Abstract
Secondary analysis of electronic health records for clinical research faces significant challenges due to known data quality issues in health data observationally collected for clinical care and the data biases caused by standard healthcare processes. In this manuscript, we contribute methodology for data quality assessment by plotting domain-level (conditions (diagnoses), drugs, and procedures) aggregate statistics and concept-level temporal frequencies (i.e., annual prevalence rates of clinical concepts). We detect common temporal patterns in concept frequencies by normalizing and clustering annual concept frequencies using K-means clustering. We apply these methods to the Columbia University Irving Medical Center Observational Medical Outcomes Partnership database. The resulting domain-aggregate and cluster plots show a variety of patterns. We review the patterns found in the condition domain and investigate the processes that shape them. We find that these patterns suggest data quality issues influenced by systemwide factors that affect individual concept frequencies.
Keywords: Electronic Health Records, Cluster Analysis, Data Accuracy
Introduction
Secondary reuse of observational health data for research is increasing in importance and popularity as electronic health data become more available and analytic methods become more powerful. However, analysis of electronic health records (EHR) continues to face many social and technical challenges, including data inaccuracy, incompleteness, and biases implicit in the healthcare recording process [1–5]. Since observational clinical data are sourced from disparate systems not designed for research purposes, researchers consuming these data must clearly understand the nuances of their data. Due to the complexity of interactions, ongoing development, and technical challenges of these systems, events that impact data integration can occur without awareness from data managers. Further, experts knowledgeable of the intricacies of the data may not be available to researchers, especially in the context of shared data in collaborative research networks, such as the Observational Health Data Sciences and Informatics (OHDSI) [6].
EHR data quality is receiving increased attention for its effects on secondary analysis for clinical research and population health informatics [7–10]. Temporal trend analysis is a common method of assessing data quality. Kahn’s harmonized data quality framework includes temporal plausibility metrics, such as comparing data value density over time against expected values [7]. Hall’s database guidelines recommend counting frequency of records and field occurrences over time to identify blocks of missing data or changes in data volume [8]. Brown et al demonstrate visually exploring trends of intrasite and intersite frequency to detect data anomalies [9]. ACHILLES performs temporal analysis of concepts in Observational Medical Outcomes Partnership (OMOP) databases to assess month-to-month stability of counts and flags changes that exceed thresholds [6]. These methods enable detection of data quality issues in individual variables but do not detect systemic issues that may affect multiple variables.
The Columbia University Irving Medical Center (CUIMC) Clinical Data Warehouse (CDW) formatted using the OMOP Common Data Model (CDM) (herein referred to as the OMOP database) contains observational data on nearly 60,000 concepts. Individual analysis of 60,000 concepts would be prohibitively time consuming. Although population prevalence of clinical concepts can change with time, we hypothesize that common patterns of temporal trends in EHRs reveal systemic factors that influence observational data and data quality.
In this manuscript, we present methods of detecting systemic, population-level data quality issues in electronic health records by analyzing domain-level aggregate statistics and identifying common temporal patterns across concepts. We apply these methods to the CUIMC OMOP database and infer and investigate system wide processes that affect the data quality.
Methods
Data Source
This study received institutional review board approval with waiver for obtaining informed consent. The CUIMC CDW contains records from multiple sites and was converted to OMOP CDM v5.1 in March 2018. Diagnoses (“conditions” in OMOP nomenclature), drugs, procedures, and visit types were collected from CUIMC’s OMOP database from the condition-occurrence, drug_exposure, procedure_occurrence, and visit_occurrence tables, respectively. We analyze conditions, drugs, and procedures to provide multiple views of patient data and visit types to indicate clinical service capacity.
