Abstract
Background
Carbapenems are β-lactam antibiotics which are used to treat severe infections caused by multidrug resistant Enterobacteriacea. The recent emergence and rapid spread of Enterobacteriaceae resistant to carbapenems is a global concern. We undertook a systematic review of the antibiotic susceptibility and genotypic characteristics of carbapenem-resistant Enterobacteriaceae in Chinese neonates.
Methods
Systematic literature reviews were conducted (PubMed/Medline, Embase, Wanfang medical online databases, China National Knowledge Infrastructure (CNKI) database) regarding sepsis caused by carbapenem-resistant Enterobacteriaceae in Chinese neonates aged 0-30 days.
Results
17 studies were identified. Eleven patients in the six studies reported the source of infection. Ten patients (10/11, 90.9%) were hospital-acquired infections. Genotypic data were available for 21 isolates in 11 studies (20 K. pneumoniae, 1 E. coli). NDM-1 was the most frequently reported carbapenem-resistant genotype (81.0%, 17/21). Carbapenem-resistant Klebsiella pneumoniae and Escherichia coli were resistant to many antibiotic classes with the exception of colistin and fosfomycin. Sequence type 105 (ST105) was the most commonly reported K. pneumoniae ST type (30.8%; 4/13), which was from the same hospital in Western China. ST17 and ST20 were the second and third most common K. pneumoniae ST type, 23.1% (3/13) and 15.4% (2/13) respectively. The three strains of ST17 are all from the same hospital in central China. The two strains of ST20, although not from the same hospital, belong to the eastern part of China.
Conclusions
Klebsiella pneumoniae with the NDM-1 genotype was the leading cause of neonatal carbapenem resistant sepsis in China. Hospital acquired infection is the main source of carbapenem resistant sepsis. There is currently no licenced antibiotic regimen available to treat such an infection in China. Improved surveillance, controlling nosocomial infection and the rational use of antibiotics are the key factors to prevent and reduce its spread.
Keywords: Klebsiella pneumoniae, Escherichia coli, Neonate, Genotype, Carbapenem-resistant
Background
According to the global reports, in 2013, 51.8% of the 6.3 million children under the age of five died of infectious diseases, while 44% (276.1 million) died during the neonatal period. Neonatal sepsis is the third leading cause of neonatal death, killing 0.421 million neonates worldwide in 2013 [1]. The overall incidence of neonatal sepsis in four Asian centres (including mainland China, Thailand, Macau, and Malaysia) was 26.1 (95% CI 24.5 to 27.8) per 1000 admissions and Klebsiella spp. was the most common Gram negative organism causing most deaths [2]. Laxminarayan et al. [3] reported that 214 000 of 690, 000 annual neonatal deaths (31%) associated with sepsis are potentially attributable to antimicrobial resistance. Carbapenems are beta-lactam antibiotics which are used to treat severe infections caused by multidrug resistant Enterobacteriaceae, such as Klebsiella pneumoniae (K. pneumoniae) and Escherichia coli (E. coli). The recent emergence and rapid spread of Enterobacteriaceae resistant to carbapenems is therefore of global concern [4].
Resistance to carbapenems includes production of carbapenemases or a combination of structural mutations and production of other β-lactamases, such as extended-spectrum β-lactamase (ESBL) and AmpC cephalosporinases. Bacteria that produce carbapenemases, enzymes that hydrolyze carbapenems, can break down other β-lactam antibiotics including penicillins, cephalosporins, and monobactams [5]. Carbapenemases can be divided into class A (e.g. K. pneumoniae carbapenemase, KPC), class B metallo-β-lactamases [MBLs, e.g. New Delhi metallo-β-lactamase (NDM), Verona integrin-encoded metallo-beta-lactamases (VIM), Imipenem-resistant Pseudomonas (IMP)] and class D β-lactamases (e.g. oxacillinases OXAs). Class C β-lactamases are rarely reported [4].
