Abstract
Three fishers (Martes pennanti), 2 gray foxes (Urocyon cinereoargenteus), 1 mink (Neovison vison), 1 skunk (Mephitis mephitis), and 1 raccoon (Procyon lotor), from Vermont and New Hampshire, had lesions on autopsy consistent with canine distemper virus (CDV) infections diagnosed in a 12-mo period in 2016–2017. Lesions of CDV infection were most commonly noted in the lungs (8 of 8 animals), urothelium (5 of 8), biliary tract (5 of 8), gastrointestinal tract (4 of 7), and brain (4 of 6). Splenic lesions were seen in 3 animals. The diagnosis was confirmed via immunohistochemistry and virus isolation. Viral genotyping indicated that all 8 animals were infected with a distinct clade of CDV that has only been reported in wildlife in New England, and this clade of viruses is distinct from vaccine strains. During the 12 mo when these cases occurred, no other CDV clade was identified in any other wildlife or domesticated animal submitted from the 2 states.
Keywords: canine distemper virus, distemper, fishers, foxes, mink, raccoons, skunks, wildlife
Canine distemper virus (CDV; family Paramyxoviridae, subfamily Orthoparamyxovirinae, genus Morbillivirus, species Canine morbillivirus) has a single-stranded, negative-sense RNA genome. The Morbillivirus genus includes measles morbillivirus, rinderpest morbillivirus, peste-des-petits-ruminant virus (species Small ruminant morbillivirus), feline morbillivirus, cetacean morbillivirus, and phocine distemper virus (species Phocine morbillivirus), among others.14 Morbilliviruses are known to cause severe, often fatal, multi-organ epitheliotropic lesions and encephalitis. Clinical signs are dictated by damage to infected epithelial cells in the respiratory tract, gastrointestinal tract, and neurons and glia. Pneumonia, conjunctivitis, and enteritis are common presenting clinical signs of CDV in dogs; encephalitis follows, after viral spread via lymphatics.17 In wildlife, these conditions occur concurrently and remain until death; advanced disease with neurologic signs can be confused with rabies.14
CDV control by vaccination of domestic animals has lowered the prevalence of infection in the United States.14 Although reports of CDV in domesticated dogs have decreased, CDV is increasingly identified in feral dogs and wild Canidae, Felidae, Mustelidae, Procyonidae, and Ursidae families, among others; infections have been reported on every continent except Australia.15,17 In the United States, CDV is a significant pathogen of raccoons (Procyon lotor), gray foxes (Urocyon cinereoargenteus), and striped skunks (Mephitis mephitis).15,17,20 There are 7 historically defined CDV lineages, based on phylogenic analysis of the hemagglutinin (H) gene sequence and named for their assumed origin: America-1, America-2, Arctic-like, Asia-1, Asia-2, Europe, and European wildlife.10 Because the H protein binds to host cells and can determine viral spread in the host, polymorphisms in the H gene can alter virus–host interaction, influencing infectivity and/or virulence.10 Based on H-gene sequencing, evidence indicates multiple circulating CDV strains in wildlife in the United States that do not fall into any of the historically described lineages.17,19,21
Previous studies have outlined lineages of CDV throughout the United States,17 with a noted dearth of data in New England wildlife. We identified no clinical isolates in the GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) using the following search terms: “distemper,” and “Hampshire” or “Vermont” or “Massachusetts” or “Connecticut.” We report herein naturally occurring infections by a distinct clade of CDV in 8 wild mesocarnivores: 3 fishers (Martes pennanti), 2 foxes (Urocyon cinereoargenteus), 1 skunk (Mephitis mephitis), 1 raccoon (Procyon lotor), and 1 mink (Neovison vison) over a 12-mo period in New Hampshire and Vermont.