Conceptual Framework
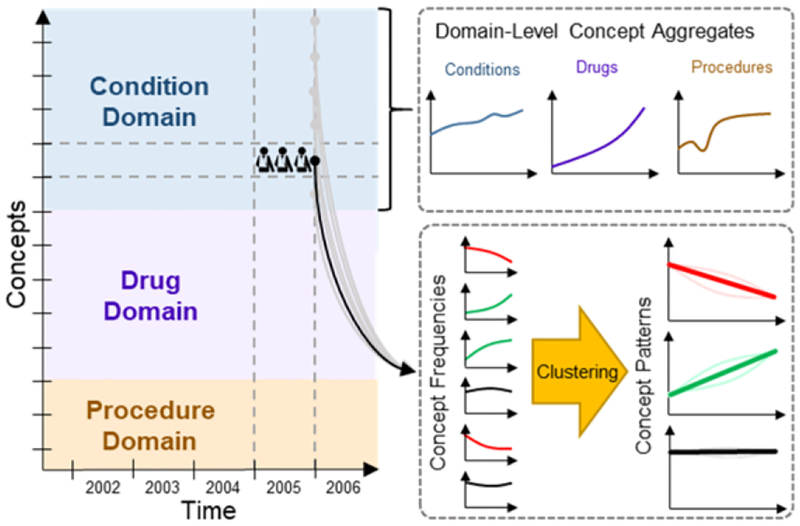
Figure 1 shows our conceptual framework for analysis. There are four entities: patients, concepts, domains, and time. Patient health is characterized in the OMOP database by individual concepts, which are categorized by domains (e.g., conditions, drugs, and procedures). For each concept, we count the number of patients in each year and plot the counts over time. Data captured in EHRs are affected by population health status, health service capacity, medical coding protocols, and operation of clinical databases. These parameters change over time, as reflected in the counts and frequency of concepts.
Figure 1–
Conceptual framework for analysis.
Concept Prevalence
We assume that population health is consistent with only minor changes annually and gradual changes over many years [11]. Correspondingly, we assume the annual frequency of most clinical concepts to be relatively stable over time if the health system is stable. Observed fluctuations in concept frequencies may have causes intrinsic to the concept (e.g., outbreaks or new interventions) or extrinsic to the concept (e.g., expansion of clinical services). We measured yearly counts and frequencies of concepts from the OMOP database. We defined counts as the number of unique patients with the concept per calendar year. We defined frequency as the count divided by number of patients in that year. We estimated the number of patients per year from the number of unique patient identifiers associated with at least one condition, drug, or procedure per year.
Domain-Level Concept Aggregates
Given a stable healthcare system, we hypothesize the domain-level aggregated metrics are highly stable over time. Fluctuations in these metrics may reflect changes to healthcare services or the EHR recording process. To identify global trends in each domain, we analyzed the yearly patient count and yearly aggregated statistics across concepts in condition, drug, and procedure domains using the domain of each concept as defined in the OMOP standard vocabulary to aggregate concepts. We analyzed four aggregated statistics across each domain per year: 1) the total patient counts across concepts; 2) the count-per-capita (total patient count divided by number of patients), indicating the average number of conditions, drugs, or procedures per patient; 3) the number of unique concepts; and 4) the mean frequency across concepts.
Clustering of Concept Patterns
Analyzing thousands of individual concept frequencies would be overwhelming and difficult to differentiate between changes in the population prevalence versus changes affecting the recorded prevalence in EHRs. To identify common behavior patterns with similar trends among concepts, we performed K-means clustering [12] within each domain. Each concept’s annual frequencies were normalized to unit magnitude and treated as vectors. We used concept frequency instead of count to normalize against population changes. We normalized frequencies to unit magnitude to match trends with similar proportional changes. Annual frequencies from calendar years 1985 to 2017 were included; we excluded 2018 since data was not yet available for the entire year. We excluded concepts with maximum frequency below 0.01% of the population to reduce noise from infrequent concepts. We performed K-means clustering on the normalized frequency vectors. We manually selected K by clustering each domain with different levels of K (4, 6, 8, 10, 12, 15, 20, and 25) and selecting the smallest K before intracluster variance ceased to improve substantially.
Post-hoc Analyses
To determine if the clustered concept patterns reveal systemic factors (e.g., changes in health service capacity or medical coding protocols) that affect concept frequency in the database, we manually reviewed the domain- and concept-level patterns, arriving at hypotheses on the causes of several patterns. We performed post-hoc analyses to validate these hypotheses.
Results
As of March 2018, the CUIMC OMOP database contains EHR records on 5,368,414 patients, covering 59,583 concepts, including 18,399 conditions, 18,691 drugs, and 22,787 procedures spanning October 1985 to March 2018.
