Abstract
Scrapie resistance or susceptibility in sheep is associated with single nucleotide polymorphisms (SNPs) at codons 136, 154, and 171 of the prion protein gene (PRNP). In addition, phenylalanine mutation at codon 141 has been recognized as a risk factor for atypical scrapie. In contrast, K222, D146, and S146 alleles confer genetic resistance to classical scrapie in goats. High-throughput genotyping technologies would provide significant benefits in scrapie eradication plans. The ability to resolve oligonucleotides varying in mass by less than a single nucleotide makes MALDI-TOF mass spectrometry (MS) a suitable platform for PRNP genotyping. We evaluated the commercial Myriapod scrapie kit (Diatech Pharmacogenetics), associated with a highly automated processing platform incorporating MALDI-TOF MS technology, to detect SNPs at codons 136, 154, 171, 141, and 222 of small ruminant PRNP. The Myriapod scrapie kit was accredited according to UNI CEI EN ISO/IEC 17025. We present the genotyping results of 10,960 sheep in Sicily and 1,822 goats in Sicily and Calabria (southern Italy) tested during 2017. We found a high frequency (43.9%) of the protective ARR allele in sheep and a promising 12.3% of the resistant K222 variant in goats. This efficient and high-throughput method is suitable for extensive PRNP genotyping, as requested in the European scrapie eradication plan.
Keywords: MALDI-TOF MS, PRNP genotyping, scrapie, single nucleotide polymorphisms, small ruminants
Scrapie is a neurodegenerative and fatal disease belonging to the group of transmissible spongiform encephalopathies (TSEs), or prion diseases, that affect sheep and goats. Scrapie in small ruminants is caused by an abnormal isoform (PrPSc) of the normal host-encoded cellular prion protein (PrPC), which accumulates in the central nervous system and peripheral tissues, causing clinical disease.17 The prion protein gene (PRNP) encoding for PrPC is highly polymorphic in sheep and goats.2 Resistance or susceptibility to scrapie in sheep has been strongly associated with single nucleotide polymorphisms (SNPs) at codons 136, 154, and 171 of the PRNP.11 PRNP alleles are conventionally reported using a 3-letter amino acid code. Five of these alleles are common and widespread among the sheep breeds (ARQ, ARR, AHQ, ARH, and VRQ); TRQ and ARK are observed more rarely. Scientific evidence indicates that the ARR allele, especially in homozygous individuals, is associated with resistance, whereas ARQ and VRQ confer highest susceptibility to classical scrapie. The other alleles have an intermediate effect, depending on PRNP genotype.3,11 Conversely, PRNP genotypes influence the resistance or susceptibility to atypical scrapie (Nor98) in a different way. The ARR allele gives no protection, whereas the AHQ and ARQ variants with phenylalanine (F) mutation at codon 141 have been associated with susceptibility.16 In contrast, K222, D146, and S146 alleles confer genetic resistance against classical scrapie strains known to occur naturally in the EU goat population.6 The protective effect for K222 has been shown to be greater than D146 and S146 in goats and ARR in sheep.6 On this basis, and with EU Decision 2003/100/EC,7 breeding programs to increase the frequency of the resistance-associated ARR allele in sheep populations have been introduced to minimize TSE risk in EU member states.
Since 2004, a large number of sheep samples have been genotyped in all European countries, and molecular genetic laboratories have applied different technologies with the aim of obtaining reliable, fast, and cost-effective methods for ovine PRNP genotyping. In Sicily, a national scrapie plan based on breeding for genetic resistance has been in force since 2004. Following our requests, Diatech Pharmacogenetics developed the Myriapod scrapie kit, a high-throughput assay based on matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) for genotyping at codons 136, 154, 171, 141, and 222 of the small ruminant PRNP gene.
