Highlights
-
•
We identified a highly conserved enhancer in the CB1 gene that contains a SNP associated with substance abuse in humans.
-
•
CRISPR genome editing to delete this enhancer caused a reduction of CB1 gene expression in hippocampus.
-
•
Deletion of this enhancer reduced alcohol intake, cannabinoid response and altered anxiety-like behaviour.
-
•
These studies provide an important platform for the future exploration of cannabinoid pharmacogenetics.
Keywords: Cannabinoid-1 receptor; CRISPR genome editing; Gene regulation; Tissue specific; Enhancer; Polymorphisms; Ethanol intake; Anxiety-related behavior; CB1 agonists; Win55,212-2; Pharmacogenetics
Abstract
The cannabinoid-1 receptor (CB1) plays a critical role in a number of biological processes including nutrient intake, addiction and anxiety-related behaviour. Numerous studies have shown that expression of the gene encoding CB1 (CNR1) is highly dynamic with changes in the tissue specific expression of CNR1 associated with brain homeostasis and disease progression. However, little is known of the mechanisms regulating this dynamic expression. To gain a better understanding of the genomic mechanisms modulating the expression of CNR1 in health and disease we characterised the role of a highly conserved regulatory sequence (ECR1) in CNR1 intron 2 that contained a polymorphism in linkage disequilibrium with disease associated SNPs. We used CRISPR/CAS9 technology to disrupt ECR1 within the mouse genome. Disruption of ECR1 significantly reduced CNR1 expression in the hippocampus but not in the hypothalamus. These mice also displayed an altered sex-specific anxiety-related behavioural profile (open field test), reduced ethanol intake and a reduced hypothermic response following CB1 agonism. However, no significant changes in feeding patterns were detected. These data suggest that, whilst not all of the expression of CNR1 is modulated by ECR1, this highly conserved enhancer is required for appropriate physiological responses to a number of stimuli. The combination of comparative genomics and CRISPR/CAS9 disruption used in our study to determine the functional effects of genetic and epigenetic changes on the activity of tissue-specific regulatory elements at the CNR1 locus represent an important first step in gaining a mechanistic understanding of cannabinoid regulatory pharmacogenetics.
1. Introduction
The cannabinoid-1 receptor (CB1) is extensively expressed in areas of the nervous system, where it plays a critical role in appetite regulation (Pomorska et al., 2016), the reward system and anxiety-related behaviour (Akirav, 2011). A number of studies have shown that expression of the gene encoding CB1 (CNR1) is highly tissue specific and subject to dynamic regulation during embryonic development, normal homeostasis as well as during the progression of various disease states (Laprairie et al., 2012). Although previous work has identified the promotor regions regulating the expression of CNR1 (Zhang et al., 2004) very little is known of the genomic mechanisms that regulate the dynamic tissue specific expression of this critical gene. Gaining an understanding of the mechanisms regulating CNR1 will not only lead to an understanding of the effects of regulatory polymorphisms and epigenetics on the changing expression of CNR1 but may also allow us to better devise personalised strategies to counter conditions such as substance abuse and anxiety disorders.
Because the CNR1 gene and its promoter region lack common polymorphisms that had been associated with disease we chose to study a region of CNR1 intron 2 which contained a linkage disequilibrium block that contained two single nucleotide polymorphisms (SNPs; rs2023239 and rs9450898) that had previously been associated with addictive behaviours (Ketcherside et al., 2017), depression (Icick et al., 2015), psychosis (Suarez-Pinilla et al., 2015), reduced hippocampal volume in cannabis abuse (Schacht et al., 2012), nicotine addiction (Chen et al., 2008), obesity (Benzinou et al., 2008) and alcohol abuse (Hutchison et al., 2008; Pava and Woodward, 2012). There is growing evidence that functional enhancer regions are often highly conserved (Montalbano et al., 2017) so we previously used a combination of comparative genomics, molecular biology and primary cell based studies to identify a highly conserved enhancer element in intron 2 of the CNR1 gene that we called evolutionary conserved region 1 (ECR1) (Nicoll et al., 2012) and which contained a SNP (rs9444584) that was in strong linkage disequilibrium (LD) with rs2023239 and rs9450898. In our previous study we had shown that polymorphic variants of ECR1 (ECR1T and C) supported different levels of marker gene expression when transfected into different primary cell types (Nicoll et al., 2012). However, we were unable to determine the role of this regulatory element in either the expression of the CNR1 gene or in physiology and behaviour. In the current study we have used CRISPR genome editing technology to disrupt the ECR1 enhancer in mice. We then went on to investigate the effects of this deletion on CNR1 gene expression in hippocampus and hypothalamus; two areas of the brain where expression of CB1 is important in modulating mood and appetite. We also analysed aspects of mouse behaviour including cannabinoid response, alcohol intake, locomoter activity and thigmotaxis; an indicator of anxiety-related behaviour. The significance of these observations is discussed in the context of cannabinoid pharmacogenomics and disease.
2. Materials and methods
2.1. Generation of gRNA molecules by a novel annealed oligo template (AOT) method
Single guide RNA (sgRNA) molecules were designed to disrupt the mouse ECR1 enhancer (ECR1; Fig. 1) using the optimised CRISPR design tool (http://CRISPR.mit.edu/). To remove the need for cloning of the guide RNA template into a plasmid as previously described (Harms et al., 2014) we devised a cloning-free technique for the production of sgRNA. This involved the annealing and PCR amplification of two oligonucleotides: the first oligonucleotide (80mer; 5′-AAAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAA-3′), represented the sgRNA scaffold and acted as a generic oligonucleotide for the generation of all subsequent templates; the second oligonucleotide, (63mer; 5′-TTAATACGACTCACTATAGGNNNNNNNNNNNNNNNNNNNGTTTTAGAGCTAGAAATAGCAAG -3′), where “N “denotes the predicted guide sequence (5′sgRNA; ATAGTAAAAGAACGATTGAA and 3′sgRNA; CTCTCCATGCAAAAATAAGG) underlined sequence represents the T7 promoter and italic sequence represents sequence complimentary to the template scaffold oligo, acted as the targeted sequence to the region of interest. These oligonucleotides were amplified using 30 cycles of a standard PCR reaction (92 °C, 30 s, 60 °C; 30 s, 72 °C 30 s) to produce a 122 bp double strand sgRNA template. 100 ng of this template was used to produce sgRNA using a mMESSAGE mMACHINE T7 in-vitro transcription kit (Ambion) described in the manufacturer’s instructions and purified using a Megaclear kit (Ambion) with modifications by Harms et al (2014).