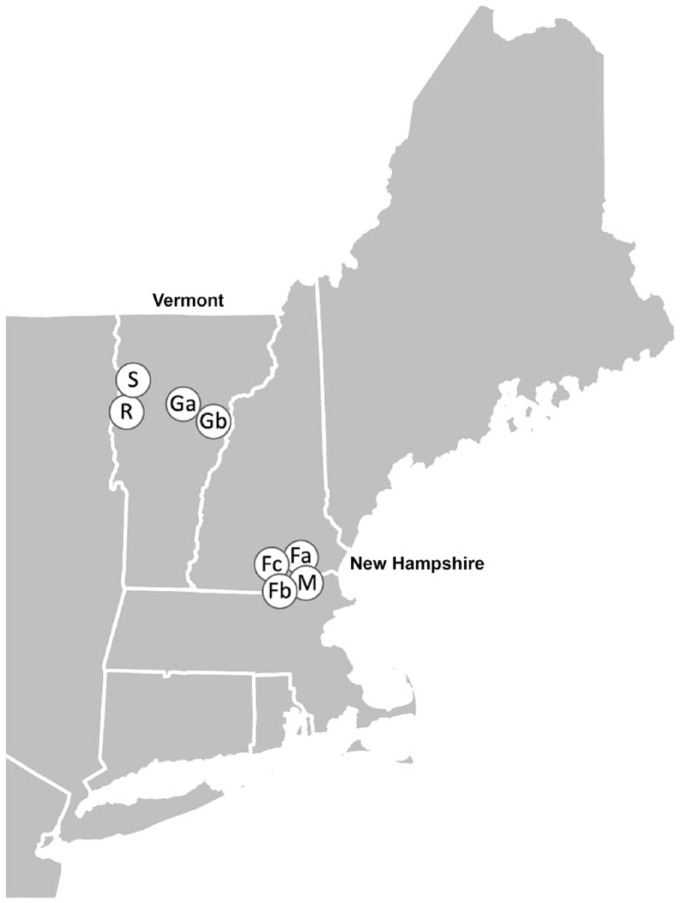
Eight free-ranging animals with signs of illness or abnormal behavior were submitted to the New Hampshire Veterinary Diagnostic Laboratory (NHVDL) by wildlife biologists from Vermont (VT) and New Hampshire (NH; Fig. 1). Animals were found dead or were euthanized prior to submission as whole carcasses; all were adults. During the same period (2016–2017), the NHVDL received 4 additional animals considered highly susceptible to CDV (1 fisher, 1 black bear, and 2 raccoons), but these animals had no lesions indicating CDV infection.
Figure 1.
Map of canine distemper virus–infected animals in New Hampshire and Vermont. Each point represents the approximate location of one animal. Capital letters indicate the species (F = fisher; G = gray fox; R = raccoon; S = striped skunk; M = mink), and when necessary, a second letter (lowercase) is provided to uniquely identify animals of the same species.
Autopsy was performed on all animals by a board-certified veterinary anatomic pathologist at the NHVDL (DB Needle or BA Stevens). Tissue samples were placed in 10% neutral-buffered formalin for 24–48 h, processed routinely, and stained with hematoxylin and eosin. Additional fresh tissue samples were stored at −20°C.
Formalin-fixed, paraffin-embedded tissues from fishers 1 and 2 were sent to the Cornell Wildlife Health Laboratory (Ithaca, NY). An automated staining system (BOND-MAX; Leica Microsystems, Buffalo Grove, IL) ran a previously validated protocol for CDV detection using an in-house mouse antibody (NP879 CDV) developed at the Baker Institute, Cornell University, for immunohistochemistry (IHC) staining, as described previously.3 Briefly, slides were dewaxed (Bond dewax solution, cat. AR9222; Leica). Heat epitope retrieval was applied for 30 min (Bond epitope retrieval solution 1, cat. AR9961; Leica). The NP879 antibody diluted at 1:5,000 was applied to slides for 15 min. A polymer (secondary antibody, PV-AP-anti-mouse IgG reagent, cat. PV6110; Leica) was applied for 10 min. Bond polymer refine red detection (cat. DS9390; Leica) was applied for 15 min. Finally, the tissue was counterstained with hematoxylin for 5 min (cat. DS9390; Leica). Nonspecific binding control slides were produced following the same method but omitting primary antibody, and no staining was noted on these slides. Positive labeling was defined as strong distinct immunoreactivity in intracytoplasmic inclusions, with absent or minimal background staining.
