Sir,
Fosfomycin is increasingly used to treat infections caused by MDR bacteria.1 Fosfomycin acts by inhibiting UDP-N-acetylglucosamine enolpyruvyl transferase (murA), which prevents the formation of N-acetylmuramic acid, an essential component of peptidoglycan.1 Although resistance to fosfomycin is still low in Escherichia coli, the acquisition of fosA may reduce future activity of fosfomycin to treat infections caused by E. coli.2 FosA is a glutathione transferase that inactivates fosfomycin through catalysing the addition of glutathione. fosA genes are often present in the chromosome of Klebsiella pneumoniae, but not in the chromosome of E. coli.2,3Klebsiella variicola is closely related and often misidentified as K. pneumoniae.4 While horizontal spread of fosA has been demonstrated in vitro,5 we here provide evidence for in vivo fosA transmission from K. variicola to E. coli, resulting in development of fosfomycin resistance.
The Medical Research Ethics Committee of the University Medical Center Utrecht confirmed that the Medical Research Involving Human Subjects Act does not apply to this study (reference number WAG/mb/18/027282). We were not able to obtain informed consent because the patient died a few years ago. All information including gender, age, dates and medical history that was not directly clinically relevant has been omitted to protect the privacy of the patient.
An aged patient had a suspicion of chronic endovascular infection of their aortic bifurcation graft, which the patient received after an acute aortic aneurysm 22 years earlier. The patient had suffered from recurrent episodes of sepsis, with blood cultures yielding Propionibacterium spp., K. variicola, Citrobacter koseri and Pseudomonas aeruginosa, as determined by MALDI-TOF MS. Positron emission tomography (PET)-CT findings were compatible with prosthetic graft infection. The patient subsequently developed septic shock with E. coli bacteraemia without a clear source of infection that was treated successfully with intravenous ceftriaxone. The isolate was resistant to amoxicillin/clavulanic acid and ciprofloxacin that had been used to suppress chronic infection, prompting the addition of oral fosfomycin at 3 g every 48 h. Seven months later, while still using fosfomycin, the patient developed spondylodiscitis. Blood cultures drawn at the time isolated E. coli with an identical resistance pattern, except being resistant to fosfomycin. Fosfomycin was discontinued and the patient received a prolonged course of ceftriaxone.
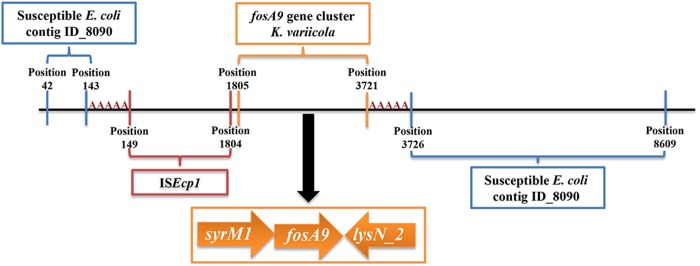
Fosfomycin susceptibility, determined by agar dilution according to CSLI guidelines,6 demonstrated a rise in the MIC from 2 mg/L for the initial E. coli isolate to >1024 mg/L for the second E. coli isolate. WGS revealed five SNP differences between E. coli isolates in the core genome, based on core genome MLST (cgMLST) analysis.7 Yet, the second E. coli isolate has a 3573 bp insertion consisting of ISEcp1, a fosA gene we named fosA9 as the next available number according to NCBI, syrM1 and lysN2. The insertion is flanked by 5 bp DRs (AAAAA) suggesting mobilization of this fosA9 gene cluster by ISEcp1 (Figure 1).8 Genes other than fosA9 responsible for fosfomycin resistance were not found. At the time of the first E. coli sepsis episode, six K. variicola had been isolated from rectum swabs and blood cultures over a period of 20 months (Table S1, available as Supplementary data at JAC Online). cgMLST analysis revealed a maximum of 16 SNP differences between K. variicola isolates.7 The same cluster as above containing fosA9, without the mobile genetic element ISEcp1, was identified in the K. variicola isolates, suggesting K. variicola to be the source of fosA9 acquired by E. coli (Figure 1). fosA genes were not identified in other clinical isolates from this patient. Sequence information of all isolates has been deposited in the European Nucleotide Archive (ENA) under project number PRJEB32329.
Figure 1.
