Abstract
Background
SWEETs (Sugar Will Eventually be Exported transporters) function as sugar efflux transporters that perform diverse physiological functions, including phloem loading, nectar secretion, seed filling, and pathogen nutrition. The SWEET gene family has been identified and characterized in a number of plant species, but little is known about in Litchi chinensis, which is an important evergreen fruit crop.
Results
In this study, 16 LcSWEET genes were identified and nominated according to its homologous genes in Arabidopsis and grapevine. Multiple sequence alignment showed that the 7 alpha-helical transmembrane domains (7-TMs) were basically conserved in LcSWEETs. The LcSWEETs were divided into four clades (Clade I to Clade IV) by phylogenetic tree analysis. A total of 8 predicted motifs were detected in the litchi LcSWEET genes. The 16 LcSWEET genes were unevenly distributed in 9 chromosomes and there was one pairs of segmental duplicated events by synteny analysis. The expression patterns of the 16 LcSWEET genes showed higher expression levels in reproductive organs. The temporal and spatial expression patterns of LcSWEET2a and LcSWEET3b indicated they play central roles during early seed development.
Conclusions
The litchi genome contained 16 SWEET genes, and most of the genes were expressed in different tissues. Gene expression suggested that LcSWEETs played important roles in the growth and development of litchi fruits. Genes that regulate early seed development were preliminarily identified. This work provides a comprehensive understanding of the SWEET gene family in litchi, laying a strong foundation for further functional studies of LcSWEET genes and improvement of litchi fruits.
Keywords: Litchi chinensis, LcSWEETs, Expression patterns, Seed development
Background
In higher plants, the major carbohydrate sucrose is transported to long-distances and regulation of its partitioning is necessary for plant development and stress responses [1, 2]. At present, three families of transporters have been characterized as key players in sugar translocation: the monosaccharide transporters (MSTs), the sucrose transporters (SUTs), and Sugar Will Eventually be Exported transporters (SWEETs) [3, 4]. The SWEET proteins function as sugar efflux transporters that transport hexose or sucrose across plasma membranes [5, 6]. They are characterized by the MtN3/saliva domain, which is also known as the PQ-loop repeat. MtN3/saliva domain comprises three alpha-helical transmembrane domains (3-TMs). Eukaryotic SWEETs typically consist of two tandemly repeated 3-TMs separated by a single TM, which constitutes a 7-TMs [7]. The distribution of carbohydrates is fundamental to crop yields. Therefore, it is important to explore the functions of SWEET proteins in plants.
SWEET proteins are considered to be involved in different sugar-efflux related processes, such as phloem loading, nectar secretion, supplying symbionts, and maternal efflux for filial tissue development [8]. Furthermore, SWEETs can be hijacked by pathogens for access to nutrition from hosts [9–12]. Indeed, SWEETs have been shown to affect various physiological processes, such as nectar secretion [13], pollen development [14, 15], modulating gibberellins response [16], senescence [17], cold tolerance [18, 19], seed and fruit development [20–22]. OsSWEET4 in rice and its ortholog ZmSWEET4c in maize have been reported to play important roles in grain filling [20].
Genome-wide identification and analysis of SWEET gene family have been reported in various plant species, such as Arabidopsis thaliana [5], Oryza sativa [23], Vitis vinifera [11], Malus domestica [24], Citrus sinensis [25], Solanum lycopersicum [26], Glycine max [27], Brassica napus [28], S. tuberosum [29], Sorghum bicolor [30], Pyrus bretschneideri [31], Cucumis sativus [32], Musa acuminate [33], Hevea brasiliensis [34], Eriobotrya japonica [35], Camellia sinensis [18], B. oleracea [19], Ananas comosus [21], Triticum aestivum [36], B. rapa [37], Phalaenopsis equestris and Dendrobium officinale [38]. In Arabidopsis, the 17 SWEET genes were classified into four clades according to the phylogenetic analysis: AtSWEET1–3 (Clade I), AtSWEET4–8 (Clade II), AtSWEET9–15 (Clade III), and AtSWEET16–17 (Clade IV) [5]. SWEETs in Clades I and II function mainly as glucose transporters [4, 5], Clade III are efficient sucrose transporters [6, 13], and Clade IV are located on tonoplast membrane and likely export fructose [4]. The SWEET genes in each clade may have similar functions, although they are versatile in different plants. At present, several SWEET genes have been identified in fruit trees [24, 25, 31, 35, 39], but their functions in fruit development are not clear.
Litchi (Litchi chinensis Sonn.) is an evergreen fruit crop, originated in China and commercially cultivated in the tropical and subtropical regions of the world. Litchi fruit has a succulent edible flesh (aril) surrounded by a red pericarp and a dark brown seed. Sugar composition in the aril of litchi fruit varies considerably among cultivars, based on different ratios of hexose/sucrose [40–42]. Moreover, the ratio of hexose/sucrose was significantly positively correlated with the weight of seeds [43]. There was no vascular tissue in the aril and the cotyledon, indicating an apoplasmic post-phloem sucrose transport from the funicle [44]. Extremely low activities of cell wall invertase (CWIN) were detected in the seed pedicel and seed coat of fruits with small seeds [45]. Silencing of LcCWIN5 or LcCWIN2 caused a reduction in the seed size [45]. It is common that CWIN and SWEETs co-expressed in the apoplasmic unloading region [46]. However, the functions of SWEETs in litchi seed development remain to be elucidated.
In this study, we comprehensively analyzed the gene structures, conserved motif compositions, chromosomal distribution of 16 SWEET genes in litchi. In addition, we studied the tissue-specific expression of LcSWEETs and their expression patterns during seed development between big-seeded and seed aborting cultivars. Moreover, the spatial expression of LcSWEET2a/3b in early seed development was analyzed. This study provides valuable information for screening important SWEET genes in litchi seed development.
Results
Identification and phylogenetic analysis of SWEET genes in litchi
Arabidopsis SWEET genes were used as queries to find putative Litchi (Litchi chinensis Sonn.) SWEET genes. A total of 16 SWEET genes with two MtN3/saliva domains (PFAM motif PF03083) were obtained in litchi (Fig. 1, Additional file 1: Figure S1). Litchi SWEET genes, hereafter referred to as LcSWEETs, were named on the basis of their percentage of identity to the 17 Arabidopsis SWEET proteins. Gene characteristics, including the complete open reading frames (ORFs), GC content, molecular weight (MW), and isoelectric point (pI) were analyzed (Table 1). Among the 16 predicted LcSWEET proteins, LcSWEET9a was identified to be the smallest protein with 229 amino acid (aa), whereas the largest one was LcSWEET15 with 300 aa. The MW of the proteins ranged from 25.6 to 33.6 kDa, and the pI ranged from 7.66 (LcSWEET15) to 9.81 (LcSWEET9b) (Table 1).
Fig. 1.
PCR amplification of full-length of the 16 LcSWEET genes in litchi. M: DNA 2000 marker; 1: LcSWEET1; 2: LcSWEET2a; 3: LcSWEET2b; 4: LcSWEET3a; 5: LcSWEET3b; 6: LcSWEET4; 7: LcSWEET5; 8: LcSWEET6; 9: LcSWEET8; 10: LcSWEET9a; 11: LcSWEET9b; 12: LcSWEET10; 13: LcSWEET11; 14: LcSWEET12; 15: LcSWEET15; 16: LcSWEET17
Table 1.