CDV was isolated from fresh frozen tissue from all animals as described previously, except that the virus was grown on Vero/SLAM (signaling lymphocyte activation molecule) cells and identified by direct fluorescent antibody technique.12 Isolated viral nucleic acid was extracted (DNeasy blood and tissue kit; Qiagen, Valencia, CA). A targeted next-generation sequencing (NGS) assay was used to amplify the entire CDV genome.2 Two pools of custom-made overlapping primers were used. Reverse-transcription PCR and library preparation were performed on the Ion Chef using the AmpliSeq library kit for Chef DL8 with incorporated barcodes (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s protocol. Libraries were templated and loaded on an Ion 314 chip using the Ion Chef instrument with the Ion PGM kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Ion Torrent machine–supplied, cloud-based bioinformatics programs and Geneious software v.11.03 (https://www.geneious.com/) were used for sequence alignment and analyses. H gene and F gene sequences obtained from the samples were aligned with additional sequences from GenBank using MAFFT, and phylogenetic trees were generated within Geneious with the Tamura–Nei model using UPGMA and 1,000 bootstrap replicates. Phylogenetic trees were also constructed using maximum likelihood with FastTree and PhyML within Geneious.3,9,11,18,22
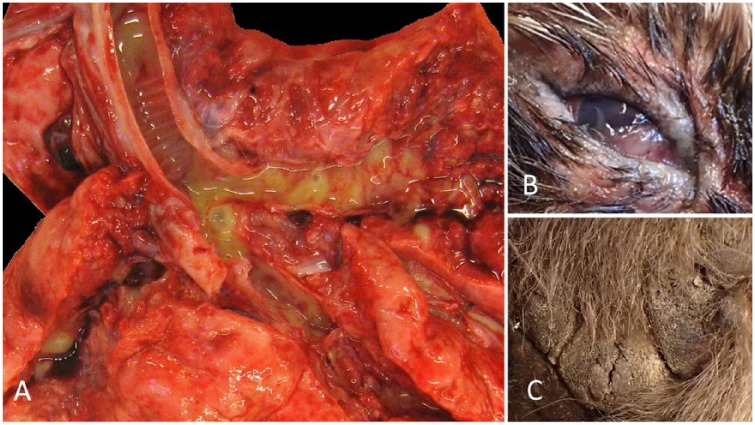
Gross anatomic lesions were noted in 6 of 8 animals. All 6 had bronchointerstitial or interstitial pneumonia (Fig. 2A). Individual animals had grossly evident bilateral conjunctivitis (raccoon; Fig. 2B), rhinitis (skunk), and proliferative footpad dermatitis (fisher C; Fig. 2C).
Figure 2.
Gross anatomic lesions of a distinct clade of canine distemper virus (CDV) infection in wildlife from New Hampshire and Vermont. A. CDV in fisher A. The terminal trachea and mainstem bronchi are opened along with the surrounding parenchyma, revealing severe, multifocal-to-coalescing areas of consolidation, and associated and surrounding erythema, with mucopurulent exudate in the airways. B. Chronic, diffuse, mucoid and proliferative conjunctivitis and palpebritis in a raccoon. C. Hyperkeratosis and degeneration of the footpads in fisher C, characterized by an irregular and pronounced surface that has multiple irregular fissures, a.k.a. “hardpad.”
Microscopic lesions attributable to CDV (Table 1) were widely distributed. Bronchointerstitial pneumonia was noted in all 8 cases (Fig. 3), lesions in the renal pelvis or urinary bladder in 5 of 8 cases, and lesions in the biliary tract. The epididymis, pancreas, thyroid gland, and lymph node each had CDV inclusions in individual animals (Table 1).
Table 1.
