Summary
Despite sharing interleukin‐4 receptor α (IL‐4Rα) in their signaling cascades, IL‐4 and IL‐13 have different functions in atopic inflammation. IL‐13 preferentially participates in the peripheral tissues because tissue‐resident group 2 innate lymphoid cells produce IL‐13 but not IL‐4. In contrast, lymph node T follicular helper cells express IL‐4 but not IL‐13 to regulate B‐cell immunity. The dominant microenvironment of IL‐13 is evident in the lesional skin of atopic dermatitis (AD). The IL‐13‐rich local milieu causes barrier dysfunction by down‐regulating the OVOL1–filaggrin (FLG) axis and up‐regulating the periostin–IL‐24 axis. Genome‐wide association studies also point to the crucial involvement of the IL‐13, OVOL1 and FLG genes in the pathogenesis of AD. Biologics targeting IL‐13, such as the anti‐IL‐4Rα antibody dupilumab and the anti‐IL‐13 antibody tralokinumab, successfully improve AD lesions and further highlight the importance of IL‐13 in the pathogenesis of AD.
Keywords: atopic dermatitis, filaggrin, interleukin‐13, OVOL1, Periostin
Introduction
Atopic dermatitis (AD) is a common eczematous skin disorder characterized by skin inflammation, barrier disruption and chronic pruritus.1 Its course involves chronic relapse with intense pruritus, which reduces quality of life and decreases treatment satisfaction of the afflicted patients.2, 3, 4 The lifetime incidence of AD is as high as 20% in the general population.5 Skin barrier dysfunction is associated with the reduced production of terminal differentiation molecules, such as filaggrin (FLG).6, 7 Abnormal skin barrier integrity also causes the increased colonization of microbes, such as Staphylococcus aureus, which further exacerbates skin inflammation.8 In addition, some autoimmune diseases are co‐morbid with AD.9, 10
The definition of ‘atopy’ is a diathesis for the overproduction of immunoglobulin E (IgE) antibodies or a personal and/or family history of asthma, allergic rhinitis, allergic conjunctivitis and AD.11 With the help of interleukin‐4 (IL‐4) and/or IL‐13 produced from type 2 helper T (Th2) cells, activated B cells produce IgE.12 The diverse activation and differentiation of multiple B‐cell subsets with significant correlation with circulating IgE levels have been reported in AD but not in psoriasis or normal controls.13 Consistent with preponderant Th2 deviation in early childhood AD,14, 15 elevated levels of total or allergen‐specific IgE have been noted in infantile and early childhood AD.16, 17 Skin barrier dysfunction with loss of function mutations of the filaggrin (FLG) gene contribute to disease progression and aberrant IgE production in AD.18
T helper type 2 cells were first reported by Mosmann et al. in 198819 and are related to allergic inflammation in AD.15, 20, 21 The gene expression of IL‐4 and IL‐13 is up‐regulated in the lesional skin of pediatric and adult AD patients compared with that in the normal skin of healthy controls.15 Type 2 predominance is likely to progress from non‐lesional to lesional skin and from acute to chronic lesions in AD.21 Type 2 predominance has been confirmed in peripheral blood T helper cells.14 The number of IL‐13‐producing Th2 cells is significantly greater in the skin‐homing cutaneous lymphocyte antigen (CLA)+ T helper cell population in both pediatric and adult AD patients than in healthy controls.14 The pathogenic importance of IL‐4 and IL‐13 has recently been reinforced by the excellent treatment response of patients with AD to the anti‐IL‐4 receptor α (IL‐4Rα, IL4R) antibody dupilumab, which inhibits both IL‐4 and IL‐13 signals.22
Notably, more recent, large‐scale transcriptomic analysis revealed the specific and dominant role of IL‐13 in the lesional skin of AD, because IL‐4 expression was almost undetectable.23 Consistent with this notion, the anti‐IL‐13 antibody tralokinumab successfully improved AD.24 In this review, we focus on IL‐13 as the major driving force of AD development.