Characteristics of the 16 LcSWEET genes in litchi
| Gene | GenBank number | Nucleotide sequence | Predicated polypeptide | |||
|---|---|---|---|---|---|---|
| ORF (bp) | GC content (%) | Length (aa) | Molecular weight (kDa) | pI | ||
| LcSWEET1 | MN480570 | 750 | 45.8 | 249 | 27.4 | 9.70 |
| LcSWEET2a | MN480571 | 708 | 39.9 | 235 | 26.0 | 8.56 |
| LcSWEET2b | MN480572 | 729 | 38.0 | 242 | 26.9 | 9.64 |
| LcSWEET3a | MN480573 | 750 | 42.0 | 249 | 28.3 | 9.47 |
| LcSWEET3b | MN480574 | 755 | 42.6 | 251 | 28.2 | 9.06 |
| LcSWEET4 | MN480575 | 726 | 43.7 | 241 | 27.2 | 9.66 |
| LcSWEET5 | MN480576 | 714 | 43.1 | 237 | 26.4 | 9.18 |
| LcSWEET6 | MN480577 | 795 | 39.6 | 264 | 29.7 | 8.93 |
| LcSWEET8 | MN480578 | 720 | 45.3 | 239 | 26.4 | 9.01 |
| LcSWEET9a | MN480579 | 690 | 40.8 | 229 | 25.6 | 9.29 |
| LcSWEET9b | MN480580 | 720 | 41.1 | 239 | 26.9 | 9.81 |
| LcSWEET10 | MN480581 | 855 | 40.3 | 284 | 32.1 | 9.55 |
| LcSWEET11 | MN480582 | 813 | 49.7 | 270 | 30.2 | 9.21 |
| LcSWEET12 | MN480583 | 901 | 40.6 | 264 | 29.8 | 9.17 |
| LcSWEET15 | MN480584 | 903 | 40.9 | 300 | 33.6 | 7.66 |
| LcSWEET17 | MN480585 | 714 | 41.0 | 237 | 25.7 | 8.73 |
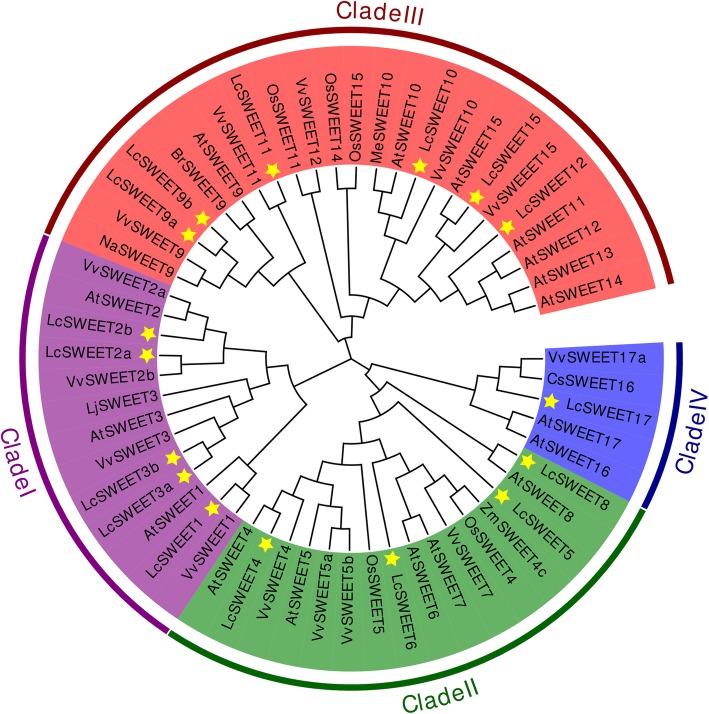
Multiple sequence alignment of the 16 LcSWEET full-length protein sequences showed that the 7-TMs were basically conserved in specific positions (Additional file 2: Figure S2). In order to investigate the evolutionary relationships between LcSWEETs and SWEETs from other plant species, a neighbor-joining phylogenetic tree was constructed using MEGA 7 software. Results showed that the SWEET proteins could be clustered into four clades (Fig. 2). Clade I contained LcSWEET1/2a/2b/3a/3b; Clade II contained LcSWEET4/5/6/8; Clade III contained LcSWEET9a/9b/10/11/12/15; Clade IV contained LcSWEET17. LcSWEET3a/3b showed high sequence identity with LjSWEET3 from Lotus japonicus [47]. LcSWEET9a/9b had high sequence identity with AtSWEET9, BrSWEET9 and NaSWEET9 [13]. And LcSWEET10 shared high sequence identity with MeSWEET10 in cassava [48].
Fig. 2.
Neighbor-joining phylogenetic tree of SWEET proteins from litchi and other plant species. The roman numbers (I-IV) labeled with various colors indicate different clades. LcSWEETs are indicated by yellow star. The amino acid sequences and their accession numbers used to generate this phylogenetic tree are listed in Additional file 3: Table S1
Gene structure and conserved motif analysis of LcSWEETs
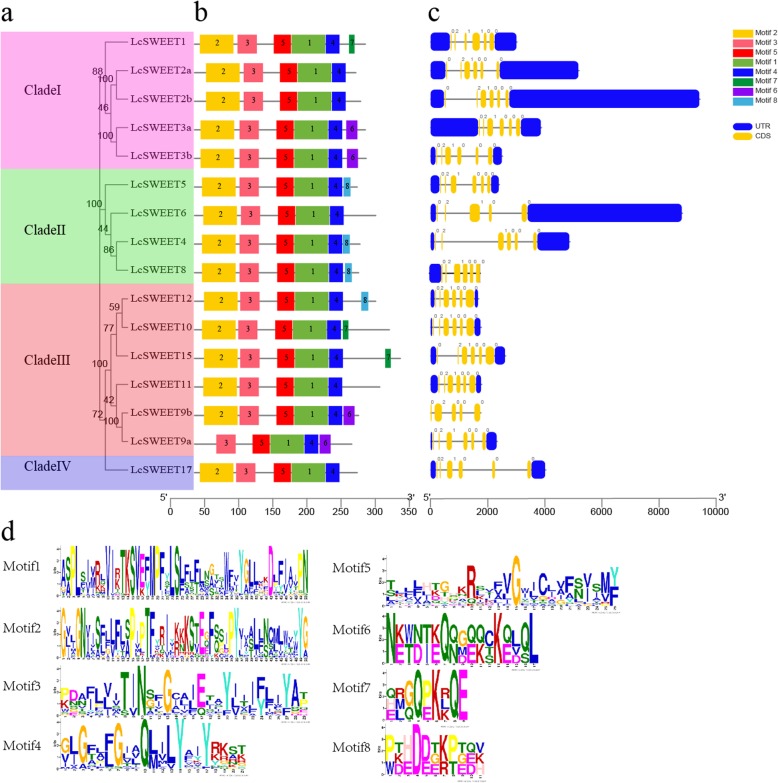
The exon-intron structures and conserved motifs were examined to gain more insight into the characteristic of LcSWEET genes. As shown in Fig. 3, most LcSWEET genes contained six exons, except LcSWEET6 and LcSWEET9b containing five exons. Moreover, LcSWEET genes in the same Clade share similar gene structures in terms of exons location and introns length, although the introns length of LcSWEET members in Clade II and Clade IV were longer.
Fig. 3.