Distribution of histologic lesions attributable to canine distemper virus (CDV) infection (with inclusions noted) in 8 wild animals from New Hampshire and Vermont, 2016–2017.
| CDV lesion locations | Fisher A | Fisher B | Fisher C | Raccoon | Striped skunk | Gray fox A | Gray fox B | Mink |
|---|---|---|---|---|---|---|---|---|
| Lung | + | + | + | + | + | + | + | + |
| Thyroid | + | NE | NE | NE | − | NE | − | NE |
| Biliary tract | + | + | − | + | − | + | + | − |
| Urinary tract | + | + | − | + | + | − | + | − |
| Epididymis | NE | + | − | NE | NE | NE | NE | − |
| Pancreas | − | − | NE | NE | NE | + | NE | − |
| Liver | − | − | − | − | − | + | − | − |
| Nasal cavity | NE | NE | NE | + | + | NE | NE | NE |
| Brain | NE | NE | + | − | + | + | + | − |
| GI tract | − | NE | − | + | + | + | + | − |
| Spleen | − | − | − | + | − | + | − | + |
| Lymph node | − | − | − | − | − | − | + | − |
| Footpad | NE | NE | + | NE | NE | NE | NE | NE |
| Conjunctiva | NE | NE | NE | + | NE | NE | NE | NE |
+ = a lesion attributed to CDV was identified in the organ; − = no lesion was noted; NE = the organ was not examined in that animal.
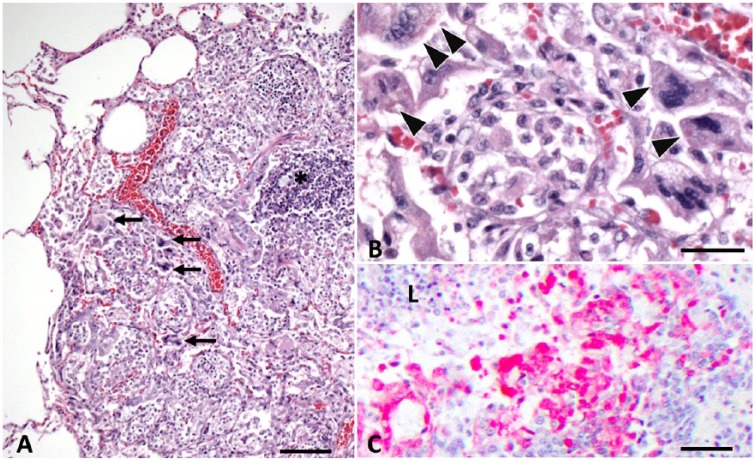
Figure 3.
Canine distemper virus (CDV) bronchointerstitial pneumonia in fisher A. A. Regional consolidation of the parenchyma caused by flooding of alveoli by inflammatory cells and amorphous debris, and expansion of the interstitium by a mixture of edema and inflammatory infiltrate. Bronchus-associated lymphoid tissue is hyperplastic (*). Multiple syncytial cells are evident (arrows). H&E. Bar = 180 µm. B. Higher magnification highlighting multiple syncytial cells that contain well-delineated, ~3–7 µm diameter intracytoplasmic inclusion bodies (arrowheads). There is a mixture of lymphohistiocytic infiltrate within the interstitium and alveolar lumina, with interspersed fibrin, amorphous debris, and rare free erythrocytes. H&E. Bar = 30 µm. C. Heavy immunohistochemical labeling of CDV antigen within the epithelial cells of a bronchiole (L = lumen) in fisher A, as well as within scattered luminal and interstitial inflammatory cells. Bar = 180 µm.
Lesions of CDV infection were consistent with classical descriptions consisting of epithelial degeneration and necrosis accompanied by neutrophilic and lymphohistiocytic infiltrate (Fig. 3A), with numerous 3–7 µm diameter, round, bright eosinophilic, intracytoplasmic inclusions (Fig. 3B), and occasional irregular polygonal multinucleated syncytial cells noted in the lungs (Fig. 3B). IHC confirmed CDV infection in the multiple tissues of both tested animals (fishers A and B; Fig. 3C).