IL‐13 signaling
Interleukin‐4 and IL‐13 are encoded by adjacent genes and share a common receptor and signaling pathway.25 However, IL‐4 and IL‐13 are differentially expressed in vivo by a number of different cells that regulate innate and adaptive immunity. For example, lymph node T follicular helper cells, which regulate B‐cell immunity, express IL‐4 but not IL‐13.25 Basophils and invariant natural killer T2 cells also express IL‐4, whereas mucosal group 2 innate lymphoid cell (ILC2s) express mostly IL‐13 and little IL‐4.25 Their differential cellular expression suggests that IL‐4 and IL‐13 have distinct and non‐redundant functions in Th2 immunity; IL‐4 plays an important role in humoral responses, whereas IL‐13 mediates tissue responses, including the recruitment of eosinophils and parasite expulsion.25 In addition to its production in Th2 cells, IL‐13 is produced in ILC2s.26 Mouse and human ILC2s are phenotypically comparable, lineage‐negative, non‐T and non‐B lymphocytes that produce high levels of IL‐13 and IL‐5.26, 27, 28 Unlike IL‐13, no or only negligible IL‐4 is produced by ILC2s.26, 27, 28, 29 The ILC2s reside in the skin and are increased in number in AD lesions.27, 28
Interleukin‐13 signaling is regulated through a complex receptor system. In non‐hematopoietic cells, IL‐13 engages a heterodimeric receptor composed of IL‐4Rα and IL‐13Rα1 (IL13RA1).25 IL‐13Rα1 binds IL‐13 with low affinity, yet when it forms a complex with IL‐4Rα, it binds with a much higher affinity, inducing the effector functions of IL‐13.25 A second receptor, IL‐13Rα2 (IL13RA2), is closely related to IL‐13Rα1; IL‐13Rα2 binds IL‐13 with high affinity but lacks any significant cytoplasmic domain and does not function as a signal mediator.25 Cells with high IL‐13Rα2 expression rapidly and efficiently deplete extracellular IL‐13.30 In parallel, IL‐13 responses were enhanced in mice lacking Il13ra2.31 These studies highlighted that IL‐13Rα2 acts as a scavenger or decoy receptor of IL‐13 that elicits antagonistic activity against IL‐13.25 Keratinocytes and fibroblasts express functional IL‐4Rα, IL‐13Rα1 and IL‐13Rα2.32, 33, 34, 35, 36
The ligation of functional heterodimeric IL‐4Rα and IL‐13Rα1 by IL‐13 induces the activation of downstream Janus kinase 2 (JAK2) and tyrosine kinase 2 (TYK2).37 JAK2 activates signal transducer and activator of transcription 3 (STAT3), and TYK2 activates STAT6 and STAT1.37 In contrast, IL‐4 signaling proceeds via the IL‐4Rα and IL‐2Rγ heterodimer.37 The ligation of IL‐4Rα and IL‐2Rγ by IL‐4 induces JAK1/JAK3 and subsequent STAT3 and STAT6 activation.37 Hence, different types of oral and topical JAK inhibitors are significantly therapeutic against AD.38, 39 As IL‐4Rα is expressed on sensory nerves, and its activation is involved in the itch sensation, the administration of a JAK inhibitor reduces itch intensity.40
IL‐13 and periostin
Periostin (POSTN) is a matricellular, non‐structural extracellular matrix protein that is highly expressed at sites of injury or inflammation.41, 42 The main source of periostin is fibroblasts. Keratinocytes produce periostin but to a much lesser extent than fibroblasts.42, 43, 44 Periostin expression is enhanced by mechanical stress or skin injury; this is indicative of the physiologically protective function of periostin, which promotes wound repair by acting on keratinocytes and fibroblasts. Along with its physiological functions, periostin plays pathogenic roles in skin fibrosis and chronic allergic inflammation.42 In normal skin, periostin is mainly distributed in the subepidermal and perifollicular compartments.42
The expression of periostin is induced by IL‐4 and IL‐13 in a STAT6‐dependent manner.41, 42 In parallel, the dermal expression of periostin is significantly up‐regulated and correlated with disease severity in AD.44 Periostin is also related to subepithelial remodeling in asthma and allergic rhinitis.41 In a mite‐induced AD mouse model, epidermal thickening and dermal fibrosis were evident with Il13 gene up‐regulation and a marked increase in periostin expression.44 All of these allergic manifestations were significantly attenuated in mice that were deficient in either Stat6 or Postn.44 Periostin stimulates the proliferation of keratinocytes and enhances their production of thymic stromal lymphopoietin (TSLP), a cardinal keratinocyte‐derived cytokine that promotes the Th2 immune response.44 Periostin signaling proceeds through integrin α v; therefore, anti‐integrin α v antibodies successfully improved mite‐induced AD.44
In addition to the mite‐induced AD model, periostin deficiency inhibited epidermal hyperplasia in an imiquimod‐induced psoriasis model without affecting the recruitment of inflammatory cells expressing IL‐17, IL‐22 and IL‐23.45 These results stress the cardinal role of the periostin–integrin α v axis in the proliferative capacity of keratinocytes and are coincident with previous findings that the integrin α v signaling axis is necessary for epidermal proliferation during cutaneous wound healing.46 As epidermal thickening (acanthosis) is a striking feature in the chronically lichenified lesional skin of AD, the increased expression of periostin may be a therapeutically important target for drug development.
AD and barrier dysfunction
Skin barrier function is disrupted in AD compared with that in normal controls.47, 48, 49 The epidermal barrier is formed by the coordinated and sequential cross‐linking of epidermal differentiation molecules such as FLG and intercellular lipids and corneocyte adhesion.47, 48, 49 The expression of FLG and the other differentiation molecules loricrin and involucrin is down‐regulated or expressed prematurely in the lesional and non‐lesional skin of AD compared with their expression in the normal skin of healthy individuals.7, 50, 51 As the daily application of moisturizer during the first 32 weeks of life reduces the risk of AD in infants,52 skin barrier dysfunction is essential to the development of AD.
