible PTP4A3 inhibitory profile (Salamoun et al., 2016; McQueeney et al., 2017). We documented that JMS-053 inhibited cancer cell migration and spheroid growth in vitro, attenuated in vivo ovarian tumor growth, and mitigated the disruption of the microvascular endothelial barrier function by vascular endothelial growth factor or lipopolysaccharide (McQueeney et al., 2017, 2018). The current project was designed to probe the structural features of JMS-053 that contribute to PTP4A3 inhibition and to investigate further the mechanism of enzyme inhibition.
The generation of reactive oxygen species (ROS) is an intrinsic component of endogenous growth factor stimulation and metabolism, which has fortified the hypothesis that protein oxidation is a post-translational modification involved in normal cell signaling and gene expression (Chakraborty et al., 2019). This suggests manipulation of ROS could be a valuable approach for the treatment of cancer. Indeed, cancer cells are known to produce more ROS than nontransformed cells (Schieber and Chandel, 2014) and at least 12 clinically used anticancer drugs either directly or indirectly generate ROS (Yokoyama et al., 2017). Recent studies have shown that many proteins become oxidized at cysteine residues under conditions that increase intracellular ROS levels (van der Reest et al., 2018). Several classes of attractive therapeutic targets are susceptible to ROS-mediated inactivation, including metalloenzymes, cysteine proteases, and PTPs, because they contain a cysteine in their active site, which is readily deprotonated as a consequence of the surrounding peptide side chain functionality (Pani et al., 2000; Johnston et al., 2008). The catalytic thiol can be oxidized to a metastable sulfenic acid and further oxidized to sulfinic or sulfonic acid, both of which are considerably more stable under physiologic conditions. Other inactivating oxidation processes have also been reported with PTPs. For example, PTEN, CDC25, LMW-PTP, DUSP12, DUSP6, SHP1, SHP2, LYP, and PTP4A phosphatases are susceptible to intramolecular disulfide bond formation (Bonham and Vacratsis, 2009; Tanner et al., 2011; Ishii et al., 2013). The creation of cyclic sulfenylamide bonds (Defelipe et al., 2015) and degradation of the catalytic cysteine to a glycine have been detected in PTPs (Orsatti et al., 2009). Such observations raise the question of whether small molecules that generate oxidative stress could promote these types of post-translational modifications, and thus inactivate PTPs.
Although in vitro PTP4A3 inhibition by JMS-053 is reversible, we previously did not formally exclude the possibility that JMS-053 forms ROS, which is a common deactivation process found for other PTP inhibitors (Johnston et al., 2008). Previous work indicates disulfide bond formation is reversible as is sulfenic acid oxidation of the catalytic thiol (Orsatti et al., 2009; Ishii et al., 2013). Thus, in the current report, we investigated the ability of JMS-053 and several recently synthesized analogs of JMS-053 to generate ROS and to inhibit PTP4A family members. We also biochemically tested our previously described computational models of the interactions between JMS-053 and PTP4A3 (McQueeney et al., 2017; Tasker et al., 2019) and interrogated the cytotoxicity of the JMS-053 analogs to human and mouse cancer cells to further clarify the actions of these compounds as chemical biology tools.
Materials and Methods
Compounds and Reagents.
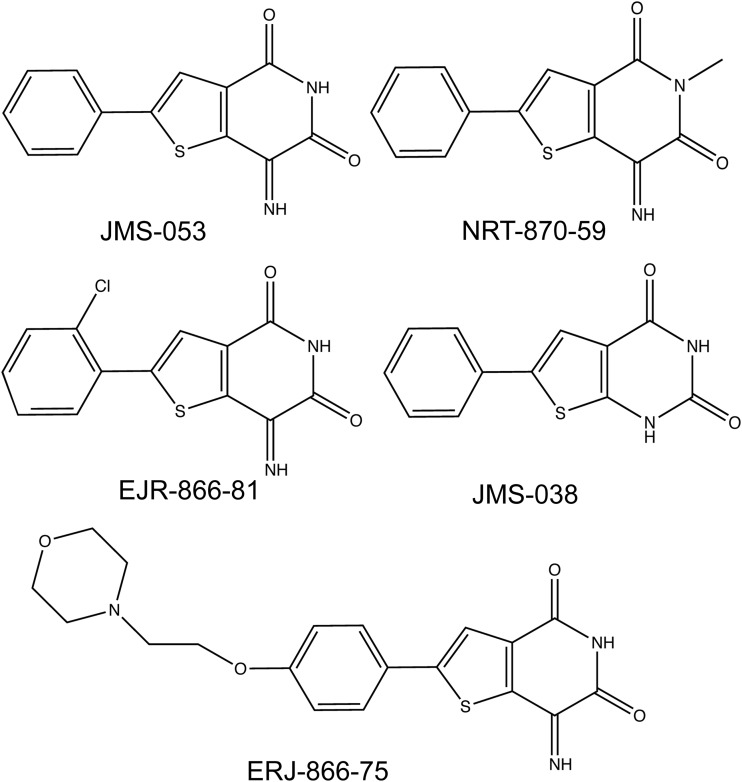
7-Amino-2-phenyl-5H-thieno[3,2-c]pyridin-4-one (thienopyridone), 6-chloro-7-(2-morpholin-4-yl-ethylamino)-quinoline-5-8-dione (DA-3003-1), JMS-053, the inactive control compound 6-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-dione (JMS-038), 7-imino-2-(4-(2-morpholinoethoxy)phenyl)thieno[3,2-c]pyridine-4,6(5H,7H)-dione (EJR-866-75), 2-(2-chlorophenyl)-7-iminothieno[3,2-c]pyridine-4,6(5H,7H)-dione (EJR-866-81), and 7-imino-5-methyl-2-phenylthieno[3,2-c]pyridine-4,6(5H,7H)-dione (NRT-870-59) (Fig. 1) were synthesized as previously described (Brisson et al., 2005; Salamoun et al., 2016; Tasker et al., 2019). 6,8-Difluoro-4-methyl-umbelliferyl phosphate (DiFMUP) was purchased from ThermoFisher Scientific (Waltham, MA). DMSO was obtained from VWR (Radnor, PA). All other reagents were obtained from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated.
Fig. 1.
Chemical structures of the thienopyridone analogs.
pET15b Bacterial Expression Constructs.
pET15b-His6-PTP4A3, -PTP4A3-K144I, -PTP4A3-C49S, -PTP4A3-A106V, -PTP4A3-A111S, -PTP4A1, -PTP4A2, -CDC25B1, and -DUSP3 bacterial expression constructs were obtained from Genscript (Piscataway, NJ). Recombinant human proteins were expressed and purified using previously described procedures (McQueeney et al., 2017, 2018).
Cancer Cell Lines.