Virus was successfully isolated and sequenced from all animals, providing partial-full gene sequences; the H and F gene sequences obtained for each sample were uploaded to GenBank (H: accessions MH644848–55; F: accessions MH651727–30 and MH665349–52). The F gene for America-3 (Edomex strain) was also added to GenBank (accession MH665353). Based on H and F gene analyses with both UPGMA and maximum likelihood methods, the sequences formed a distinct clade containing only the viruses isolated from our 8 animals and a single virus isolated from a raccoon in Rhode Island in 2012 (GenBank accession KU666057; Supplementary Figs. 1, 2; only UPGMA results shown). The viral gene encoding the H protein sequence was assessed at position 549 (Supplementary Fig. 1): the raccoon, mink, fox A, and fishers A–C encoded a tyrosine (the Y549H mutation), whereas the skunk and fox B had the genetic code for a histidine (wild type).
Sequence analysis of H and F genes found this clade to have just a single previously reported isolate from a raccoon in Rhode Island in 2012 (GenBank entry www.ncbi.nlm.nih.gov/nuccore/992370942; Supplementary Figs. 1, 2). A study based on the variation of H gene sequences in field isolates from North America and Europe showed “pronounced genetic diversity” among field isolates and “substantial genetic difference” between field and vaccine strains.4 To date, CDV vaccines are mainly composed of America-1 lineage, although no field strains from this lineage are reported, as of 2017, as circulating in dogs in the United States.14 Vaccinated dogs with active CDV infections in North America have had isolates related to an isolate from Missouri in the Edomex/America-3 clade, and the Arctic-like clade.10,16 Thus, either the classically described lineages appear more widespread than initially thought, or are fluid in their geographic distribution.
We found only one previous report of CDV in fishers, describing infection of animals from a remote area of California with a strain in the emerging Edomex/America-3 clade.12,19 Our fisher isolates are not in this lineage. All 4 California fishers had pneumonia, just as ours did (Figs. 2, 3); 2 of 4 California fishers had pododermatitis, as noted in fisher C (Fig. 2); and none of the California fishers had CDV-related encephalitis, whereas the only fisher we report with brain available for examination (fisher C) had CDV encephalitis (Table 1). There is a report of 4 wild mink with naturally occurring CDV infection in Florida in 2004, wherein the most commonly affected organ was footpad, followed by stomach and lung—the mink in our study had lesions in the lung and spleen only (Table 1).6
Contrary to fishers and mink, the other mesocarnivores in our study have widely reported CDV infection, with lymphoid depletion and pneumonia common and encephalitis rare in a study of 22 raccoons; our raccoon had those common lesions with the addition of encephalitis, conjunctivitis, rhinitis, and enteritis (Fig. 2B, Table 1).5 As we report, striped skunks often have pneumonia and meningoencephalitis, as well as rhinitis, enteritis, and cystitis (Table 1).8,24,25 CD is perhaps the most common disease of gray foxes, given that a study found CDV in 123 of 157 (78%) gray foxes.7 In general, the lesions we noted were similar to those reported previously; the few unique lesions or CDV distribution such as epididymitis and pancreatitis may be a result of broader tissue sampling and examination, or a result of wider tissue tropism or infectivity of this distinct clade.
Although aerosol is the most common mode of CDV transmission, virus in urine, feces, or cutaneous exudates can infect wildlife in close quarters.13 Clinically and subclinically infected animals can shed CDV up to 90 d after infection.13 The virus is not thought to survive more than transiently outside of a host, thus animal interaction and biological reservoirs appear key to its perpetuation.13 This need for interaction to ensure transmission indicates that the animals we describe likely had some significant contact with other animals infected with this distinct clade of CDV, although the nature of this contact is speculative. Modes of infection less likely than direct contact include ectoparasite vectors or vertical transmission, as demonstrated in farmed mink and a wild ferret, respectively, as well as potentially through infected rodent prey.23
We assessed our samples for the Y549H mutation that has been purported to allow for non-canid CDV infectivity.23 In the cases we present, all 4 animals from NH (all 3 fishers, the mink), and gray fox A and the raccoon from VT, encoded for Y at position 549, as did the historical Rhode Island isolate; the striped skunk and gray fox B had RNA encoding H at 549 (Supplementary Fig. 1). The 2 viruses encoding 549H were most like each other and had split from all of the other members of this new clade in a relatively early evolutionary event resulting in one branch giving rise to just the 549H isolates (striped skunk, gray fox B), and a second branch giving rise to all other isolates we describe and the historical isolate (Supplementary Figs. 1, 2). Our isolates include a wild canid-origin CDV that has the mutation (gray fox A), and a non-canid origin CDV that has the wild-type 549H. Thus, our data indicate Y549H may be more widely and randomly distributed among wildlife-origin CDV, infecting canids and non-canids; it may be that Y549H is more common and thus helpful to delineate evolutionary changes within clades as they emerge or are identified.