Domain-Level Concept Aggregates
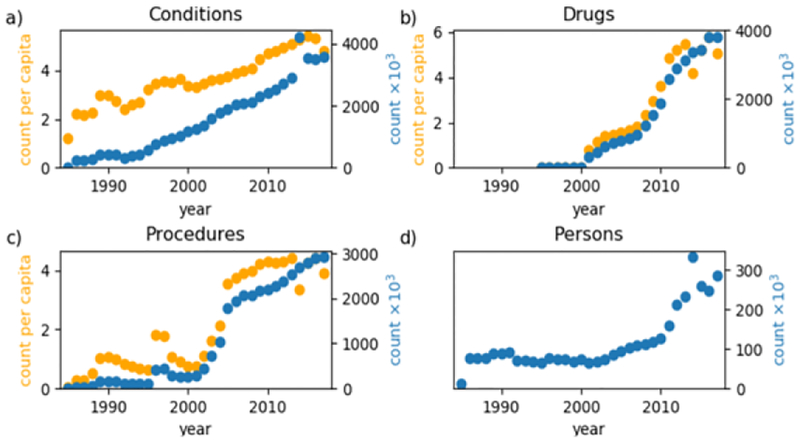
Figure 2 shows plots of the domain-level count-per-capita and total counts of a) conditions, b) drugs, c) procedures, and d) people per year. Patient counts are relatively flat between 1986–2001, except for a slight depression from 1992–1995, and grows steadily from 2001–2010 and rapidly after 2010, with a spike in 2014. The condition domain total count and count-per-capita increase nearly linearly over time. The condition count-per-capita is slightly unstable before 2000 but steadily increases after 2000, peaking in 2015 and dropping in 2016–2017. The total count of conditions spikes in 2014. Drug data is nearly non-existent before 2001 but steadily increases until it peaks in 2013. The drug count-per-capita drops substantially in 2014 and fluctuates through 2017. The procedure domain is the most unstable. Procedures display notable growth in 1989, a large spike in 1996–1997, rapid increase from 2001–2005 and steady increase thereafter. Similar to drug count-per-capita, procedure count-per-capita drops in 2014 and fluctuates through 2017.
Figure 2–
Total count (blue) and count-per-capita (orange) of a) conditions, b) drugs, c) procedures and d) people per year.
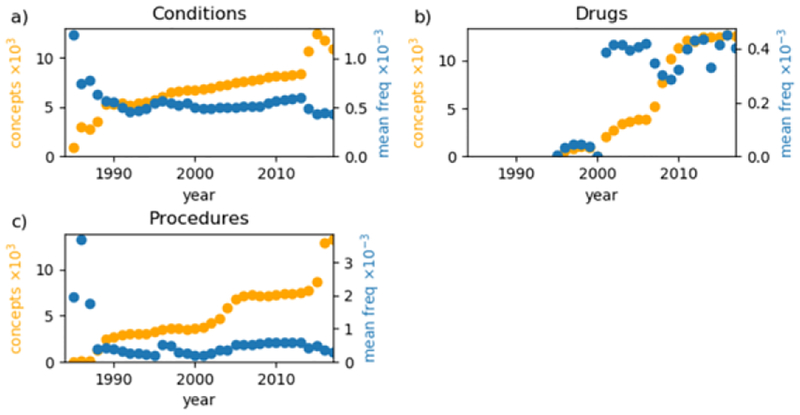
Figure 3 shows the annual number of distinct concepts and the mean frequency of patients across concepts for each domain. The condition domain stably grows from 1989–2013 but fluctuates thereafter. The drug domain is relatively stable from 2001–2006 but fluctuates thereafter. In the procedure domain, the number of concepts stably grows from 1989 to 2001, grows quickly to 2005, slowly grows through 2014, and quickly grows again. The mean frequency of procedure concepts has similar periods of behavior with an additional sharp increase in 1996.
Figure 3–
Count of unique concepts (orange) and the mean frequency of concepts (blue) per year for a) conditions, b) drugs, and c) procedures.