Authenticated OVCAR4 cells were obtained from Charles River Laboratories (New York, NY) and Kuramochi from (Sekisui XenoTech, LLC, Kansas City, KS). Authenticated MDA-MB-231, Hs578T, and IMR90 cells were obtained from America Type Culture Collection (Manassas, VA). Nonmalignant human H10-180 ovarian epithelial cells were a gift from Charles N. Landen (University of Virginia) and have been previously described (McQueeney et al., 2017, 2018). OVCAR4, Kuramochi, and MDA-MB-231 cells were maintained in RPMI 1640 (Life Technologies, Grand Island, NY) supplemented with 10% FBS, with no antibiotics, and were passaged <20 times. Hs578T cells were grown in Dulbecco’s minimum essential medium supplemented with 10% FBS and 0.01% bovine insulin, with no antibiotics, and were likewise passaged <20 times. The four human cell lines are valuable because they are genetically diverse and have been fully annotated for mutations and mRNA expression by the Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle). All four cell lines have mutations in the DNA binding domain of TP53 (Supplemental Table 1), which encodes the direct target of PTP4A3: p53 (Basak et al., 2008). All All of the cell lines also have missense mutations in HRAS, KRAS, or IL6R, which could influence the actions of PTP4A3 (Supplemental Table 1). Mixed mouse PTP4A3 wild-type and null colon cancer cells and the colony formation assay conditions have been previously described (McQueeney et al., 2018). Exponentially growing ovarian and breast cancer cells were harvested and seeded (250 cells/22 µl) in each well of a 384-well ultralow attachment U-bottom microtiter plate (Corning, Corning, NY). Plates were incubated for 24 hours (37°C at 5% CO2) to allow for spheroid formation. Compounds (3 μl) were then added in final DMSO concentrations of 0.5%, as was the vehicle control (0.5% DMSO). Positive control wells contained 10% DMSO. The microtiter plates were incubated for 48 hours (37°C, 5% CO2) and 25 μl of CellTiterGlo 3D Reagent (Promega, Fitchburg, WI) was added. Plates were incubated with shaking for 30 minutes at room temperature. Luminescence data were captured on a SpectraMax M5 multimode plate reader (San Jose, CA). The effects of NRT-870-059 on colon cancer colony formation were performed as previously described (McQueeney et al., 2018).
In Vitro Biochemical Analysis of PTP4A Inhibition.
Enzyme activity assays were performed in triplicate in 384-well Greiner Bio-One black small volume microtiter plates, as previously described (Salamoun et al., 2016; McQueeney et al., 2017, 2018), using recombinant human His6-tagged PTP4A1, PTP4A2, PTP4A3, PTP4A3 mutants, CDC25B, or DUSP3 and substrate DiFMUP (12 μM) incubated at 25°C for 25 minutes in 40 mM Tris-HCl (pH 7.0), 75 mM NaCl, 2 mM EDTA, and 4 mM dithiothreitol (DTT) buffer. The assays were fully automated using an Agilent Bravo Liquid Handling Platform and miniaturized to 15 μl total volume. Dilutional reversibility assays were performed in a 100 μl total reaction volume using the same assay conditions (McQueeney et al., 2017, 2018). His6-tagged PTP4A3 (1 μg) was preincubated for 30 minutes with 0, 86, or 860 nM compound and then diluted 10-fold. Reactions were initiated with the addition of 45 μl of substrate for a final DiFMUP concentration of 12 μM and incubated at room temperature for 25 minutes. Preincubation studies with NRT-870-59 were performed by incubating the compound with PTP4A3 for 2 hours with continuous shaking, after which time substrate was added and the standard assay conditions were followed. Fluorescence data were captured on a SpectraMax M5 and phosphatase activity was expressed as a percentage of maximal activity.
In Vitro and Cell-Based Reactive Oxygen Species Assessments.
In vitro generation of ROS by JMS-053 and it analogs was determined by two orthogonal fluorescence- and absorbance-based assays using resazurin and phenol red as substrates, respectively, as described previously (Lor et al., 2007; Johnston, 2011). Cellular ROS generation was determined using a ROS-Glo H2O2 Assay (Promega, Madison, WI) according to the manufacturer’s instructions. In brief, 1,000 OVCAR4 cells were seeded per well of a 384-well microtiter plate and incubated until they were ∼70% confluent. Cells were exposed to compounds or control vehicle for 10 minutes, and then ROS-Glo Detection Solution was added to each well. Plates were incubated for 20 minutes and relative luminescence units were captured on a SpectraMax M5.
Filter-Aided Sample Preparation of Peptides for Liquid Chromatography–Tandem Mass Spectrometry Analysis.
Samples were prepared in duplicate using a modified filter-aided sample preparation of peptides protocol (Wiśniewski et al., 2009) to compare between reducing and nonreducing conditions. Purified PTP4A3 (30 µg) was incubated with 200 nM of JMS-053, NRT-870-59, or water for 10 minutes at room temperature. Once the incubation was complete, samples were diluted in either 10 M urea/25 mM ammonium bicarbonate (ABC) or 2 mM urea in Tris-HCl, transferred to 10 kDa filters (Sartorious, AG, Göttingen, Germany), and centrifuged at 14,000g for 15 minutes. Samples were diluted in buffer and centrifuged two more times before further treatment, and all flow through was discarded. Reduction with 12.5 mM DTT was performed in samples denatured by the urea/ABC buffer for 45 minutes at 56°C followed by alkylation at 37°C with 50 mM iodoacetic acid in the dark, while corresponding samples were alkylated with 2 mM iodoacetic acid at 37°C in the dark for 2 hours. All filters were subsequently washed with urea/ABC buffer and centrifuged three times at 14,000g for 15 minutes. This step was repeated another three times with 25 mM ABC before the proteolytic digest. Filters were transferred to new tubes before the addition of trypsin/lys-c (Promega) at a 1:100 ratio of protease to PTP4A3 and incubated in a shaker at 37°C for 12 hours. Filter units were transferred to new, low-bind tubes washed with 25 mM ABC and centrifuged three times at 14,000g for 10 minutes. The flow through was collected and acidified with 0.1% formic acid, desalted on C18 StageTips, and lyophilized. Peptides were resuspended in 50 µl of 0.1% formic acid for analysis.
Electrospray Ionization Liquid Chromatography–Tandem Mass Spectrometry Acquisition.
An integrated liquid chromatography autosampler (Easy-nLC 1200; ThermoFisher Scientific) was used to load peptides onto a trap column (2 cm Nano-Trap, 5 µm C18; Thermo Fisher Scientific) and washed for 2 minutes with 1% B (80% acetonitrile and 1% formic acid). The peptides were eluted from the trap column and through a homemade 10-cm nanocapillary analytical column (5 µm C18 packed in 360 µm o.d. × 75 µm i.d. fused silica), with an integrated electrospray tip, using a 65-minute 1%–95% reverse-phase liquid chromatography gradient (A: 0.1% formic acid; B: 80% acetonitrile and 0.1% formic acid) with the following parameters: 0–2 minutes, 1% B at 400 nl/min; 2–21 minutes, 45% B at 300 nl/min; 21–25 minutes, 95% B at 300 nl/min; and 25–35 minutes, 1% B at 400 nl/min. The eluting peptides were analyzed using a linear ion trap mass spectrometer (ThermoFisher Scientific), which was operated with a top five data-dependent acquisition method that consisted of one full MS1 scan (375–1800 mass-to-charge ratio (m/z)) followed by five MS2 scans of the most abundant ions with a normalized collision energy of 35. Peptide identification was accomplished by matching observed peptide m/z values to predicted average peptide masses for PTP4A3 by using an ExPASy in silico digest (PeptideMass) with a 500 ppm mass tolerance (Gasteiger et al., 2005). Peptides were further verified by matching fragment ions predicted from the sequence using ProteinProspector MS-Product (version 5.22.1).
Gene Expression.
Total RNA was isolated as previously described (McQueeney et al., 2018) from wild-type PTP4A3 mouse colon cancer cells treated with 1 μM JMS-053 or vehicle control (1% DMSO) for 24 hours or isogenic PTP4A3 null mouse colon cancer cells. Next-generation sequencing was conducted as previously described (McQueeney et al., 2018) and pathway analyses were conducted with transcripts that were either significantly increased or decreased using Reactome software (https://reactome.org). Total RNA was extracted from human and mouse cells to measure PTP4A3 mRNA levels by real-time quantitative polymerase chain reaction and normalized to human glyceraldehyde-3-phosphate dehydrogenase, actin, and hypoxanthine phosphoribosyltransferase as previously described (McQueeney et al., 2018).