Recent studies suggest that carbapenem resistance is increasing in China. A national report using data from CHINET (a Chinese antimicrobial resistance surveillance network) has shown that the overall prevalence of imipenem-resistant K. pneumoniae increased from 3.0% to 20.9% and meropenem-resistance from 2.9% to 24.0% between 2005 and 2017. These data included both children and adults and most of the samples were from sputum and urine. Among the five children’s hospitals, the resistance rate of K. pneumoniae isolated from one hospital to imipenem was 2.5%, while from the other four hospitals resistance rates ranged from 32.1% to 45.5%. Little information was available on age ranges and types of samples [6]. This systematic review aimed to summarize the current data from both English and Chinese language sources on the antibiotic susceptibility and genotypic characteristics of carbapenem-resistant Enterobacteriaceae (K. pneumoniae and E. coli) causing neonatal sepsis in China.
Methods
Definitions
Carbapenem-resistance was defined as resistance to any one of meropenem, imipenem, or ertapenem according to the US Central Laboratory Standards Institute (CLSI). In 2015 the breakpoint was changed from 2010. Laboratories using Enterobacteriaceae minimal inhibitory concentration (MIC) interpretive criteria for carbapenems described in M100-S20 (January 2010) performed the modified Hodge test (MHT), Carba NP test and/or a molecular assay when isolates of Enterobacteriaceae were suspicious for carbapenemase production based on impipenem or meropenem MICs of 2–4 ug/ml or ertapenem MIC of 2 ug/ml in 2015 [7]. Carbapenem-resistant K. pneumoniae or E. coli sepsis was defined as a laboratory confirmed culture of K. pneumoniae or E. coli obtained from the blood accompanied with signs and symptoms of infection [8]. Neonates were defined as age 0-30 days [9].
Search strategy and selection criteria
This systematic review was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidance (PRISMA) [10]. We searched the published literature from PubMed/Medline, Embase, China National Knowledge Infrastructure (CNKI) and Wanfang med online databases between January 1, 2000, and June 28, 2018. We used the search terms (“beta-lactamases/or carbapenemase, or carbapenem resistance/resistant or drug resistance or carbapenemase* or carbapenem adj1 resist* or MBL or metallo-b-lactamase or VIM or NDM or OXA or oxacillinase or IMP or KPC or Klebsiella pneumoniae carbapenemase or OmpK”) AND (“Enterobacteriaceae/or enterobacteriaceae or Escherichia/or Escherichia or Escherichia coli or Klebsiella or Klebsiella/Klebsiella pneumoniae/Klebsiella oxytoca”) AND (“China or Chinese”) AND (“neonate or newborn or infant”) for English databases. We used search terms (“Carbapenems or Carbapenem”) AND (“antibiotic resistance”) AND (“infant”, OR “neonatal”) for Chinese databases. We limited the searches to Chinese territories, including Taiwan, Hong Kong, and Macau. The full search strategy is available in Additional file 1: Table S1.
Inclusion and exclusion criteria
We include studies with original data on carbapenem-resistant K. pneumoniae or E. coli sepsis in neonates, which contained any antimicrobial resistance (AMR) or genotype data, or showed the proportion of carbapenem resistant isolates of all Gram negative isolates, or clinical data (including patient demographics, underlying conditions, and antibiotic treatment). We only included blood stream infections. The full details of inclusion and exclusion criteria are presented in Additional file 2: Table S2.
Statistical analysis
Descriptive analysis was performed to investigate the distribution of genotype and MLST typing. Antimicrobial resistance rates were reported by median with interquartile interval (IQI).