We evaluated the performance of the kit by analyzing 10,960 healthy sheep within the genotyping program for scrapie resistance from Sicily, and 1,822 healthy goat samples from Sicily and Calabria (southern Italy) within the Italian Ministry of Health project “Scrapie control in goats through genetics.” All samples were collected during 2017. Peripheral blood samples were collected in vacuum vials with EDTA and stored at 4°C until DNA extraction. Genomic DNA (gDNA) was isolated (QIAamp 96 DNA, QIAcube HT kit, QIAcube HT; Qiagen, Hilden, Germany) according to the manufacturer’s protocol.
The Myriapod scrapie kit allows detection and identification, by MALDI-TOF MS, of the SNPs at codon 136, 141, 154, 171, and 222 of the PRNP gene associated with resistance or susceptibility to scrapie in small ruminants. This kit uses the MassARRAY system (Agena Bioscience, San Diego, CA) associated with single-base extension technology. The MassARRAY platform is simple, requires little operator input, and has high-throughput, thus allowing analysis of all of the mutations in a single run.
A 406-bp PRNP fragment encompassing codons 136, 141, 154, 171, and 222 was amplified from gDNA with specific primers included in the Myriapod scrapie kit. Amplification was performed in 5-µL reactions containing: 10× PCR buffer, 2 mM MgCl2 (final concentration), 0.5 mM of each dNTP, 1 U/reaction enzyme, 0.1 µM of each primer, and 2 µL of gDNA (1–100 ng/µL). Thermocycling conditions were as follows: hold 2 min at 95°C; heat step 30 s at 95°C, 30 s at 56°C, 60 s at 72°C for 45 cycles, and an elongation step of 5 min at 72°C. Unincorporated dNTPs were dephosphorylated by treatment with shrimp alkaline phosphatase (SAP), adding to each PCR reaction 2 µL of SAP mix, containing 1.53 µL of water, 0.17 µL of buffer, and 0.3 µL of SAP enzyme. The plate was incubated for 40 min at 37°C and subsequently for 5 min at 85°C to inactivate the enzyme. All reactions were prepared in 96-well plates and manually sealed with a cover film.
The iPLEX assay (Agena Bioscience) consists of a single-base extension reaction allowing the determination of, according to the mass revealed, the specific nucleotide added. The master mix for the iPLEX reaction was carried out starting from 0.56 µL of reaction buffer, 7–14 µM of each extension primer depending on the molecular mass, 0.2 µL of termination mix (Agena Bioscience), and 0.04 µL of Thermo Sequenase (Agena Bioscience). A total of 2 µL of master mix was added to the SAP-treated PCR products. The termination mix contained chemically modified dideoxynucleosides, which are chain-elongating inhibitors of DNA polymerase with specific known mass. This ensures single-base extension, avoiding nonspecific products derived from multiple base additions that can cause multiple peaks in the final spectrum. The multiplex reactions were incubated at 94°C for 30 s followed by 40 cycles of 5 s at 94°C, (5×) 5 s at 52°C, and 5 min at 80°C. The masses of the primers and the predicted products of their extension were then obtained (Table 1).
Table 1.
Mass of the extension primer and the expected masses of each assay included in the Myriapod scrapie kit (Diatech Pharmacogenetics).
| Codon | Assay | UEP mass | EXT1 |
EXT2 |
||
|---|---|---|---|---|---|---|
| Nucleotide | Expected primer mass | Nucleotide | Expected primer mass | |||
| 136 pos. 1 | Z01 | 4,657 | C | 4,904 | T | 4,984 |
| 136 pos. 2 | Z02 | 4,921 | G | 5,208 | A | 5,192 |
| 141 | Z03 | 5,491 | T | 5,762 | C | 5,778 |
| 154 | Z04 | 5,818 | G | 6,065 | A | 6,145 |
| 171 | Z05 | 5,179 | A | 5,451 | G | 5,467 |
| 171 | Z06 | 7,077 | G | 7,324 | T | 7,348 |
| 171 pos. 1 | Z07 | 6,463 | C | 6,710 | A | 6,734 |
| 171 pos. 2 | Z08 | 6,426 | G | 6,673 | A | 6,753 |
| 222 | Z09 | 4,510 | A | 4,837 | C | 4,797 |
pos. = position; UEP = unextended primer. Each SNP is identified by an assay name in order to be distinguished during the analysis. The expected nucleotides in each position are indicated by EXT1 and EXT2, and the respective masses are indicated in the “expected primer mass” column.