Statistical Analysis.
All statistical analyses were performed with GraphPad Prism 7.0. Data are presented as average (mean) ± S.D. or S.E. The P values were calculated with Student’s t test for comparisons involving two groups and one- or two-way ANOVA for comparisons involving more than two groups. A value of P < 0.05 was considered statistically significant. Each experiment is represented by at least three biologic replicates and three technical replicates (per independent experiment) unless otherwise indicated.
Results
Inhibition of PTP4A3 by JMS-053 and New Analogs.
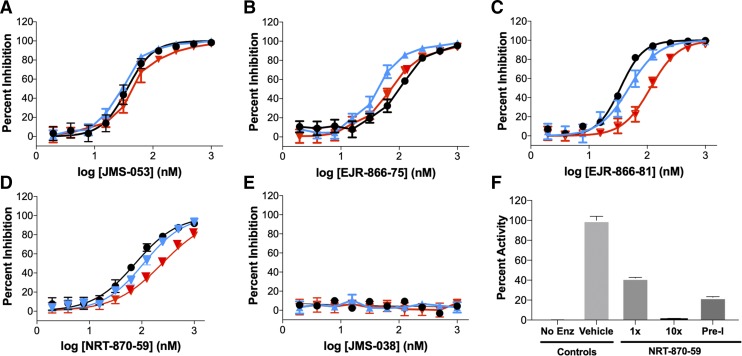
We recently synthesized a series of 19 analogs of the novel iminothienopyridinedione JMS-053, which is a reversible, allosteric, and cell-active small molecule PTP4A3 inhibitor with an in vitro IC50 value of ∼30 nM for recombinant human PTP4A3 (Salamoun et al., 2016; McQueeney et al., 2017, 2018; Tasker et al., 2019). From the series, three JMS-053 analogs, EJR-866-75, EJR-866-81, and NRT-870-59 (Fig. 1), were selected for further study because they retained the ability to potently inhibit PTP4A3 in vitro and had structural features that potentially should reduce metabolism or increase water solubility (Tasker et al., 2019). We also synthesized the valuable inactive congener JMS-053 (Fig. 1) as a control compound. Since the endogenous substrate for PTP4A3 has not yet been firmly established, previous enzymatic studies employed artificial small molecule substrates, such as DiFMUP. With this substrate, PTP4A3 utilizes a two-step in vitro kinetic cycle, which involves a long-lived phosphocysteine intermediate (Gulerez et al., 2016). The first step is rapid followed by slower steady-state conversion, which is likely to represent the kinetically more meaningful parameter within cells. Using DiFMUP and a 25-minute incubation that reflects the steady-state rate, we found that the PTP4A3 IC50 values for EJR-866-75, ERJ-866-81, and NRT-870-59 were 98.2, 36.1, and 86.0 nM, respectively (Supplemental Fig. 1; Table 1). The inactive control congener, JMS-038, failed to inhibit PTP4A3 (Supplemental Fig. 1; Table 1). JMS-053 was equipotent against PTP4A3 and its close family members PTP4A1 and PTP4A2, confirming our previous results (McQueeney et al., 2017) (Fig. 2A; Table 1). This pan-PTP4A phosphatase inhibition was also observed with a less potent parent compound, thienopyridone (Daouti et al., 2008; Hoeger et al., 2014). ERJ-866-75 showed little preference among the PTP4A family members (Fig. 2B; Table 1). Surprisingly, EJR-866-81 and the thienopyridone core–modified NRT-870-59 displayed a pronounced preference for PTP4A3 compared with PTP4A2 (Fig. 2, C and D; Table 1). This is the first demonstration of tractable selectivity by a potent small molecule inhibitor for PTP4A3 versus PTP4A2, which is rather remarkable considering there is ∼80% identity in the overall amino acid composition between these two phosphatases and 100% identity in the catalytic P loop (HCVAGLGRA) (Rios et al., 2013). However, it is noteworthy that a 10-fold selectivity for inhibition of PTP4A1 over PTP4A3 was previously reported for the chemically unrelated procyanidin B3 (Stadlbauer et al., 2015). JMS-038 was inactive against all three PTP4A3 family members (Fig. 2E).
TABLE 1.
IC50 values for PTP4A3 inhibitors
The in vitro IC50 values for the iminopyridinediones were determined with recombinant human protein using the artificial substrate DiFMUP, an automated liquid handling platform, and 25-minute incubation. N = 3 independent experiments unless otherwise marked.
| Compound | Mean (nM) ± S.E.M. | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| PTP4A1 | PTP4A2 | PTP4A3 | PTP4A3 C49S | PTP4A3 K144I | PTP4A3 A106V | PTP4A3 A111S | CDC25B | DUSP3 | |
| JMS-053 | 29.1 ± 2.7 | 48.0 ± 14.5 | 34.7 ± 2.5a | 156.5 ± 20.1b | 42.7 ± 8.0 | 26.3 ± 2.5 | 104.4 ± 7.7b,c | 92.6 ± 10.2 | 207.6 ± 46.3 |
| EJR-866-81 | 49.0 ± 5.0 | 112.9 ±10.0b | 36.1 ± 1.4 | 390.0 ± 72.5b | 59.1 ± 5.9 | 54.5 ± 2.4 | 61.0 ± 2.8b,c | 65.5 ± 16.6 | 240.8 ± 30.9 |
| EJR-866-75 | 43.2 ± 8.5 | 73.9 ± 10.0 | 98.2 ± 2.5d | 229.7 ±54.6b | 68.5 ± 1.0 | 57.5 ± 18.6 | 172.0 ± 46.0b,c,d | 122.6 ± 22.6 | 521.0 ± 128.4 |
| NRT-870-59e | 133.2 ± 31.7a | 264.4 ± 37.4a,b | 86.0 ± 12.6a | 332.2 ± 52.5b | 180.2 ± 64.5e | 57.7 ± 18.6 | >1000c | >1000 | 411.8 ± 75.3 |
| JMS-038 | >1000 | >1000 | >1000 | ND | >1000 | ND | ND | ND | >1000 |
ND, not determined.
N = 8.
P < 0.05 compared with PTP4A3.
N = 2.
N = 6.
N = 5.
Fig. 2.
Small molecule inhibitor PTP4A1, PTP4A2, and PTP4A3 concentration-response curves and reversibility studies. PTP4A family inhibition: (A) JMS-053; (B) EJR-866-75; (C) EJR-866-81; (D) NRT-870-59; and (E) JMS-038 (black symbols, PTP4A3; blue symbols, PTP4A1; red symbols, PTP4A2). (F) Reversible PTP4A3 inhibition by NRT-870-59. Full-length recombinant PTP4A3 was treated with a concentration of NRT-870-59 equal to the IC50 value or 10-fold higher, namely, 86 or 860 nM, respectively. A preincubated (Pre-I) sample was exposed to a concentration of NRT-870-59 that was 10-fold higher than the IC50 value for 30 minutes and then diluted to 86 nM. Enz, enzyme. N = 3. Bars = S.E.M. unless smaller than the symbol.
Inhibition Specificity with Other PTPs.
JMS-053 inhibited the oncogenic dual specificity phosphatase CDC25B with an IC50 value of 92.6 nM (Supplemental Fig. 2A; Table 1), which was consistent with our previous report (McQueeney et al., 2017). EJR-866-81 and EJR-866-75 also were potent in vitro inhibitors of CDC25B with IC50 values of 65.5 and 122.6 nM, respectively (Supplemental Fig. 2, B and C; Table 1). In contrast, NRT-870-59 did not inhibit CDC25B at concentrations ≤1 μM (Supplemental Fig. 2D; Table 1). All of the compounds were 5- to 6-fold less potent as inhibitors of the dual specificity phosphatase DUSP3 (Table 1). Like JMS-053 (McQueeney et al., 2017), NRT-870-59 inhibition was reversible as measured with a dilutional assay (Fig. 2F).