Results
Literature search and study selection
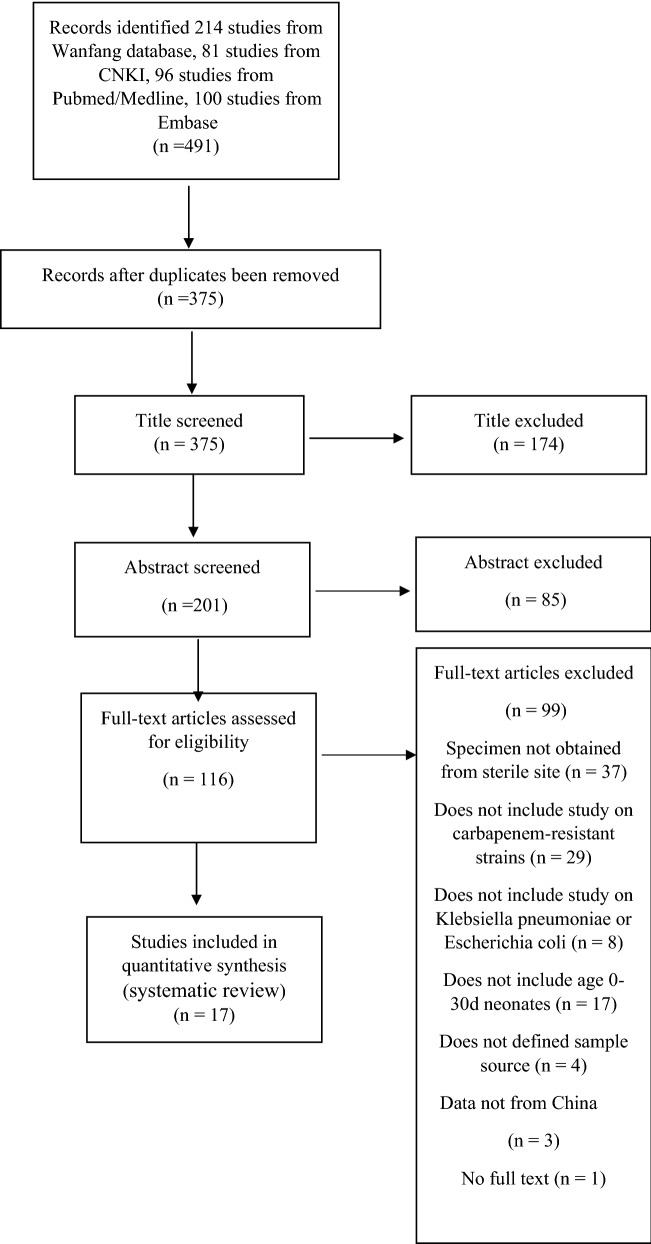
We identified 491 studies from Chinese and English database searches: 81 from CNKI, 214 from Wanfang med database, 96 from Pubmed/Medline and 100 from Embase (the flow chart is shown in Fig. 1). A total of 17 studies met the inclusion criteria and were included for final review, of these 11 (64.7%) reported genotype (including carbapenemase, β-lactamase genes and AmpC cephalosporinases genes) distribution and 9 (52.9%) reported AMR and clinical data. Only 6 studies reported treatment outcomes and gave the proportion of carbapenem-resistant isolates relative to all Gram-negative isolates. The full list of studies included in the review is available in Additional file 3: Table S3.
Fig. 1.
Search strategy and process of study selection
Demographics and clinical presentations of K. pneumoniae or E. coli infections
All 17 studies were from tertiary hospitals. Based on the Government economic divisions of China, 7 studies were from Eastern China, 6 studies from Central China, and 4 studies from Western China. Only 9 of 17 studies reported clinical data, including patient demographics, underlying conditions, and antibiotic treatment. A total of 16 infants were included in these 9 studies. Eight of 16 patients were reported to have underlying conditions, including 6 with lung disease, 2 with necrotizing enterocolitis (NEC) and 2 with recent surgery. Ten patients in these 9 studies reported antibiotic treatment: 5 received meropenem alone, 1 ceftazidime alone, 3 patients had received two antibiotics (piperacillin/sulbactam and ceftazidime; imipenem and amikacin; meropenem and ciprofloxacin) and 1 patient had received more than three antibiotics. Eight patients in these 4 studies had received antibiotics prior to the onset of the relevant infection: 5 received meropenem, 1 received panipenem and 2 didn’t report the type the antibiotics. Clinical treatment outcomes were reported in 13 patients from 6 studies; 3 died and their deaths were attributed to the infection. Eleven patients of the six studies reported the source of infection. Ten patients (10/11, 90.9%) were hospital-acquired infections, while only one was considered to be a vertical transmission.