Concept-Level Analyses
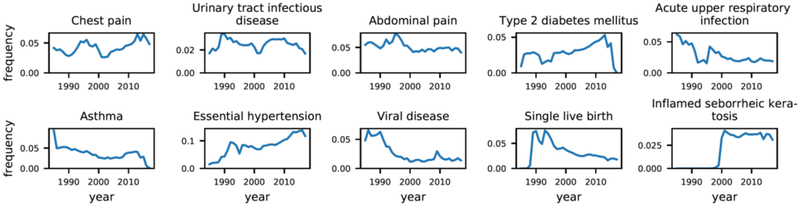
Due to space limitations, we primarily present and discuss the results for concept-level analyses in the condition domain and exclude drug and procedure results. Figure 4 plots annual frequencies for the ten conditions with the highest total counts across all years, showing multiple remarkable behaviors. Type 2 diabetes mellitus and asthma both begin to drop in 2014 and are nearly zero by 2017. Type 2 diabetes mellitus and acute upper respiratory infection both exhibit abrupt depressions from 1992–1995. Inflamed seborrheic keratosis is nearly zero until 1999 but rapidly increases and plateaus after 2001. Chest pain and abdominal pain both increase in frequency from roughly 1990–1995 and decrease from 1995–2000.
Figure 4–
Example annual concept trends for the ten most prevalent conditions.
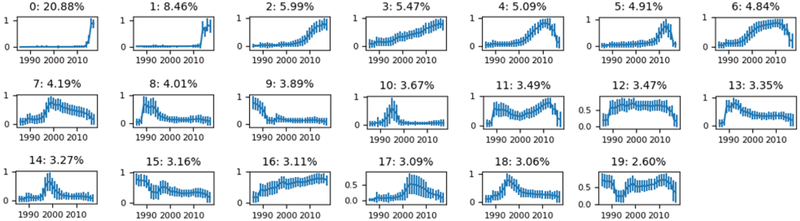
K-means clustering was performed for conditions (K=20). Figure 5 shows plots of cluster centroids (i.e., the temporal pattern) for condition concepts. The clusters are sorted in descending order of the number of concepts belonging to each cluster. The two largest condition clusters have nearly zero frequency until a rapid increase in 2014. Clusters 4, 5, 6, 11, 12, and 19 drop in frequency starting around 2014. Cluster 12 is the only cluster with constant frequency over a large time period but only contains 3.47% of condition concepts. Clusters 3 and 16 increase almost linearly with time. Other clusters display patterns that are more dynamic. Clusters 7, 8, 10, 13, 14, 17, and 18 exhibit peaks at different times. Condition cluster 11 decreases from 1989–1999 and then increases until 2014.
Figure 5–
K-means clusters for conditions. Plots show cluster centroids (the cluster trend over time) with standard deviation across concepts as error bars (intracluster vaiance). Subplot titles show cluster labels and the percent of concepts belonging to each cluster.
Post-hoc Analyses of Trends
To demonstrate that the above methods reveal systemic data quality issues, we manually reviewed the domain- and concept-level patterns, hypothesized the causes of several patterns, and performed post-hoc analyses to test these hypotheses.
Condition clusters 0 and 1 behave like step functions with a rapid increase in 2014 while many other clusters begin to decrease around the same time. The number of condition concepts (Fig. 2a) also suddenly increases in 2014. We hypothesized that these patterns were caused when the health systems converted from using one medical coding system to another, e.g., from ICD9CM to ICD10CM. To investigate this, we analyzed the composition of medical coding systems (i.e., OMOP source vocabularies) contributing to these clusters. Table 1 shows the source vocabulary composition of condition clusters 0 and 1 compared against the aggregate of all other trends. Condition clusters 0 and 1 are composed of 93.9% and 92.0% ICD10CM source concepts, respectively. The remaining clusters are composed of 83.2% ICD9CM concepts.
Table 1–
Source vocabulary composition of condition clusters.
| Vocabulary ID | Cluster 0 | Cluster 1 | Remaining clusters |
|---|---|---|---|
| SNOMED | 0.2% | 1.6% | 0.0% |
| ICD10CM | 93.9% | 92.0% | 16.8% |
| ICD9CM | 6.0% | 6.4% | 83.2% |
The domain-level count of condition concepts displays an abrupt depression from 1992–1995 (Fig. 1a). Condition clusters 9, 15, and 19 also display similar depressions from 1992–1995, but this behavior is not noticeable in other clusters. Since the behavior was isolated to a few trends, we hypothesized that this depression was caused by changes affecting specific service types (e.g., temporary loss of data or reduction of services) which cover the conditions in these clusters.