Phylogenetic relationship, gene structure and conserved motif analysis of LcSWEET genes. a The neighbor-joining phylogenetic tree was constructed with MEGA7 using amino acid sequences of LcSWEETs, and the bootstrap test replicate was set as 1000 times. b The motif composition of LcSWEET proteins. Five motifs were displayed in different colored rectangles. c Exon-intron structure of 16 LcSWEET genes. Yellow roundrectangles represent exons and black lines with same length represent introns. Blue roundrectangles indicate the UTR region. d The amino acid sequences of 8 motifs of LcSWEET proteins were showed
Furthermore, sequence motifs in 16 LcSWEETs were predicted using the MEME program. A total of 8 motifs were identified and named motif 1 to 8 (Fig. 3). Motif 1–5 were detected in all LcSWEET proteins except LcSWEET9a lacking Motif 2. Motif 6 was detected in LcSWEET3a, LcSWEET3b, LcSWEET9a, and LcSWEET9b. Motif 7 was detected in LcSWEET1, LcSWEET10, and LcSWEET15. Motif 8 was only detected in LcSWEET4, LcSWEET8, LcSWEET5 and LcSWEET12. Overall, the conserved motif in the N-terminus of all LcSWEET proteins was similar.
Chromosomal localization and synteny analysis of LcSWEETs
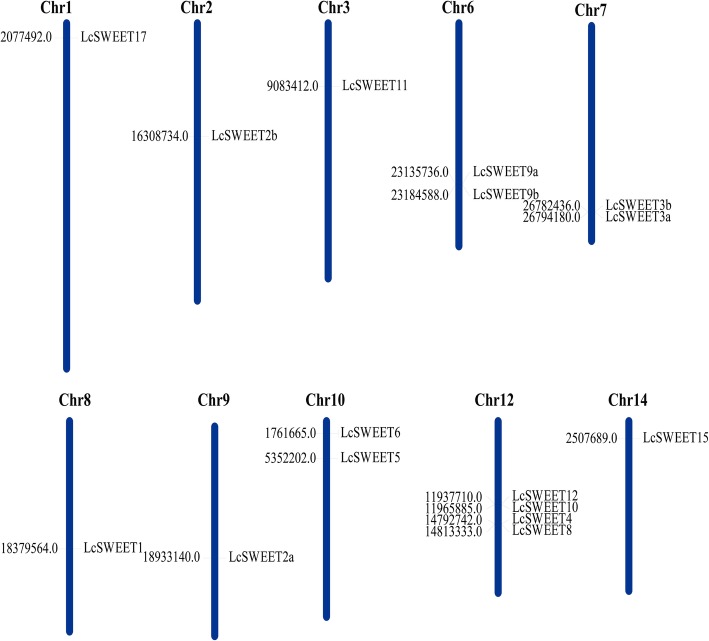
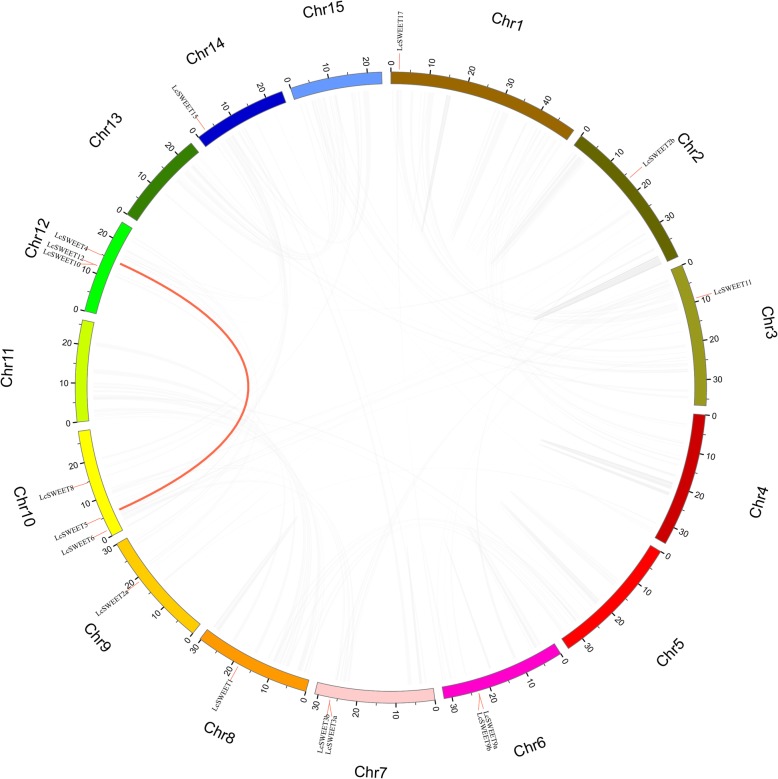
According to the gene loci information, the 16 LcSWEET genes were unevenly distributed in 9 chromosomes and the detailed chromosomal locations were shown in the Fig. 4. Most of LcSWEET members were dispersed to different chromosomes. However, there were 4 LcSWEET genes in Chromosome 12. After the synteny analysis of LcSWEETs, there was one pairs of segmental duplicated events (Fig. 5). LcSWEET4 and LcSWEET5 may be generated by fragment duplication. LcSWEET3a/3b, LcSWEET9a/9b and LcSEET10/12 were clustered into 3 tandem duplication events with BLASTP and MCScanX methods. Based on the above results, some LcSWEET genes were probability generated by gene segmental or tandem duplication.
Fig. 4.
Schematic representations of the chromosomal location of the LcSWEET genes. The chromosome number is indicated on the top of each chromosome
Fig. 5.
The synteny analysis of LcSWEET genes. Fifteen chromosomes were drawn in different colors. The chromosome location of LcSWEET genes were shown by short red lines on the circle. Gray lines indicate all synteny blocks in the litchi genome, and the red lines indicate the duplication of LcSWEET gene pairs
Tissue-specific expression of LcSWEET genes
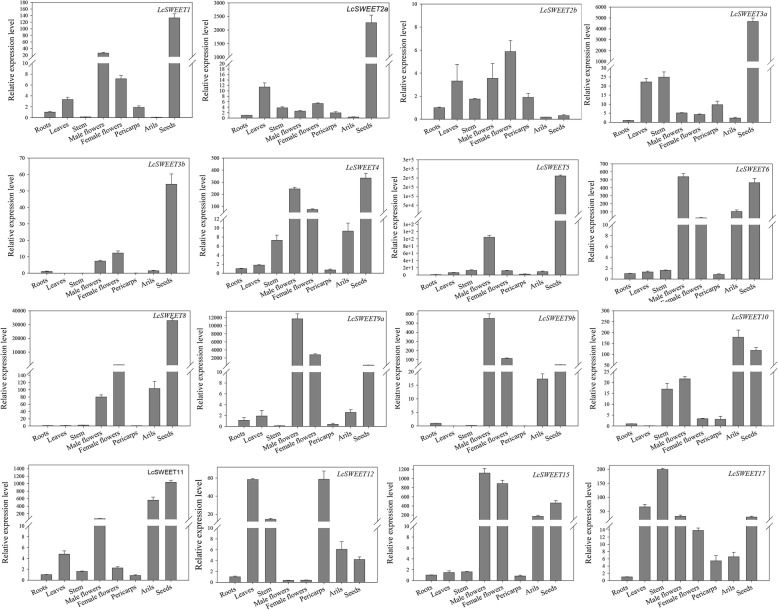
To obtain insights into the physiological functions of the LcSWEET genes, the expression patterns of each LcSWEET gene in different tissues (leaves, stems, roots, male flowers, female flowers, pericarps, arils, and seeds) were measured by RT-qPCR assay. The results showed that the expression pattern differed among the 16 LcSWEET genes (Fig. 6). Most of the LcSWEET genes showed higher expression levels in reproductive organs. However, LcSWEET2b was almost not detected in the arils and seeds, and LcSWEET12/17 had high expression levels in leaves and stems. The expression levels of LcSWEET1/2a/3a/3b/4/5/8/11 in the seeds were the highest among different tissues. Moreover, LcSWEET2a/3a/3b/5 were mainly expressed in seeds. LcSWEET6/9a/9b/15 showed higher expression levels in flowers, arils and seeds. Interestingly, the expression levels of these genes in male flowers were higher than those in female flowers. LcSWEET10 had the highest expression level in arils and LcSWEET17 showed the highest expression level in stems. LcSWEET12 was expressed at high levels in leaves and pericarps.