The proportion of carbapenem resistant strains of all Gram negative strains
Only 6 studies (35%; 6/17) reported the proportion of carbapenem resistant isolates relative to all Gram-negative isolates causing sepsis. Overall, 39 (5.3%) carbapenem resistant K. pneumoniae and E. coli isolates were reported of out of a total of 740 Gram-negative isolates.
Antimicrobial resistance genotype and Multilocus Sequence Type (MLST)
Genotypic data were available for 21 isolates in 11 studies (20 K. pneumoniae, 1 E coli). The most commonly reported genotype was NDM-1 (81.0%, 17/21), followed by KPC-2 (9.5%, 2/21) and IMP-4 (9.5%, 2/21). 15 isolates from 9 studies were tested β-lactamase genes, 66.7% (10/15) isolates carried TEM and SHV genotypes, and 80.0% (12/15)carried CTX-M. 7 isolates from 5 studies were Amp C gene positive, and more than half of them were CMY-4/30 (57.1%; 4/7).
Antibiotic susceptibility results were reported from 19 isolates in 9 studies. The resistance rates of carbapenem-resistant Klebsiella pneumoniae (CRKP) and carbapenem-resistant Escherichia coli (CREC) to second-, third-, and fourth-generation cephalosporins were 100% (IQI 100%–100%). All isolates were susceptible to colistin and fosfomycin (Table 1). MLST was identified for 14 isolates (13 K. pneumoniae and 1 E.coli) from 8 studies. ST105 was the most common K. pneumoniae ST type (30.8%; 4/13), followed by ST17 and ST 20 with 23.1% (3/13) and 15.4% (2/13), respectively (Table 2).
Table 1.
The proportion of isolates demonstrating antimicrobial resistance
| First author | Publication year | Sample | Sample size (number) | Aztreonam % | Levofloxacin % | Ciprofloxacin % | Gentamicin % | Amikacin % | Tigecyclin % | Imipenem % | Meropenem % | Ertapenem | Cefatriaxone % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| He et al. | 2017 | KP | 5 | 100 | 100 | 100 | 100 | ||||||
| Jiang et al. | 2012 | KP | 1 | 0 | 0 | 0 | 0 | 0 | 100 | 100 | |||
| Jin et al. | 2015 | KP | 1 | 100 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | ||
| Zheng et al. | 2016 | KP | 4 | 0 | 0 | 0 | 0 | 0 | 100 | 100 | |||
| Liu et al. | 2013 | KP | 1 | 100 | 100 | 100 | 100 | 100 | |||||
| Zhang et al. | 2015 | KP | 3 | 0 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | ||
| Zhang et al. | 2015 | KP | 1 | 100 | 100 | 0 | 100 | 100 | 100 | 100 | |||
| Qin et al. | 2014 | E.coli | 1 | 100 | 0 | 100 | 100 | 0 | 0 | 100 | 100 | ||
| KP | 1 | 100 | 0 | 0 | 100 | 0 | 100 | 100 | 100 | ||||
| Jin et al. | 2017 | KP | 1 | 100 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | |
| Meidan | 100 | 0 | 50 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | |||
| IQI 25% | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 100 | 100 | 100 | |||
| IQI 75% | 100 | 0 | 100 | 100 | 0 | 0 | 100 | 100 | 100 | 100 |
| First author | Publication year | Sample | Sample size (number) | Cefotaxime % | Ceftazidim % | Cefepime % | Cefoxitin % | Fosfomycin % | Piperacillin % |
PIP/TZB % |
Colsitin |
|---|---|---|---|---|---|---|---|---|---|---|---|
| He et al. | 2017 | KP | 5 | 100 | |||||||