Fig. 1.
Bioinformatic analysis of the human CNR1 locus using the UCSC genome browser. A. Genetic linkage analysis within intron 2 of the human CNR1 locus showing degrees of linkage disequilibrium between disease associated SNPs (rs2023239 and rs9450898; highlighted by red boxes) and rs9444584 (Green box). Deep red triangles indicate linkage disequilibrium expressed as r2>0.95 and pink r2> 0.9. B. Sequence analysis of the mouse ECR1 enhancer region showing levels of mammalian conservation (blue peaks) and depth of conservation (green bars). Long black bars labelled with numbers (41,39,32,36,28,29,38,31,30,37) reflect the alignment of sequences derived from PCR products amplified from ear plug DNA of CRISPR genome edited mice. Deletions are indicated by thin lines covered in chevrons. Short black bars (gRNAECR1a and gRNAECR1b) reflect the regions targeted by the guide RNA used to target the ECR1 locus. The red line highlights where the rs9444584 polymorphisms within the ECR1 enhancer lies within the equivalent human locus. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
2.2. Production of transgenic and genome edited mice
RNA (10 ng/μl each gRNA as produced above) and mRNA encoding a wild type CAS9 with a nuclear localisation signal (Life Technologies) was microinjected into the cytoplasm (RNA) of 1-cell C57/BL6 embryos as described (Harms et al., 2014). Surviving two-cell embryos were introduced into host CD1 mothers using oviduct transfer as previously described (Nagy et al., 2003). Once weaned, ear-clip biopsies were recovered and analysed by PCR using ECR1 flanking primers (mECR1 test for; TGTGTGCAGAGAGGGGAGAC; mECR1 test rev; CTTTAGGAGTGGACAAGGGGTC) as previously described (Harms et al., 2014).
2.3. Quantitative reverse transcriptase-PCR
Brain tissues (hypothalamus and hippocampus) were dissected and snap frozen on dry ice. Total RNA was extracted using the isolate II RNA minikit and converted to cDNA using reverse transcriptase (Bioline). Quantitative reverse transcriptase-PCR (QrtPCR) to determine mRNA expression levels of mouse CNR1 was undertaken using gene specific primers (CNR1for. 5′-GGATGCGAAGGGGTTCCCTC-3′; CNR1rev. 5′-AACCAACGGGGAGTTGTCTC-3′) on derived cDNA using mouse Qrt-PCR primers as previously described (Shanley et al., 2010, 2011) using a Roche Light Cycler 480 with Roche SYBR green. All QrtPCR analyses were normalised using mouse primers specific to the Nono housekeeping gene (Nonofor. 5′-GCCAGAATGAAGGCTTGACTAT-3′; Nonorev. 5′-TATCAGGGGGAAGATTGCCCA-3′) that gave the most stable expression in all neuronal tissues analysed compared to the βactin, HPRT or GAPDH genes.
2.4. Animal maintenance
All mice were housed under standard UK Home Office approved laboratory conditions (12 h light/12 h dark cycle and constant temperature (22 ± 2 °C) and humidity (55 ± 10%)), with food and water available ad libitum.
2.5. Ethanol intake studies
Oral ethanol self-administration and preference were examined using a two-bottle choice protocol as described previously (Hungund et al., 2003). Mice were housed in TSE home cage systems (TSE, Bad Homburg, Germany) that record liquid intake automatically using weight sensors attached to suspended liquid containers. Initially mice were group housed (˜4 per cage) and habituated for 5 days to allow for adaptation to the monitored bottles, then they were single housed for 3 days prior to introduction of the ethanol solution. Intake of water and 10% ethanol solution (both containing 0.2% saccharine to normalise preference to ethanol) were monitored at 60 min intervals over a 3-week period and recorded by TSE PhenoMaster Software. These mice were not used for subsequent tests.
2.6. Hypothermic response studies
Win55,212-2 was dissolved to 1 mg/ml in ethanol and mixed with 2 mg/ml tween 80 in ethanol in a 1:1 ratio. Ethanol was evaporated off and the drug/tween 80 mixture was reconstituted in 0.9% saline. Homozygous wildtype and ECR1KO mice male and female age matched littermates (6–12 weeks old) were injected i.p. with 5 mg/kg of Win55,212-2 and core temperature measurements taken at 0 min, 30 min and 60 min using a rectal thermometer inserted 2 cm (Rawls et al., 2002). These mice were not used for subsequent tests.
2.7. Food intake analysis
For food intake studies, singly housed male and females litter and age matched (12–20 weeks) ECR1KO and WT animals were provided with a choice of low fat diet (LFD) or high fat diet (HFD) (LFD; 22.03 kcal% protein, 68.9 kcal% carbohydrate and 9.08 kcal% fat. HFD; 20 kcal% protein, 20 kcal% carbohydrate and 60 kcal % fat; Research Diets Inc.) in different hoppers. The position of each hopper was changed regularly to rule out the possibility of position effects. Animals were weighed daily over a period of 23 days and LFD and HFD were also weighed daily to determine intake of each diet. These mice were not used for subsequent tests
2.8. Body fat mass composition
Body fat mass versus lean mass composition of each mouse in feeding studies was analysed at the beginning of the food intake study and at the end using an Echo MRI-3-in-1 scanner (Echo Medical Systems, Houston, TX, USA). Mice were placed individually into a plastic tube and inserted into the scanner where total body fat and lean mass were determined at the beginning and at the end of each study.