Before MALDI–TOF MS analysis, the product of reactions was diluted with 41 µL of water (MS plus; Diatech Pharmacogenetics) and then purified by incubating for 15 min (MassARRAY clean resin; Agena Bioscience) at room temperature on a rotating shaker. Volumes of the purified reactions (in nL) were automatically dispensed in a 96-well chip (SpectroCHIP, MassARRAY RS-1000 nanodispenser 6-pin; Agena Bioscience). Finally, the chip was read (MassARRAY Analyzer 4 (96) mass spectrometer; Agena Bioscience) and analyzed (Typer 4.0 software; Agena Bioscience). Both PCR and extension assays of the Myriapod scrapie kit were designed with MassARRAY Typer 4 assay designer software (Agena Bioscience).
The design was based on a proprietary algorithm of Agena Bioscience. The reference sequence of the PRNP (GenBank JX187539.1) and the relevant SNPs to be detected were introduced into the software. Taking into consideration the melting temperature, % cytosine (C) and guanine (G), and duplex formation, the software produces the best combination of PCR and EXT primers, which were used to produce the commercial Myriapod scrapie kit. The validation of each assay of the Myriapod scrapie kit was performed by testing 55 DNA samples. Sheep samples were previously genotyped at codons A136V, R154H, R171H, K171Q, and L141F using the accredited RT-PCR method (GenoScrapie Plus v.1.0; Real Gene Biotech, Reggio Calabria, Italy); goat samples underwent sequencing analysis. Genomic DNA was extracted from peripheral blood of sheep with suitable starting quantity (>10 ng/reaction).
Samples were previously characterized through quantitative PCR. Twenty-one samples were tested in order to define the functionality of each assay; the other 34 were characterized in a blind test in order to validate the Myriapod scrapie kit. The results obtained showed a concordance of 100% of the genotype with the PCR results.
The analytical sensitivity of the assay was defined by determining the minimum amount of DNA needed to detect the mutations. We used decreasing amounts of DNA (50 ng, 10 ng, 5 ng, and 1 ng) extracted from different blood samples. All of the samples tested confirmed the expected genotype regardless of the starting DNA amount, indicating that the sensitivity of the assay is 1 ng of template DNA. System reproducibility and repeatability have been evaluated from the data derived from 3 independent runs with the same amount of DNA. Genotyping results were reproducible for all assays and samples analyzed. Two different batches of PCR and extension primers were tested with the same DNA samples. Genotype results from the different lots were comparable. The Myriapod