Although the strongest genetic risk factors for AD are loss of function mutations in the FLG gene,53 FLG mutations were not found in all AD patients; they were less common in southern Europeans with AD54 and were even absent in patients with AD from some African countries,55 suggesting that FLG mutations only partly explain FLG protein down‐regulation in AD. The FLG mutation was also not related to the development of AD in patients from a subtropical island in Japan.56
IL‐13 and OVOL1–FLG axis
The coordinated expression of FLG and other epidermal differentiation proteins is crucial for skin barrier protection.1, 7 OVOL1 is an upstream transcription factor that regulates FLG expression.6 It is intriguing that FLG, OVOL1 and IL13 were the three genes most significantly associated with AD among 31 susceptible gene loci reported in a meta‐analysis of genome‐wide association studies.52 IL‐4Rα signaling by IL‐14 and IL‐13 significantly reduced the expression of FLG, loricrin and involucrin in keratinocytes.7, 57, 58, 59, 60 At least two mechanisms mediate the IL‐4/IL‐13‐induced FLG down‐regulation: OVOL1 inactivation and the periostin–IL‐24 axis (Fig. 1).43, 60 The activation of OVOL1 induces its cytoplasmic‐to‐nuclear translocation and up‐regulates FLG and loricrin expression.6, 61 Notably, IL‐4 and IL‐13 consistently inhibit FLG expression by interfering with OVOL1 signaling.6 Interleukin‐13 also inhibits the involucrin expression but in an OVOL1‐independent manner61 and exacerbates barrier dysfunction. Epidermal keratinocytes in barrier‐disrupted skin produce large amounts of TSLP, IL‐25 and IL‐33, which promote the differentiation of Th2 cells and ILC2s and stimulate the production of IL‐13.29, 62 Hence, a vicious cycle is formed to develop atopic dry skin. These results suggest the crucial involvement of the IL‐13–OVOL1–FLG axis in the pathogenesis of AD.
Figure 1.
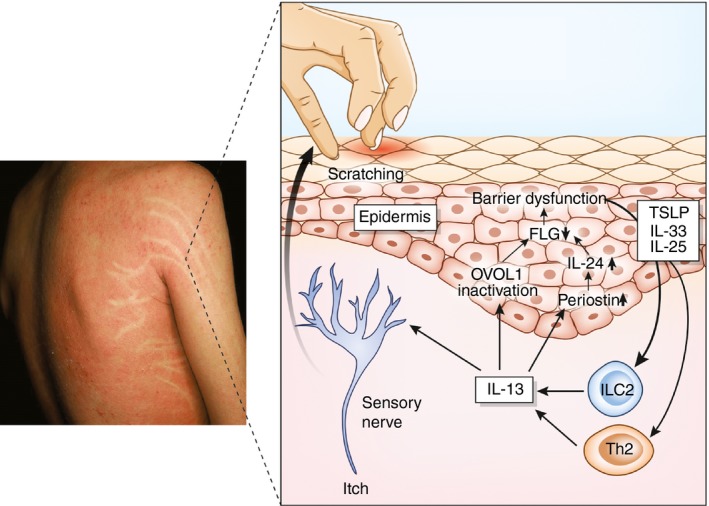
Simplified pathogenesis of atopic dermatitis. In the lesional skin of atopic dermatitis, T helper type 2 (Th2) cells and innate lymphoid cells (ILCs) produce high amounts of interleukin‐13 (IL‐13). IL‐13 induces OVOL1 inactivation and up‐regulates the periostin–IL‐24 axis, which down‐regulates filaggrin (FLG) and induces subsequent barrier dysfunction. Barrier dysfunction augments the epidermal production of thymic stromal lymphopoietin (TSLP), IL‐25 and IL‐33. These cytokines promote the differentiation of Th2 cells and group 2 ILCs (ILC2) and facilitate their production of IL‐13. IL‐13 also stimulates the sensory nerve and evokes the itch sensation. Itch‐induced scratching further deteriorates barrier dysfunction.