2.9. Open Field test
Movement and thigmotaxis in ECR1 deletion and WT littermate male and female age matched (6–12weeks) mice was tested using the open field test (Crawley, 1985). Briefly, the open field test consisted of a square 30 cm (30 cm high) PVC open field arena positioned on a white base with overhead lighting applying 100 lx at the base. Animals were transported individually to the testing room, habituated (30 min.) and released into the corner of the arena. Ambulatory activity was recorded in the open field for 600 s using an overhead video camera and the ANY-maze tracking software. The software was used to define a 35 cm2 mid zone and 30 cm2 peripheral zone. Travel between these zones was referred to as “line crossings”. Distance travelled, mean speed, time in peripheral zone, entries into centre zone and time in centre zone, were determined by the software. Many of these mice were included in subsequent food intake studies.
2.10. Data analysis
Statistical significance of data sets was analysed using two-tailed unpaired parametric Student t-test using GraphPad PRISM version 5.02 (GraphPad Software, La Jolla, CA, USA) except for data in Fig. 4D which was analysed using 2-way ANOVA. p-values, numbers used, and t-values are displayed for each graph in the figure legends for significant. ANOVA analysis for Fig. 4D includes the p-value and F-value.
Fig. 4.
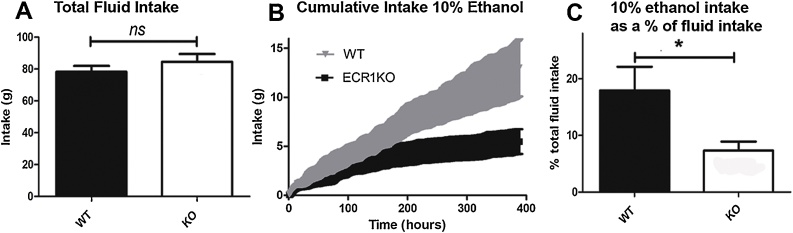
A Comparison of total fluid intake over 400 h by wild type (WT) and ECR1KO animals given a choice of drinking water or water and 10% ethanol (n = 11–16; error bars = SDM, ns = no significance, p = 0.33, t = 1.008). B, a time plot comparing cumulative hourly intake of 10% ethanol between WT (grey triangles) and ECR1KO (black squares) age matched littermates over a 400 -h period (n = 11–16; error bars = SDM). C, a comparison of total intake of 10% ethanol expressed as a percentage of total intake comparing wild type and ECR1KO animals over the 400 h of the study (n = 11–16; error bars = SDM, p = 0.02, t = 2.454).
2.11. Ethics
All animal experiments were conducted in full accordance with UK Home Office guidelines as stipulated in the Animals in Scientific Research act of 1986. This study was undertaken under a current UK Home Office 5-year project licence (PPL number: P5D8BAA7D) which was subjected to ethical review by the University of Aberdeen ethical review board prior to submission to the Home Office in 2016. Animals were euthanised using UK Home Office approved schedule-1 techniques.
3. Results
Our previous studies used a combination of comparative genomics and linkage disequilibrium analysis to identify a highly conserved polymorphic sequence (ECR1; rs9444584) within intron 2 of the CNR1 gene that was in strong LD with two other SNPs (Fig. 1A, rs2023239 and rs9450898) that were previously associated with various pathologies (Benzinou et al., 2008; Chen et al., 2008; Hutchison et al., 2008; Icick et al., 2015; Ketcherside et al., 2017; Pava and Woodward, 2012; Schacht et al., 2012; Suarez-Pinilla et al., 2015) and which displayed allele specific differential enhancer activity in different tissues (Nicoll et al., 2012). In the current study we asked whether this enhancer could be disrupted using CRISPR genome editing and what effects disrupting this enhancer would have on the expression of the endogenous CNR1 gene in different tissues as well as a broad analysis of food and alcohol intake, cannabinoid response, movement and anxiety-related behaviour.
3.1. Cytoplasmic microinjection of gRNA; produced using the AOL protocol, and Cas9 mRNA efficiently disrupts the ECR1 enhancer in mice
All animal experiments complied with current U.K. Home Office Animals (Scientific Procedures) Act (1986) guidelines. To determine the possible function of the ECR1 sequence we used CRISPR genome editing to disrupt the most highly conserved core region of ECR1 from the mouse genome (Fig. 1A). This was done by microinjecting guide RNA molecules (gRNA; gRNAECR1a and gRNAECR1b; Fig. 1B), produced using our AOL method (see materials and methods), and designed to disrupt the core region of ECR1, centred on a sequence homologous to that containing the human rs9444584 polymorphism (Fig. 1B), together with CAS9 mRNA into the cytoplasm of fertilised single cell mouse embryos. We injected 120 1-cell embryos with gRNA and Cas9 mRNA and transferred 90 surviving 2 cell embryos into 4 pseudopregnant female mice. These mice gave birth to 23 pups of which 10 proved to be homozygous for the ECR1 disruption (Fig. 2A). Sequence analysis of earclip DNA demonstrated that 90% of deletions were within +/- 20 base pairs of the PAM sequences associated with each gRNA (Fig. 1B). We derived 3 lines containing nearly identical deletions of ECR1 (Fig. 1B; ECR1KO lines 29, 30 and 31) that closely matched the PAM sequence of our guides. Further analysis of earclip DNA explored the top 5 likely off target sequences (Sequences within the mouse genome that differed by one or two base pairs) from the founder line but could not identify any evidence of off-target changes to the genomes of any of the lines chosen for further analysis (Hay et al., 2016).
Fig. 2.
Bar graphs showing QrtPCR analysis of mouse CNR1 expression in total RNA derived from A, hippocampus (p = 0.0182, t = 2.142) and B, hypothalamus (p = 0.8132, t = 0.2379) comparing CNR1 mRNA expression in wild type (WT) and ECR1KO (KO) animals normalised to the Nono reference gene (n = 19–31; error bars = SDM, *=p<0.05, ns = not significant).