Inhibition of PTP4A3 Mutants.
We previously proposed a binding site for JMS-053 based on computational docking studies using an energy refined structure (McQueeney et al., 2017). In agreement with a noncompetitive mechanism, the putative allosteric binding site occupied a pocket that was flanked by the α3, α4, and α6 helices on PTP4A3 along with the tryptophan-proline-aspartate (WPD) loop. The well-desolvated inhibitor binding mode was predicted to hold the WPD loop in the closed conformation in part via hydrogen bonds between the compound’s pyridinedione oxygens and the K144 side chain amine (McQueeney et al., 2017). We tested this model by generating the PTP4A3 K144I mutant, which we predicted would be resistant to inhibition by JMS-053. The K144I mutant shared a similar steady-state Michaelis constant (Km) and catalytic rate constant (Kcat) for the artificial substrate DiFMUP (4.73 μM and 6.47 minute−1) with wild-type PTP4A3 (4.04 μM and 5.77 minute−1) (Supplemental Fig. 3A; Table 2) and was inhibited with a similar IC50 value by JMS-053 (42.7 ± 8.0 nM) (Supplemental Fig. 3B; Table 1). These results suggested that K144 was not essential for JMS-053 binding to PTP4A3. The IC50 values for EJR-866-81, EJR-866-75, and NRT-870-59 were also similar for the K144I mutant and wild type, while JMS-038 did not inhibit the mutant form of PTP4A3 (Supplemental Fig. 3, C–F; Table 1). Thus, we further modified our exploratory docking model with the A chain of PTP4A3 Protein Data Bank entry 5TSR, which represents the reduced form of the enzyme with a closed WPD loop and proposed strong hydrogen bond interactions with JMS-053 to A106 and A111 (Tasker et al., 2019). We created the A106V and A111S mutants to test this hypothesis. The A106V mutant exhibited a markedly higher Km value for DiFMUP and a significantly lower Kcat/Km value compared with the wild-type PTP4A3 (Table 2). This finding would be consistent with the proposed role of the flexible WPD loop. However, JMS-053 and all three analogs retained similar IC50 values compared with the wild-type enzyme. These results suggested an interaction with A106 was not essential for JMS-053 inhibition of PTP4A3. In contrast, the A111S mutant had a Kcat/Km value that was similar to the wild-type enzyme, although the mutant Km and Kcat values were both higher than that of the wild-type enzyme. The IC50 values for JMS-053, EJR-866-81, EJR-866-75, and NRT-870-59 were higher for the A111S mutant compared with the wild-type phosphatase (Supplemental Fig. 4; Table 1), supporting a role for this amino acid in the binding and inhibition of this class of compounds.
TABLE 2.
Kinetic determinations for wild-type and mutant forms of PTP4A3
The kinetic parameters were determined with wild-type and mutant forms of recombinant human PTP4A3 and the artificial substrate DiFMUP. N = 3, mean ± S.E.M.
| Parameter | PTP4A3 | PTP4A3 C49S | PTP4A3 K144I | PTP4A3 A106V | PTP4A3 A111S |
|---|---|---|---|---|---|
| Km (μM) | 4.04 ± 0.59 | 41.10 ± 9.01 | 4.73 ± 0.71 | 32.04 ± 6.88 | 18.94 ± 6.16 |
| Kcat (min−1) | 5.77 ± 0.53 | 4.93 ± 0.24 | 6.47 ± 0.69 | 8.51 ± 2.14 | 38.72 ± 6.78 |
| Kcat/Km (min−1/μM−1) | 1.50 ± 0.53 | 0.13 ± 0.03 | 1.40 ± 0.24 | 0.26 ± 0.01 | 2.21 ± 0.46 |
Many PTPs are inhibited by intramolecular disulfide bond formation (Buhrman et al., 2005; Ishii et al., 2013) and C49 has been reported to be the only cysteine involved in an intramolecular disulfide bond formation with the catalytic C104 of PTP4A3 (Kozlov et al., 2004; Orsatti et al., 2009). Therefore, we generated the conservative C49S PTP4A3 mutant, which had impaired catalytic activity consistent with previous published results (McParland et al., 2011; Zhang et al., 2017) (Supplemental Fig. 5; Table 2). PTP4A3 C49S also had a 10-fold higher steady-state Km value for DiFMUP, in agreement with previous results with a different artificial substrate (Kozlov et al., 2004), but a similar Kcat value compared with wild-type PTP4A3 (Table 2). The higher Km value likely reflects the importance of the CXnE motif in PTP4A3 for DiFMUP dephosphorylation, although it is interesting to note that previous work has demonstrated mutation of the glycine adjacent to C49, namely E50R, does not alter the Km value of PTP4A3 for DiFMUP but increases the Kcat value (Hoeger et al., 2017). JMS-053 and all of the analogs, with the exception of the inactive JMS-038 control compound, retained partial inhibition of PTP4A3 C49S (Supplemental Fig. 5; Table 1). Thus, C49-C104 disulfide bond formation is unlikely to be the primary inhibitory mode of JMS-053 or the analogs, although these data did not formally exclude the involvement of C49. We previously hypothesized (McQueeney et al., 2017) that JMS-053 interacted with an allosteric site flanked by the highly flexible WPD loop and the α3 helix, which are located close to C49. This binding mode may account for the partial inhibition of the C49S mutant by JMS-053 and its analogs.
Reactive Oxygen Species Generation.
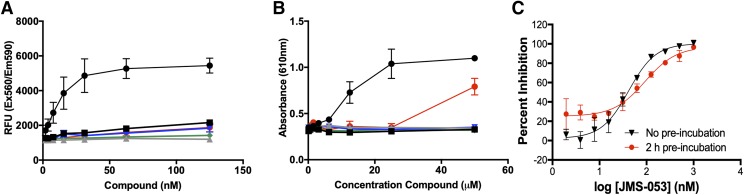
Many small molecule PTP inhibitors have been found to be redox cycling compounds, generating H2O2, superoxide, hydroxyl radicals, and singlet oxygen in the presence of strong reducing agents such as DTT, which are common in many PTP assay buffers (Soares et al., 2010; Johnston, 2011). Therefore, we formally examined JMS-053 and its analogs for their ability to generate ROS in the in vitro buffer conditions (Fig. 3). DTT can cause a net 2e− reduction of resazurin to create the highly fluorescent product resorufin, and this reaction forms the basis of a successfully used assay to identify redox cycling compounds, particularly those that generate H2O2 in high-throughput screening assays (Lor et al., 2007; Johnston, 2011). A positive control quinolinedione (DA-3003-1), which we previously found generates ROS in vitro with PTP assay buffers (Vogt et al., 2008), yielded a concentration-dependent increase in the relative fluorescence units. Concentrations of DA-3003-1 as low as 15 nM caused a significant increase in oxidized product (Fig. 3A). In contrast, the JMS-053 series showed little or no ability to form ROS at concentrations that were 2-fold higher than the IC50 value, i.e., <62 nM, when compared with the inactive compound JMS-038. We also evaluated the redox liabilities of JMS-053 and its analogs using a phenol red/horseradish peroxidase assay, which can readily detect H2O2 generated in vitro by redox cycling compounds coincubated with DTT and is based on absorbance not fluorescence (Vogt et al., 2008). The positive control compound DA3003-1 (Vogt et al., 2008) produced a pronounced concentration-dependent increase in absorbance, while no change in absorbance was detected with JMS-053, EJR-866-75, NRT-870-59, and JMS-038 at concentrations ≤50 μM (Fig. 3B). EJR-866-81 showed no evidence for H2O2 generation at concentrations of ≤25 μM but increased absorbance at 50 μM (Fig. 3B). Moreover, preincubation of PTP4A3 with JMS-053 for 2 hours did not markedly alter PTP4A3 inhibition (Fig. 3C), which was consistent with our previous observation of reversibility (McQueeney et al., 2017). Addition of 1 U/ml of catalase decreased by 19-fold the IC50 value of the control ROS-generating compound DA-3003-1 from 60.2 nM to 1.1 μM, while the same catalase concentration only decreased the IC50 value of JMS-053 by 3-fold to 108.5 nM. The modest reduction in the JMS-053 IC50 value in the presence of catalase might reflect the production of low levels of ROS that were not detected with the surrogate assays or simply be due to binding of the compound to catalase. This aspect of the work is worthy of further investigation.