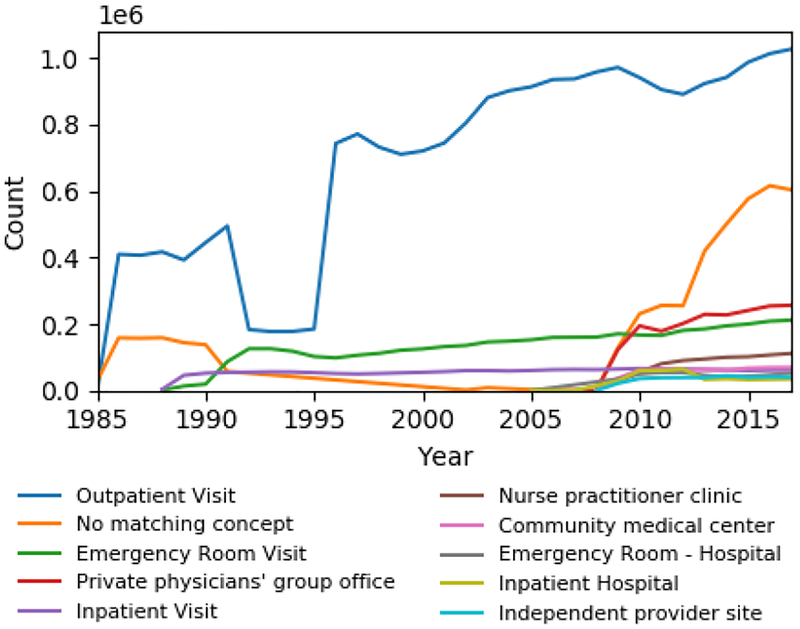
To investigate whether changes in health services occurred, we analyzed the number of patient visits per year stratified by visit type (e.g., inpatient, outpatient, etc.) from the OMOP visit-occurrence table. Figure 6 shows plots of the annual counts for the 10 most frequent visit types, which account for 97.3% of visits. We excluded 17 less frequent visit types, including cardiology clinic, general surgery clinic, community clinic, etc., from the figure for visual clarity. Outpatient visit is the most frequent type and exhibits a large depression from 1992–1995. The other visit types do not display similar depressions. Inpatient visits appear in 1989 and are very stable throughout the years. Emergency room visits appear in 1989 with unstable growth until 1995 and stable growth thereafter. 11.7% of visits did not have a specified visit type (No matching concept).
Figure 6–
Annual number of visits for the 10 most common visit concepts.
Discussion
Concept Prevalence
Reviewing plots of annual frequencies of individual concepts, we found that the large majority of concepts exhibited a variety of unstable behaviors. Figure 1 shows a sample of plots, including the ten most prevalent conditions. We show the most frequent conditions because we assumed they would generally behave more stable than less frequent conditions. If frequencies of clinical terms in the EHR are representative of general population prevalence, then the behavior of terms in the EHR should reflect the behavior of the clinical concepts in the population. We should expect relative stability from the conditions in Figure 1, but from this small sample, it is immediately apparent that concepts exhibit a wide variety of behaviors in the EHR. Most of these fluctuations are too dramatic to be reflective of real health changes in the population. For example, Type 2 diabetes mellitus and asthma fall to nearly 0% frequency in 2015 while inflamed seborrheic keratosis was nearly 0% until 1999 but plateaus from 2000 onward. These patterns suggest underlying changes in the usage of medical coding systems (e.g., changing from ICD9CM to ICD10CM). Type 2 diabetes mellitus and acute upper respiratory infection both have abruptly depressed frequencies from 1992–1995. Chest pain and abdominal pain have similarly shaped growth and decline patterns in the 1990s.