3.2. Disruption of the CR1 sequence reduced the expression of CNR1 in hippocampal cells but not the hypothalamus
To determine the effects of disrupting ECR1 in mice we used QrtPCR to detect tissue changes in the expression of CNR1 mRNA in tissues derived from the hippocampus and the hypothalamus of ECR1KO and age and sex matched wild type littermates. ECR1KO animals had a 17% reduction in CNR1 mRNA in the hippocampus (Fig. 2A) but not in the hypothalamus (Fig. 2B). These observations suggest a role for ECR1 as a tissue specific regulatory sequence with greater relevance for CNR1 expression in the hippocampus.
3.3. Disruption of ECR1 reduces the CB1 activated hypothermic response
Mice lacking CB1 have a reduced hypothermic response to CB1 agonists (Ledent et al., 1999). In order to assess the effects of ECR1 deletion of CB1 activity we tested the well-known hypothermic response of wild type and ECR1KO animals treated with the CB1 agonist Win55,212-2(Grim et al., 2016). We injected adult male and female ECR1KO and wild-type mice with the CB1 agonist Win55,212-2 (5 mg/kg; i.p) (Irving et al., 2000). We were unable to detect any sex specific difference in Win55,212-2 response (data not shown) but we observed that, 30 min after injection, ECR1KO mice of both sexes displayed a significantly reduced hypothermic response compared to wild types (WT) consistent with a change in CB1 receptor expression (Fig. 3A). The difference in effect between ECR1KO and WT animals, although still observable, became non-significant at 60 min after injection (Fig. 3B).
Fig. 3.
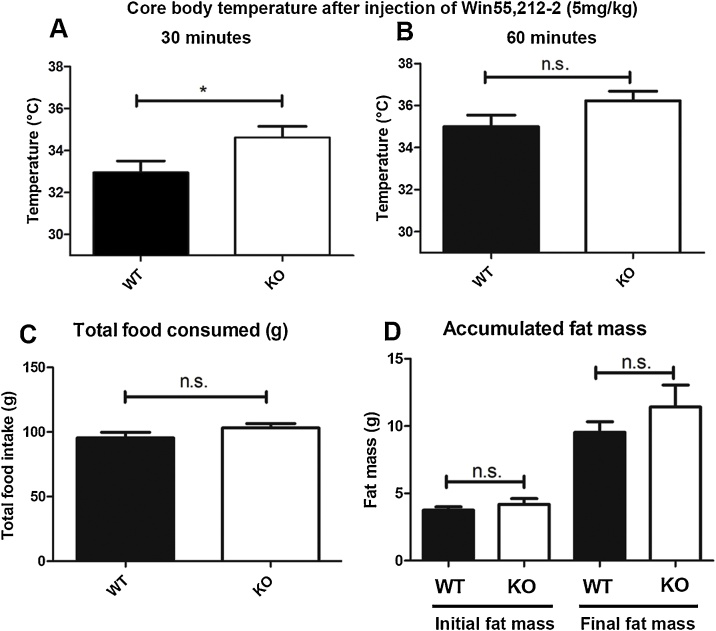
A and B Comparison of core temperature measurements from wild type (WT) and ECR1KO (KO) animals taken (A) 30 min (p = 0.04, t = 2.131) and (B) 60 min (t-test, p = 0.09, t = 1.744) after intraperitoneal (i.p.) injection of 5 mg/kg of Win55,212-2 (n = 17–19; error bars = SDM). C Comparisons of total food intake (g) in ECR1KO (KO) and wild type (WT) animals allowed ad libitum access to a choice of low fat and high fat diet over 29 days (n = 8; p = 0.17, t = 1.419). D. Comparisons of fat mass deposition in ECR1KO (KO) and wild type (WT) animals assessed using Echo-MRI scanning before and after 29 days of ad libitum access to a choice of low fat and high fat diet (n = 8; ns = no significance, 2-way ANOVA, p = 0.48, F = 0.85).
3.4. Disruption of ECR1 did not affect food intake or weight gain
Previous analysis had shown that deletion of the CNR1 gene (Cota et al., 2003) or treatment of mice with CB1 antagonists (Viveros et al., 2008) significantly decreased food intake and fat deposition in mice. To determine whether ECR1 played a role in food intake or fat deposition we provided age matched ECR1KO and WT male and female littermate mice (12–18 weeks old) with free access to two hoppers containing high fat diet (HFD; 60% of calories from fat) or low-fat diet (4% of calories from fat). No significant change in preference for HFD over LFD was detected (data not shown). Furthermore, no significant change in overall food intake was observed because of ECR1 deletion after 29 days exposure to ad libitum HFD/LFD exposure (Fig. 3C). To detect possible changes in fat/lean mass each animal was assessed using echo MRI scanning at the beginning of the trial and at the end (29 days) and no significant difference was observed in the deposition of fat between these groups of animals (Fig. 3D) suggesting that ECR1 did not play an important role in appetite or weight gain.
3.5. Disruption of ECR1 reduced alcohol intake in mice
Mice lacking the CB1 gene have reduced ethanol intake (Hungund et al., 2003) and pharmacological blockade of CB1 also has an effect on alcohol intake (Femenia et al., 2010; Lallemand and De Witte, 2006) demonstrating an important mechanistic link between CB1 expression and alcohol consumption. We therefore explored the effects of the ECR1 knockout on the ethanol intake of ECR1KO mice in comparison to wild type littermates by exposing singly housed ECR1KO and WT mice to a choice of either water or water and 10% ethanol. We observed that, despite having the same total fluid intake (Fig. 4A), ECR1KO mice drank significantly less 10% ethanol than WT animals (Fig. 4B and C) over the 16 days that they were exposed to the choice. Overall ECR1KO animals drank less than half of the 10% ethanol drunk by their wild type littermates.
