Abstract
Post-translational modifications (PTMs) of proteins play key roles in cellular processes. Hence, PTM identification is crucial for elucidating the mechanism of complex cellular processes and disease. Here we present a method for PTM detection at the single-molecule level using FraC biological nanopores. We focus on two major PTMs, phosphorylation and glycosylation, that mutually compete for protein modification sites, an important regulatory process that has been implicated in the pathogenic pathways of many diseases. We show that phosphorylated and glycosylated peptides can be clearly differentiated from nonmodified peptides by differences in the relative current blockade and dwell time in nanopore translocations. Furthermore, we show that these PTM modifications can be mutually differentiated, demonstrating the identification of phosphorylation and glycosylation in a label-free manner. The results represent an important step for the single-molecule, label-free identification of proteoforms, which have tremendous potential for disease diagnosis and cell biology.
Keywords: Nanopores, post-translational modifications, label-free detection, glycosylation, phosphorylation
In the pursuit of a comprehensive understanding of the molecular mechanisms at work in living organisms, one inevitably arrives at the study of proteins. These organic polymers are the molecular machines of the cell that enable a spectacularly wide array of functionalities. Cells employ proteins for a host of different processes such as signaling, recognition, differentiation, gene regulation, structuring, and many more.1 To realize such a diverse set of functionalities, cells need a way to diversify a wide array of different proteins. Post-translational modifications, or PTMs, are one of the methods by which the protein pool of a cell can be expanded by orders of magnitude.2,3 This collective term refers to all modifications occurring during and after the translational synthesis of proteins, usually involving the addition of chemical groups to specific amino acids along the protein. PTMs play vital roles in protein (in)activation, stability, and recognition, among other things.4−6 In the medical field in particular, PTMs have been found to play key roles in pathogenic pathways for cancer, Parkinson’s disease, Alzheimer’s disease, and diabetes.7,8 Current detection methods face challenges in detecting low-copy number variants within the vast mixture of proteins in biological samples. Consequently, it is pivotal to detect protein PTM variants with high sensitivity.
Mass spectrometry is the current standard method for PTM detection.9,10 The technique, however, has its limitations, mainly brought about its narrow dynamic range (104–105) compared to the immense dynamic range of protein concentrations that are present in in vivo samples (108–109).3,11−13 Challenges arise when low-copy number proteoforms are of interest but get swamped in a sea of others. Furthermore, a protein can have different kinds and quantities of PTMs. In practice, sparsely occurring proteins are invisible in the noise of the diverse mixture of abundant proteins.11 For this very reason, many biomarkers for cancer and other diseases escape timely detection.14−16 To alleviate this problem, prepurification steps based on chromatography or antibodies are often used to enrich samples of interest.9,15 Other approaches include the PTM-specific removal and subsequent chemical labeling, enabling the researcher to detect it through characteristic mass-shifts in a mass spectrum.15,17 These enrichment and labeling methods are, however, time-consuming and highly PTM specific which discourages its use in the blanket analysis approaches that are sought after in clinical research.
Single-molecule techniques, such as nanopores, open the door for new approaches for PTM detection with ultimate sensitivity. In conventional MS, thousands of peptides are ionized and measured at the same time. As a consequence, low-copy number species are often impossible to distinguish from background noise in the measurements. Through the use of nanopores, single molecules can be analyzed and therefore in principle low-copy-number species can be detected. In a nanopore setup, an insulating membrane with a nanometer-sized aperture separates two electrolyte-filled compartments. Upon applying an electric potential, an ionic current flows through the nanopore and a decrease in the current is measured when a protein translocates through the pore.18,19 Several groups have previously reported the study of peptides20−31 or unfolded proteins32−40 translocating through a nanopore. In addition to that, others have reported the capabilities of nanopore sensors as mass detectors.22,29,30,41 Features in the protein structure, such as PTMs, can result in slightly different current blockage characteristics, allowing for label-free detection. In recent years, this principle was used for the detection of large PTMs such as a large N-glycosylation or ubiquitination.42,43 The detection of small PTMs (<500 Da) has remained a challenge and so far was only demonstrated for a phosphorylated protein.44
It has been recently shown that phosphorylation and O-glycosylation GlcNAc (N-acetylglucosamine) are intricately connected and exhibit a complex interplay within the cell.7 These PTMs compete for serine and threonine modification sites and therefore mutually regulate each other. Malfunctions of this regulatory network have been associated with chronic diseases such as diabetes, cancer, and Alzheimer’s.7,8,10 For example, in Alzheimer’s disease a decrease in the levels of O-GlcNAc in the tau protein has been associated with its hyper-phosphorylated state and the formation of the intraneuronal tangles characteristic of the disease. Consequently, finding efficient mechanisms for the label-free detection of these PTMs is of direct medical relevance.
Here, we demonstrate the proof of concept of label-free detection of both phosphorylation and O-glycosylation and their discrimination from unmodified peptides. This is achieved using a biological nanopore (Fragaceatoxin C, or FraC)45 and a model peptide system in which a serine is modified with either of these PTMs. We first show that phosphorylated and nonphosphorylated peptides can be distinguished by their difference in the relative current blockade. Subsequently, the same is done for glycosylated and nonglycosylated peptides. Finally, we demonstrate for the first time that phosphorylated and O-glycosylated peptides can be differentiated from each other in a label-free manner.