Domain-Level Analysis
The domain-level aggregate plots show high-level changes observable in the EHR. Figure 1 shows the annual number of patients, total patient count across concepts, and the count of concepts per capita for each domain. Some of the patterns observed in individual concepts also exist at the domain-level. From 1992–1995, the number of patients abruptly drops relative to surrounding years, corresponding to decreased condition counts. This suggests that the dips in type 2 diabetes mellitus and acute upper respiratory infection were caused by system level events, potentially affecting many conditions. Drug counts were practically non-existent before 2001 and almost linearly increased after 2001, indicating that drug data was not available before 2001. Changes in the annual number of unique concepts and the mean patient frequency across concepts (Fig. 2) may suggest changes in the usage of medical coding systems. The increased number of concepts in the condition domain from 2014–2015, in the drug domain from 2007–2011, and in the procedure domain from 2002–2005 and 2015–2017 all suggest that changes occurred in the medical coding system.
Concept-Level Analysis
As discussed above, many concept frequency curves display fluctuations too dramatic to be caused by changes in population prevalence. We applied K-means clustering to find groups of concepts that have similar behavioral patterns. Clusters contain hundreds to thousands of concepts, so their average frequency curves are minimally influenced by true changes in individual concepts’ population prevalences. These cluster patterns may suggest changes in the medical system that influence the clinical recording process, including service capacity changes (e.g., expansion of medical system) or events that affect patient data records (e.g., deployment of new clinical databases). In Figure 5, various patterns among condition concepts can be easily identified along with the proportion of concepts that follow the pattern. A small number of concepts remain stable and constant over time (e.g., condition cluster 12, 3.47% of conditions), and the stability only lasts for a period (1989–2013). The majority of concepts are stable only during certain time windows, and these windows vary by cluster.
Several clusters have sharp increases that correspond with decreases in other clusters. Clusters 0 and 1 increase while clusters 4, 5, 6, 11, 12, and 19 drop around 2014. We hypothesized that these patterns were caused by changes in the medical coding systems in 2014. To test this hypothesis, we analyzed the composition of source vocabularies among these clusters to determine if these clusters are primarily represented by different vocabularies (Table 1). Indeed, clusters 0 and 1 are composed of over 90% ICD10CM concepts, while all other clusters are cumulatively composed of 83.2% ICD9CM concepts, supporting the hypothesis above. Similar patterns were observed in the procedure domain associated with changes from ICD9Proc to ICD10PCS in 2016 (data not shown).
Condition clusters 9, 15, and 19 exhibit a marked drop in frequency between 1992–1995, corresponding with dips in domain-level counts (Fig. 2a). Since this behavior was not observed among most other clusters, we hypothesized that this depression was isolated to specific sites or services. To test this, we analyzed annual visit type occurrences (Fig. 5). Outpatient visits display a similar depression between 1992–1995 while the other visit types behave stably during this time, suggesting that clusters 9, 15, and 19 are primarily composed of conditions observed in outpatient settings. Inpatient visit and emergency room visit types both begin in 1989, corresponding with the opening of the Milstein Hospital Building of Presbyterian Hospital. A flurry of other visit types, including private physicians’ group office, nurse practitioner clinic, and community medical center, begin to appear in 2009, suggesting that the network of clinical data sources connected to the clinical data warehouse was expanding to more sites.
Limitations
There are a number of limitations to our methods. While clustering the annual concept frequencies reveals patterns of behavior among the concepts, the clusters on their own do not provide meaningful insight for the causes of these patterns. For some patterns, we were able to generate hypotheses and corroborate the causes by investigating other information sources (e.g., visit types and source vocabularies) to reveal systemic forces behind these patterns. However, many patterns are unexplained, and we cannot infer causality for the hypothesized causes of patterns as we did not apply statistical analyses. Even without causal explanations, these methods still allow researchers to identify time frames of data instability to exclude from their analyses or interpret with caution. These methods do not produce quantitative, scalar, and invariant results, which are desired characteristics of data quality metrics to allow comparison of data quality statistics across data sets and time [13]. The domain- and concept-level analyses were performed at annual intervals, which may be too coarse to detect short data aberrations. However, the method should perform well with shorter intervals.
The selected number of clusters for K-means clustering was arbitrary. The choice of K influences the resulting clusters and, thus, the interpretation of results. We attempted to use the elbow method [14] to guide our choice, but the intercluster sum of squared differences had broad-shouldered changes over K in all domains (data not shown). This indicates the presence of many small clusters or many concepts may not follow common patterns, thus, there may not exist an optimal choice for K.