| Jiang et al. | 2012 | KP | 1 | 100 | 100 | 100 | 100 | 0 | |||
| Jin et al. | 2015 | KP | 1 | 100 | 100 | 100 | 100 | 0 | 100 | 0 | |
| Zheng et al. | 2016 | KP | 4 | 100 | 100 | 100 | 100 | ||||
| Liu et al. | 2013 | KP | 1 | 100 | 100 | 100 | |||||
| Zhang et al. | 2015 | KP | 3 | 100 | 100 | 100 | 100 | ||||
| Zhang et al. | 2015 | KP | 1 | 100 | 100 | 100 | 100 | 100 | |||
| Qin et al. | 2014 | E.coli | 1 | 100 | 100 | 0 | 100 | 0 | |||
| KP | 1 | 100 | 100 | 0 | 100 | 0 | |||||
| Jin et al. | 2017 | KP | 1 | 100 | 100 | 100 | 0 | 100 | 0 | ||
| Meidan | 100 | 100 | 100 | 100 | 0 | 100 | 100 | 0 | |||
| IQI 25% | 100 | 100 | 100 | 100 | 0 | 100 | 100 | 0 | |||
| IQI 75% | 100 | 100 | 100 | 100 | 0 | 100 | 100 | 0 |
E. coli, Escherichia coli; KP, Klebsiella pneumoniae; PIP/TZB, piperacillin/tazobactam
Table 2.
Distribution of antimicrobial resistance genotypes and MLSTs among carbapenem-resistant isolates
| First Author | Economic division | Hospital level | Year of publication | Year data collection | Sample source | CLSI Criteria (year) |
Studies type | Community acquired or hospital acquired infection | Organisms and sample size (n) | Resistance gene | MLST | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carbapenemase (n) | β-lactamase genes | Amp C | |||||||||||
| He JR et al. | Central China | Tertiary hospitals | 2017 | 2016.9–2016.10 | Blood | 2015 | Case reports | UNK | KP (n = 5) | bla NDM-1 (n = 5) | – | – | – |
| Jiang MJ et al. | Eastern China | Tertiary hospitals | 2012 | 2009.7 | Blood | 2011 | Case reports | UNK | KP (n = 1) | bla KPC-2 (n = 1) | bla CTX-M-14 (n = 1), bla SHV-2 (n = 1) | bla DHA-1 (n = 1) | – |
| Xu C et al. | Eastern China | Tertiary hospitals | 2015 | 2013.4–2013.5 | Blood | 2013 | Case reports | Hospital acquired | KP (n = 1) | bla NDM-1 (n = 1) | bla TEM-1 (n = 1), | – | ST22 (n = 1) |
| Yao MZ et al. | Eastern China | Tertiary hospitals | 2003 | 1997.1–2002.8 | Blood | UNK | Cross-sectional study | UNK | KP (n = 1) | – | – | – | – |
| Jiang DQ et al. | Western China | Tertiary hospitals | 2017 | 2013.1–2016.12 | Blood | UNK | Cross-sectional study | UNK | KP (n = 2) | – | – | – | – |
| Song HY, et al. | Eastern China | Tertiary hospitals | 2012 | 2009.1–2010.12 | Blood | UNK | Cross-sectional study | UNK | KP (n = 1) | – | – | – | – |
| Zhang ZM et al. | Central China | Tertiary hospitals | 2014 | 2011–2013 | Blood | UNK | Cross-sectional study | UNK |
KP (n = 18) E. coli (n = 9) |
– | – | – | – |
| Tai SH, et al. | Central China | Tertiary hospitals | 2017 | 2014.1–2016.6 | Blood | 2013 | Cross-sectional study | UNK | KP (n = 1) | – | – | – | – |
| Tian HR, et al. | Western China | Tertiary hospitals | 2016 | 2013.1–2014.12 | Blood | UNK | Cross-sectional study | UNK | KP (n = 7) | – | – | – | – |
| Chen S et al. | Western China | Tertiary hospitals | 2014 | 2009.1–2010.12 | Blood | 2010 | Cross-sectional study | Hospital acquired infection | KP (n = 1) | bla IMP-4 (n = 1) | – | – | – |