Fig. 6.
Quantitative real-time PCR analysis of the expression levels of 16 LcSWEET genes in in different litchi tissues. LcActin (HQ615689) was used as an internal control. The vertical bars indicate the standard error of three replicates
Expression patterns of LcSWEET genes during seed development
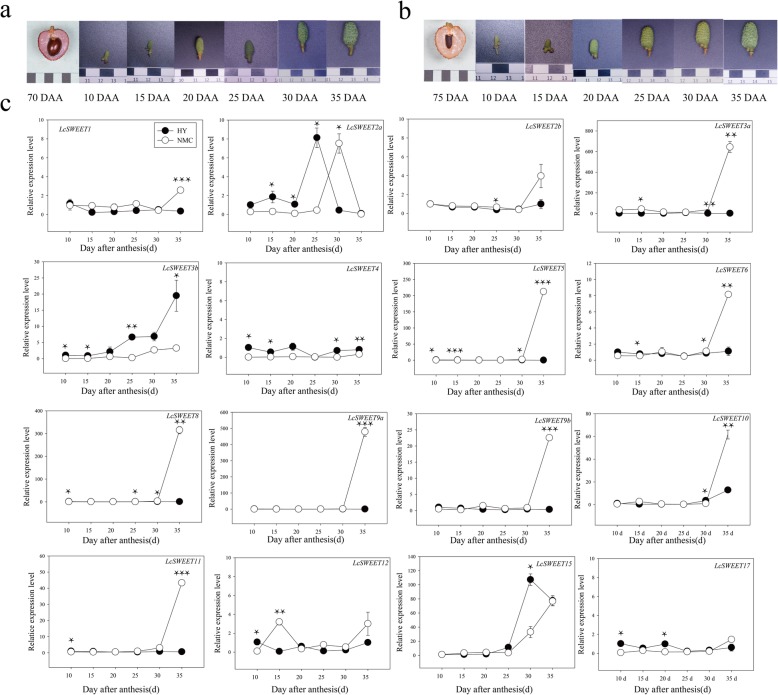
Fruit with a small seed is an economically desirable trait of litchi fruits, and seed size has been found to be associated with the expression levels of LcCWINs, which might affect seed development through sugar import and/or sugar signaling [45]. According to our previous study [45], 28 days after anthesis (DAA) represents a transition point between the cell division stage and the filling stage during litchi seed development. As the seed size has been determined at the cell division stage. We systematically studied the expression of LcSWEET genes at early stages of seed development. The expression patterns of LcSWEET genes at six developmental stages were different between big-seeded cultivar ‘Heiye’ (HY) and seed-aborting cultivar ‘Nuomici’ (NMC) (Fig. 7). The expression of LcSWEET1/4/12/17 remained low in both big-seeded and seed-aborting cultivars through the six developmental stages. A significant increasing was observed in the expression of LcSWEET2b/3a/5/6/8/9a/9b/11 at 35 DAA in NMC, which was not observed in HY. The expression of LcSWEET10/15 increased at 30 DAA in both HY and NMC. Two LcSWEET genes, LcSWEET2a and LcSWEET3b, were found to be correlated with early seed development. The expression of LcSWEET2a peaked at 25 DAA in HY, which was earlier than that in NMC. The expression of LcSWEET3b was higher in HY than that in NMC. With the seed development, a noticeable increased expression of LcSWEET3b was observed since 20 DAA in HY, however, the levels of this gene in NMC were just slightly elevated.
Fig. 7.
Changes in the fruit development of HY (a), NMC (b), and the expression of LcSWEET genes during seed development of HY and NMC as determined by quantitative real-time PCR (c). LcActin (HQ615689) was used to normalize gene expression. Asterisks denote a significant difference during seed development between HY and NMC samples (*p < 0.05, ** p < 0.01, *** p < 0.001). The error bars represent the standard error of three replicates
Spatial patterning of LcSWEET2a and LcSWEET3b in early seed development
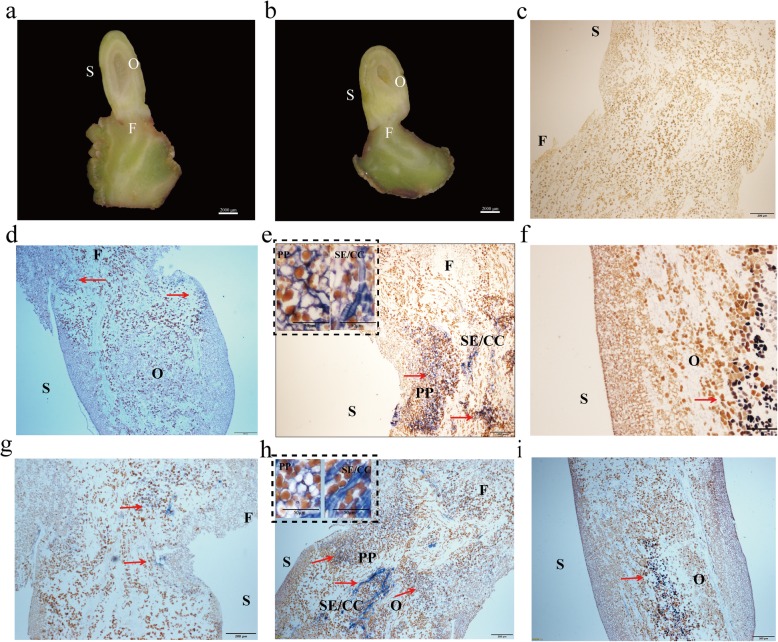
According to Lü et al. [49], the seed at 15 DAA was the critical stage of early embryonic development in litchi. Our previous study indicated funicle was the site of sucrose unloading [44]. Therefore, we conducted in situ hybridization experiments on the funicle and seed at 15 DAA to further explore the roles of LcSWEET2a and LcSWEET3b in early seed development. The results indicated that the spatial distribution of LcSWEET2a and LcSWEET3b transcripts appeared more restrictive to specific areas (Fig. 8). In HY, abundant LcSWEET2a and LcSWEET3b were expressed in the funicle and ovule in comparison with the sense control. However, only little LcSWEET2a and LcSWEET3b transcripts were detected in the funicle and no transcript signals were found in the ovule of NMC. Thus, the different spatial patterning of LcSWEET3b and LcSWEET2a between NMC and HY indicated their involvement in the early seed development in litchi.
Fig. 8.
