Abstract
Peroxisomes are multifunctional organelles with roles in cellular metabolism, cytotoxicity, and signaling. The plastic nature of these organelles allows them to respond to diverse biological processes, such as virus infections, by remodeling their biogenesis, morphology, and composition to enhance specific functions. During virus infections in humans, peroxisomes act as important immune signaling organelles, aiding the host by orchestrating antiviral signaling. However, more recently it was discovered that peroxisomes can also benefit the virus, facilitating virus–host interactions that rewire peroxisomes to support cellular processes for virus replication and spread. Here, we describe recent studies that uncovered this double-edged character of peroxisomes during infection, highlighting mechanisms that viruses have coevolved to take advantage of peroxisome plasticity. We also provide a perspective for future studies by comparing the established roles of peroxisomes in plant infections and discussing the promise of virology studies as a venue to reveal the uncharted biology of peroxisomes.
Peroxisome Biology and Virus Infections: A Changing Outlook
Nearly a decade ago, peroxisomes were identified as important immune signaling organelles in humans [1]. This discovery prompted much effort towards characterizing the peroxisome-mediated immune response [2,3], redefining the scientific community’s view of peroxisomes as major cellular organelles. In fact, peroxisomes are multifunctional and dynamic, and are known to play a variety of critical roles in development [4-6], cellular stress [7,8], cancer [9], and virus infection in plants [10]. Despite this, and the knowledge that human viruses cause extensive changes in other host organelles [11], peroxisomal characteristics beyond immune signaling have not been deeply investigated in cells under invasion by pathogens.
Several recent studies have tackled this broader question of peroxisome biology in infection, and have uncovered diverse virus–host interactions that rewire peroxisomal metabolism, structure, and biogenesis to support virus replication and spread. These investigations also discovered new links between peroxisome dynamics and functions, revealing relationships at the core of peroxisome biology that have long been challenging to study. The peroxisome is emerging as a double-edged sword of infection, an organelle that can be hijacked to serve either the virus or the host. The findings we review here have contributed to a changing perspective of peroxisomes in infectious diseases, identifying new possibilities for antiviral therapeutics in addition to establishing a venue to examine the fundamentals of peroxisome biology.
Peroxisomes Are Plastic Organelles with Diverse Roles in Health and Disease
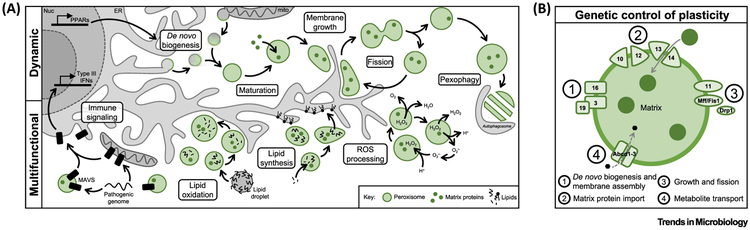
Among the last membrane-bound organelles to be defined, peroxisomes are found in nearly all eukaryotic organisms, acting as the subcellular hubs of reactive oxygen species (ROS) regulation and lipid and amino acid β-oxidation [12] (Figure 1A). They are also plastic organelles; peroxisome numbers, shape, and composition are fluid properties, often changing to tailor peroxisome functions in response to cellular demands. Peroxisome biogenesis is primarily controlled by peroxin (PEX) genes, which regulate self-sustaining cycles of maturation, fission, and pexophagy [13-16] (Figure 1B). However, peroxisomes can also form de novo by the fusion of pre-peroxisomal vesicles derived from the endoplasmic reticulum (ER) and mitochondria [17-19], a process initiated by transcription factors known as peroxisome proliferator-activated receptors (PPARs). The morphology of peroxisomes is likely regulated by a combination of molecular motor activity [20] and ER-mediated lipid transfer [21,22], which allows for peroxisome membrane growth during maturation and prior to fission. Additionally, peroxisome composition is dictated by the expression and import of PEX proteins, peroxisome membrane proteins (PMPs), cellular metabolites, and metabolic enzymes in the peroxisome matrix [12,13].
Figure 1. Peroxisomes Are Dynamic and Multifunctional Organelles.
(A) The peroxisome biogenesis cycle in mammalian cells, including de novo biogenesis, peroxisome growth, fission, and pexophagy. Primary peroxisome functions in human cells are depicted below. (B) Major peroxisome proteins involved in peroxisome formation and maintenance. PEX proteins are indicated by their respective gene number. Abbreviations: IFN, interferon; ROS, reactive oxygen species.