Our cases include multiple species from distinct regions fatally infected by highly homologous CDV isolates. The phylogenetic clustering of these viruses is significant because the 2 states are separated along virtually their entire border by the Connecticut River (Fig. 1). The only previously identified member of the clade is from a state at least 160 km (100 mi) away. Homologous members of a distinct clade of virus infecting multiple species of wildlife on 2 sides of the largest river in New England suggests potential for wide and rapid spread, or previously unrecognized widespread distribution in the region. The latter may be favored as there is separate clustering of the 4 VT-origin isolates and the 4 NH-origin isolates based on the H and F gene sequences (Supplementary Figs. 1, 2). It is possible that many such smaller geographically related clusters occur within distinct clades of CDV and other viruses circulating in wildlife, and that increased surveillance could expose these trends.
It is also likely important that this small group of 8 initial animals infected with this distinct clade includes 3 fishers; this species has only been noted with CDV infection once prior on the opposite coast of the continent.12 Although the lesions we describe in our 3 fisher are similar to those described in California, we have the additional finding of neurotropism, which may indicate increased infectivity or pathogenicity in the distinct strain we described compared to others. The emerging distinct strain of CDV in the southeastern United States (including Tennessee) has diverged enough from vaccine strains that they elicit significantly different antibody titers, and could potentiate vaccine breakthrough.1,19 There is the additional potential for spillover into non-traditional hosts, locally threatened wild populations, and zoological collections.1 The 8 animals we report and the distinct clade of CDV infecting them highlight the importance of prospective observational diagnostic pathology studies of wildlife to identify emerging pathogens, and the value of advanced molecular techniques in assessing pathogen genealogy as cases emerge.
Supplemental Material
Supplemental material, DS1_JVDI_10.1177_1040638719847510 for Infection of eight mesocarnivores in New Hampshire and Vermont with a distinct clade of canine distemper virus in 2016–2017 by David B. Needle, Vivien C. Burnell, Marίa J. Forzán, Edward J. Dubovi, Krysten L. Schuler, Chris Bernier, Nicholas A. Hollingshead, Julie C. Ellis, Brian A. Stevens, Patrick Tate, Eman Anis and Rebecca P. Wilkes in Journal of Veterinary Diagnostic Investigation
Acknowledgments
We thank W Cottrell, Northeast Wildlife Disease Cooperative (NWDC) Field Veterinarian, for facilitating NWDC members (NH Fish and Game, VT Fish and Wildlife) submission of cases for diagnosis.
Footnotes
Declaration of conflicting interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
Supplementary Material: Supplementary material for this article is available online.