Fig. 3.
Redox activity of JMS-053 analogs and preincubation studies with JMS-053. (A) Detection of ROS using resazurin in the presence of DTT. (B) Detection of ROS using a phenol red/horseradish peroxidase assay in the presence of DTT (black circles, DA-3003-1; black squares, JMS-053; gray triangles, JMS-038; inverted blue triangles, EJR-866-75; green diamonds, NRT-870-59; red circles, EJR-866-81). (C) Preincubation of PTP4A3 with JMS-053 for 2 hours prior to addition of substrate. Black symbols represent no preincubation and red symbols represent 2-hour preincubation. Bars = S.E.M., N = 3.
Lack of PTP4A3 Oxidation as Measured by Mass Spectrometry.
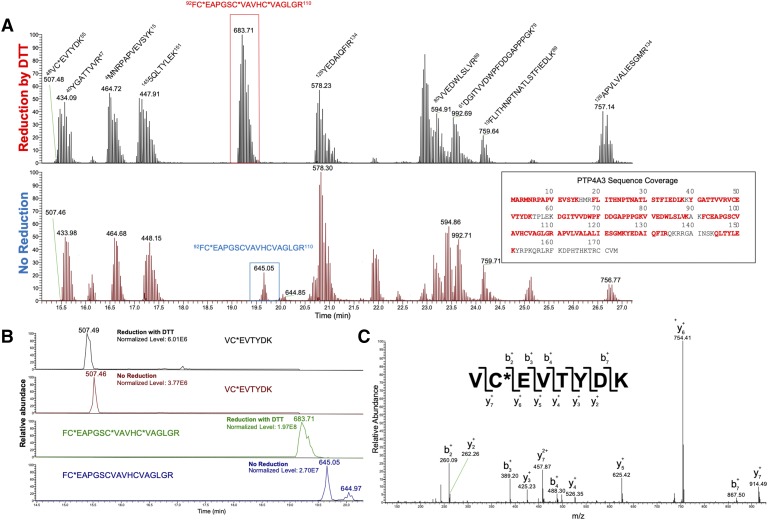
Although the bulk of our assays provided no support for significant ROS generation by JMS-053 and its analogs, the kinetics and chemistry of ROS are notoriously complex. Therefore, we used liquid chromatography–tandem mass spectrometry (LC-MS/MS) combined with selective Cys alkylation to examine the status of the disulfide formation between C104 and C49, which forms the functionally important CXnE motif. PTP4A3 was incubated in the presence or absence of 200 nM JMS-053 for 30 minutes and then labeled at reduced Cys residues, using the alkylating reagent iodoacetamide. After alkylation, the samples were digested with tosyl phenylalanyl chloromethyl ketone–modified trypsin/lys-c and the trypic digests were analyzed by electrospray ionization LC-MS/MS as previously described (Orsatti et al., 2009). Prominent peaks for each of the Cys-containing peptides were observed, and there was no evidence for C49-C104 dimer formation in the absence or presence of JMS-053 (Fig. 4). Moreover, JMS-053 did not appear to oxidize C49, C93, or C104; convert the catalytic C104 to glycine; or generate a sulfenyl-amide species (Fig. 4). No cysteine oxidation was observed with preincubation of PTP4A3 and NRT-870-59 in the absence or presence of DTT (Supplemental Fig. 6). Collectively, these data further support an inhibitory mechanism that is independent of the generation of ROS.
Fig. 4.
(A) Base peak chromatograms of wild-type PTP4A3 tryptic peptides after treatment of the protein with JMS-053. A comparison was made between samples that were treated with DTT to reduce disulfide bonds before alkylation with iodoacetic acid (IAA), and those that received only IAA treatment. PTP4A3 contains two tryptic peptides with Cys. While little to no change was observed in the peptide containing C49 [mass-to-charge ratio (m/z) = 507.4], the second Cys containing peptide (m/z = 683.7) disappeared in the absence of DTT. A new peak for this peptide was found with an m/z = 645.0 corresponding to a single alkylation of C93. No additional peaks were found to indicate the formation of a disulfide bond. (B) Base peaks for Cys containing peptides at m/z = 507.4, m/z = 683.7, and m/z = 645.0. The single Cys on peptide VCEVTYDK allowed for quick interpretation for the presence of possible disulfide bonds at C49 by measuring their relative abundances between reduced and nonreduced samples. No significant change was found in this peptide’s relative abundance. In the absence of DTT, the peak found at m/z = 645.0 contributed significantly to the relative abundance of the second peptide of interest with a single alkylation. A second peak was found with a retention time ∼21.2 minutes and is likely a structural isomer of the peptide with an alkylation on the other cysteine residues. The sum of these peaks accounted for a majority of the FCEAPGSCVAVHCVAGLGR peptide. (C) MS2 fragment spectra of m/z 507.4. The y and b ion coverage provided the amino acid sequence of VC (+57.02) EVTYDK and confirmed the identity of the peptide found in both reduced and nonreduced samples.
Cytotoxicity of JMS-053 and Analogs to Cancer Cells.
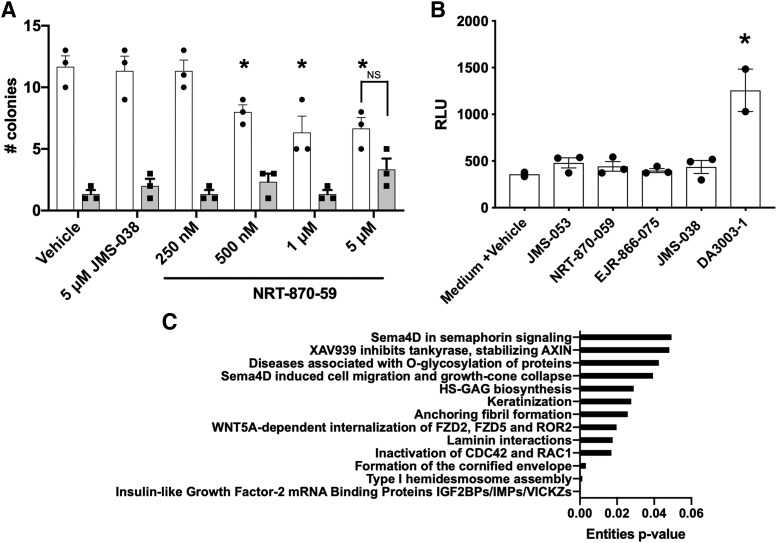
We next investigated the ability of the JMS-053 series to kill human tumor cells grown as three-dimensional spheroids, which retain many of the critical intercellular signaling pathways that are modified by PTP4A3 (McQueeney et al., 2018). We used four human cancer cell lines with diverse genetic backgrounds that have been well annotated in the publicly available Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle). All four cell lines have higher PTP4A3 expression levels compared with a nonmalignant counterpart (Supplemental Fig. 7, A and B). Human triple negative breast cancer Hs578T and high-grade serous ovarian cancer OVCAR4 cells were the most sensitive to a brief 48- or 72-hour exposure to JMS-053 with values of IC50 < 9 μM, while the high-grade serous ovarian Kuramochi cells, which have a long doubling time (∼72 hours), had an intermediate potency and triple negative MDA-MB-231 breast cancer cells were the least sensitive (Supplemental Fig. 8; Table 3). This profile of Hs578T and OVCAR4 being most responsive was seen with all analogs. EJR-855-81, EJR-866-75, and NRT-870-59 were cytotoxic to human Hs578T breast cancer and OVCAR4 ovarian cancer spheroid tumor cells grown in vitro with IC50 values below 20 μM. Spheroid colony formation is an alternative method to evaluate extracellular matrix–dependent cytotoxicity. Similar to our previous results with JMS-053 (McQueeney et al., 2017), wild-type mouse colon cancer cells, which express considerable PTP4A3 (Supplemental Fig. 7C) (McQueeney et al., 2017), exhibited a statistically significant decrease in colony formation after exposure to 500 nM NRT-870-59 with a somewhat flat concentration-response curve up to 5 μM. However, importantly, no statistically significant decrease in colony formation was observed with the PTP4A3 null cells treated at any concentration of NRT-870-59 and no difference in colony formation was seen between wild-type and null cells treated with 5 μM NRT-870-59 (Fig. 5A), indicating a dependence of target expression in the cellular effects. This is similar to what we previously observed with JMS-053 (McQueeney et al., 2017). However, the low colony numbers with our vehicle-treated PTP4A3 null cells does limit the dynamic range of the assay.