scrapie kit was accredited according to DM 25.11.2015 of the Italian Ministry of Health, within our official TSE laboratory according to UNI CEI EN ISO/IEC 17025. The method showed a sensitivity of 100%, a specificity of 100%, an accuracy of 100%, and a concordance (kappa) of 1.00 (95% confidence interval: 0.98–1.00) during the annual national proficiency tests of the Italian Public Health Institute performed in 2016.5
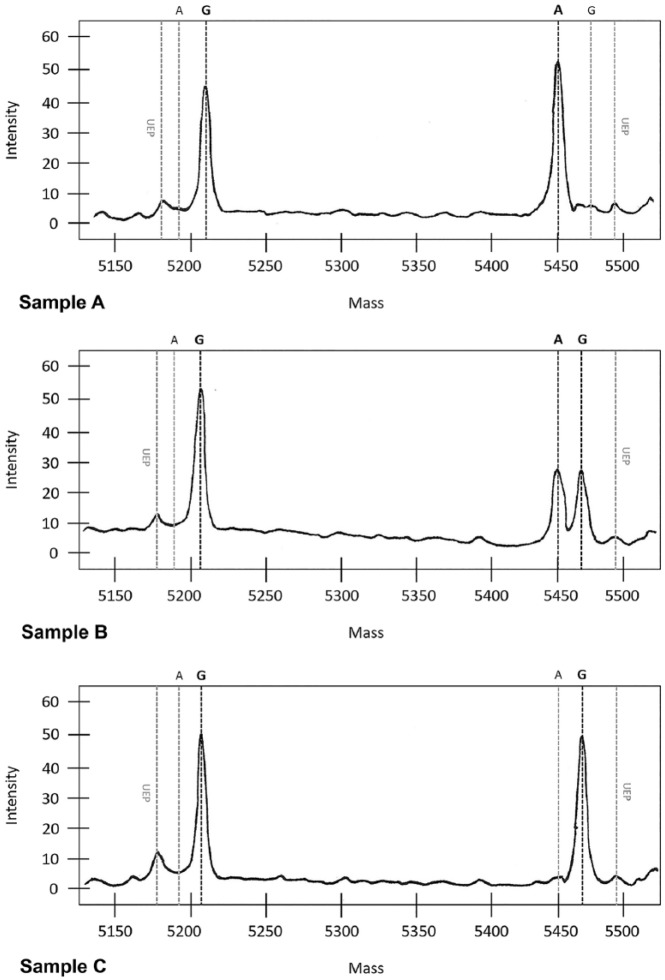
The primer extension reaction produced clearly interpretable spectra with balanced peak heights (Fig. 1). This allowed reliable identification of the PRNP genotypes corresponding to the SNPs interrogated by the corresponding primers. Approximately 2% of the samples needed to be retested. The main reason for retesting was poor gDNA quality or quantity, resulting in insufficient or null amplification; DNA was re-extracted.
Figure 1.
Spectra of codon 171. Samples A and C are homozygous samples (G and A, respectively); sample B is a heterozygous sample. In each spectrum, the unused primers remaining after the extension reaction (iPLEX) are indicated by UEP (unextended primer). The molecular mass, and hence the mass of the extension product, depends on the length of the primers (see Table 1 for details).
The method used allowed the identification of 7 different alleles: AHQ, ARH, VRQ, ARQ, ARR, ARK, and AFRQ, combined in 17 different genotypes (data not shown); the TRQ allele was not found. Allele frequencies ranged from 0.02% (ARK) to 47.3% (ARQ; Table 2). We found a low frequency (2.1%) of the very susceptible VRQ allele and a high frequency (43.9%) of the protective ARR. The AHQ and ARQ variants with phenylalanine (F) mutation at codon 141, associated with susceptibility to atypical scrapie (Nor98), had frequencies of 4.0% and 1.0%, respectively. The 3 most representative genotypes were ARR/ARQ (40.1%), ARQ/ARQ (24.7%), and ARR/ARR (20.6%; data not shown).
Table 2.
PRNP alleles and their frequencies (%) in sheep in Sicily.
| Allele | n | % |
|---|---|---|
| AHQ | 433 | 4.0 |
| ARH | 188 | 1.7 |
| VRQ | 228 | 2.1 |
| ARQ | 5,188 | 47.3 |
| ARR | 4,813 | 43.9 |
| ARK | 2 | 0.02 |
| AFRQ | 108 | 1 |
| Total | 10,960 | 100.0 |
Codon 222 had the expected mutation with lysine (K) instead of glutamine (Q) in 398 goats with a frequency of 12.3%. We found 348 (19.1%) Q/K heterozygous and 50 (2.8%) K/K homozygous goats (Table 3).
Table 3.
PRNP polymorphism on codon 222 and their frequencies (%) in goats in Sicily and Calabria, Italy.
| Codon 222 | n | % |
|---|---|---|
| Q/Q | 1,424 | 78.1 |
| Q/K | 348 | 19.1 |
| K/K | 50 | 2.8 |
| Total | 1,822 | 100.0 |
K222 frequency is 12.3%.