Spatial distribution of LcSWEET2a and LcSWEET3b mRNA in 15 DAA funicle and ovule of HY and NMC. Longitudinal sections of 15 DAA seed of HY (a) and NMC (b) were viewed under bright field. Longitudinal sections of 15 DAA funicle and ovule with sense RNA probes (c). Longitudinal sections of 15 DAA funicle and ovule of NMC (d) and HY (e, f) hybridized with antisense RNA probes for LcSWEET2a. Longitudinal sections of 15 DAA funicle and ovule of NMC (g) and HY (h, i) hybridized with antisense RNA probes for LcSWEET3b. Magnified views of the SE/CC and PP were inside the black virtual frame on the left top corner of (e and h). Red arrows indicate the LcSWEET2a and LcSWEET3b signals. S, seed; O, ovule; F, funicle. SE/CC, Sieve element-companion cell. PP, parenchyma cells. Bars = 2000 μm (a, b), 200 μm (c-i), 50 μm (magnified views of the SE/CC and PP)
Discussion
SWEET gene family in litchi
Plant SWEET proteins play critical roles in growth, development, and stress responses by regulation of sugar transport and distribution [4, 8]. Recently, SWEET gene families have been analyzed from over 20 plant species. Increasing evidence indicates SWEETs play important roles in fruit development and ripening [21, 22]. However, the SWEET gene family has not been studied in litchi. In this study, we identified and characterized SWEET gene family in litchi through genome-wide analyses, and further investigated their expression patterns during early seed development.
In higher plants, the number of reported SWEET genes varies from 7 to 108, with 17 in Arabidopsis [5], 17 in grapevine [11], 21 in rice [23], and 108 in wheat [50]. In the present study, 16 LcSWEET genes were isolated from litchi. The varied number of SWEET genes in different plant species may be evolved from tandem or segmental duplication [27, 50, 51]. Indeed, one segmental duplicated event (LcSWEET4 and LcSWEET5) and 3 tandem duplication events (LcSWEET3a/3b, LcSWEET9a/9b and LcSEET10/12) were identified in the present study (Figs. 4, 5). The length of LcSWEET proteins ranged from 229 aa to 300 aa, which was similar to that has been reported in other plants, such as 233–308 aa in tomato [26], 171–333 aa in banana [33], and 215–340 aa in apple [39].
Genomic structural analysis showed that majority of LcSWEET genes consisted of six exons except LcSWEET6 and LcSWEET9b (Fig. 3). Similar results were earlier reported in Chinese white pear [31], banana [33], and apple [39]. Chen et al. [5] carried out phylogenetic analysis of Arabidopsis SWEET genes and divided them into four clades. Our results support LcSWEET genes could be classified into four clades. Clade I, II, III, and IV contained 4, 4, 6, and 1 LcSWEET genes, respectively. In the previous studies, it has been shown that phylogenetic analysis also supports the results of conserved motif analysis [33, 50]. Our results indicated that gene members in each clade harbored some unique conserved motif (Fig. 3), indicating they might have different functions in litchi.
Expression and function diversity of SWEET genes in litchi
Plant SWEET genes are found to be differentially expressed in tissues and are involved in different sugar transport [52]. Differential expression analysis of SWEET genes in litchi has implement us to find out specialized functions of each SWEET protein in sugar transport. In this study, most of the LcSWEET genes were highly expressed in flowers and seeds (Fig. 6), which are strong carbon sinks. However, the expression patterns of the pairs of duplicated genes differed in litchi (Figs. 6, 7). For example, LcSWEET3a/3b had different expression levels during early seed development, whereas LcSWEET9a/9b had similar expression patterns in flowers and seeds. Our results are consistent with results reported for apple [39] and wheat [50]. These results suggest that the SWEET genes in litchi have undergone duplication and some duplicated SWEET genes have diverged in functions.
LcSWEET2a and LcSWEET3b were mainly expressed in seeds, and may play important roles in sugar partitioning during seed development. The physiological roles of LcSWEET genes in seed development were further discussed below in detail. SWEET9 has been identified as nectary-specific in Arabidopsis, Brassica, and Nicotiana, and function as a sucrose transporter for nectar production [13]. In this study, LcSWEET9a/9b showed high expression levels in flower. And LcSWEET9a/9b was shown to share high sequence identity with AtSWEET9, BrSWEET9 and NaSWEET9 (Fig. 2). These results suggested that LcSWEET9a/9b appeared to be responsible for nectar secretion in litchi. The transcript levels of LcSWEET6/15 were higher in the flowers, arils and seeds than in other tissues, indicating the function of these genes in specialized organs. Similar studies found that the expression levels of Arabidopsis AtSWEET15 and tea CsSWEET15 were high in flowers and seeds [18, 53]. LcSWEET10 was expressed at high levels in aril. A previous work revealed that grapevine VvSWEET10 was strongly expressed at the onset of ripening and can improve grape sugar content [22]. Overexpression of VvSWEET10 in grapevine callus and tomato increased the sugar levels significantly [22]. The potential role of LcSWEET10 in litchi fruit sugar accumulation is worthy to make further studies.
LcSWEET2a/3b are involved in early seed development in litchi
SWEET transporters are known to transfer assimilate from maternal tissues to developing seeds [4], supplying nutritional and signaling sugars for seed development. The roles of SWEET transporters in seeds were demonstrated during seed filling stage. In Arabidopsis, AtSWEET11, AtSWEET12 and AtSWEET15 were expressed in both seed coat and endosperm, and the AtSWEET11, AtSWEET12 and AtSWEET15 triple mutant showed delayed embryo development and seed shrinkage [53]. Notably, the rice OsSWEET11 and OsSWEET15 transporters both contribute to seed filling with seemingly redundant roles [54]. Overall, SWEET transporters in Clade III responsible for importing sucrose play central roles in seed filling. However, the function of SWEET transporters in early seed development was unclear. In our previous study, the seed development in litchi could be clearly divided into the cell division stage and the filling stage around 28 DAA [45]. Here we performed the temporal and spatial expression patterns of LcSWEET2a and LcSWEET3b during early seed development in litchi. Clearly, the big-seeded cultivar HY showed higher expression levels of LcSWEET2a and LcSWEET3b than those in seed-aborting cultivar NMC. Moreover, the expression sites of LcSWEET2a and LcSWEET3b in HY concentrated in the funicle and ovule, which are the sites of apoplasmic phloem unloading of sucrose [44]. Therefore, we speculate that LcSWEET2a and LcSWEET3b may cooperate with CWIN in hexose import into liquid endosperm. Indeed, we previously found significant lower levels of CWIN protein and activity associated with seed abortion in NMC [45]. During early seed development, CWIN hydrolyze incoming sucrose into hexoses, and the adjacent expression of hexose transporters will be predicted. SWEETs in Clade I and II as hexose transporters may be involved in importation of CWIN-derived hexoses into endosperm. In maize, ZmSWEET4c was expressed in the basal endosperm transfer layer (BETL) and necessary for BETL differentiation [20]. Because OsSWEET4 was predominantly expressed at early stages of seed development, it appears to have similar roles as ZmSWEET4c [20]. The hexoses produced from CWIN and SWEET transporters are thought to stimulate mitotic activity to increase cell number [55]. Taken together, our finding strongly indicates that LcSWEET2a and LcSWEET3b function in determining the seed abortion or seed size by direct translocation of carbohydrates.
Conclusions
In summary, our work firstly analyzed the SWEET gene family in litchi. Sixteen LcSWEET genes were isolated and comprehensively characterized, including gene structures, conserved motifs, chromosomal distribution, gene duplication, evolutionary relationships, spatial and temporal expression patterns. The phylogenetic tree divided the LcSWEET gene into four clades, each of which had similar gene structures and motif compositions. Tissue-specific expression patterns suggested the function diversity of these LcSWEET genes. The temporal and spatial expression patterns of LcSWEET2a and LcSWEET3b suggested their central roles during early seed development. Our research lays a foundation for the elaboration of the functions of the LcSWEET genes in the growth and development of litchi fruits.