3.6. Disruption of the ECR1 enhancer affected thigmotaxis in a sex specific manner but not locomotor activity
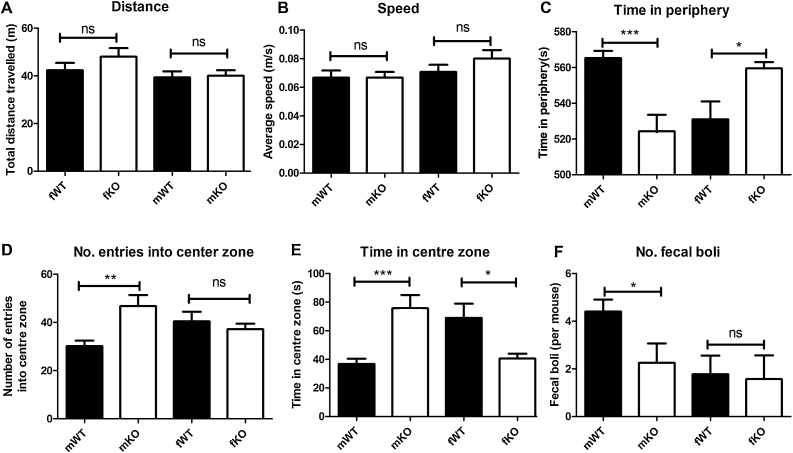
Previous studies of CNR1 knockout animals have indicated a complex role for CB1 in modulating anxiety-related behaviour (Patel and Hillard, 2009). CB1 expression in the hippocampus plays a role in anxiety-related behaviour (Bambico et al., 2009; Viveros et al., 2007) and we demonstrate that disruption of the ECR1 enhancer reduced expression of CNR1 mRNA in the hippocampus by 17%. To determine whether disruption of ECR1 influenced levels locomotor activity or anxiety-related behaviour we compared the behaviour of male and female wild type and ECR1KO animals using the open field test (OFT); an accepted model of locomotor activity, and thigmotaxis; an anxiety-related behaviour that increases the need for an animal to make contact with a solid object such as a wall, in mice (Crawley, 1985) (Seibenhener and Wooten, 2015). We were unable to detect any significant difference in the speed or distance travelled by male or female ECR1KO and WT littermates within the arena of the OFT suggesting that disruption of ECR1 had no effect on male or female locomotor activity (Fig. 5A and B). However, we observed increased thigmotaxis in female ECR1KO mice; reflected by their need to stay at the periphery of the arena, compared to wild type mice, and a corresponding reduction in centre entries and time spent there (Fig. 5C–E). In contrast, ECR1 KO mice displayed significantly reduced thigmotaxis and a corresponding increase in centre entries and time spent there (Fig. 5 C–E). Interestingly, male wild type mice deposited more fecal boli than male ECR1KO mice (Fig. 5F) which also represents an accepted measure of anxiety-related behaviour in mice (Crawley, 1985). Although more tests are required to make a certain conclusion regarding the anxiety status of these animals the changes in thigmotaxis and centre entry times displayed by our ECR1KO male and female animals suggest that ECR1 may play a role in modulating anxiety-related behaviour in mice.
Fig. 5.
An open field test analysis comparing wild type (WT) and ECR1KO (KO) female and male mice demonstrating the total distance travelled (A), the average speed of the animals (B), the time in the periphery (C), The number of entries into the centre of the test arena (D) the time spent in the centre zone of the arena (E) and numbers of faecal boli deposited (F). n = 8–10, error bars = SDM, ns = not significant, *; p ≤ 0.05, **; p ≤ 0.01. (F-values; A, p = 0.2, F = 1.524; B, p = 0.3, F = 1.099; C, p = 0.0001, f = 9.647, D, p = 0.0048, F = 5.082; E, p = 0.0001, F = 9.092).
4. Discussion
We had previously demonstrated the presence of a highly conserved and polymorphic region of DNA within intron 2 of the CNR1 gene that demonstrated an ability to upregulate promoter function in different primary neuronal cell types (Nicoll et al., 2012). In addition, different polymorphic variants of ECR1 induced altered levels of TATA box promoter activity suggesting that polymorphic variation within ECR1 may play a functional role in determining the activity of the CNR1 promoter (Zhang et al., 2004). Following our discovery that ECR1 had enhancer-like properties we asked whether ECR1 played a role in modulating CNR1 gene expression in vivo. We also asked whether disrupting ECR1 would have any effects on physiologies associated with changes in CB1 expression including cannabinoid response, food and alcohol intake, movement or anxiety-related behaviour.
Analysis of tissues derived from animals containing the CAS9-driven ECR1 disruption demonstrated a significant reduction of CNR1 mRNA within the hippocampus of these animals compared to wild type littermates but did not seem to affect gross levels of CNR1 mRNA expression in the hypothalamus. These observations suggest that the mouse ECR1 enhancer; that is represented by the ancestral T-allele in all mammalian species except humans, has tissue specific properties and is only required to support the expression of the CNR1 gene in specific tissues and at specific times. The observation that ECR1 only supports CNR1 mRNA expression in the hippocampus is not surprising as many genes depend on the combined input of many enhancers to ensure that they are expressed in appropriate levels in different cell types and in response to the correct cues (Rickels and Shilatifard, 2018). Moreover, expression of CNR1 and CB1 in hippocampus has been previously shown to be markedly different to that found in the hypothalamus providing further evidence of the existence of different regulatory mechanisms governing CNR1 expression in these regions (Howlett et al., 2002). Although we have identified a reduction in gross CNR1 expression in our ECR1KO mice, more localised comparisons of CNR1 mRNA expression remains to be done in more localised regions of the brain such as the nucleus accumbens, amygdala, ventral tegmental area and pre-frontal cortex.
The observation that disruption of ECR1 reduces the hypothermic response of CB1 agonism was consistent with a role for ECR1 in controlling CB1 receptor expression. The modulation of body temperature by CB1 agonism is regulated by a part of the hypothalamus known as the preoptic anterior hypothalamus (POAH) (Rawls et al., 2002). However, as described above, we were unable to detect any change in the expression of the CNR1 mRNA in hypothalamic tissues by QrtPCR. This suggests that ECR1 only controls CNR1 expression in a small region of the hypothalamus and, judging by the effects of ECR1 disruption on thermoregulation following CB1 agonism, within groups of cells that include the POAH. Fine dissection of the POAH followed by QrtPCR would conclusively address this hypothesis.