Many methods have been used for PRNP genotyping in small ruminants, including: PCR–restriction fragment length polymorphism typing,10 DNA strand conformation polymorphism detection by denaturing gradient gel electrophoresis,3 hybridization with allele-specific oligonucleotides,12 primer extension assay,20 amplification refractory mutation system,4 and MALDI-TOF MS.18 Among these techniques, MALDI-TOF MS nucleic acid analysis has several benefits. The technique is faster compared to conventional electrophoresis (a final mass spectrum can be obtained in <10 s), the results are absolute, thanks to the intrinsic property of the mass-to-charge ratio (m/z), and it is also automated; therefore, data can be obtained with little operator input.9 The entire analytical process of this multiplex assay (from DNA amplification to extension products) is performed in a single well, hence the risk of cross-contamination and volume loss is reduced and the number of samples per run is increased. For these reasons, the Myriapod scrapie kit has great potential for large-scale PRNP genotyping in sheep. In addition, it allows efficient screening of large numbers of goats for identification of animals carrying the codon K222 variant. Moreover, the possibility to expand the assay panels based on presence or absence of resistant alleles in the various European goat populations (e.g., codons 142, 146, and 211) makes the Myriapod scrapie kit suitable for future breeding programs for TSE resistance in all European goats. A universal large-scale small ruminant assay for scrapie resistance detection would include all PRNP polymorphisms linked with the disease according to breed and/or geographic regions.
A limitation to the practical application of this technique is that all of the implicated SNPs are located in the PRNP and, in the case of close SNPs, the extension primers designed to interrogate neighboring positions would overlap and likely interfere with each other.1 MALDI-TOF MS nucleic acid analysis has already been used successfully for high-throughput genotyping at codons 136, 154, and 171 in sheep in New Zealand,13 and for genotyping at codon 146 of caprine PRNP in Cyprus.1 A previous survey in Sicilian sheep on a limited number of reproductive animals showed an average frequency of 35.5%, 55.3%, and 4.6% of the ARR, ARQ, and VRQ haplotypes respectively.19 In our study, we found a higher frequency of the ARR resistant allele (43.9%) and a lower frequency of the susceptible ARQ and VRQ alleles (47.3% and 2.1%, respectively) in a large number of reproductive males within the Sicilian sheep population. In goats, we found a promising 12.3% of the resistant K222 variant, consistent with previous studies in Sicily14,15,19 and Calabria.8
The commercial Myriapod scrapie kit has been used within our TSE official laboratory since 2016. Our experience confirmed high accuracy, reproducibility, and repeatability of the results obtained, with reduced hands-on time. The hands-on time is calculated considering the operator turnaround time: PCR 30 min, SAP 10 min, and iPLEX 20 min (in total 1 h for a 96-well plate with 96 samples). On the other hand, considering the instrument time [2 h 20 min for PCR, 50 min for SAP, and 3 h for iPLEX performed on a thermocycler; 10 min to automatically transfer the 96-well plate (MassARRAY RS-1000 Nanodispenser 6-pin) on the SpectroCHIP, and 10 min to read the SpectroCHIP (MassARRAY Analyzer 4, 96)], the total time needed to obtain the complete profile for 96 samples with the Myriapod scrapie kit is 7 h 30 min. The estimated price for sample genotyping assay in our laboratory is ~15.00 euros or 16.96 USD and includes reagents, plastic ware, and SpectroCHIPs required for the PCR amplification and EXT assay preparation and analysis. Genomic DNA extraction cost is not included.
The sheep genotyping method used previously in our laboratory was based on reverse-transcription PCR (“ready for use” commercial kit) and, at the same cost per sample, it provided 5 independent assays (A136V, R154H, R171H, K171Q, and L141F) for each sample (1 per well), with an estimated hands-on time of 2 h for a plate with a maximum of 17 samples. Based on our experience, the Myriapod scrapie kit is confirmed to be an efficient and high-throughput method, suitable for extensive PRNP genotyping in the Italian small ruminant population, in accordance with the national scrapie eradication plan.