Methods
Plant materials
Litchi (Litchi chinensis Sonn.) cultivars ‘Heiye’ (HY), and ‘Nuomici’ (NMC) were grown in the orchard of South China Agricultural University, Guangzhou, China. Trees used for samples collection were under the same integrated orchard management practices. Leaves, stems, roots, male flowers, female flowers, pericarps, arils, and seeds of FZX were collected for gene cloning and tissue-specific gene expression. For gene expression in early seed development, 6 stages of seeds of HY and NMC from 10 DAA until 35 DAA at 5 days interval were sampled. For in situ hybridization experiments, HY and NMC seeds at 15 DAA were collected. Ten uniformly sized fruits were sampled at every stage (one replicate). Three replicates were collected. All samples were immediately frozen in liquid nitrogen and kept at − 80 °C before use.
Identification and molecular cloning of SWEET gene family in litchi
Litchi SWEET gene family were identified by protein Blast of the 17 Arabidopsis SWEET proteins against Litchi genome database (unpublished). The conserved domains of LcSWEET proteins were identified by using NCBI-CCD (https://www.ncbi.nlm.nih.gov/cdd).
Total RNA was extracted using the Quick RNA isolation Kit (Huayueyang, Beijing, China) and the first-strand cDNA was synthesized using Scientific RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA). The coding sequences of LcSWEET genes were amplified from cDNA using gene-specific primers (Additional file 4: Table S2). Phanta Max Super-Fidelity DNA Polymerase (Vazyme, Nanjing, China) was used for PCR amplification. The purified PCR products were ligated into the pEASY-Blunt Cloning Vector (TransGen, Beijing, China) for sequencing.
Sequence analysis
Multiple sequence alignments of the amino acid sequences were generated using DNAMAN software and TMHMM server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The number of amino acids, molecular weights, and theoretical pI were analyzed on the ExPASy website (http://web.expasy.org/potparam/).
Phylogenetic analysis
The full-length amino acid sequences of SWEETs for phylogenetic tree were downloaded from TAIR (https://www.arabidopsis.org/) and NCBI database (https://www.ncbi.nlm.nih.gov), respectively. A neighbor-joining phylogenetic tree were constructed using MEGA7 software with bootstrap test of 1000 times [56]. Then, the phylogenetic tree was annotated with EVOLVIEW (http://120.202.110.254:8280/evolview) [57].
Gene structure analysis and identification of conserved motifs
The exon-intron structures were analyzed by TBtools software (http://www.tbtools.com/) and the MEME (http://meme-suite.org/) website was used for conserved protein motif analysis.
Chromosomal distribution and gene synteny analysis
The location information of LcSWEET genes were obtained from the Litchi genome database (unpublished). The gene location map was constructed using MapChart [58]. The synteny analysis was constructed using the MCScanX and CRCOS [59].
RT-qPCR analysis
RT-qPCR was conducted to determine the expression profile for each member of the LcSWEET genes using various tissues and developmental stages seeds. RT-qPCR expression analysis was carried out using our established protocol [60]. All reactions were performed in triplicate with three biological replicates. Gene primer sequences for RT-qPCR were listed in Additional file 5: Table S3.
Statistical analysis
All the reported values were expressed as mean ± standard error (SE). Significant differences were determined with a T-TEST using IBM SPSS Statistics software 19.0 (SPSS Inc., USA) for Windows.
In situ hybridizations
In situ hybridization was carried out using the protocol of Jackson [61] with some modifications. The funicle and seed of 15 DAA of HY and NMC were fixed in 10% Neutral Formalin Fix Solution (NBF). The samples were rinsed twice each in 1× PBS on ice for 15 min. The samples were dehydrated in a graded ethanol series. To transparent, we replaced the ethanol gradually with xylene. The samples were infiltrated with Paraplast Plus at Paraplat:xylene volume ratio of 2:1, 1:1, 2:1 for 1 h in a 60 °C oven at each step. Lastly, the samples were embedded in 100% Paraplast at 60 °C overnight and stored at 4 °C.
Probes for in situ hybridization were amplified using cDNA from funicle and seed at 15 DAA as template. Sense and antisense RNA probes were synthesized using a DIG RNA labeling mix (Roche Diagnostics, USA). To generate gene-specific probes of LcSWEET2a, a 140 bp fragment, was amplified using LcSWEET2aF, 5′-GAAAGGCAGATTTTTGTTGG-3′, and LcSWEET2aR, 5′ -AAGAAGGTTGAAAGCGAGAG-3′. To generate gene-specific probes of LcSWEET3b, a 172 bp fragment, was amplified using the primer set of LcSWEET3bF, 5′-ATGACCTATGGACTACTGG-3′, and LcSWEET3bR, 5′ -GCTGTTTGGACTTCTCCAT-3′. Fluorescent microscope (ZEISS) was used to observe the samples.
Supplementary information
Additional file 1: Figure S1. The MtN3/saliva domain of LcSWEETs based on the entire protein sequences using NCBI CCD (https://www.ncbi.nlm.nih.gov/cdd).
Additional file 2: Figure S2. Alignment of 16 LcSWEET protein sequences. Highly conserved residues are indicated in color and the seven transmenbrane domains are indicated with TM.
Additional file 3: Table S1. The accession number and amino acid sequences used to generate phylogenetic tree.
Additional file 4: Table S2. Primers used for SWEET genes isolation.
Additional file 5: Table S3. Primers used for RT-qPCR.
Acknowledgements
We thank Dr. Rui Xia for providing the litchi genome sequence. We are very grateful to the editor and reviewers for critically evaluating the manuscript and providing constructive comments for its improvement.
Abbreviations
- aa
Amino acid
- CWIN
Cell wall invertase
- DAA
Days after anthesis
- MSTs
Monosaccharide transporters
- MW
Molecular weight
- NBF
Neutral Formalin Fix Solution
- ORFs
Open reading frames
- pI
Isoelectric point
- RT-qPCR
Reverse transcription quantitative real-time polymerase chain reaction
- SUTs
Sucrose transporters
- SWEETs
Sugar Will Eventually be Exported transporters
- TMs
Alpha-helical transmembrane domains
Authors’ contributions
JZ conceived and designed the experiments. HX, DW, YQ, and AM performed the experiments. HX and JZ analyzed the data. HX, JF, YQ, GH, and JZ wrote the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Fund of China (31501734), China Litchi and Longan Industry Technology Research System (CARS-32-05), the Key Areas of Science and Technology Planning Project of Guangdong Province (2018B020202011), and YangFan Innovative & Entrepreneurial Research Team Project (2014YT02H013). The funders had no role in study design, data collection, analysis and interpretation, or preparation of the manuscript.
Availability of data and materials
The data that support the results are included within the article and its additional file.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-019-2120-4.