Results
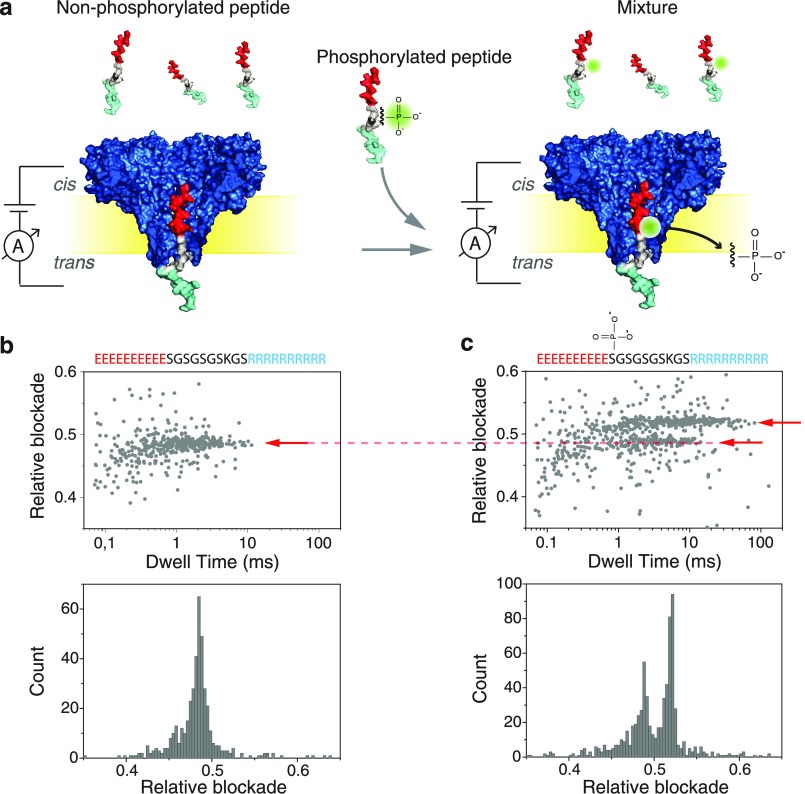
For the detection of phosphorylation and O-glycosylation, we used a model peptide (N′-EEEEEEEEEESGSGSGSKGSRRRRRRRRRR-C′) and the FraC nanopore. Our approach is presented in Figure 1. This peptide contains a stretch of 10 negatively charged glutamic acid residues at the N-terminus and a stretch of 10 positively charged arginine residues at the C-termimus. The two charged regions are connected by a flexible sequence of glycine (G) and serine (S) residues. Upon applying a negative bias to the trans compartment, the positive R-stretch enters the pore first, thus orienting the peptide in a defined way. Subsequently, when the negative E-stretch enters the proximity of the pore constriction, the positive bias of the cis compartment exerts an opposing force.46 A similar peptide was used by Asandei et al. to study peptide–nanopore interactions at different pH values and to show the differentiation between alanine and tryptophan residues.47,48 The peptide essentially gets stalled at an equilibrium position where the forces in both directions cancel out. This tug-of-war mechanism gives rise to long read lengths (>1 ms), enabling one to extensively probe a particular region of the peptide until it is thermally disturbed and translocated. Moreover, the pulling mechanism stretches the peptide, allowing the analysis of a linearized molecule.49
Figure 1.
(a) Schematic representation of the measurement setup, where peptides are driven through a FraC nanopore. Experiments were done with a nonphosphorylated peptide (left) and with an equimolar-concentration mixture of phosphorylated and nonphosphorylated peptides (right). (b) Scatter plot of relative blockade versus dwell time (top), and relative-blockade histogram (bottom) for the nonphosphorylated peptide. (c) Scatter plot of relative blockade versus dwell time for the mixture of phosphorylated and nonphosphorylated peptides. A second population with higher relative blockade is now visible, as shown by the arrows.
Substantial evidence was found for the existence of such an equilibrium position by tagging this model peptide with large chemical moieties at various locations along the peptide.46 These previous experiments reported that the peptide is stalled in the FraC nanopore at the position where amino acid in position 11 (from the N-terminus) is closest to the pore constriction. For PTM analysis, we therefore placed a serine in position 11 and evaluated variants of the peptide containing either phosphorylation or O-glycosylation in this position.
Measurements were performed using a wild type FraC nanopore in a buffer containing 10 mM Tris, 1 mM EDTA, and either 1 M NaCl or 0.8 M NaCl as specified at pH 7.5. If necessary, all the required sample preparation steps previous to the measurement can be done at physiological conditions, and the salt concentration can be increased right before the nanopore measurements. Peptides were added to the cis side of the flow cell at concentrations between 100–300 nM (Figure 1a). A negative bias of −90 mV was applied to the trans compartment to avoid gating that is observed in FraC under positive bias.
Identification of Phosphorylated Peptides with a FraC Nanopore
After the addition of peptides containing no PTM modifications to the cis compartment, we observed well-defined and consistent current blockades. Figure 1b shows a scatter diagram of the relative current blockade versus the dwell time observed for N = 449 translocation events, and the histogram for the relative blockade. We define the relative blockade as the current blockade (ΔI = Iblockade – Iopenpore) divided by the open pore current (Iopenpore). For the control peptide without PTMs, a relative blockade of 0.48 ± 0.01 was observed with a mean translocation time of 1.6 ± 1.1 ms. Error bars represent the standard deviation over at least three independent experiments.