ORCID iDs: David B. Needle  https://orcid.org/0000-0002-7215-5306
https://orcid.org/0000-0002-7215-5306
Marίa J. Forzán  https://orcid.org/0000-0002-0810-9911
https://orcid.org/0000-0002-0810-9911
References
- 1. Anis E, et al. Antigenic analysis of genetic variants of canine distemper virus. Vet Microbiol 2018;19:154–160. [DOI] [PubMed] [Google Scholar]
- 2. Anis E, et al. Phylogenetic analysis of the wild-type strains of canine distemper virus circulating in the United States. Virol J 2018;15:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Appel MJG, et al. Canine distemper epizootic in lions, tigers, and leopards in North America. J Vet Diagn Invest 1994;6:227–228. [DOI] [PubMed] [Google Scholar]
- 4. Bolt G, et al. Genetic diversity of the attachment (H) protein gene of current field isolates of canine distemper virus. J Gen Virol 1997;78:367–372. [DOI] [PubMed] [Google Scholar]
- 5. Cranfield MR, et al. Canine distemper in wild raccoons (Procyon lotor) at the Metropolitan Toronto Zoo. Can Vet J 1984;25:63–66. [PMC free article] [PubMed] [Google Scholar]
- 6. Cunningham MW, et al. Canine distemper epizootic in Everglades mink. J Wildl Dis 2009;45:1150–1157. [DOI] [PubMed] [Google Scholar]
- 7. Davidson WR, et al. Diseases diagnosed in gray foxes (Urocyon cinereoargenteus) from the Southeastern United States. J Wildl Dis 1992;28:28–33. [DOI] [PubMed] [Google Scholar]
- 8. Gehrt SD, et al. Pathogen dynamics and morbidity of striped skunks in the absence of rabies. J Wildl Dis 2010;46:335–347. [DOI] [PubMed] [Google Scholar]
- 9. Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010;59:307–321. [DOI] [PubMed] [Google Scholar]
- 10. Kapil S, et al. Canine distemper virus strains circulating among North American dogs. Clin Vaccine Immunol 2008;15:707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Katoh K, et al. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 2002;30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keller SM, et al. Canine distemper in an isolated population of fishers (Martes pennanti) from California. J Wildl Dis 2012;48:1035–1041. [DOI] [PubMed] [Google Scholar]
- 13. Loots AK, et al. Advances in canine distemper virus pathogenesis research: a wildlife perspective. J Gen Virol 2017;98:311–321. [DOI] [PubMed] [Google Scholar]
- 14. MacLachlan JN, Dubovi EJ, eds. Paramyxoviridae and pneumoviridae. In: Fenner’s Veterinary Virology. Boston, MA: Elsevier, 2017:327–356. [Google Scholar]
- 15. Martinez-Gutierrez M, Ruiz-Saenz J. Diversity of susceptible hosts in canine distemper virus infection: a systematic review and data synthesis. BMC Vet Res 2016;12:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pardo ID, et al. Phylogenetic characterization of canine distemper viruses detected in naturally infected dogs in North America. J Clin Microbiol 2005;43:5009–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pope JP, et al. Characterization of a novel canine distemper virus causing disease in wildlife. J Vet Diagn Invest 2016;28:506–513. [DOI] [PubMed] [Google Scholar]
- 18. Price MN, et al. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 2009;26:1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Riley MC, Wilkes RP. Sequencing of emerging canine distemper virus strain reveals new distinct genetic lineage in the United States associated with disease in wildlife and domestic canine populations. Virol J 2015;12:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roscoe DE. Epizootiology of canine distemper in New Jersey raccoons. J Wildl Dis 1993;29:390–395. [DOI] [PubMed] [Google Scholar]
- 21. Schumaker BA, et al. Canine distemper outbreak in pet store puppies linked to a high-volume dog breeder. J Vet Diagn Invest 2012;24:1094–1098. [DOI] [PubMed] [Google Scholar]
- 22. Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 1993;10:512–526. [DOI] [PubMed] [Google Scholar]
- 23. Trebbien R, et al. Wildlife reservoirs of canine distemper virus resulted in a major outbreak in Danish farmed mink (Neovison vison). PLoS One. 2014;9:e85598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Woolf A, Gremillion-Smith C. Pathologic findings in rabies-suspect, random-source, and accidentally killed skunks. J Am Vet Med Assoc 1986;189:1089–1091. [PubMed] [Google Scholar]
- 25. Woolf A, et al. Evidence of canine distemper virus infection in skunks negative for antibody against rabies virus. J Am Vet Med Assoc 1986;189:1086–1088. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS1_JVDI_10.1177_1040638719847510 for Infection of eight mesocarnivores in New Hampshire and Vermont with a distinct clade of canine distemper virus in 2016–2017 by David B. Needle, Vivien C. Burnell, Marίa J. Forzán, Edward J. Dubovi, Krysten L. Schuler, Chris Bernier, Nicholas A. Hollingshead, Julie C. Ellis, Brian A. Stevens, Patrick Tate, Eman Anis and Rebecca P. Wilkes in Journal of Veterinary Diagnostic Investigation