IL‐13 and periostin–IL‐24 axis
In addition to the IL‐13–OVOL1–FLG axis, IL‐13‐induced FLG down‐regulation is partly mediated by the IL‐13–periostin–IL‐24 axis.43 Interleukin‐24 belongs to an IL‐20 subfamily that includes IL‐19, IL‐20, IL‐22 and IL‐26.63 Although gene and protein expression of IL‐24 is up‐regulated in the lesional skin of AD,23, 43 its implication remains elusive. Interleukin‐13 up‐regulates the expression of periostin in keratinocytes through STAT6 activation.43 Periostin stimulates keratinocyte to produce IL‐24 and IL‐24 down‐regulates the FLG expression via STAT3 activation.43 In a mite antigen‐induced AD model, the Flg expression was decreased in the lesional murine skin. However, the decrease of Flg expression was not observed in mice deficient for Stat6 or Postn.43 Moreover, the phosphorylated forms of both STAT6 and STAT3 were abundantly expressed in the lesional epidermal keratinocytes of individuals with AD.43 Like IL‐13, IL‐31 is a pruritogenic cytokine produced from Th2 cells.64 Interleukin‐31 stimulates keratinocytes to produce IL‐24. Then, IL‐24 contributes to down‐regulate the expression of FLG.65
Anti‐IL‐13 biologics for the treatment of AD
Standard therapeutics for AD include topical emollients for barrier dysfunction and topical steroids and calcineurin inhibitors for skin inflammation.11, 66 These conventional treatments are more or less effective in reducing atopic inflammation and itch; however, patient satisfaction and adherence to conventional treatments are generally low, as reported in daily clinics.67, 68, 69, 70
The anti‐IL‐4Rα antibody dupilumab inhibits the binding of IL‐4 and IL‐13 to IL‐4Rα and blocks IL‐4Rα signaling.22 Dupilumab significantly improved skin lesions and pruritus in patients with moderate to severe AD in two randomized, placebo‐controlled phase 3 clinical trials.22 The severity of AD was authentically evaluated using the Eczema Area and Severity Index (EASI). A reduction in the EASI score of at least 75% (EASI75) was observed in 51% and 44% of patients in dupilumab monotherapy groups and only 15% and 12% of the patients in placebo groups, respectively, at 16 weeks post‐treatment.22 Dupilumab also provided a clinically meaningful improvement in patient‐reported outcome measures.71
Dupilumab therapy significantly reduced the lesional expression of IL‐13 and IL‐13‐regulated Th2 signature genes, such as CCL17, CCL18 and CCL26, in AD patients.72 It also restored the down‐regulation of FLG and loricrin and reduced epidermal acanthosis in the lesional skin of AD patients.72 In addition, dupilumab normalized the type 2 serum biomarkers CCL17, CCL18 and periostin.72
Tralokinumab is a fully human monoclonal antibody that potently and specifically neutralizes IL‐13.24 In a phase 2b study, 42·5% of patients injected with 300 mg tralokinumab every 2 weeks for 12 weeks achieved EASI75, which was significantly higher than that in the placebo control group (15·5%).24 Tralokinumab inhibits serum type 2 signature biomarkers such as periostin, CCL17 and IgE, but only the periostin level is a predictive biomarker for a good treatment response to tralokinumab in AD.24 Dipeptidyl peptidase‐4 (DPP‐4), a regulator of glucose metabolism, is also a type 2 signature biomarker.24, 73 Similar to the results of periostin treatment, patients with AD with high serum levels of DPP‐4 showed a greater treatment response to tralokinumab.24 In a phase 2b clinical trial of tralokinumab for severe asthma, high serum periostin and DPP‐4 levels were also significant biomarkers to predict a good treatment response to tralokinumab.73
Conclusion
Interleukin‐4 and IL‐13 share IL‐4Rα in their signaling cascades and induce similar biological responses. However, IL‐13 likely preferentially participates in peripheral tissues, including the skin, because tissue‐residing ILC2s produce IL‐13 but not IL‐4.26, 27, 28 An IL‐13‐dominant microenvironment is evident in the lesional skin of AD,23 and the IL‐13‐rich local milieu causes barrier dysfunction by the down‐regulation of the OVOL1–FLG axis.6, 7 The linkages among IL‐13–OVOL1–FLG may be particularly crucial for the development of AD, because genes encoding these three molecules are the three most susceptible genes for the development of AD.53 The successful improvement of AD by treatment with the anti‐IL‐4Rα antibody dupilumab and the anti‐IL‐13 antibody tralokinumab further highlights the importance of IL‐13 in the pathogenesis of AD.
Funding
This work was partly supported by grants from The Ministry of Health, Labour, and Welfare in Japan (H30‐Shokuhin‐Shitei‐005) and The Leading Advanced Projects for Medical Innovation in Japan (LEAP).
Disclosures
The authors declare that they have no conflict of interest regarding the publication of this article.