We also demonstrated that disruption of ECR1 reduces alcohol intake in both male and female mice. This observation is consistent with a role for ECR1 in controlling CNR1 expression as mice lacking the CB1 gene have reduced ethanol intake (Hungund et al., 2003). Critically, humans containing specific haplotypes of an LD block containing the ECR1 enhancer are more susceptible to addictive behaviours including addiction to alcohol (Hutchison et al., 2008). Thus, the identification of a polymorphic enhancer within the CNR1 locus that modulates ethanol intake is important to our understanding of how polymorphisms within the human genome contribute to increased alcohol intake. Previous analyses of the regions of the brain where CB1 is known to affect alcohol intake suggest the involvement of CB1 expression in the nucleus accumbens (NAc) (Hungund and Basavarajappa, 2004). Future studies will involve a more focussed analysis of CNR1 mRNA from the NAc to determine whether the reduction in alcohol intake observed was linked to a reduction in the expression of CNR1 mRNA in this region of the brain. In contrast to our observed decrease in ethanol intake we did not observe any difference in food intake or fat deposition between ECR1KO or WT animals suggesting that the role played by ECR1 in fat and ethanol intake differed and provides clues suggesting that the mechanisms modulating expression of the CB1 receptor in ethanol and food intake also differ.
We showed that disruption of ECR1 caused a significant decrease in the expression of CNR1 in the hippocampus where CB1 gene expression plays a critical role in protecting the nervous system against the depression associated effects of stress (Scarante et al., 2017). We therefore explored the effects of the disruption of ECR1 on levels of anxiety-related behaviour in ECR1KO mice using the open field test which assesses locomoter movement and thigmotaxis; the need to stay close to the walls of the test arena. The value of using thigmotaxis as a measure of anxiety like behaviour was demonstrated when animals given anxiolytic drugs such as diazepan or chlordiazepoxide displayed significantly reduced thigmotaxis (Choleris et al., 2001). Female mice displayed increased levels of thigmotaxis and spent less time in the centre of the OFT arena suggesting increased anxiety levels in female mice lacking ECR1 which is consistent with previous observations of increased anxiety-related behaviour in CNR1 gene disrupted mice (Patel and Hillard, 2009). However, in male mice we show that disruption of ECR1 appears to decrease anxiety related behaviours as these mice spend a significantly longer time in the centre of the OFT and less time at the periphery. Although this observation is compelling and consistent with a role for ECR1 in governing the manifestation of anxiety-related behaviour, more work needs to be done using tests such as the elevated zero maze, light-dark experiments or novelty suppressed feeding test before a conclusive role for ECR1 in mood modulation can be asserted. These observations also suggest that the role of ECR1 in anxiety-related behaviour modulation is sex specific. Whilst a great deal of work remains to be undertaken to determine the mechanisms governing ECR1 activity with respect to the generation of anxiety-related behaviour it is likely that the direct or indirect effects of testosterone or oestrogens on ECR1 activity, or the CNR1 promoter, would be a good place to start.
Another possibility that requires further exploration is that ECR1 may play a dual role in modulating the tissue specific expression at the transcriptional level whilst also modulating the identity and amount of different splice forms that are produced from the human CNR1 gene in different tissues. Fine dissection of neuronal tissues in mice lacking ECR1 combined with mRNA isoform mRNA analysis using splice-form specific PCR primers would allow us to address this possibility.
As mentioned above, it is likely that ECR1 only represents a small component of the regulatory machinery required to modulate the dynamic expression of the CNR1 gene in the brain. Although we initially chose to explore ECR1, due to the disease association of its SNP, we also observed that the CNR1 locus is bracketed by many other regions of even higher non-coding conservation, many of which have been as highly conserved as the CNR1 coding region, that may also represent enhancers involved in modulating CNR1 expression. Therefore, our analysis of the ECR1 enhancer only represents the start of a larger study to identify the core enhancer elements that modulate the expression of the CNR1 locus.
5. Conclusions
We have used comparative genomics and CRISPR/CAS9 genome editing to demonstrate that the ECR1 enhancer sequence plays a role in several physiologies including anxiety-related behaviour, alcohol intake and cannabinoid response. Moreover, we demonstrate that disruption of ECR1 has a significant effect on the expression of the CNR1 gene in the hippocampus. The functional analysis of the ECR1 enhancer sequence using the combination of comparative genomics and CRISPR genome editing described in the current manuscript lays the foundation for further studies on the effects of polymorphic variation on the activity of ECR1 and how these polymorphisms may contribute to anxiety-related disorders, substance abuse disorders. Furthermore, establishing a functional role for ECR1 in cannabinoid response may provide a platform for exploring a role for ECR1 polymorphisms in stratified response to drugs targeted against the CB1 receptor and in contributing to our understanding of cannabinoid pharmacogenetics. The exciting combination of comparative genomics and CRISPR genome editing described in the current manuscript is a blueprint for further studies to identify and characterise the functional non-coding elements essential for health well as permitting novel insights into identifying the genetic and epigenetic mechanisms contributing to disease susceptibility.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgements
EH was funded by Medical Research Scotland (PhD-719-2013) and GW Pharmaceuticals. AMcE was funded by BBSRC project grant (BB/N017544/1). PB and DW are funded by the Scottish Government Rural and Environment Science and Analytical Services Division to the Rowett Institute.