Footnotes
Declaration of conflicting interests: Enrico Gagliostro is an employee of Diatech Pharmacogenetics. The remainder of the authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Sergio Migliore  https://orcid.org/0000-0002-6685-7109
https://orcid.org/0000-0002-6685-7109
References
- 1. Arrizubieta MJ, et al. Design and validation of a high-throughput assay to detect codon 146 polymorphisms in the caprine prion protein gene. Anal Biochem 2009;393:229–233. [DOI] [PubMed] [Google Scholar]
- 2. Baylis M, McIntyre KM. Transmissible spongiform encephalopathies: scrapie control under new strain. Nature 2004;432:810–811. [DOI] [PubMed] [Google Scholar]
- 3. Belt PB, et al. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J Gen Virol 1995;76(Pt 3):509–517. [DOI] [PubMed] [Google Scholar]
- 4. Buitkamp J, Semmer J. A robust, low- to medium-throughput prnp genotyping system in sheep. BMC Infect Dis 2004;4:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chiappini B, et al. Identification of prion protein genotype in sheep: 11 years of proficiency tests in Italy. Accred Qual Assur 2019;24:49–55. [Google Scholar]
- 6. EFSA Panel on Biological Hazards. Genetic resistance to transmissible spongiform encephalopathies (TSE) in goats. EFSA J 2017;15:4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. European Commission. Commission Decision (2003/100/EC) of 13 February 2003 laying down minimum requirements for the establishment of breeding programmes for resistance to transmissible spongiform encephalopathies in sheep. Off J Eur Commun 2003; L041:41–45. [Google Scholar]
- 8. Fantazi K, et al. Analysis of differences in prion protein gene (PRNP) polymorphisms between Algerian and Southern Italy’s goats. Ital J Anim Sci 2018;17:578–585. [Google Scholar]
- 9. Griffin TJ, Smith LM. Single-nucleotide polymorphism analysis by MALDI-TOF mass spectrometry. Trends Biotechnol 2000;18:77–84. [DOI] [PubMed] [Google Scholar]
- 10. Hunter N. Natural transmission and genetic control of susceptibility of sheep to scrapie. Curr Top Microbiol Immunol 1991;172:165–180. [DOI] [PubMed] [Google Scholar]
- 11. Hunter N. Molecular biology and genetics of scrapie in sheep. In: Piper L, Ruvinsky A. eds. The Genetics of Sheep. UK: CAB International, 1997;225–240. [Google Scholar]
- 12. Ishiguro N, et al. Rapid analysis of allelic variants of the sheep PrP gene by oligonucleotide probes. Microbiol Immunol 1998;42:579–582. [DOI] [PubMed] [Google Scholar]
- 13. Lee MA, et al. Distribution of prion protein genotypes in breeds of sheep in New Zealand. N Z Vet J 2007;55:222–227. [DOI] [PubMed] [Google Scholar]
- 14. Migliore S, et al. Biodiversity and selection for scrapie resistance in goats: genetic polymorphism in “Girgentana” breed in Sicily, Italy. Small Rumin Res 2015;125:137–141. [Google Scholar]
- 15. Migliore S, et al. Cross-sectional study of PRNP gene in two native Sicilian goat populations in Italy; a relation between prion gene polymorphisms and scrapie incidence. J Genet 2017; 96:319–325. [DOI] [PubMed] [Google Scholar]
- 16. Moum T, et al. Polymorphisms at codons 141 and 154 in the ovine prion protein gene are associated with scrapie Nor98 cases. J Gen Virol 2005;86:231–235. [DOI] [PubMed] [Google Scholar]
- 17. Prusiner SB. Molecular biology of prion diseases. Science 1991;252:1515–1522. [DOI] [PubMed] [Google Scholar]
- 18. Storm N, et al. MALDI-TOF mass spectrometry-based SNP genotyping. Methods Mol Biol 2003;212:241–262. [DOI] [PubMed] [Google Scholar]
- 19. Vitale M, et al. Scrapie incidence and PRNP polymorphisms: rare small ruminant breeds of Sicily with TSE protecting genetic reservoirs. BMC Vet Res 2016;12:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zsolnai A, et al. Detection of single-nucleotide polymorphisms coding for three ovine prion protein variants by primer extension assay and capillary electrophoresis. Electrophoresis 2003;24:634–638. [DOI] [PubMed] [Google Scholar]