References
- 1. Furue M, Chiba T, Tsuji G, Ulzii D, Kido‐Nakahara M, Nakahara T et al Atopic dermatitis: immune deviation, barrier dysfunction, IgE autoreactivity and new therapies. Allergol Int 2017; 66:398–403. [DOI] [PubMed] [Google Scholar]
- 2. Arima K, Gupta S, Gadkari A, Hiragun T, Kono T, Katayama I et al Burden of atopic dermatitis in Japanese adults: analysis of data from the 2013 National Health and Wellness Survey. J Dermatol 2018; 45:390–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jung HJ, Bae JY, Kim JE, Na CH, Park GH, Bae YI et al Survey of disease awareness, treatment behavior and treatment satisfaction in patients with atopic dermatitis in Korea: a multicenter study. J Dermatol 2018; 45:1172–80. [DOI] [PubMed] [Google Scholar]
- 4. Komura Y, Kogure T, Kawahara K, Yokozeki H. Economic assessment of actual prescription of drugs for treatment of atopic dermatitis: differences between dermatology and pediatrics in large‐scale receipt data. J Dermatol 2018; 45:165–74. [DOI] [PubMed] [Google Scholar]
- 5. Williams H, Stewart A, von Mutius E, Cookson W, Anderson HR. Is eczema really on the increase worldwide? J Allergy Clin Immunol 2008; 121:947–54. [DOI] [PubMed] [Google Scholar]
- 6. Tsuji G, Hashimoto‐Hachiya A, Kiyomatsu‐Oda M, Takemura M, Ohno F, Ito T et al Aryl hydrocarbon receptor activation restores filaggrin expression via OVOL1 in atopic dermatitis. Cell Death Dis 2017; 8:e2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van den Bogaard EH, Bergboer JG, Vonk‐Bergers M, van Vlijmen‐Willems IM, Hato SV, van der Valk PG et al Coal tar induces AHR‐dependent skin barrier repair in atopic dermatitis. J Clin Invest 2013; 123:917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Furue M, Iida K, Imaji M, Nakahara T. Microbiome analysis of forehead skin in patients with atopic dermatitis and healthy subjects: implication of Staphylococcus and Corynebacterium . J Dermatol 2018; 45:876–7. [DOI] [PubMed] [Google Scholar]
- 9. Cipriani F, Marzatico A, Ricci G. Autoimmune diseases involving skin and intestinal mucosa are more frequent in adolescents and young adults suffering from atopic dermatitis. J Dermatol 2017; 44:1341–8. [DOI] [PubMed] [Google Scholar]
- 10. Furue M, Kadono T. “Inflammatory skin march” in atopic dermatitis and psoriasis. Inflamm Res 2017; 66:833–42. [DOI] [PubMed] [Google Scholar]
- 11. Saeki H, Nakahara T, Tanaka A, Kabashima K, Sugaya M, Murota H et al Clinical practice guidelines for the management of atopic dermatitis 2016. J Dermatol 2016; 43:1117–45. [DOI] [PubMed] [Google Scholar]
- 12. Poulsen LK, Hummelshoj L. Triggers of IgE class switching and allergy development. Ann Med 2007; 39:440–56. [DOI] [PubMed] [Google Scholar]
- 13. Czarnowicki T, Esaki H, Gonzalez J, Renert‐Yuval Y, Brunner P, Oliva M et al Alterations in B‐cell subsets in pediatric patients with early atopic dermatitis. J Allergy Clin Immunol 2017; 140:134–44. [DOI] [PubMed] [Google Scholar]
- 14. Czarnowicki T, Esaki H, Gonzalez J, Malajian D, Shemer A, Noda S et al Early pediatric atopic dermatitis shows only a cutaneous lymphocyte antigen (CLA)+ TH2/TH1 cell imbalance, whereas adults acquire CLA+ TH22/TC22 cell subsets. J Allergy Clin Immunol 2015; 136:941–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Esaki H, Brunner PM, Renert‐Yuval Y, Czarnowicki T, Huynh T, Tran G et al Early‐onset pediatric atopic dermatitis is TH2 but also TH17 polarized in skin. J Allergy Clin Immunol 2016; 138:1639–51. [DOI] [PubMed] [Google Scholar]
- 16. Esaki H, Takeuchi S, Furusyo N, Yamamura K, Hayashida S, Tsuji G et al Levels of immunoglobulin E specific to the major food allergen and chemokine (C‐C motif) ligand (CCL)17/thymus and activation regulated chemokine and CCL22/macrophage‐derived chemokine in infantile atopic dermatitis on Ishigaki Island. J Dermatol 2016; 43:1278–82. [DOI] [PubMed] [Google Scholar]
- 17. Takeuchi S, Esaki H, Furusyo N, Hayashida S, Yamamura K, Tsuji G et al Incidence, serum IgE and TARC/CCL17 levels in atopic dermatitis associated with other allergic diseases: an update from the Ishigaki cohort. Acta Derm Venereol 2015; 95:480–4. [DOI] [PubMed] [Google Scholar]
- 18. Weidinger S, Rodríguez E, Stahl C, Wagenpfeil S, Klopp N, Illig T et al Filaggrin mutations strongly predispose to early‐onset and extrinsic atopic dermatitis. J Invest Dermatol 2007; 127:724–6. [DOI] [PubMed] [Google Scholar]