| Jin Y, et al. | Eastern China | Tertiary hospitals | 2015 | 2012.8–2013.9 | Blood | 2013 | Cross-sectional study | Hospital acquired infection | KP (n = 1) | bla NDM-1 (n = 1) | bla TEM-1 (n = 1), bla CTX-M -14 (n = 1), | bla DHA-1 (n = 1) | ST20 (n = 1) |
| Zheng R, et al. | Western China | Tertiary hospitals | 2016 | 2014.1–2014.3 | Blood | 2013 | Cross-sectional study | Hospital acquired infection | KP (n = 4) | bla NDM-1 (n = 4), bla IMP-4 (n = 1) | bla CTX-M-15 (n = 4), bla SHV-1 (n = 4) | – |
ST105 (n = 4) |
| Liu Y, et al. | Eastern China | Tertiary hospitals | 2013 | 2010.6–2010.9 | Blood | 2009 | Cross-sectional study | UNK | KP (n = 1) | bla KPC-2 (n = 1) | bla SHV-12 (n = 1), bla TEM-1 (n = 1), bla CTX-M -14 (n = 1), | – | UD (n = 1) |
| Zhang XY, et al. | Central China | Tertiary hospitals | 2015 | 2012.8–2013.3 | Blood | 2012 | Case report |
Hospital acquired infection (n = 2) vertical transmission infection (n = 1) |
KP (n = 3) | bla NDM-1 (n = 3), | TEM-1 (n = 3), bla CTX-M-15 (n = 3), bla SHV-1(n = 3), |
bla CMY-4 (n = 3) |
ST17 (n = 3) |
| Zhang Y, et al. | Central China | Tertiary hospitals | 2015 | 2013.2.18 | Blood | 2014 | Case report | Hospital acquired infection (n = 1) | KP (n = 1) | 0 | SHV-11 (n = 1), TEM-53 (n = 1), | 0 | ST65 (n = 1) |
| Qin SS, et al. | Central China | Tertiary hospitals | 2014 | 2011.6–2012.6 | Blood | 2012 | Cross-sectional study | UNK | KP (n = 1) | bla NDM-1 (n = 1) | bla TEM-1 (n = 1), CTX-M-15(n = 1) | – | ST966 (n = 1) |
| E.coli (n = 1) | bla NDM-1 (n = 1) | bla TEM-1(n = 1) | bla CMY-30 (n = 1) | ST40 (n = 1) | |||||||||
| Jin Y, et al. | Eastern China | Tertiary hospitals | 2017 | 2013.7.29 | Blood | 2014 | Cross-sectional study | UNK | KP (n = 1) | bla NDM-1 (n = 1) | bla TEM-1(n = 1), bla CTX-M-15 (n = 1) | bla DHA-1 (n = 1) | ST20 (n = 1) |
CRE, carbapenem resistant Enterobacteriaceae; E coli, Escherichia coli; KP, Klebsiella pneumoniae; NDM, New Delhi Metallo-beta-lactamase-1; Amp C, AmpC cephalosporinases; MLST, Multilocus sequence types; UD, unidentified
Discussion
This is the first review of carbapenem-resistant Enterobacteriaceae (CRE) sepsis in Chinese neonates. Although the prevalence of adults and children with infections resistant to imipenem and meropenem reported by CHINET in 2017 increased significantly, the samples were mainly derived from non-sterile body fluids, and the data for children were not broken down by age. This review has demonstrated that there are very limited recent data on carbapenem resistant isolates in neonates in China. CRKP is reported more than CREC. NDM-1 was the most commonly reported carbapenemase genotype, consistent with previous reports from Asia [11], but different to reports from the United States, where KPC is the most common genotype identified in children [12]. It is worth noting that the CLSI breakpoints for carbapenem changed in 2010 and in 2015, the CDC revised the definition for CRE. In this review, 12 studies provided CLSI reference standards. Among the 12 studies, only one adopted the CLSI standard of 2015, and the others adopted the CLSI standard of before 2015.