References
- 1.Wind J, Smeekens S, Hanson J. Sucrose: metabolite and signaling molecule. Phytochemistry. 2010;71:1610–1614. doi: 10.1016/j.phytochem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Braun DM. SWEET! The pathway is complete. Science. 2012;335:173–174. doi: 10.1126/science.1216828. [DOI] [PubMed] [Google Scholar]
- 3.Chen LQ, Cheung LS, Feng L, Tanner W, Frommer WB. Transport of sugars. Annu Rev Biochem. 2015;84:865–894. doi: 10.1146/annurev-biochem-060614-033904. [DOI] [PubMed] [Google Scholar]
- 4.Eom JS, Chen LQ, Sosso D, Julius BT, Lin IW, Qu XQ, et al. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr Opin Plant Biol. 2015;25:53–62. doi: 10.1016/j.pbi.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, et al. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science. 2012;335:207–211. doi: 10.1126/science.1213351. [DOI] [PubMed] [Google Scholar]
- 7.Xuan YH, Hu YB, Chen LQ, Sosso D, Ducat DC, Hou BH, et al. Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc Natl Acad Sci U S A. 2013;110:E3685–E3694. doi: 10.1073/pnas.1311244110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen LQ. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 2014;201:1150–1155. doi: 10.1111/nph.12445. [DOI] [PubMed] [Google Scholar]
- 9.Antony G, Zhou J, Huang S, Li T, Liu B, White F, et al. Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell. 2010;22:3864–3876. doi: 10.1105/tpc.110.078964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Streubel J, Pesce C, Hutin M, Koebnik R, Boch J, Szurek B. Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. Oryzae. New Phytol. 2013;200:808–819. doi: 10.1111/nph.12411. [DOI] [PubMed] [Google Scholar]
- 11.Chong J, Piron MC, Meyer S, Merdinoglu D, Bertsch C, Mestre P. The SWEET family of sugar transporters in grapevine: VvSWEET4 is involved in the interaction with Botrytis cinerea. J Exp Bot. 2014;22:6589–6601. doi: 10.1093/jxb/eru375. [DOI] [PubMed] [Google Scholar]
- 12.Cox KL, Meng F, Wilkins KE, Li F, Wang P, Booher NJ, et al. TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton. Nat Commun. 2017;8:15588. doi: 10.1038/ncomms15588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin IW, Sosso D, Chen LQ, Gase K, Kim SG, Kessler D, et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature. 2014;508:546–549. doi: 10.1038/nature13082. [DOI] [PubMed] [Google Scholar]
- 14.Guan YF, Huang XY, Zhu J, Gao JF, Zhang HX, Yang ZN. Ruptured pollen grain1, a member of the mtn3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol. 2008;147:852–863. doi: 10.1104/pp.108.118026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun MX, Huang XY, Yang J, Guan YF, Yang ZN. Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod. 2013;26:83–91. doi: 10.1007/s00497-012-0208-1. [DOI] [PubMed] [Google Scholar]
- 16.Kanno Y, Oikawa T, Chiba Y, Ishimaru Y, Shimizu T, Sano N, et al. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat Commun. 2016;7:13245. doi: 10.1038/ncomms13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Liu L, Huang W, Yuan M, Zhou F, Li X, et al. Overexpression of OsSWEET5 in rice causes growth retardation and precocious senescence. PLoS One. 2014;9:e94210. doi: 10.1371/journal.pone.0094210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Yao L, Hao X, Li N, Qian W, Yue C, et al. Tea plant SWEET transporters: expression profling, sugar transport, and the involvement of CsSWEET16 in modifying cold tolerance in Arabidopsis. Plant Mol Biol. 2018;96:577–592. doi: 10.1007/s11103-018-0716-y. [DOI] [PubMed] [Google Scholar]
- 19.Zhang W, Wang S, Yu F, Tang J, Shan X, Bao K, et al. Genome-wide characterization and expression profling of SWEET genes in cabbage (Brassica oleracea var. capitata L.) reveal their roles in chilling and clubroot disease responses. BMC Genomics. 2019;20:93. doi: 10.1186/s12864-019-5454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sosso D, Luo D, Li QB, Sasse J, Yang J, Gendrot G, et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat Genet. 2015;47:1489–1493. doi: 10.1038/ng.3422. [DOI] [PubMed] [Google Scholar]
- 21.Guo C, Li H, Xia X, Liu X, Yang L. Functional and evolution characterization of SWEET sugar transporters in Ananas comosus. Biochem Biophys Res Commun. 2018;496:407–414. doi: 10.1016/j.bbrc.2018.01.024. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Zou L, Ren C, Ren F, Wang Y, Fan P, et al. VvSWEET10 mediates sugar accumulation in grapes. Genes. 2019;10:255. doi: 10.3390/genes10040255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan M, Wang SP. Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol Plant. 2013;6(3):665–674. doi: 10.1093/mp/sst035. [DOI] [PubMed] [Google Scholar]
- 24.Wei X, Liu F, Chen C, Ma F, Li M. The Malus domestica sugar transporter gene family: identifications based on genome and expression profiling related to the accumulation of fruit sugars. Front Plant Sci. 2014;5:569. doi: 10.3389/fpls.2014.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Q, Tang Z, Xu Q, Deng X. Isolation, phylogenetic relationship and expression profiling of sugar transporter genes in sweet orange (Citrus sinensis) Plant Cell Tissue Organ Cult. 2014;119:609–624. doi: 10.1007/s11240-014-0560-y. [DOI] [Google Scholar]
- 26.Feng C, Han J, Han X, Jiang J. Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene. 2015;573:261–272. doi: 10.1016/j.gene.2015.07.055. [DOI] [PubMed] [Google Scholar]
- 27.Patil G, Valliyodan B, Deshmukh RK, Prince SJ, Nicander B, Zhao M, et al. Soybean (Glycine max) SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genomics. 2015;16(1):520. doi: 10.1186/s12864-015-1730-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jian H, Lu K, Yang B, Wang T, Zhang L, Zhang A, et al. Genome-wide analysis and expression profiling of the SUC and SWEET gene families of sucrose transporters in oilseed rape (Brassica napus L.) Front Plant Sci. 2016;7:1464. doi: 10.3389/fpls.2016.01464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manck-Götzenberger J, Requena N. Arbuscular mycorrhiza symbiosis induces a major transcriptional reprogramming of the potato SWEET sugar transporter family. Front Plant Sci. 2016;7:487. doi: 10.3389/fpls.2016.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizuno H, Kasuga S, Kawahigashi H. The sorghum SWEET gene family: stem sucrose accumulation as revealed through transcriptome profiling. Biotechnol Biofuels. 2016;9(1):127. doi: 10.1186/s13068-016-0546-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Qin M, Qiao X, Cheng Y, Li X, Zhang H, et al. A new insight into the evolution and functional divergence of sweet transporters in Chinese white pear (Pyrus bretschneideri) Plant Cell Physiol. 2017;58:839–850. doi: 10.1093/pcp/pcx025. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Feng S, Ma S, Sui X, Zhang Z. Spatiotemporal expression and substrate specificity analysis of the cucumber SWEET gene family. Front Plant Sci. 2017;8:1855. doi: 10.3389/fpls.2017.01855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miao H, Sun P, Liu Q, Miao Y, Liu J, Zhang K, et al. Genomewide analyses of SWEET family proteins reveal involvement in fruit development and abiotic/biotic stress responses in banana. Sci Rep. 2017;7:3536. doi: 10.1038/s41598-017-03872-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sui JL, Xiao XH, Qi JY, Fang YJ, Tang CR. The SWEET gene family in Hevea brasiliensis – its evolution and expression compared with four other plant species. FEBS Open Bio. 2017;7:1943–1959. doi: 10.1002/2211-5463.12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y, Wang Y, Shan Y, Qin Q. Characterization of SWEET family members from loquat and their responses to exogenous induction. Tree Genet Genomes. 2017;13:123. doi: 10.1007/s11295-017-1200-6. [DOI] [Google Scholar]