- 19. Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol 1986; 136:2348–57. [PubMed] [Google Scholar]
- 20. Esaki H, Ewald DA, Ungar B, Rozenblit M, Zheng X, Xu H et al Identification of novel immune and barrier genes in atopic dermatitis by means of laser capture microdissection. J Allergy Clin Immunol 2015; 135:153–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gittler JK, Shemer A, Suárez‐Fariñas M, Fuentes‐Duculan J, Gulewicz KJ, Wang CQ et al Progressive activation of TH2/TH22 cytokines and selective epidermal proteins characterizes acute and chronic atopic dermatitis. J Allergy Clin Immunol 2012; 130:1344–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simpson EL, Bieber T, Guttman‐Yassky E, Beck LA, Blauvelt A, Cork MJ et al Two phase 3 trials of dupilumab versus placebo in atopic dermatitis. N Engl J Med 2016; 375:2335–48. [DOI] [PubMed] [Google Scholar]
- 23. Tsoi LC, Rodriguez E, Degenhardt F, Baurecht H, Wehkamp U, Volks N et al Atopic dermatitis is an IL‐13 dominant disease with greater molecular heterogeneity compared to psoriasis. J Invest Dermatol 2019; 139:1480–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wollenberg A, Howell MD, Guttman‐Yassky E, Silverberg JI, Kell C, Ranade K et al Treatment of atopic dermatitis with tralokinumab, an anti‐IL‐13 mAb. J Allergy Clin Immunol 2019; 143:135–41. [DOI] [PubMed] [Google Scholar]
- 25. Ranasinghe C, Trivedi S, Wijesundara DK, Jackson RJ. IL‐4 and IL‐13 receptors: roles in immunity and powerful vaccine adjuvants. Cytokine Growth Factor Rev 2014; 25:437–42. [DOI] [PubMed] [Google Scholar]
- 26. Hurrell BP, Shafiei Jahani P, Akbari O. Social networking of group two innate lymphoid cells in allergy and asthma. Front Immunol 2018; 9:2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mashiko S, Mehta H, Bissonnette R, Sarfati M. Increased frequencies of basophils, type 2 innate lymphoid cells and Th2 cells in skin of patients with atopic dermatitis but not psoriasis. J Dermatol Sci 2017; 88:167–74. [DOI] [PubMed] [Google Scholar]
- 28. Salimi M, Barlow JL, Saunders SP, Xue L, Gutowska‐Owsiak D, Wang X et al A role for IL‐25 and IL‐33‐driven type‐2 innate lymphoid cells in atopic dermatitis. J Exp Med 2013; 210:2939–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Camelo A, Rosignoli G, Ohne Y, Stewart RA, Overed‐Sayer C, Sleeman MA et al IL‐33, IL‐25, and TSLP induce a distinct phenotypic and activation profile in human type 2 innate lymphoid cells. Blood Adv 2017; 1:577–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kasaian MT, Raible D, Marquette K, Cook TA, Zhou S, Tan XY et al IL‐13 antibodies influence IL‐13 clearance in humans by modulating scavenger activity of IL‐13Rα2. J Immunol 2011; 187:561–9. [DOI] [PubMed] [Google Scholar]
- 31. Wood N, Whitters MJ, Jacobson BA, Witek J, Sypek JP, Kasaian M et al Enhanced interleukin (IL)‐13 responses in mice lacking IL‐13 receptor α 2. J Exp Med 2003; 197:703–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Akaiwa M, Yu B, Umeshita‐Suyama R, Terada N, Suto H, Koga T et al Localization of human interleukin 13 receptor in non‐haematopoietic cells. Cytokine 2001; 13:75–84. [DOI] [PubMed] [Google Scholar]
- 33. Campbell‐Harding G, Sawkins H, Bedke N, Holgate ST, Davies DE, Andrews AL. The innate antiviral response upregulates IL‐13 receptor α2 in bronchial fibroblasts. J Allergy Clin Immunol 2013; 131:849–55. [DOI] [PubMed] [Google Scholar]
- 34. Mitamura Y, Murai M, Mitoma C, Furue M. NRF2 activation inhibits both TGF‐β1‐ and IL‐13‐mediated periostin expression in fibroblasts: benefit of cinnamaldehyde for antifibrotic treatment. Oxid Med Cell Longev 2018; 2018:2475047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mitamura Y, Nunomura S, Nanri Y, Arima K, Yoshihara T, Komiya K et al Hierarchical control of interleukin 13 (IL‐13) signals in lung fibroblasts by STAT6 and SOX11. J Biol Chem 2018; 293:14646–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sivaprasad U, Warrier MR, Gibson AM, Chen W, Tabata Y, Bass SA et al IL‐13Rα2 has a protective role in a mouse model of cutaneous inflammation. J Immunol 2010; 185:6802–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bhattacharjee A, Shukla M, Yakubenko VP, Mulya A, Kundu S, Cathcart MK. IL‐4 and IL‐13 employ discrete signaling pathways for target gene expression in alternatively activated monocytes/macrophages. Free Radic Biol Med 2013; 54:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nakagawa H, Nemoto O, Yamada H, Nagata T, Ninomiya N. Phase 1 studies to assess the safety, tolerability and pharmacokinetics of JTE‐052 (a novel Janus kinase inhibitor) ointment in Japanese healthy volunteers and patients with atopic dermatitis. J Dermatol 2018; 45:701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rodrigues MA, Torres T. JAK/STAT inhibitors for the treatment of atopic dermatitis. J Dermatolog Treat 2019; 31:1–8. [DOI] [PubMed] [Google Scholar]