TABLE 3.
Cellular IC50 values for loss of spheroid viability after 48-hour exposure
Exponentially growing human breast and ovarian cancer cells were plated in ultralow attachment U-bottom microtiter plates and cultured for 24 hours to allow spheroid formation. Compounds were added to the preexisting spheroids and microtiter plates were incubated for 48 hours. Cell viability was determined with CellTiterGlo. N = 3, mean ± S.E.M.
| Compound | (μM) ± S.E.M. | |||
|---|---|---|---|---|
| MDA-MB-231 | Hs578T | OVCAR4 | Kuramochi | |
| JMS-053 | 32.67 ± 7.02 | 8.48 ± 2.38 | 4.42 ± 1.04 | 13.25 ± 0.65 |
| EJR-866-75 | >50 μM | 12.01 ± 3.84 | 19.64 ± 4.66a | >50 μM |
| EJR-866-81 | >50 μM | 14.39 ± 4.91 | 12.35 ± 2.26a | >50 μM |
| NRT-870-59 | 61.54 ± 9.66a | 10.07 ± 1.93 | 11.50 ± 2.38a | 34.14 ± 8.60a |
| JMS-038 | >50 μM | >50 μM | >50 μM | >50 μM |
P < 0.05 compared with JMS-053.
Fig. 5.
Inhibition of colony formation, lack of intracellular reactive oxygen species generation, and gene expression profiling. (A) Mouse colon cancer cells (white columns) and an isogenic PTP4A3 null cell line (gray columns) were exposed to NRT-870-59, JMS-038, or vehicle and colony formation wasdetermined after 2-week incubation. The mean value from three wells and error bars = S.E.M. Representative of two independent assays with similar results. N = 3. *P < 0.05. Error bars = S.E.M. NS = difference not statistically significant. (B) Formation of ROS was determined in confluent OVCAR4 ovarian cancer cells using the ROS-Glo H2O2 assay. N = 3. *P < 0.05. Error bars = S.E.M. (C) Commonly altered pathways in PTP4A3 wild-type and null cells and the JMS-053–treated PTP4A3 wild-type cells.
Lack of Cellular ROS Formation with JMS-053.
We next probed the potential of JMS-053 and its analogs to generate significant ROS bursts within cells. A robust elevation of ROS was observed 10 minutes after exposure to the positive control DA-3003-1 (Fig. 5B). In contrast, neither JMS-053 nor any of the analogs produced detectable ROS in cells (Fig. 5B). This lack of intracellular ROS generation coupled with the known endogenous reductants of the PTP4A3 disulfide bond, such as thioredoxin-related protein 23 (Ishii et al., 2013), offers further support for an oxidant-independent mechanism of inhibition by JMS-053 and its analogs. The oxidant independence of JMS-053 was further supported by examining the changes in gene expression, which have been extensively analyzed by others using H2O2 (Harris et al., 2019). When we compared the mRNA expression of 31,037 genes in a previously described isogenic pair of colorectal cancer cells that are wild type or null for PTP4A3 (McQueeney et al., 2018), we detected 3289 transcripts that were differentially expressed in wild-type and null cells. A pathway analysis of these upregulated and downregulated gene transcripts implicated extracellular matrix and extracellular signal–regulated kinase pathway alterations but not oxidative stress (Supplemental Fig. 9A). None of the gene transcripts commonly associated with oxidative stress, such as NRF2, NQO1, HMOX1, HSPA6, GADD34, or CHOP (Harris et al., 2019), were elevated in the PTP4A3 null cells. Treatment of the wild-type colorectal cancer cells for 24 hours with 1 μM JMS-053 did not elevate the transcripts commonly associated with oxidative stress, such as NRF2, NQO1, HMOX1, HSPA6, GADD34, or CHOP. A pathway analysis of the upregulated and downregulated gene transcripts did not demonstrate any prominent changes in the canonical oxidative stress pathways (Supplemental Fig. 9B). The pathways common to the expression of the 241 gene transcripts altered in both the null cells and the JMS-053–treated wild-type cells were frequently related to extracellular matrix and migration including hemidesmosome assembly, laminin interactions, anchoring fibril formation, keratinization, glycosaminoglycan synthesis, and O-glycosylation (Fig. 5C). This is consistent with our previous observations about the importance of the extracellular matrix and adhesion in the cellular actions of PTP4A3 (McQueeney et al., 2018). These changes could be related to the proposed role of PTP4A3 in regulating epithelial mesenchymal transition (Wang et al., 2007) and the keratins in colon cancer (Karantza, 2011). These subjects warrant further investigation.
Discussion
A number of cellular functions have been attributed to PTP4A3 family members (Rios et al., 2013; Yu and Zhang, 2017; Hardy et al., 2018). Most, but not all, of the evidence supporting the oncogenic roles of PTP4A3 have been generated using genetic approaches, which are not easily titratable and frequently irreversible. Therefore, rigorously validated small molecules that inhibit PTP4A phosphatases should be valuable pharmacological reagents. While this paper was being prepared work was published suggesting thienopyridone and JMS-053 generated ROS, and the authors of that study concluded these compounds were inappropriate for therapeutic development or studies of PTP4A function (Zhang et al., 2019). However, based on our results we must demur. We found reversible inhibition with dilution assays, which would be inconsistent with the formation of a sulfinic or sulfonic acid at cysteines and would be expected from extreme oxidation by ROS. While those authors reported NMR results that indicated a conformation change consistent with disulfide bond formation between C49 and C104 in PTP4A3 (Zhang et al., 2019), we observed no evidence for disulfide bond formation by LC-MS/MS with either JMS-053 or NRT-870-059 (Fig. 4; Supplemental Fig. 6). Importantly, we did not observe ROS formation in cells treated with JMS-053 (Fig. 5B), nor did we find a change in the gene expression profile consistent with oxidative stress (Fig. 5C).