The dynamic regulation of peroxisomes is an essential aspect of peroxisome biology, illustrated by the high variation in peroxisome properties across cell types, organisms, and even temporally within an individual cell. Numerous studies have linked peroxisome dynamics to condition- or organism-specific peroxisome functions. For example, PEX genes are upregulated [23] and peroxisomes are elongated [8,24,25] during cell stress, and peroxisome numbers are increased in cell types with elevated lipid catabolism [26,27], during mitosis [6], in cancerous cells [9], and during cell differentiation [4,28]. These changes may enhance metabolic output for the high energy consumption required for these processes. In addition, peroxisome content can be markedly different between organisms, leading to additional organelle functions such as the glyoxylate cycle in plants and plasmalogen synthesis in animals [12,29]. Alternatively, an absence of peroxisomes, such as in peroxisome biogenesis disorders (PBDs, e.g., Zellweger syndrome), inhibits processes that often overlap with mitochondrial, lysosomal, and ER-mediated functions [30]. As investigations were long limited to characterizing peroxisomes in single conditions or cell types, many peroxisome characteristics and functions have only recently been elucidated, and it is becoming increasingly evident that peroxisome plasticity is critical for human development and disease.
Peroxisome-Driven Immune Signaling at the Virus–Host Interface
The peroxisome was first recognized as a key subcellular signaling center upon the discovery that the RIG-I-like receptor (RLR) adaptor protein MAVS, an innate immune signaling factor, localizes to the peroxisome in human cells [1]. MAVS, previously thought to be exclusive to the mitochondria, mounts a rapid antiviral response upon the RIG-I-mediated sensing of pathogenic genomes, specifically controlling the type III interferon response when activated at the peroxisome [2]. Combined with the known cellular detoxification functions of peroxisomes, the identification of peroxisomal MAVS lead to the idea that peroxisomes primarily act in host defense during infection as antiviral signaling organelles.
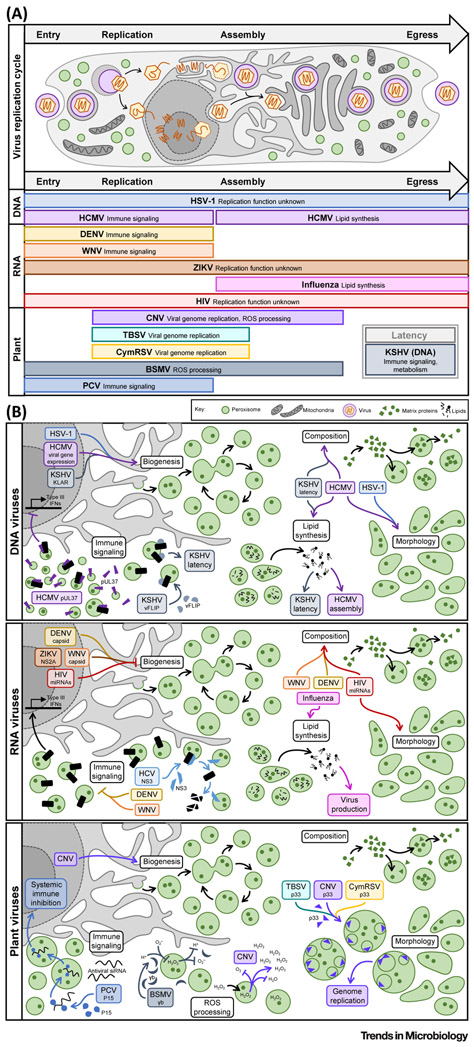
Further evidence for the antiviral function of peroxisomes comes from the finding that viruses have acquired several mechanisms to inhibit peroxisome-mediated immune signaling (Figure 2A). Some, including hepatitis C virus (HCV) and human cytomegalovirus (HCMV), use viral proteins to directly target peroxisomal MAVS and inhibit its function. HCV encodes the multifunctional serine protease NS3-4A, which localizes to peroxisome membranes and cleaves MAVS, resulting in its degradation [31,32] (Figure 2B). Similarly, the HCMV protein pUL37 (also known as vMIA) localizes to both mitochondria and peroxisomes to sequester MAVS, drive organelle fragmentation, and disrupt antiviral signaling [33]. In a different twist of fate, Kaposi’s sarcoma-associated herpesvirus (KSHV) repurposes peroxisomal MAVS to facilitate latency, using it to stabilize expression of the viral protein vFLIP and maintain oncogenic activity [34].
Figure 2. Viruses Manipulate Peroxisome Functions in a Temporal Manner during the Viral Replication Cycle.