- 36.Gao Y, Wang ZY, Kumar V, Xu XF, Yuan DP, Zhu XF, et al. Genome-wide identification of the SWEET gene family in wheat. Gene. 2018;642:284–292. doi: 10.1016/j.gene.2017.11.044. [DOI] [PubMed] [Google Scholar]
- 37.Li H, Li X, Xuan Y, Jiang J, Wei Y, Piao Z. Genome wide identifcation and expression profling of SWEET genes family reveals its role during Plasmodiophora brassicae-induced formation of clubroot in Brassica rapa. Front Plant Sci. 2018;9:207. doi: 10.3389/fpls.2018.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang T, Song Z, Meng WL, Li LB. Identification, characterization, and expression of the SWEET gene family in Phalaenopsis equestris and Dendrobium officinale. Biol Plant. 2018;62:24–32. doi: 10.1007/s10535-017-0750-7. [DOI] [Google Scholar]
- 39.Zhen Q, Fang T, Peng Q, Liao L, Zhao L, Owiti A, et al. Developing gene-tagged molecular markers for evaluation of genetic association of apple SWEET genes with fruit sugar accumulation. Hortic Res. 2018;5:14. doi: 10.1038/s41438-018-0024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang HC, Huang HB, Huang XM, Hu ZQ. Sugar and acid compositions in the arils of Litchi chinensis Sonn.: cultivar differences and evidence for the absence of succinic acid. J Hortic Sci Biotechnol. 2006;81:57–62. doi: 10.1080/14620316.2006.11512029. [DOI] [Google Scholar]
- 41.Yang Z, Wang T, Wang H, Huang X, Qin Y, Hu G. Patterns of enzyme activities and gene expressions in sucrose metabolism in relation to sugar accumulation and composition in the aril of Litchi chinensis Sonn. J Plant Physiol. 2013;170:731–740. doi: 10.1016/j.jplph.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 42.Wang D, Zhao J, Hu B, Li J, Qin Y, Chen L, et al. Identification and expression profile analysis of the sucrose phosphate synthase gene family in Litchi chinensis Sonn. PeerJ. 2018;6:e4379. doi: 10.7717/peerj.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang ZY, Zhang JQ, Wang TD, Huang XM, Hu GB, Wang HC. Does acid invertase regulate the seed development of Litchi chinensis? Acta Hort. 2014;1029:301–307. doi: 10.17660/ActaHortic.2014.1029.37. [DOI] [Google Scholar]
- 44.Wang TD, Zhang HF, Wu ZC, Li JG, Huang XM, Wang HC. Sugar uptake in the aril of litchi fruit depends on the apoplasmic post-phloem transport and the activity of proton pumps and the putative transporter LcSUT4. Plant Cell Physiol. 2015;56:377–387. doi: 10.1093/pcp/pcu173. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Wu Z, Hu F, Liu L, Huang X, Zhao J, et al. Aberrant seed development in Litchi chinensis is associated with the impaired expression of cell wall invertase genes. Hortic Res. 2018;5:39. doi: 10.1038/s41438-018-0042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen S, Ma S, Liu Y, Liao S, Li J, Wu L, et al. Cell wall invertase and sugar transporters are differentially activated in tomato styles and ovaries during pollination and fertilization. Front Plant Sci. 2019;10:506. doi: 10.3389/fpls.2019.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugiyama A, Saida Y, Yoshimizu M, Takanashi K, Sosso D, Frommer WB, et al. Molecular characterization of LjSWEET3, a sugar transporter in nodules of Lotus japonicus. Plant Cell Physiol. 2017;58:298–306. doi: 10.1093/pcp/pcw190. [DOI] [PubMed] [Google Scholar]
- 48.Cohn M, Bart RS, Shybut M, Dahlbeck D, Gomez M, Morbitzer R, et al. Xanthomonas axonopodis virulence is promoted by a transcription activator-like Efector-mediated induction of a SWEET sugar transporter in cassava. Mol Plant-Microbe Interact. 2014;27:1186–1198. doi: 10.1094/MPMI-06-14-0161-R. [DOI] [PubMed] [Google Scholar]
- 49.Lü LX, Chen JL, Chen XJ. An observation on the process of embryo development in litchi. Subtrop Plant Res Comm. 1985;1:1–5. [Google Scholar]
- 50.Gautam T, Saripalli G, Gahlaut V, Kumar A, Sharma PK, Balyan HS, et al. Further studies on sugar transporter (SWEET) genes in wheat (Triticum aestivum L.) Mol Biol Rep. 2019;46:2327–2353. doi: 10.1007/s11033-019-04691-0. [DOI] [PubMed] [Google Scholar]
- 51.Jia B, Zhu XF, Pu ZJ, Duan YX, Hao LJ, Zhang J, et al. Integrative view of the diversity and evolution of SWEET and SemiSWEET sugar transporters. Front Plant Sci. 2017;8:2178. doi: 10.3389/fpls.2017.02178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeena GS, Kumar S, Shukla RK. Structure, evolution and diverse physiological roles of SWEET sugar transporters in plants. Plant Mol Biol. 2019;100:351–365. doi: 10.1007/s11103-019-00872-4. [DOI] [PubMed] [Google Scholar]
- 53.Chen LQ, Lin IW, Qu XQ, Sosso D, McFarlane HE, Londoño A, Samuels AL, Frommer WB. A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell. 2015;27:607–619. doi: 10.1105/tpc.114.134585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang J, Luo D, Yang B, Frommer WB, Eom JS. SWEET11 and 15 as key players in seed filling in rice. New Phytol. 2018;218:604–615. doi: 10.1111/nph.15004. [DOI] [PubMed] [Google Scholar]
- 55.Weber H, Borisjuk L, Wobus U. Controlling seed development and seed size in Vicia faba: a role for seed coat-associated invertases and carbohydrate state. Plant J. 1996;10:823–834. doi: 10.1046/j.1365-313X.1996.10050823.x. [DOI] [Google Scholar]
- 56.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H, Gao S, Lercher MJ, Hu S, Chen WH. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012;40:W569–W572. doi: 10.1093/nar/gks576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu B, Lai B, Wang D, Li J, Chen L, Qin Y, et al. Three LcABFs are involved in the regulation of chlorophyll degradation and anthocyanin biosynthesis during fruit ripening in Litchi chinensis. Plant Cell Physiol. 2019;60:448–461. doi: 10.1093/pcp/pcy219. [DOI] [PubMed] [Google Scholar]
- 61.Jackson AC. Detection of rabies virus mRNA in mouse brain by using in situ hybridization with digoxigenin-labelled RNA probes. Mol Cell Probes. 1992;6:131–136. doi: 10.1016/0890-8508(92)90057-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. The MtN3/saliva domain of LcSWEETs based on the entire protein sequences using NCBI CCD (https://www.ncbi.nlm.nih.gov/cdd).
Additional file 2: Figure S2. Alignment of 16 LcSWEET protein sequences. Highly conserved residues are indicated in color and the seven transmenbrane domains are indicated with TM.
Additional file 3: Table S1. The accession number and amino acid sequences used to generate phylogenetic tree.
Additional file 4: Table S2. Primers used for SWEET genes isolation.
Additional file 5: Table S3. Primers used for RT-qPCR.
Data Availability Statement
The data that support the results are included within the article and its additional file.