There are several significant differences between our study and that of Zhang et al. (2019), which might explain the disagreement in the conclusions. First, we examined steady-state kinetics, which we believe are more pharmacologically relevant, rather than burst kinetics with an artificial substrate. Second, we employed control compounds—the inactive JMS-038 and the ROS-generating compound DA-3003-1—in most of our studies. Third, we examined the reduction of resazurin in vitro with JMS-053 at ≤125 nM or 5× the IC50 values rather than with 5 μM, which is 160-fold greater than the IC50 value for PTP4A3. Fourth, we avoided using high concentrations of the solvent DMSO, which we have found facilitates PTP4A3 oxidation in vitro. Fifth, we used full-length recombinant PTP4A3 rather than a truncated version of the phosphatase. Finally, we extensively interrogated the purity of our compounds with 1H and 13C NMR, crystallography, LC-MS/MS, and elemental analyses and we did not rely solely on 1H NMR to document compound purity. Indeed, a comparison between our published 1H NMR results (Salamoun et al., 2016; Tasker et al., 2019) and that used in the recently published work (Zhang et al., 2019) suggests considerable impurities or decomposition of the compounds used in the Zhang et al. (2019) study. PTP4A3 is notoriously susceptible to oxidation from cations and oxidized DMSO (Orsatti et al., 2009); therefore, great care needs to be taken in the preparation of PTP4A3, the buffers, and the compound being tested.
Thus, we propose that the iminothienopyridones can inhibit at pharmacologically relevant concentrations the phosphatase activity of PTP4A3 independent of prominent ROS generation. While PTPs, in general, are suspectable to oxidation in vitro, a recent proteome-wide analysis of intracellular cysteine oxidation with exogenous H2O2 exposure did not show any evidence of PTP4A3 oxidation (van der Reest et al., 2018). Even if the iminothienopyridones produced ROS at very high concentrations, they could be potentially useful since at least 12 clinically used anticancer drugs have been shown to produce ROS (Yokoyama et al., 2017). This may be due in part to an altered redox homeostasis in cancer cells that preferentially sensitizes them to ROS (Yang et al., 2018). Recent work has highlighted the close integration of processes that regulate protein homeostasis and emphasize the unique dependence of cancer cells on thiols and deubiquitinases for survival (Harris et al., 2019). Loss of intracellular thiol-based reductants led to endoplasmic reticulum and proteotoxic stress and altered cancer cell sensitivity to therapeutic agents (Harris et al., 2019). Thus, the generation of intracellular ROS by compounds can selectively alter cancer cell redox status. Therefore, collectively, our results should stimulate further investigation of the pharmacological actions of the iminothienopyridone chemotype.
Acknowledgments
We are grateful for the assistance from the University of Virginia Advanced Microscopy Facility and the Bioinformatics Core. We thank Awais Paracha, who was supported by the Summer Undergraduate Research Fellow Award to the University of Virginia from the American Society for Pharmacology and Experimental Therapeutics, for assistance with the initial ROS studies. We appreciate the cancer cell line data available from the Cancer Cell Line Encyclopedia.
Abbreviations
- ABC
ammonium bicarbonate
- DA-3003-1
6-chloro-7-(2-morpholin-4-yl-ethylamino)-quinoline-5-8-dione
- DiFMUP
6,8-difluoro-4-methyl-umbelliferyl phosphate
- DTT
dithiothreitol
- EJR-866-75
7-imino-2-(4-(2-morpholinoethoxy)phenyl)thieno[3,2-c]pyridine-4,6(5H,7H)-dione
- EJR-866-81
2-(2-chlorophenyl)-7-iminothieno[3,2-c]pyridine-4,6(5H,7H)-dione
- JMS-038
6-phenylthieno[2,3-d]pyrimidine-2,4(1H,3H)-dione
- JMS-053
7-imino-2-phenylthieno[3,2-c]pyridine-4,6(5H,7H)-dione
- Kcat
catalytic rate constant
- Km
Michaelis constant
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- m/z mass-to-charge ratio NRT-870-59
7-imino-5-methyl-2-phenylthieno[3,2-c]pyridine-4,6(5H,7H)-dione
- PTP
protein tyrosine phosphatase
- ROS
reactive oxygen species
- thienopyridone
7-amino-2-phenyl-5H-thieno[3,2-c]pyridin-4-one
- WPD
tryptophan-proline-aspartate
Authorship Contributions
Participated in the research design: Lazo, Wipf, Sharlow, Hsu.
Conducted experiments: Blanco, Garrott, Hart, Tasker, Rastelli.
Contributed new reagents or analytic tools: McCloud, Hsu, Burnett, Wipf.
Performed data analysis: Hsu, McCloud, Blanco, Tasker, Rastelli, Burnett, Wipf.
Wrote or contributed to the writing of the manuscript: Lazo, Wipf, Sharlow.
Footnotes
The work was supported by grants from the Department of Defense [Grant W81XWH-18-1-0012], the National Institutes of Health [Grant S10 OD021723], the Fiske Drug Discovery Fund, the Owens Foundation, the Ivy Foundation, and the National Science Foundation Graduate Research Fellowship Program [Grant 2018255830].
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
This article is dedicated to the memory of Nancy Ann Lazo Higgins, who courageously fought her ovarian cancer.
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Basak S, Jacobs SB, Krieg AJ, Pathak N, Zeng Q, Kaldis P, Giaccia AJ, Attardi LD. (2008) The metastasis-associated gene Prl-3 is a p53 target involved in cell-cycle regulation. Mol Cell 30:303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonham CA, Vacratsis PO. (2009) Redox regulation of the human dual specificity phosphatase YVH1 through disulfide bond formation. J Biol Chem 284:22853–22864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson M, Nguyen T, Wipf P, Joo B, Day BW, Skoko JS, Schreiber EM, Foster C, Bansal P, Lazo JS. (2005) Redox regulation of Cdc25B by cell-active quinolinediones. Mol Pharmacol 68:1810–1820. [DOI] [PubMed] [Google Scholar]
- Buhrman G, Parker B, Sohn J, Rudolph J, Mattos C. (2005) Structural mechanism of oxidative regulation of the phosphatase Cdc25B via an intramolecular disulfide bond. Biochemistry 44:5307–5316. [DOI] [PubMed] [Google Scholar]
- Chakraborty AA, Laukka T, Myllykoski M, Ringel AE, Booker MA, Tolstorukov MY, Meng YJ, Meier SR, Jennings RB, Creech AL, et al. (2019) Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science 363:1217–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daouti S, Li WH, Qian H, Huang KS, Holmgren J, Levin W, Reik L, McGady DL, Gillespie P, Perrotta A, et al. (2008) A selective phosphatase of regenerating liver phosphatase inhibitor suppresses tumor cell anchorage-independent growth by a novel mechanism involving p130Cas cleavage. Cancer Res 68:1162–1169. [DOI] [PubMed] [Google Scholar]
- Defelipe LA, Lanzarotti E, Gauto D, Marti MA, Turjanski AG. (2015) Protein topology determines cysteine oxidation fate: the case of sulfenyl amide formation among protein families. PLOS Comput Biol 11:e1004051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander P, Rawls K, Tsimelzon A, Shepherd J, Mazumdar A, Hill J, Fuqua SA, Chang JC, Osborne CK, Hilsenbeck SG, et al. (2016) Phosphatase PTP4A3 promotes triple-negative breast cancer growth and predicts poor patient survival. Cancer Res 76:1942–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. (2005) Protein Identification and Analysis Tools on the Expasy Server, Humana Press. [DOI] [PubMed] [Google Scholar]