In 2017, the World Health Organization published a list of priority pathogens in order to inform global AMR research. CRE is one of the highest priority pathogens for the development of new antibiotics [13], but there are few new antibiotics available. Cefiderocol, is a novel catechol-substituted siderophore cephalosporin with potent activity against meropenem-non susceptible Enterobacteriaceae [14], including metallo-β-lactamases (NDM-1, VIM, IMP). This is the most clinically advanced drug active against NDM carbapenem resistant organisms (CROs) infections [15], but no paediatric studies have yet commenced recruitment. The current standard treatment for NDM CRE infections is polymyxin based combination therapy [16]. However, polymixin E has complex pharmacokinetics requiring hydrolysis of the prodrug colistimethate sodium to colistin, making this less suitable for neonates and infants, and there are no pharmacokinetics data for polymixin B in neonates [17]. Other older, off patent drugs that have potential activity against CROs include fosfomycin and tigecycline, but again, these have no published PK data in neonates. In our study, we found that the currently reported carbapenem-resistant Enterobacteriaceae sepsis in neonate is mainly nosocomial infection. In view of the fact that there is no appropriate antibiotics to treat carbapenem resistant bacteria infection in neonates, it is very important to strengthen epidemiological surveillance, stringent standard infection control practices in healthcare settings, and to enhance the rational use of antibiotics.
Supplementary information
Additional file 1: Table S1. Search terms.
Additional file 2: Table S2. Inclusion and exclusion criteria.
Additional file 3: Table S3. Characteristics of studies included and data type extracted for neonatal sepsis caused by carbapenem-resistant isolates.
Acknowledgements
Not applicable.
Abbreviations
- K. pneumonia
Klebsiella pneumonia
- E. coli
Escherichia coli
- ESBL
extended-spectrum β-lactamase
- KPC
K. pneumoniae carbapenemase
- MBLs
metallo-β-lactamases
- NDM
New Delhi metallo-β-lactamase
- VIM
verona integrin-encoded metallo-beta-lactamases
- IMP
imipenem-resistant Pseudomonas
- OXAs
oxacillinases
- CHINET
chinese antimicrobial resistance surveillance network
- CLSI
Central Laboratory Standards Institute
- MIC
minimal inhibitory concentration
- MHT
Modified Hodge test
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidance
- CNKI
China National Knowledge Infrastructure
- AMR
antimicrobial resistance
- NEC
necrotizing enterocolitis
- CRKP
carbapenem-resistant Klebsiella pneumonia
- CREC
carbapenem-resistant Escherichia coli
- CRE
carbapenem-resistant Enterobacteriaceae
- CROs
carbapenem resistant organisms
Authors’ contributions
The concept of the estimates and the technical oversight of the paper was P.H, Y.W and M. S. The reviews, analyses, and first draft of the manuscript were undertaken by Y.D. Other specific contributions were made by YH, MS, PH, YD and YH undertook the data abstraction. YD and YH undertook the statistical analyses. All authors read and approved the final manuscript.
Funding
This work was supported by Beijing Hospitals Authority Youth Programme (Grant Number: QML 20181207), National Natural Science Foundation of China (Grant Number: 81872676), Beijing Natural Science Foundation (Grant Number: 7192063), Special Fund of the Pediatric Medical Coordinated Development Center of Beijing Municipal Administration of Hospitals (Grant Number: XTYB201806).
Availability of data and materials
All the data for this paper can be found in the additional files. All data analyzed during this study are included in this published article.
Ethics approval and consent to participate
This paper is a systematic review, so ethical approval and consent is not required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12941-019-0334-9.