Subsequently, peptides containing a single phosphorylation (i.e., a single PO32– group) in position 11 were added to the cis side of the chamber at a same concentration, thus leading to a one-to-one mixture of unmodified and PTMed peptides. As shown in Figure 1c, this led to two clear populations with comparable densities in the scatter plot of relative blockade versus dwell time. This indicates that the phosphorylation PTM can be clearly detected with the FraC nanopore. The well-defined difference in current blockade is also apparent in the histogram displayed in Figure 1c which shows two clearly separated peaks. The relative blockade of the bottom population (0.48 ± 0.02) corresponded to the unmodified control peptide, whereas a larger relative blockade of 0.52 ± 0.01 was found for the phosphorylated peptide. This represents an 8.7% increase compared to the control peptide.
Identification of Glycosylated Peptides with a FraC Nanopore
We expanded our nanopore detection to the analysis of the O-glycosylated peptide. O-glycosylated peptides have the glycan group attached to the amino acid residue through an oxygen group. O-glycans such as GlcNAc (N-acetylglucosamine) occur naturally in serines and threonines and are of growing importance in proteomics because of their close connection to phosphorylation regulation which plays a key role in several chronic diseases.
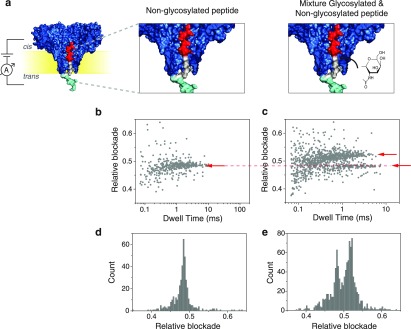
Here, we analyze a peptide containing an O-GlcNAc glycosylation in position 11. GlcNAc is a small PTM comprising a single sugar moiety with a total mass of 203 Da. Figure 2 shows the result of the nonglycosylated peptide as well as that of a mixture containing the O-glycosylated peptide together with the control peptide at equimolar concentrations. In Figure 2c,e, two distinct populations are clearly discernible in their relative blockade values, leading us to conclude that the nanopore is able to distinguish peptides containing a O-GlcNAc PTM from the unmodified peptide. Analysis of repeat measurements show that the O-GlcNAc variant has an average relative blockade of 0.52 ± 0.01. This represents an 8.3% increase compared to the unmodified peptide.
Figure 2.
(a) Schematic representation of the measurements for the non-glycosylated peptides (left) and for an equimolar concentration mixture of glycosylated and non-glycosylated peptides (right). (b) Scatter plot of relative blockade versus dwell time for the non-glycosylated peptide. (c) Scatter plot of relative blockade versus dwell time for the mixture of glycosylated and non-glycosylated peptides. A second population with higher relative blockade is visible as shown by the arrows. (d) Relative blockade histogram of the non-glycosylated peptide. (e) Relative blockade histogram of the mixture of non-glycosylated and glycosylated peptide.
We note that the increase in relative blockade observed by the phosphorylated and glycosylated variants are almost identical (8.7% versus 8.3%) despite their molecular weight difference (80 Da for phosphorylation versus 203 Da for glycosylation). We hypothesize that this relates to the negative charge of the phosphoryl group.
Distinguishing Different PTM Modifications with a FraC Nanopore
As observed in Figures 1 and 2, the phosphorylated and glycosylated peptides cannot be mutually differentiated based on the relative blockade as measured at 1 M NaCl because the increase in relative blockade caused by the phosphoryl group (8.7%) and the O-GlcNAc group (8.3%) is nearly identical under these measurements conditions. This was also confirmed by measuring a mixture containing three different peptides, the phosphorylated peptide, the O-glycosylated peptide, and the control peptide (SI; Figure S1), where only two populations were observed.
Despite the similarity in relative blockade observed for both PTMs, we noted a difference between the phosphorylated and glycosylated peptides in other translocation characteristics, notably the translocation time (Figure 3). For the phosphorylated peptide, an average dwell time of 3.2 ± 2.1 ms is observed, indicating an increase in translocation time compared to the control peptide that has an average dwell time of 1.6 ± 1.1 ms. For the glycosylated peptide, on the other hand, a decrease in dwell time is observed leading to an average dwell time of 0.4 ± 0.2 ms. We attribute the increase in dwell time observed with the phosphorylated peptide to a charge imbalance in the construct caused by the PTM. The addition of a phosphoryl group with a net negative charge shifts the force equilibrium brought about by the positive and negative tail regions of peptide and as a result the dwell time increases. We hypothesize that the decrease in dwell time for the GlcNAc group is of entropic nature. Confining the O-GlcNAc moiety into the pore constriction limits its degrees of freedom. As a result, the equilibrium state of the peptide is less energetically favorable for the glycosylated variant than for the unmodified peptide. This increases the probability to thermally disturb the force equilibrium, leading to shorter dwell times.
Figure 3.
Dwell time histograms for the unmodified peptide (top), the glycosylated peptide (middle), and the phosphorylated peptide (bottom).
Although the average dwell time for the two PTMs is significantly different, the dwell time of each population is quite widely spread and therefore this characteristic is not ideal for a proper real-time differentiation between the two PTM modifications. Hence, we looked for other ways to differentiate the two PTM variants. The relative blockades are nearly identical in 1 M NaCl, but if the salt conditions were changed the relative blockade of each modification would be altered to a different degree. Unlike the O-GlcNAc group, which is essentially uncharged, the phosphorylation contains two negative charges at pH = 7.5. The ionic strength of our buffer should therefore particularly affect the charge screening of the phosphoryl group, altering its translocation characteristics.