Schematic representation of the contig (ECO-BAB-IMI-103297_P-ACH-BAB-IMI-103242_1528359160_131_length_8653_cov_18.1163_ID_8928, 8653 bp) in the fosfomycin-resistant E. coli isolate containing a fosA9 gene cluster originating from a K. variicola isolate. The ISEcp1-syrM1-fosA9-lysN2 region is flanked by 5 bp DRs (AAAAA), suggesting mobilization from K. variicola by ISEcp1. Upstream and downstream sequences of the insertion region align to contig ECO-BAB-IMI-103298_P-ACH-BAB-IMI-103242_1528359160_92_length_16411_cov_29.2905_ID_8090 from the first susceptible E. coli isolate. Sequence information of complete genomes of all isolates and separate sequences of the relevant contigs (containing fosA9 in E. coli and K. variicola, and ECO-BAB-IMI-103298_P-ACH-BAB-IMI-103242_1528359160_92_length_16411_cov_29.2905_ID_8090 from the susceptible E. coli) have been deposited in the ENA under project number PRJEB32329. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
fosA transfer from Klebsiella spp. to E. coli, leading to fosfomycin resistance, has been demonstrated in vitro.3 Based on publicly available genomes, fosA and adjacent genes are well conserved in K. variicola (minimum 98% identity to fosA9) and K. pneumoniae (minimum 94% identity to fosA9) isolates. According to mlplasmids, PlasmidFinder and contig coverage, fosA9 was predicted to be located in the chromosome of the second E. coli and all K. variicola isolates.9,10 However, based on BLASTn, the contig containing fosA9 aligns to plasmid sequences. The localization of fosA9 in E. coli can thus only be confirmed by completely assembling its genome using long-read sequencing, as the mobilization of the fosA9 gene cluster by an IS element might switch its genomic background. We postulate that fosA9 transfer from K. variicola to E. coli occurred in the gastrointestinal tract, as K. variicola was not co-cultured in the blood at the time of E. coli bacteraemia. We hypothesize that fosfomycin pressure played a role in this transfer; however, this has to be confirmed with further experiments in vitro. Acquisition of fosA9 was associated with an 8-fold increase in the MIC for E. coli (from 2 to 1024 mg/L) while, despite the presence of fosA9 in the chromosome of the K. variicola isolates, the fosfomycin MICs were below the EUCAST susceptibility breakpoint of ≤32 mg/L (Table S1).6 This could suggest either higher dependency of E. coli growth on glutathione or a difference in fosA9 expression or metabolism, i.e. higher expression by the ISEcp1 promoter present upstream of the fosA9 gene cluster.8
In conclusion, our case illustrates the potential of long-term use of oral fosfomycin to promote horizontal gene transfer of fosA9 from commensal gut flora to potential pathogenic microorganisms, such as E. coli.
Supplementary Material
Acknowledgements
We are grateful to Dr Ad C. Fluit and Dr Anita Schurch from the University Medical Center Utrecht for the critical appraisal of this case report prior to submission.
Funding
This study was carried out as part of our routine work.
Transparency declarations
None to declare.
References
- 1. Karageorgopoulos DE, Wang R, Yu XH. et al. Fosfomycin: evaluation of the published evidence on the emergence of antimicrobial resistance in Gram-negative pathogens. J Antimicrob Chemother 2012; 67: 255–68. [DOI] [PubMed] [Google Scholar]
- 2. Ito R, Mustapha MM, Tomich AD. et al. Widespread fosfomycin resistance in Gram-negative bacteria attributable to the chromosomal fosA gene. MBio 2017; 8: e00749–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guo Q, Tomich AD, McElheny CL. et al. Glutathione-S-transferase FosA6 of Klebsiella pneumoniae origin conferring fosfomycin resistance in ESBL-producing Escherichia coli. J Antimicrob Chemother 2016; 71: 2460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Linson SE, Long SW, Ojeda Saavedra M. et al. Whole-genome sequencing of human clinical Klebsiella pneumoniae isolates reveals misidentification and misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere 2017; 2: e00290–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Klontz EH, Tomich AD, Günther S. et al. Structure and dynamics of FosA-mediated fosfomycin resistance in Klebsiella pneumoniae and Escherichia coli. Antimicrob Agents Chemother 2017; 61: e01572–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically—Eleventh Edition: M07. CLSI, Wayne, PA, USA, 2018. [Google Scholar]
- 7. De Been M, Pinholt M, Top J. et al. Core genome multilocus sequence typing scheme for high-resolution typing of Enterococcus faecium. J Clin Microbiol 2015; 53: 3788–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poirel L, Decousser JW, Nordmann P.. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob Agents Chemother 2003; 47: 2938–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arredondo-Alonso S, Rogers MRC, Braat JC. et al. mlplasmids: a user-friendly tool to predict plasmid- and chromosome-derived sequences for single species. Microb Genom 2018; 4: e000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carattoli A, Zankari E, Garcìa FA. et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 2014; 58: 3895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.