- 40. Oetjen LK, Mack MR, Feng J, Whelan TM, Niu H, Guo CJ et al Sensory neurons co‐opt classical immune signaling pathways to mediate chronic itch. Cell 2017; 171:217–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Izuhara K, Conway SJ, Moore BB, Matsumoto H, Holweg CT, Matthews JG et al Roles of periostin in respiratory disorders. Am J Respir Crit Care Med 2016; 193:949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamaguchi Y. Periostin in skin tissue and skin‐related diseases. Allergol Int 2014; 63:161–70. [DOI] [PubMed] [Google Scholar]
- 43. Mitamura Y, Nunomura S, Nanri Y, Ogawa M, Yoshihara T, Masuoka M et al The IL‐13/periostin/IL‐24 pathway causes epidermal barrier dysfunction in allergic skin inflammation. Allergy 2018; 73:1881–91. [DOI] [PubMed] [Google Scholar]
- 44. Masuoka M, Shiraishi H, Ohta S, Suzuki S, Arima K, Aoki S et al Periostin promotes chronic allergic inflammation in response to Th2 cytokines. J Clin Invest 2012; 122:2590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arima K, Ohta S, Takagi A, Shiraishi H, Masuoka M, Ontsuka K et al Periostin contributes to epidermal hyperplasia in psoriasis common to atopic dermatitis. Allergol Int 2015; 64:41–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Duperret EK, Natale CA, Monteleon C, Dahal A, Ridky TW. The integrin αv–TGFβ signaling axis is necessary for epidermal proliferation during cutaneous wound healing. Cell Cycle 2016; 15:2077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chiba T, Nakahara T, Kohda F, Ichiki T, Manabe M, Furue M. Measurement of trihydroxy‐linoleic acids in stratum corneum by tape‐stripping: possible biomarker of barrier function in atopic dermatitis. PLoS One 2019; 14:e0210013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Elias PM. Primary role of barrier dysfunction in the pathogenesis of atopic dermatitis. Exp Dermatol 2018; 27:847–51. [DOI] [PubMed] [Google Scholar]
- 49. Kim BE, Leung DYM. Significance of skin barrier dysfunction in atopic dermatitis. Allergy Asthma Immunol Res 2018; 10:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jensen JM, Fölster‐Holst R, Baranowsky A, Schunck M, Winoto‐Morbach S, Neumann C et al Impaired sphingomyelinase activity and epidermal differentiation in atopic dermatitis. J Invest Dermatol 2004; 122:1423–31. [DOI] [PubMed] [Google Scholar]
- 51. Seguchi T, Cui CY, Kusuda S, Takahashi M, Aisu K, Tezuka T. Decreased expression of filaggrin in atopic skin. Arch Dermatol Res 1996; 288:442–6. [DOI] [PubMed] [Google Scholar]
- 52. Horimukai K, Morita K, Narita M, Kondo M, Kitazawa H, Nozaki M et al Application of moisturizer to neonates prevents development of atopic dermatitis. J Allergy Clin Immunol 2014; 134:824–30. [DOI] [PubMed] [Google Scholar]
- 53. Paternoster L, Standl M, Waage J, Baurecht H, Hotze M, Strachan DP et al Multi‐ancestry genome‐wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet 2015; 47:1449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cascella R, Foti Cuzzola V, Lepre T, Galli E, Moschese V, Chini L et al Full sequencing of the FLG gene in Italian patients with atopic eczema: evidence of new mutations, but lack of an association. J Invest Dermatol 2011; 131:982–4. [DOI] [PubMed] [Google Scholar]
- 55. Thawer‐Esmail F, Jakasa I, Todd G, Wen Y, Brown SJ, Kroboth K et al South African amaXhosa patients with atopic dermatitis have decreased levels of filaggrin breakdown products but no loss‐of‐function mutations in filaggrin. J Allergy Clin Immunol 2014; 133:280–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sasaki T, Furusyo N, Shiohama A, Takeuchi S, Nakahara T, Uchi H et al Filaggrin loss‐of‐function mutations are not a predisposing factor for atopic dermatitis in an Ishigaki Island under subtropical climate. J Dermatol Sci 2014; 76:10–5. [DOI] [PubMed] [Google Scholar]