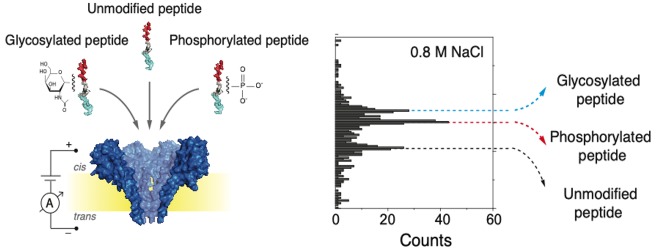
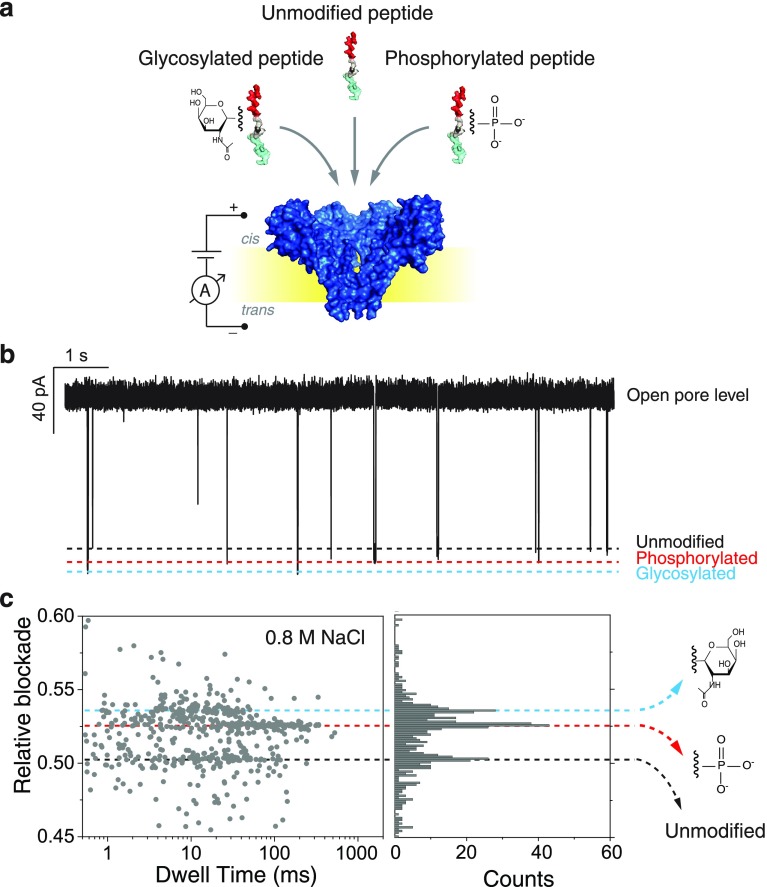
To test this, we measured a mixture containing the three peptide variants (the two modified peptides and the control) in equimolar concentrations in a buffer containing a lower salt concentration of 0.8 M NaCl at pH 7.5. Measurements at other salt concentrations such as 0.6 and 2 M NaCl were also performed (example traces can be found in Figure S2). The lowest concentration, 0.6 M, resulted in near-stalled translocations, obstructing data acquisition. The higher concentrations, for example 2 M, gave rise to very fast translocations, hampering the signal-to-noise ratio. As shown in Figure 4, at 0.8 M NaCl three different populations can be clearly observed in the scatter plot and histogram. Importantly, these conditions enable us to detect and differentiate the three different peptide variants. The control peptide had a mean value of 0.502 ± 0.004; the phosphorylated peptide had a mean value of 0.526 ± 0.002; and the glycosylated peptide had a mean value of 0.536 ± 0.003 (Figure S3). The results were confirmed by three independent experiments containing the peptide mixture (Figure S4) and by independent measurements of each PTM (Figure S5). Example event traces can be found in Figure S6. From the data, we can establish that at 0.8 M NaCl the increase in relative blockade of the glycosylated peptide compared to the control peptide remained largely unchanged (7.6%), whereas for the phosphorylated peptide, an increase in relative blockade of 4.8% compared to the control peptide is observed at 0.8 M NaCl, which is reduced as compared to 8.7% at 1 M NaCl. The mean values of the different populations are well-defined and separated from each other by values equivalent to at least two times sigma (Figure S2 and S3). We do not observe an overlap between the peptides containing PTMs and the control peptide. The overlap between the phosphorylated population and the glycosylated population is equivalent to approximately 5% (Figures S2 and S3).
Figure 4.
(a) Schematic representation of the measurement approach and (b) an example current trace obtained for a measurement on a mixture of the three peptides: the unmodified control peptide, the phosphorylated peptide and the glycosylated peptide. Data are taken at 0.8 M NaCl with pH 7.5. (c) Scatter plot of relative blockade vs dwell time and a relative blockade histogram of the mixture of the three peptides. Three different current blockade levels are observed. The first peak has a mean value of 0.502 (sd = 0.004), the second peak has a mean value of 0.526 (sd = 0.002), and the third peak has a mean value of 0.536 (sd = 0.003).