References
- 1.Li L, Shefali O, Daniel H, et al. Global, regional, and national causes of child mortality in 2000–13, with projections to inform post-2015 priorities: an updated systematic analysis. 2015. p. 430-440. [DOI] [PubMed]
- 2.Al-Taiar A, Hammoud MS, Cuiqing L, et al. Neonatal infections in China, Malaysia, Hong Kong and Thailand. Arch Dis Child Fetal Neonatal Ed. 2013;98(3):F249–F255. doi: 10.1136/archdischild-2012-301767. [DOI] [PubMed] [Google Scholar]
- 3.Laxminarayan R, Matsoso P, Pant S, Brower C, Røttingen JA, Klugman K, Davies S. Access to effective antimicrobials: a worldwide challenge. Lancet. 2016;387(10014):168–175. doi: 10.1016/S0140-6736(15)00474-2. [DOI] [PubMed] [Google Scholar]
- 4.Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Logan LK. Carbapenem-resistant enterobacteriaceae: an emerging problem in children. Clin Infect Dis. 2012;55(6):852–859. doi: 10.1093/cid/cis543. [DOI] [PubMed] [Google Scholar]
- 6.Hu F, Guo Y, Zhu D, Wang F, Jiang X, Xu Y. Antimicrobial resistance profile of clinical isolates in hospitals across China: report from the CHINET surveillance program. Chin J Infect Chemother. 2017;2018:241–251. [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. M100-S25. Performance standards for antimicrobial susceptibility testing: twenty-fifth informational supplement [S]. Wayne: CLSI. 2015.
- 8.Group CM Protocol for diagnosis and treatment of neonatal septicemia. Chin J Pediatr. 2003;41(12):897–899. [PubMed] [Google Scholar]
- 9.FDA. General Clinical Pharmacology Considerations for Pediatric studies for Drugs and Biological products. 2014.
- 10.Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700. doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zmarlicka MT, Nailor MD, Nicolau DP. Impact of the New Delhi metallo-beta-lactamase on beta-lactam antibiotics. Infect Drug Resist. 2015;8:297–309. doi: 10.2147/IDR.S39186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pannaraj PS, Bard JD, Cerini C, Weissman SJ. Pediatric carbapenem-resistant Enterobacteriaceae in Los Angeles, California, a high-prevalence region in the United States. Pediatr Infect Dis J. 2015;34(1):11–16. doi: 10.1097/INF.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO . Essential medicines and health products: Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. Geneva: WHO; 2018. [Google Scholar]
- 14.Hackel MA, Tsuji M, Yamano Y, Echols R, Karlowsky JA, Sahm DF. In vitro activity of the siderophore cephalosporin, cefiderocol, against a recent collection of clinically relevant gram-negative bacilli from north America and Europe, including carbapenem-nonsusceptible isolates (SIDERO-WT-2014 Study) Antimicrob Agents Chemother. 2017;61(9):e00093. doi: 10.1128/AAC.00093-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrei S, Valeanu L, Chirvasuta R, Stefan M-G. New FDA approved antibacterial drugs: 2015–2017. Discoveries. 2018;6(1):e81. doi: 10.15190/d.2018.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papst L, Beović B, Pulcini C, Durante-Mangoni E, Rodríguez-Baño J, Kaye KS. Antibiotic treatment of infections caused by carbapenem-resistant Gram-negative bacilli: an international ESCMID cross-sectional survey among infectious diseases specialists practicing in large hospitals. Clin Microbiol Infect. 2018;24(10):1070–1076. doi: 10.1016/j.cmi.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Thomas R, Velaphi S, Ellis S, et al. The use of polymyxins to treat carbapenem resistant infections in neonates and children. 2019;20(4):415–422. doi: 10.1080/14656566.2018.1559817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Search terms.
Additional file 2: Table S2. Inclusion and exclusion criteria.
Additional file 3: Table S3. Characteristics of studies included and data type extracted for neonatal sepsis caused by carbapenem-resistant isolates.
Data Availability Statement
All the data for this paper can be found in the additional files. All data analyzed during this study are included in this published article.