- 57. Nakahara T, Mitoma C, Hashimoto‐Hachiya A, Takahara M, Tsuji G, Uchi H et al Antioxidant Opuntia ficus‐indica extract activates AHR‐NRF2 signaling and upregulates filaggrin and loricrin expression in human keratinocytes. J Med Food 2015; 18:1143–9. [DOI] [PubMed] [Google Scholar]
- 58. Takei K, Mitoma C, Hashimoto‐Hachiya A, Takahara M, Tsuji G, Nakahara T et al Galactomyces fermentation filtrate prevents T helper 2‐mediated reduction of filaggrin in an aryl hydrocarbon receptor‐dependent manner. Clin Exp Dermatol 2015; 40:786–93. [DOI] [PubMed] [Google Scholar]
- 59. Takei K, Mitoma C, Hashimoto‐Hachiya A, Uchi H, Takahara M, Tsuji G et al Antioxidant soybean tar Glyteer rescues T‐helper‐mediated downregulation of filaggrin expression via aryl hydrocarbon receptor. J Dermatol 2015; 42:171–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tsuji G, Ito T, Chiba T, Mitoma C, Nakahara T, Uchi H et al The role of the OVOL1–OVOL2 axis in normal and diseased human skin. J Dermatol Sci 2018; 90:227–31. [DOI] [PubMed] [Google Scholar]
- 61. Hashimoto‐Hachiya A, Tsuji G, Murai M, Yan X, Furue M. Upregulation of FLG, LOR, and IVL expression by Rhodiola crenulata root extract via aryl hydrocarbon receptor: differential involvement of OVOL1. Int J Mol Sci 2018; 19:E1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Han H, Roan F, Ziegler SF. The atopic march: current insights into skin barrier dysfunction and epithelial cell‐derived cytokines. Immunol Rev 2017; 278:116–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Niess JH, Hruz P, Kaymak T. The interleukin‐20 cytokines in intestinal diseases. Front Immunol 2018; 9:1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Furue M, Yamamura K, Kido‐Nakahara M, Nakahara T, Fukui Y. Emerging role of interleukin‐31 and interleukin‐31 receptor in pruritus in atopic dermatitis. Allergy 2018; 73:29–36. [DOI] [PubMed] [Google Scholar]
- 65. Cornelissen C, Marquardt Y, Czaja K, Wenzel J, Frank J, Lüscher‐Firzlaff J et al IL‐31 regulates differentiation and filaggrin expression in human organotypic skin models. J Allergy Clin Immunol 2012; 129:426–33. [DOI] [PubMed] [Google Scholar]
- 66. Ohtsuki M, Morimoto H, Nakagawa H. Tacrolimus ointment for the treatment of adult and pediatric atopic dermatitis: review on safety and benefits. J Dermatol 2018; 45:936–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nakahara T, Fujita H, Arima K, Taguchi Y, Motoyama S, Furue M. Treatment satisfaction in atopic dermatitis relates to patient‐reported severity: a cross‐sectional study. Allergy 2019; 74:1179–81. [DOI] [PubMed] [Google Scholar]
- 68. Saeki H, Imafuku S, Abe M, Shintani Y, Onozuka D, Hagihara A et al Poor adherence to medication as assessed by the Morisky Medication Adherence Scale‐8 and low satisfaction with treatment in 237 psoriasis patients. J Dermatol 2015; 42:367–72. [DOI] [PubMed] [Google Scholar]
- 69. Takeuchi S, Oba J, Esaki H, Furue M. Non‐corticosteroid adherence and itch severity influence perception of itch in atopic dermatitis. J Dermatol 2018; 45:158–64. [DOI] [PubMed] [Google Scholar]
- 70. Wei W, Anderson P, Gadkari A, Blackburn S, Moon R, Piercy J et al Extent and consequences of inadequate disease control among adults with a history of moderate to severe atopic dermatitis. J Dermatol 2018; 45:150–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Simpson EL, Gadkari A, Worm M, Soong W, Blauvelt A, Eckert L et al Dupilumab therapy provides clinically meaningful improvement in patient‐reported outcomes (PROs): a phase IIb, randomized, placebo‐controlled, clinical trial in adult patients with moderate to severe atopic dermatitis (AD). J Am Acad Dermatol 2016; 75:506–15. [DOI] [PubMed] [Google Scholar]
- 72. Guttman‐Yassky E, Bissonnette R, Ungar B, Suárez‐Fariñas M, Ardeleanu M, Esaki H et al Dupilumab progressively improves systemic and cutaneous abnormalities in atopic dermatitis patients. J Allergy Clin Immunol 2019; 143:155–72. [DOI] [PubMed] [Google Scholar]
- 73. Brightling CE, Chanez P, Leigh R, O'Byrne PM, Korn S, She D et al Efficacy and safety of tralokinumab in patients with severe uncontrolled asthma: a randomised, double‐blind, placebo‐controlled, phase 2b trial. Lancet Respir Med 2015; 3:692–701. [DOI] [PubMed] [Google Scholar]