We hypothesize that the lower increase in relative blockade observed for the phosphoryl moiety at lower salt has the same origin as the inverted current rectification previously observed in FraC (Figure S7).49 Previous studies have reported that nanopores with strong ion selectivity and a diameter comparable to the Debye screening length exhibit a reversion in current rectification.50−52 The exact mechanism is still a matter of debate but it is speculated that it originates from the distribution of ions in the pore vestibule and at the trans region at the narrow tip of the pore. Our previous molecular dynamics simulations support this hypothesis and show that an accumulation of cations is observed at the cis opening of FraC, while an accumulation of anions is observed at the trans exit of the pore.46 In line with this reasoning, the presence of a stronger negatively charged phosphoryl group in the FraC constriction has an effect on this mechanism leading to an increase in current and thus a decrease in relative blockade compared to the 1 M NaCl case.
Discussion
In this work, we have shown that phosphorylation and glycosylation post-translational modifications can be detected using the wild type FraC nanopore. We use a bipolar peptide that contains a stretch of negative charges at the N-terminus and positive charges at the C-terminus. The peptide is stalled upon translocating the nanopore, allowing us to probe a particular region of the peptide with great sensitivity.49 Upon placing a phosphorylation or a small O-glycosylation group in this peptide, both PTM peptide variants could be distinguished from the control peptide with an unmodified serine residue. To the best of our knowledge, this constitutes the first reported detection of a O-glycan PTM using nanopores. O-glycans play a crucial role for protein conformation, solubility, and stability but, compared to N-glycans, their detection is challenging in mass spectrometry.10 The main reasons for the latter difficulties are the lack of a consensus amino acid sequence where the PTM is present, which hinders the determination of the PTM site, and the lack of universal enzymes to release O-glycans from the protein substrates. Nanopores thus present an attractive alternative for O-glycosylation characterization.
Next to the change in relative blockade detected for the PTM variants, we also observed changes in the translocation time of the peptides containing PTMs. Longer dwell times were observed for the phosphorylated peptide, presumably because of the negative charges in the phosphoryl group, which increase the forces in the peptide toward the cis compartment. Shorter dwell times were observed for the O-GlcNAc variant, which may be due to the entropic penalty associated with confining the sugar moiety in the pore constriction.
Upon changing the NaCl salt concentration from 1 to 0.8 M, an interesting effect was observed for the phosphorylation. A reduced blockade was observed at lower salt concentration. The result is counterintuitive, as one might expect an increase in the size of the electrical double layer at lower salt and thus an increase of the current blockade caused by the phosphoryl group. We hypothesize that the observed result has the same origin as the inversion of current rectification phenomena observed in FraC, where an accumulation or depletion of ions in the cis and trans opening of the pore has an important effect on the pore conductance.49 The presence of an effectively larger negative charge in the pore constriction generates an increased current.
In conclusion, our results show, for the first time, nanopore-based differentiation between phosphorylated and glycosylated peptide variants in a label-free manner. These results bear potential for PTM detection that is important for the study of diseases such as cancer or diabetes, where the intricate regulation of phosphorylation and glycosylation plays an important role in the pathogenesis of the disease.
In future work, the proof-of-principle nanopore-based detection of PTMs on peptides shown here may be expanded to PTM detection in full-length proteins upon using an enzyme or another mechanism to slow down the linear translocation of a full-length protein through the nanopore. As proteins of interest will likely be in a complex mixture containing other proteins, antibody-based methods or chromatography might be required to isolate particular proteins for efficient analysis. PTM detection on proteins would allow the identification of the variety of proteoforms in a sample, providing a radical improvement to current proteomics. Present day proteomics methods typically rely on approaches in which proteins are digested and peptide fragments are analyzed with their PTMs, which is inherently problematic because proteoforms cannot be correctly identified from the fragmented information on these analytes. Nanopore methods that sequentially read full-length proteins could thus allow the sensitive detection of the proteoforms present in a sample, bringing important improvements for PTM-based biomarker detection and the correct identification of proteoforms against the “protein inference problem”. Our current results present a promising step into this direction.
Materials and Methods
Peptide Design and Synthesis
The peptides used in this work were the control peptide with sequence EEEEEEEEEESGSGSGSKGSRRRRRRRRRR (HPLC purity = 96.33%, MW = 3662.77 Da), the phosphorylated peptide with sequence EEEEEEEEEES(p-S)GSGSGSKGSRRRRRRRRRR (HPLC purity = 96.41%, MW = 3743.07 Da), and the glycosylated peptide with sequence EEEEEEEEEES(β-GlcNAc-S)GSGSGSKGCRRRRRRRRRR (HPLC purity = 99%, MW = 3865.38 Da). The control and phosphorylated peptides were synthesized by Biomatik Corporation (Cambridge, CA). The glycosylated peptide was synthesized by Thermo Fisher Scientific. Synthesis was performed using standard solid-phase methods and the peptides were further purified using reverse-phase HPLC and analyzed by mass spectrometry. Peptides were kept lyophilized or, when necessary, aliquoted to a final concentration of 10 mg/mL at −20 °C.
Electrical Recording in Planar Lipid Membranes
Electrical recording was performed using planar lipid membranes, as described before.53,54 Briefly, a 25 μm thick Teflon film (Goodfellow Corporation, Pennsylvania U.S.A.) containing an orifice of approximately 70 μm width separates the cis and trans compartments. To form the membranes, 10 μL of 5% hexadecane in pentane was added to the Teflon film and the pentane was allowed to evaporate. The reservoirs were filled with buffer and 10 μl of 10 mg/mL 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) in pentane. Membranes were spontaneously formed using the Montal-Mueller method. Ag/AgCl electrodes were placed in each compartment with the ground electrode in the cis side. Wild type FraC oligomers were added to the cis side of the chamber. Upon pore insertion, the pore was characterized by measuring traces at different voltages and taking an I–V curve. The substrate was added to the cis side of the chamber and current recordings were done at −90 mV.