We determined the annual number of patients based on observations in the condition, drug, and procedure tables and subsequently used this patient count in calculating concept frequencies of those conditions, drugs, and procedures. This introduces bias into the calculation of concept frequency. Ideally, the patient population should be identified externally from the variables undergoing analysis. The OMOP CDM contains observation_periods which should identify the start and end dates when the patient was observed in the database. However, in our database, 66.6% of patients have missing start dates, which may indicate an error in the extract-transform-load process, which converts data from the clinical data warehouse to the OMOP CDM. Also, we assume that each patient has a single unique identifier. This may not be true if patients have duplicate registrations within the CDW.
Developing this analysis on an OMOP database confers both advantages and disadvantages. By analyzing the transformed database, we lose some ability to investigate data provenance and sources of data quality issues. However, developing this analysis on a widely adopted CDM allows other institutions to immediately benefit and learn from these results. Although other data models will not be able to replicate this analysis directly, the methods are simple to implement in other systems.
Conclusions
We contribute methods for analyzing EHR data quality aspects (e.g., temporal plausibility, consistency, etc.) for secondary analysis using domain-level aggregate statistics and clustering the annual concept frequency trends to find common temporal patterns across concepts. These patterns may indicate EHR data quality issues caused by operational or system-wide factors that affect multiple concepts simultaneously.
Acknowledgements
This study was supported in part by grants OT3TR002027 from NCATS and R01LM009886-08A1 from NLM.
References
- [1].Hripcsak G, and Albers DJ, Next-generation pheno-typing of electronic health records, J Am Med Inform Assoc 20 (2013), 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Platt R, and Lieu T, Data enclaves for sharing information derived from clinical and administrative data, JAMA (2018), E1–E2. [DOI] [PubMed] [Google Scholar]
- [3].Botsis T, Hartvigsen G, Chen F, and Weng C, Secondary use of EHR: data quality issues and informatics opportunities, Summit on Translat Bioinforma 2010 (2010), 1–5. [PMC free article] [PubMed] [Google Scholar]
- [4].Kharrazi H, Wang C, and Scharfstein D, Prospective EHR-based clinical trials: the challenge of missing data, J Gen Intern Med 29 (2014), 976–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kharrazi H et al. , Comparing population-based risk-stratification model performance using demographic, diagnosis and medication data extracted from outpatient electronic health records versus administrative claims, Med Care 55 (2017), 789–796. [DOI] [PubMed] [Google Scholar]
- [6].Hripcsak G et al. , Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers, Stud Health Technol Inform 216 (2015), 574–578. [PMC free article] [PubMed] [Google Scholar]
- [7].Kahn MG et al. , A harmonized data quality assessment terminology and framework for the secondary use of electronic health record data, EGEMS (Wash DC) 4 (2016), 1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hall GC et al. , Guidelines for good database selection and use in pharmacoepidemiology research, Pharmacoepidemiology and Drug Safety 21 (2012), 1–10. [DOI] [PubMed] [Google Scholar]
- [9].Brown J, Kahn M, and Toh S, Data quality assessment for comparative effectiveness research in distributed data networks, Med Care 51 (2013), S22–S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kharrazi H et al. , A proposed national research and development agenda for population health informatics: summary recommendations from a national expert workshop, J Am Med Inform Assoc 24 (2017), 2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].van Oostrom SH et al. , Time trends in prevalence of chronic diseases and multimorbidity not only due to aging: data from general practices and health surveys, PLoS One 11 (2016), e0160264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hartigan JA, and Wong MA, Algorithm AS 136: a K-means clustering algorithm, Journal of the Royal Statistical Society. Series C (Applied Statistics) 28 (1979), 100–108. [Google Scholar]
- [13].Johnson SG, Speedie S, Simon G, Kumar V, and Westra BL, Application of an Ontology for Characterizing Data Quality for a Secondary Use of EHR Data, Appl Clin Inform 07 (2016), 69–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tibshirani R, Walther G, and Hastie T, Estimating the number of clusters in a data set via the gap statistic, Journal of the Royal Statistical Society: Series B (Statistical Methodology) 63 (2001), 411–423. [Google Scholar]