References
- Akirav I. The role of cannabinoids in modulating emotional and non-emotional memory processes in the hippocampus. Front. Behav. Neurosci. 2011;5:34. doi: 10.3389/fnbeh.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambico F.R., Duranti A., Tontini A., Tarzia G., Gobbi G. Endocannabinoids in the treatment of mood disorders: evidence from animal models. Curr. Pharm. Des. 2009;15:1623–1646. doi: 10.2174/138161209788168029. [DOI] [PubMed] [Google Scholar]
- Benzinou M., Chevre J.C., Ward K.J., Lecoeur C., Dina C., Lobbens S., Durand E., Delplanque J., Horber F.F., Heude B., Balkau B., Borch-Johnsen K., Jorgensen T., Hansen T., Pedersen O., Meyre D., Froguel P. Endocannabinoid receptor 1 gene variations increase risk for obesity and modulate body mass index in European populations. Hum. Mol. Genet. 2008;17:1916–1921. doi: 10.1093/hmg/ddn089. [DOI] [PubMed] [Google Scholar]
- Chen X., Williamson V.S., An S.S., Hettema J.M., Aggen S.H., Neale M.C., Kendler K.S. Cannabinoid receptor 1 gene association with nicotine dependence. Arch. Gen. Psychiatry. 2008;65:816–824. doi: 10.1001/archpsyc.65.7.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choleris E., Thomas A.W., Kavaliers M., Prato F.S. A detailed ethological analysis of the mouse open field test: effects of diazepam, chlordiazepoxide and an extremely low frequency pulsed magnetic field. Neurosci. Biobehav. Rev. 2001;25:235–260. doi: 10.1016/s0149-7634(01)00011-2. [DOI] [PubMed] [Google Scholar]
- Cota D., Marsicano G., Tschop M., Grubler Y., Flachskamm C., Schubert M., Auer D., Yassouridis A., Thone-Reineke C., Ortmann S., Tomassoni F., Cervino C., Nisoli E., Linthorst A.C., Pasquali R., Lutz B., Stalla G.K., Pagotto U. The endogenous cannabinoid system affects energy balance via central orexigenic drive and peripheral lipogenesis. J. Clin. Invest. 2003;112:423–431. doi: 10.1172/JCI17725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley J.N. Exploratory behavior models of anxiety in mice. Neurosci. Biobehav. Rev. 1985;9:37–44. doi: 10.1016/0149-7634(85)90030-2. [DOI] [PubMed] [Google Scholar]
- Femenia T., Garcia-Gutierrez M.S., Manzanares J. CB1 receptor blockade decreases ethanol intake and associated neurochemical changes in fawn-hooded rats. Alcohol. Clin. Exp. Res. 2010;34:131–141. doi: 10.1111/j.1530-0277.2009.01074.x. [DOI] [PubMed] [Google Scholar]
- Grim T.W., Morales A.J., Gonek M.M., Wiley J.L., Thomas B.F., Endres G.W., Sim-Selley L.J., Selley D.E., Negus S.S., Lichtman A.H. Stratification of cannabinoid 1 receptor (CB1R) agonist efficacy: manipulation of CB1R density through use of transgenic mice reveals congruence between in vivo and in vitro assays. J. Pharmacol. Exp. Ther. 2016;359:329–339. doi: 10.1124/jpet.116.233163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms D.W., Quadros R.M., Seruggia D., Ohtsuka M., Takahashi G., Montoliu L., Gurumurthy C.B. Mouse genome editing using the CRISPR/Cas system. Curr. Protoc. Hum. Genet. 2014;83(15):11–27. doi: 10.1002/0471142905.hg1507s83. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay E.A., Khalaf A.R., Marini P., Brown A., Heath K., Sheppard D., MacKenzie A. An analysis of possible off target effects following CAS9/CRISPR targeted deletions of neuropeptide gene enhancers from the mouse genome. Neuropeptides. 2016 doi: 10.1016/j.npep.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett A.C., Barth F., Bonner T.I., Cabral G., Casellas P., Devane W.A., Felder C.C., Herkenham M., Mackie K., Martin B.R., Mechoulam R., Pertwee R.G. International union of pharmacology. XXVII. Classification of cannabinoid receptors. Pharmacol. Rev. 2002;54:161–202. doi: 10.1124/pr.54.2.161. [DOI] [PubMed] [Google Scholar]
- Hungund B.L., Basavarajappa B.S. Role of endocannabinoids and cannabinoid CB1 receptors in alcohol-related behaviors. Ann. N. Y. Acad. Sci. 2004;1025:515–527. doi: 10.1196/annals.1316.064. [DOI] [PubMed] [Google Scholar]
- Hungund B.L., Szakall I., Adam A., Basavarajappa B.S., Vadasz C. Cannabinoid CB1 receptor knockout mice exhibit markedly reduced voluntary alcohol consumption and lack alcohol-induced dopamine release in the nucleus accumbens. J. Neurochem. 2003;84:698–704. doi: 10.1046/j.1471-4159.2003.01576.x. [DOI] [PubMed] [Google Scholar]
- Hutchison K.E., Haughey H., Niculescu M., Schacht J., Kaiser A., Stitzel J., Horton W.J., Filbey F. The incentive salience of alcohol: translating the effects of genetic variant in CNR1. Arch. Gen. Psychiatry. 2008;65:841–850. doi: 10.1001/archpsyc.65.7.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icick R., Peoc’h K., Karsinti E., Ksouda K., Hajj A., Bloch V., Prince N., Mouly S., Bellivier F., Lepine J.P., Laplanche J.L., Vorspan F. A cannabinoid receptor 1 polymorphism is protective against major depressive disorder in methadone-maintained outpatients. Am. J. Addict. 2015;24:613–620. doi: 10.1111/ajad.12273. [DOI] [PubMed] [Google Scholar]
- Irving A.J., Coutts A.A., Harvey J., Rae M.G., Mackie K., Bewick G.S., Pertwee R.G. Functional expression of cell surface cannabinoid CB(1) receptors on presynaptic inhibitory terminals in cultured rat hippocampal neurons. Neuroscience. 2000;98:253–262. doi: 10.1016/s0306-4522(00)00120-2. [DOI] [PubMed] [Google Scholar]