Data Acquisition and Analysis
Nanopore recording was collected using a patch-clamp amplifier (Axopatch 200B, molecular devices, U.S.A.) at a filtering frequency of 100 kHz. The data was digitized using an Axon Digidata 1550B digitizer at a sampling frequency of 500 kHz. The signal was low-pass filtered at 10 kHz and was processed using a Matlab script (Transalyzer).55
Acknowledgments
We would like to thank Peggy Bohländer, Rienk Eelkema, Martin Pabs, Robert Cordfunke, Sonja Schmid, and Mike Filius for fruitful discussions. This work was supported by the Foundation for Fundamental Research on Matter (Vrije Programma SMPS) to C.J., C.D., and G.M., and by the European Research Council Advanced Grant SynDiv (No. 669598), The Netherlands Organisation for Scientific Research (NWO/OCW) through the NanoFront and BaSyC grants
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nanolett.9b03134.
Data of the measurement of a mixture containing glycosylated, phosphorylated, and control peptide at 1 M NaCl, example traces of peptide translocations at different salt concentrations, three independent measurements of the peptide mixture at 0.8 M NaCl, data of independent measurements of glycosylated and phosphorylated peptide at 0.8 M NaCl, example traces of the different peptides, and current versus voltage plots of the FraC nanopore (PDF)
The authors declare the following competing financial interest(s): L.R.P., C.D., and C.J. are co-founders and shareholders of Bluemics, a company engaged in the development of nanopore sensors for protein analysis.
Supplementary Material
References
- Alberts; Johnson; Lewis; Raff; Roberts; Walter. Molecular Biology of the Cell; Garland Science, 2002. 10.1091/mbc.E14-10-1437 [DOI] [Google Scholar]
- Aebersold R.; Agar J. N.; Amster I. J.; Baker M. S.; Bertozzi C. R.; Boja E. S.; Costello C. E.; Cravatt B. F.; Fenselau C.; Garcia B. A. How many human proteoforms are there?. Nat. Chem. Biol. 2018, 14 (3), 206–214. 10.1038/nchembio.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarenko E. A. ; Poverennaya E. V.; Ilgisonis E. V.; Pyatnitskiy M. A.; Kopylov A. T.; Zgoda V. G.; Lisitsa A. V.; Archakov A. I. The Size of the Human Proteome: The Width and Depth. Int. J. Anal. Chem. 2016, 2016, 1–6. 10.1155/2016/7436849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan G.; Walther D. The Roles of Post-translational Modifications in the Context of Protein Interaction Networks. PLoS Comput. Biol. 2015, 11, e1004049 10.1371/journal.pcbi.1004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorre D. G.; Kudryashova N. V.; Godovikova T. S. Chemical and functional aspects of posttranslational modification of proteins. Acta Naturae 2009, 1, 29–51. 10.32607/20758251-2009-1-3-29-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle H. A.; Mamula M. J. Post-translational protein modifications in antigen recognition and autoimmunity. Trends Immunol. 2001, 22, 443–449. 10.1016/S1471-4906(01)01976-7. [DOI] [PubMed] [Google Scholar]
- Hart G. W.; Akimoto Y.. The O-GlcNAc Modification. Essentials of Glycobiology; Cold Spring Harbor Laboratory Press, 2009. [PubMed] [Google Scholar]
- Liu F.; Iqbal K.; Grundke-Iqbal I.; Hart G. W.; Gong C.-X. O-GlcNAcylation regulates phosphorylation of tau: a mechanism involved in Alzheimer’s disease. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 10804–9. 10.1073/pnas.0400348101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel O.; Loroch S.; Sickmann A.; Zahedi R. P. Current strategies and findings in clinically relevant post-translational modification-specific proteomics. Expert Rev. Proteomics 2015, 12, 235–253. 10.1586/14789450.2015.1042867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulagapati S.; Koppolu V.; Shantha Raju T. Decoding of O-Linked Glycosylation by Mass Spectrometry. Biochemistry 2017, 56, 1218–1226. 10.1021/acs.biochem.6b01244. [DOI] [PubMed] [Google Scholar]
- Zubarev R. A. The challenge of the proteome dynamic range and its implications for in-depth proteomics. Proteomics 2013, 13, 723–726. 10.1002/pmic.201200451. [DOI] [PubMed] [Google Scholar]
- Anderson N. L. The Human Plasma Proteome: History, Character, and Diagnostic Prospects. Mol. Cell. Proteomics 2002, 1, 845–867. 10.1074/mcp.R200007-MCP200. [DOI] [PubMed] [Google Scholar]
- Marshall A. A cast of thousands. Nat. Biotechnol. 2003, 21, 213. 10.1038/nbt0303-213. [DOI] [PubMed] [Google Scholar]