- Gulerez I, Funato Y, Wu H, Yang M, Kozlov G, Miki H, Gehring K. (2016) Phosphocysteine in the PRL-CNNM pathway mediates magnesium homeostasis. EMBO Rep 17:1890–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy S, Kostantin E, Hatzihristidis T, Zolotarov Y, Uetani N, Tremblay ML. (2018) Physiological and oncogenic roles of the PRL phosphatases. FEBS J 285:3886–3908. [DOI] [PubMed] [Google Scholar]
- Harris IS, Endress JE, Coloff JL, Selfors LM, McBrayer SK, Rosenbluth JM, Takahashi N, Dhakal S, Koduri V, Oser MG, et al. (2019) Deubiquitinases maintain protein homeostasis and survival of cancer cells upon glutathione depletion. Cell Metab 29:1166–1181.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeger B, Diether M, Ballester PJ, Köhn M. (2014) Biochemical evaluation of virtual screening methods reveals a cell-active inhibitor of the cancer-promoting phosphatases of regenerating liver. Eur J Med Chem 88:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeger B, Rios P, Berteotti A, Hoermann B, Duan G, Köhn M. (2017) Mutational analysis of a conserved glutamate reveals unique mechanistic and structural features of the phosphatase PRL-3. ACS Omega 2:9171–9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Funato Y, Miki H. (2013) Thioredoxin-related protein 32 (TRP32) specifically reduces oxidized phosphatase of regenerating liver (PRL). J Biol Chem 288:7263–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston PA. (2011) Redox cycling compounds generate H2O2 in HTS buffers containing strong reducing reagents—real hits or promiscuous artifacts? Curr Opin Chem Biol 15:174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston PA, Soares KM, Shinde SN, Foster CA, Shun TY, Takyi HK, Wipf P, Lazo JS. (2008) Development of a 384-well colorimetric assay to quantify hydrogen peroxide generated by the redox cycling of compounds in the presence of reducing agents. Assay Drug Dev Technol 6:505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza V. (2011) Keratins in health and cancer: more than mere epithelial cell markers. Oncogene 30:127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Cheng J, Ziomek E, Banville D, Gehring K, Ekiel I. (2004) Structural insights into molecular function of the metastasis-associated phosphatase PRL-3. J Biol Chem 279:11882–11889. [DOI] [PubMed] [Google Scholar]
- Lazo JS, Sharlow ER. (2016) Drugging undruggable molecular cancer targets. Annu Rev Pharmacol Toxicol 56:23–40. [DOI] [PubMed] [Google Scholar]
- Lor LA, Schneck J, McNulty DE, Diaz E, Brandt M, Thrall SH, Schwartz B. (2007) A simple assay for detection of small-molecule redox activity. J Biomol Screen 12:881–890. [DOI] [PubMed] [Google Scholar]
- McParland V, Varsano G, Li X, Thornton J, Baby J, Aravind A, Meyer C, Pavic K, Rios P, Köhn M. (2011) The metastasis-promoting phosphatase PRL-3 shows activity toward phosphoinositides. Biochemistry 50:7579–7590. [DOI] [PubMed] [Google Scholar]
- McQueeney KE, Salamoun JM, Ahn JG, Pekic P, Blanco IK, Struckman HL, Sharlow ER, Wipf P, Lazo JS. (2018) A chemical genetics approach identifies PTP4A3 as a regulator of colon cancer cell adhesion. FASEB J 32:5661–5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueeney KE, Salamoun JM, Burnett JC, Barabutis N, Pekic P, Lewandowski SL, Llaneza DC, Cornelison R, Bai Y, Zhang ZY, et al. (2017) Targeting ovarian cancer and endothelium with an allosteric PTP4A3 phosphatase inhibitor. Oncotarget 9:8223–8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsatti L, Innocenti F, Lo Surdo P, Talamo F, Barbato G. (2009) Mass spectrometry study of PRL-3 phosphatase inactivation by disulfide bond formation and cysteine into glycine conversion. Rapid Commun Mass Spectrom 23:2733–2740. [DOI] [PubMed] [Google Scholar]
- Pani G, Colavitti R, Bedogni B, Anzevino R, Borrello S, Galeotti T. (2000) A redox signaling mechanism for density-dependent inhibition of cell growth. J Biol Chem 275:38891–38899. [DOI] [PubMed] [Google Scholar]
- Rios P, Li X, Köhn M. (2013) Molecular mechanisms of the PRL phosphatases. FEBS J 280:505–524. [DOI] [PubMed] [Google Scholar]
- Salamoun JM, McQueeney KE, Patil K, Geib SJ, Sharlow ER, Lazo JS, Wipf P. (2016) Photooxygenation of an amino-thienopyridone yields a more potent PTP4A3 inhibitor. Org Biomol Chem 14:6398–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber M, Chandel NS. (2014) ROS function in redox signaling and oxidative stress. Curr Biol 24:R453–R462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares KM, Blackmon N, Shun TY, Shinde SN, Takyi HK, Wipf P, Lazo JS, Johnston PA. (2010) Profiling the NIH Small Molecule Repository for compounds that generate H2O2 by redox cycling in reducing environments. Assay Drug Dev Technol 8:152–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlbauer S, Rios P, Ohmori K, Suzuki K, Köhn M. (2015) Procyanidins negatively affect the activity of the phosphatases of regenerating liver. PLoS One 10:e0134336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford SM, Bottini N. (2017) Targeting tyrosine phosphatases: time to end the stigma. Trends Pharmacol Sci 38:524–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner JJ, Parsons ZD, Cummings AH, Zhou H, Gates KS. (2011) Redox regulation of protein tyrosine phosphatases: structural and chemical aspects. Antioxid Redox Signal 15:77–97. [DOI] [PubMed] [Google Scholar]
- Tasker NR, Rastelli EJ, Blanco IK, Burnett JC, Sharlow ER, Lazo JS, Wipf P. (2019) In-flow photooxygenation of aminothienopyridinones generates iminopyridinedione PTP4A3 phosphatase inhibitors. Org Biomol Chem 17:2448–2466. [DOI] [PubMed] [Google Scholar]
- van der Reest J, Lilla S, Zheng L, Zanivan S, Gottlieb E. (2018) Proteome-wide analysis of cysteine oxidation reveals metabolic sensitivity to redox stress. Nat Commun 9:1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt A, McDonald PR, Tamewitz A, Sikorski RP, Wipf P, Skoko JJ, III, Lazo JS. (2008) A cell-active inhibitor of mitogen-activated protein kinase phosphatases restores paclitaxel-induced apoptosis in dexamethasone-protected cancer cells. Mol Cancer Ther 7:330–340. [DOI] [PubMed] [Google Scholar]
- Wang H, Quah SY, Dong JM, Manser E, Tang JP, Zeng Q. (2007) PRL-3 down-regulates PTEN expression and signals through PI3K to promote epithelial-mesenchymal transition. Cancer Res 67:2922–2926. [DOI] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M. (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. [DOI] [PubMed] [Google Scholar]
- Yang H, Villani RM, Wang H, Simpson MJ, Roberts MS, Tang M, Liang X. (2018) The role of cellular reactive oxygen species in cancer chemotherapy. J Exp Clin Cancer Res 37:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama C, Sueyoshi Y, Ema M, Mori Y, Takaishi K, Hisatomi H. (2017) Induction of oxidative stress by anticancer drugs in the presence and absence of cells. Oncol Lett 14:6066–6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZH, Zhang ZY. (2017) Regulatory mechanisms and novel therapeutic targeting stragegies for protein tyrosine phosphatases. Chem Rev 50:122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kozlov G, Li X, Wu H, Gulerez I, Gehring K. (2017) PRL3 phosphatase active site is required for binding the putative magnesium transporter CNNM3. Sci Rep 7:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kozlov G, Chen YS, Gehring K. (2019) Mechanism of thienopyridone and iminothienopyridinedione inhibition of protein phosphatases. MedChemComm 10:791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]