- Ketcherside A., Noble L.J., McIntyre C.K., Filbey F.M. Cannabinoid receptor 1 gene by cannabis use interaction on CB1 receptor density. Cannabis Cannabinoid Res. 2017;2:202–209. doi: 10.1089/can.2017.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand F., De Witte P. SR147778, a CB1 cannabinoid receptor antagonist, suppresses ethanol preference in chronically alcoholized Wistar rats. Alcohol. 2006;39:125–134. doi: 10.1016/j.alcohol.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Laprairie R.B., Kelly M.E., Denovan-Wright E.M. The dynamic nature of type 1 cannabinoid receptor (CB(1)) gene transcription. Br. J. Pharmacol. 2012;167:1583–1595. doi: 10.1111/j.1476-5381.2012.02175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledent C., Valverde O., Cossu G., Petitet F., Aubert J.F., Beslot F., Bohme G.A., Imperato A., Pedrazzini T., Roques B.P., Vassart G., Fratta W., Parmentier M. Unresponsiveness to cannabinoids and reduced addictive effects of opiates in CB1 receptor knockout mice. Science. 1999;283:401–404. doi: 10.1126/science.283.5400.401. [DOI] [PubMed] [Google Scholar]
- Montalbano A., Canver M.C., Sanjana N.E. High-throughput approaches to pinpoint function within the noncoding genome. Mol. Cell. 2017;68:44–59. doi: 10.1016/j.molcel.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A.G.M., Vinterstein K., Behringer R. third edition ed. Cold Spring Harbor laboratory Press, Cold Spring Harbor; 2003. Manipulating the Mouse Embryo. [Google Scholar]
- Nicoll G., Davidson S., Shanley L., Hing B., Lear M., McGuffin P., Ross R., MacKenzie A. Allele-specific differences in activity of a novel cannabinoid receptor 1 (CNR1) gene intronic enhancer in hypothalamus, dorsal root ganglia, and hippocampus. J. Biol. Chem. 2012;287:12828–12834. doi: 10.1074/jbc.M111.336750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S., Hillard C.J. Role of endocannabinoid signaling in anxiety and depression. Curr. Top. Behav. Neurosci. 2009;1:347–371. doi: 10.1007/978-3-540-88955-7_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pava M.J., Woodward J.J. A review of the interactions between alcohol and the endocannabinoid system: implications for alcohol dependence and future directions for research. Alcohol. 2012;46:185–204. doi: 10.1016/j.alcohol.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomorska D.K., do-Rego J.C., do-Rego J.L., Zubrzycka M., Janecka A. Opioid and cannabinoid system in food intake. Curr. Pharm. Des. 2016;22:1361–1370. doi: 10.2174/1381612822666160125114144. [DOI] [PubMed] [Google Scholar]
- Rawls S.M., Cabassa J., Geller E.B., Adler M.W. CB1 receptors in the preoptic anterior hypothalamus regulate WIN 55212-2 [(4,5-dihydro-2-methyl-4(4-morpholinylmethyl)-1-(1-naphthalenyl-carbonyl)-6H-pyrr olo[3,2,1ij]quinolin-6-one]-induced hypothermia. J. Pharmacol. Exp. Ther. 2002;301:963–968. doi: 10.1124/jpet.301.3.963. [DOI] [PubMed] [Google Scholar]
- Rickels R., Shilatifard A. Enhancer logic and mechanics in development and disease. Trends Cell Biol. 2018;28:608–630. doi: 10.1016/j.tcb.2018.04.003. [DOI] [PubMed] [Google Scholar]
- Scarante F.F., Vila-Verde C., Detoni V.L., Ferreira-Junior N.C., Guimaraes F.S., Campos A.C. Cannabinoid modulation of the stressed hippocampus. Front. Mol. Neurosci. 2017;10:411. doi: 10.3389/fnmol.2017.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacht J.P., Hutchison K.E., Filbey F.M. Associations between cannabinoid receptor-1 (CNR1) variation and hippocampus and amygdala volumes in heavy cannabis users. Neuropsychopharmacology. 2012;37:2368–2376. doi: 10.1038/npp.2012.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener M.L., Wooten M.C. Use of the Open Field Maze to measure locomotor and anxiety-like behavior in mice. J. Vis. Exp. 2015 doi: 10.3791/52434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanley L., Davidson S., Lear M., Thotakura A.K., McEwan I.J., Ross R.A., Mackenzie A. Long-range regulatory synergy is required to allow control of the TAC1 locus by MEK/ERK signalling in sensory neurones. Neurosignals. 2010;18:173–185. doi: 10.1159/000322010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanley L., Lear M., Davidson S., Ross R., Mackenzie A. Evidence for regulatory diversity and auto-regulation at the TAC1 locus in sensory neurones. J. Neuroinflammation. 2011;8:10. doi: 10.1186/1742-2094-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Pinilla P., Roiz-Santianez R., Ortiz-Garcia de la Foz V., Guest P.C., Ayesa-Arriola R., Cordova-Palomera A., Tordesillas-Gutierrez D., Crespo-Facorro B. Brain structural and clinical changes after first episode psychosis: focus on cannabinoid receptor 1 polymorphisms. Psychiatry Res. 2015;233:112–119. doi: 10.1016/j.pscychresns.2015.05.005. [DOI] [PubMed] [Google Scholar]
- Viveros M.P., de Fonseca F.R., Bermudez-Silva F.J., McPartland J.M. Critical role of the endocannabinoid system in the regulation of food intake and energy metabolism, with phylogenetic, developmental, and pathophysiological implications. Endocr. Metab. Immune Disord. Drug Targets. 2008;8:220–230. doi: 10.2174/187153008785700082. [DOI] [PubMed] [Google Scholar]
- Viveros M.P., Marco E.M., Llorente R., Lamota L. The role of the hippocampus in mediating emotional responses to nicotine and cannabinoids: a possible neural substrate for functional interactions. Behav. Pharmacol. 2007;18:375–389. doi: 10.1097/FBP.0b013e3282d28fb4. [DOI] [PubMed] [Google Scholar]
- Zhang P.W., Ishiguro H., Ohtsuki T., Hess J., Carillo F., Walther D., Onaivi E.S., Arinami T., Uhl G.R. Human cannabinoid receptor 1: 5′ exons, candidate regulatory regions, polymorphisms, haplotypes and association with polysubstance abuse. Mol. Psychiatry. 2004;9:916–931. doi: 10.1038/sj.mp.4001560. [DOI] [PubMed] [Google Scholar]