- Rusling J. F.; Kumar C. V.; Gutkind J. S.; Patel V. Measurement of biomarker proteins for point-of-care early detection and monitoring of cancer. Analyst 2010, 135, 2496. 10.1039/c0an00204f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanash S. M.; Pitteri S. J.; Faca V. M. Mining the plasma proteome for cancer biomarkers. Nature 2008, 452, 571–579. 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- Rifai N.; Gillette M. A.; Carr S. A. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 2006, 24, 971–983. 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- Nørregaard Jensen O. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr. Opin. Chem. Biol. 2004, 8, 33–41. 10.1016/j.cbpa.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Dekker C. Solid-state nanopores. Nat. Nanotechnol. 2007, 2, 209–215. 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- Albrecht T.; Gibb T.; Nuttall P.. Ion Transport in Nanopores. In Engineered Nanopores for Bioanalytical Applications; Elsevier Inc., 2013; Chapter 1. [Google Scholar]
- Goodrich C. P. Single-Molecule Electrophoresis of β-Hairpin Peptides by Electrical Recordings and Langevin Dynamics Simulations. J. Phys. Chem. B 2007, 111, 3332–3335. 10.1021/jp071364h. [DOI] [PubMed] [Google Scholar]
- Movileanu L.; Schmittschmitt J. P.; Martin Scholtz J.; Bayley H. Interactions of Peptides with a Protein Pore. Biophys. J. 2005, 89, 1030–1045. 10.1529/biophysj.104.057406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piguet F.; Ouldali H.; Pastoriza-Gallego M.; Manivet P.; Pelta J.; Oukhaled A. Identification of Single Amino Acid Differences in Uniformly Charged Homopolymeric Peptides with Aerolysin Nanopore. Nat. Commun. 2018, 9 (1), 966. 10.1038/s41467-018-03418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G.; Willems K.; Soskine M.; Wloka C.; Maglia G. Electro-osmotic capture and ionic discrimination of peptide and protein biomarkers with FraC nanopores. Nat. Commun. 2017, 8, 935. 10.1038/s41467-017-01006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefureac R.; Long Y.-T.; Kraatz H.-B.; Howard P.; Lee J. S. Transport of α-Helical Peptides through α-Hemolysin and Aerolysin Pores †. Biochemistry 2006, 45, 9172–9179. 10.1021/bi0604835. [DOI] [PubMed] [Google Scholar]
- Mohammad M. M.; Movileanu L. Excursion of a single polypeptide into a protein pore: simple physics, but complicated biology. Eur. Biophys. J. 2008, 37, 913–925. 10.1007/s00249-008-0309-9. [DOI] [PubMed] [Google Scholar]
- Sutherland T. C.; Long Y.-T.; Stefureac R.-I.; Bediako-Amoa I.; Kraatz H.-B.; Lee J. S. Structure of Peptides Investigated by Nanopore Analysis. Nano Lett. 2004, 4 (7), 1273–1277. 10.1021/nl049413e. [DOI] [Google Scholar]
- Mahendran K. R.; Romero-Ruiz M.; Schlösinger A.; Winterhalter M.; Nussberger S. Protein Translocation through Tom40: Kinetics of Peptide Release. Biophys. J. 2012, 102, 39–47. 10.1016/j.bpj.2011.11.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z.; Wang S.; Zhao Z.; Haque F.; Guo P. Fingerprinting of Peptides with a Large Channel of Bacteriophage Phi29 DNA Packaging Motor. Small 2016, 12 (33), 4572–4578. 10.1002/smll.201601157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G.; Voet A.; Maglia G. FraC nanopores with adjustable diameter identify the mass of opposite-charge peptides with 44 Da resolution. Nat. Commun. 2019, 10, 835. 10.1038/s41467-019-08761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavis A. E.; Brady K. T.; Hatmaker G. A.; Angevine C. E.; Kothalawala N.; Dass A.; Robertson J. W. F.; Reiner J. E. Single Molecule Nanopore Spectrometry for Peptide Detection. ACS Sensors 2017, 2 (9), 1319–1328. 10.1021/acssensors.7b00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asandei A.; Rossini A. E.; Chinappi M.; Park Y.; Luchian T. Protein Nanopore-Based Discrimination between Selected Neutral Amino Acids from Polypeptides. Langmuir 2017, 33, 14451–14459. 10.1021/acs.langmuir.7b03163. [DOI] [PubMed] [Google Scholar]
- Payet L.; Martinho M.; Pastoriza-Gallego M.; Betton J. M.; Auvray L.; Pelta J.; Mathé J. Thermal Unfolding of Proteins Probed at the Single Molecule Level Using Nanopores. Anal. Chem. 2012, 84 (9), 4071–4076. 10.1021/ac300129e. [DOI] [PubMed] [Google Scholar]
- Pastoriza-Gallego M.; Rabah L.; Gibrat G.; Thiebot B.; van der Goot F. G.; Auvray L; Betton J.-M.; Pelta J. Dynamics of Unfolded Protein Transport through an Aerolysin Pore. J. Am. Chem. Soc. 2011, 133 (9), 2923–2931. 10.1021/ja1073245. [DOI] [PubMed] [Google Scholar]
- Oukhaled G.; Mathé J.; Biance A.-L.; Bacri L.; Lairez D.; Pelta J.; Auvray L. Unfolding of Proteins and Long Transient Conformations Detected by Single Nanopore Recording. Phys. Rev. Lett. 2007, 98 (15), 158101. 10.1103/PhysRevLett.98.158101. [DOI] [PubMed] [Google Scholar]
- Restrepo-Pérez L.; John S.; Aksimentiev A.; Joo C.; Dekker C. SDS-assisted protein transport through solid-state nanopores. Nanoscale 2017, 9, 11685–11693. 10.1039/C7NR02450A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaga D. S.; Li J. Single-Molecule Protein Unfolding in Solid State Nanopores. J. Am. Chem. Soc. 2009, 131, 9287–9297. 10.1021/ja901088b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Fologea D.; Rollings R.; Ledden B. Characterization of Protein Unfolding with Solid-state Nanopores. Protein Pept. Lett. 2014, 21, 256–265. 10.2174/09298665113209990077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman K. J.; Haq S. R.; Edel J. B.; Jemth P.; Kim M. J. Single molecule unfolding and stretching of protein domains inside a solid-state nanopore by electric field. Sci. Rep. 2013, 3, 1638. 10.1038/srep01638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z.; Kennedy E.; Hokmabadi M.; Timp G. Discriminating Residue Substitutions in a Single Protein Molecule Using a Sub-nanopore. ACS Nano 2017, 11, 5440–5452. 10.1021/acsnano.6b08452. [DOI] [PubMed] [Google Scholar]
- Kennedy E.; Dong Z.; Tennant C.; Timp G. Reading the primary structure of a protein with 0.07 nm3 resolution using a subnanometre-diameter pore. Nat. Nanotechnol. 2016, 11, 968–976. 10.1038/nnano.2016.120. [DOI] [PubMed] [Google Scholar]
- Robertson J. W. F.; Rodrigues C. G.; Stanford V. M.; Rubinson K. A.; Krasilnikov O. V.; Kasianowicz J. J. Single-molecule mass spectrometry in solution using a solitary nanopore. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (20), 8207–11. 10.1073/pnas.0611085104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahie M. A.; Chen M. Electrostatic Interactions between OmpG Nanopore and Analyte Protein Surface Can Distinguish between Glycosylated Isoforms. J. Phys. Chem. B 2015, 119, 10198–10206. 10.1021/acs.jpcb.5b06435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C.; Van Meervelt V.; van Gelder D.; Danda N.; Jager N.; Williams C. P.; Maglia G. Label-Free and Real-Time Detection of Protein Ubiquitination with a Biological Nanopore. ACS Nano 2017, 11 (5), 4387–4394. 10.1021/acsnano.6b07760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen C. B.; Rodriguez-Larrea D.; Bayley H. Single-molecule site-specific detection of protein phosphorylation with a nanopore. Nat. Biotechnol. 2014, 32, 179–181. 10.1038/nbt.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K.; Caaveiro J. M. M.; Morante K.; González-Mañas J. M.; Tsumoto K. Structural basis for self-assembly of a cytolytic pore lined by protein and lipid. Nat. Commun. 2015, 6, 6337. 10.1038/ncomms7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo-Pérez L. et al. Resolving Chemical Modifications to a Single Amino Acid within a Peptide Using a Biological Nanopore. ACS Nano 2019; doi: 10.1021/acsnano.9b05156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asandei A.; Chinappi M.; Kang H. K.; Seo C. H.; Mereuta L.; Park Y.; Luchian T. Acidity-Mediated, Electrostatic Tuning of Asymmetrically Charged Peptides Interactions with Protein Nanopores. ACS Appl. Mater. Interfaces 2015, 7 (30), 16706–16714. 10.1021/acsami.5b04406. [DOI] [PubMed] [Google Scholar]
- Asandei A.; Rossini A. E.; Chinappi M.; Park Y.; Luchian T. Protein Nanopore-Based Discrimination between Selected Neutral Amino Acids from Polypeptides. Langmuir 2017, 33, 14451–14459. 10.1021/acs.langmuir.7b03163. [DOI] [PubMed] [Google Scholar]
- Zhao S.; Restrepo-Pérez L.; Soskine M.; Maglia G.; Joo C.; Dekker C.; Aksimentiev A. Electro-Mechanical Conductance Modulation of a Nanopore Using a Removable Gate. ACS Nano 2019, 13 (2), 2398–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momotenko D.; Cortés-Salazar F.; Josserand J.; Liu S.; Shao Y.; Girault H. H. Ion current rectification and rectification inversion in conical nanopores: a perm-selective view. Phys. Chem. Chem. Phys. 2011, 13 (12), 5430. 10.1039/c0cp02595j. [DOI] [PubMed] [Google Scholar]
- Yan Y.; Wang L.; Xue J.; Chang H.-C. Ion current rectification inversion in conic nanopores: Nonequilibrium ion transport biased by ion selectivity and spatial asymmetry. J. Chem. Phys. 2013, 138, 044706 10.1063/1.4776216. [DOI] [PubMed] [Google Scholar]
- Wloka C.; Mutter N. L.; Soskine M.; Maglia G. Alpha-Helical Fragaceatoxin C Nanopore Engineered for Double-Stranded and Single-Stranded Nucleic Acid Analysis. Angew. Chem., Int. Ed. 2016, 55, 12494–12498. 10.1002/anie.201606742. [DOI] [PubMed] [Google Scholar]
- Maglia G.; Heron A. J.; Stoddart D.; Japrung D.; Bayley H. Analysis of Single Nucleic Acid Molecules with Protein Nanopores. Methods Enzymol. 2010, 475, 591–623. 10.1016/S0076-6879(10)75022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsmann T.; Heimburg T.; Keyser U.; Mahendran K. R.; Winterhalter M. Protein reconstitution into freestanding planar lipid membranes for electrophysiological characterization. Nat. Protoc. 2015, 10, 188–198. 10.1038/nprot.2015.003. [DOI] [PubMed] [Google Scholar]
- Plesa C.; Dekker C. Data analysis methods for solid-state nanopores. Nanotechnology 2015, 26, 084003. 10.1088/0957-4484/26/8/084003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.