(A) Upper panel. General schematic of the cellular life cycle of an enveloped virus, depicting both a nuclear and cytoplasmic genome replication route. Lower panel. Virus-induced peroxisome modulations occur in accordance with each stage (entry, replication, assembly, egress, or latency) of the virus replication cycle. For each virus, the rectangle length corresponds to the stages of infection known to correspond with peroxisome remodeling, with the virus name in bold and beside the regulated function. Virus type is indicated to the left. (B) Diverse aspects of peroxisome biology are regulated during infection with human DNA (upper), human RNA (middle), and plant (lower) viruses. In each case, the virus is indicated with the respective viral factor or process (if known) in italics. Abbreviations: BSMV, barley stripe mosaic virus; CNV, cucumber necrosis virus; CymRSV, Cymbidium ringspot virus; DENV, dengue virus; HCMV, human cytomegalovirus; HCV, hepatitis C virus; HIV, human immunodeficiency virus; HSV-1, herpes simplex virus type 1; KSHV, Kaposi’s sarcoma-associated herpesvirus; PCV, peanut clump virus; ROS, reactive oxygen species; TBSV, tomato bushy stunt virus; WNV, West Nile virus; ZIKV, Zika virus.
Other aspects of peroxisome plasticity, beyond MAVS localization, are also important in the regulation of immune signaling. By examining peroxisome-mediated immune signaling under exposure to a range of cellular conditions and pathogens, Odendall et al. showed that an increase in peroxisome numbers enhances the type III interferon response [2]. This suggested that peroxisome dynamics are tightly linked to the magnitude of peroxisome-dependent antiviral signaling, and recent studies of virus infections have further demonstrated this relationship (Figure 2B). For example, the flaviviruses dengue (DENV) and West Nile (WNV) target their capsid proteins to peroxisome membranes, where they interact with the biogenesis factor PEX19, cause a nearly 40% decrease in peroxisome numbers, and consequently inhibit type III interferon expression [35]. Zika virus (ZIKV), another member of the Flavivirus family, alters peroxisome numbers through a different mechanism: the viral protein NS2A localizes at peroxisomes and interacts with both PEX19 and PEX3 to inhibit peroxisome biogenesis [36]. While this has not been fully linked to immune modulation, it is likely that ZIKV inhibits peroxisomal signaling in a similar manner to its DENV and WNV family members.
While these examples support a correlative – and rather intuitive – link between the number of peroxisomes in a cell and the magnitude of peroxisome-mediated antiviral signaling, the relationship between peroxisome dynamics and immune signaling seems more complex. For example, Bender et al. observed that cells lacking peroxisomes still mounted an immune response during HCV infection [31]. This finding could be explained, in part, by the presence of ER- and mitochondrial-derived pre-peroxisomal vesicles during de novo biogenesis (Figure 1A), which may be capable of antiviral signaling. However, the flavivirus studies further found that, surprisingly, permanently reducing peroxisome numbers (e.g., via PEX gene knockdowns or in PBD patient samples) negatively impacts virus production [35,36]. This was also illustrated in HCMV and herpes simplex type 1 (HSV-1) infections, along with the discovery that these viruses increase peroxisome biogenesis during infection [37] (Figure 2B). Moreover, cells from patients with Zellweger’s syndrome, which lack peroxisomes entirely, were shown to be resistant to infection by these herpesviruses [37]. These results muddle what is expected of peroxisomes as antiviral organelles, and suggest that other peroxisome functions may be required for virus replication.
Going Proviral: Peroxisome Metabolism and Virus Replication
Certain cellular lipids, including very-long-chain fatty acids (e.g., docosahexaenoate, DHA) and ether lipids (e.g., plasmalogens), can only be synthesized at the peroxisome, often in sync with β-oxidation pathways (Figure 1A) [26,27]. These lipids are thought to be particularly important for membrane integrity and fluidity, highlighted by their involvement in PBD and neurodegeneration [38,39], and plasmalogens specifically are key participants in membrane fission and fusion events [40]. Due to these features, peroxisome-synthesized lipids are intriguing candidates for a variety of infection-induced processes, especially formation of viral envelopes, modulation of cellular trafficking, and maintenance of host and virus fitness. Indeed, plasmalogens were shown to be components of infectious particles for several enveloped viruses, including HCMV [41], WNV [42], and influenza [43].
Several recent studies have identified peroxisomal lipid synthesis as a proviral peroxisomal function (Figure 2A). For example, Jean Beltran et al. uncovered elevated peroxisome-mediated plasmalogen synthesis as a requirement for secondary envelopment during the assembly of HCMV [37]. Given their finding that HSV-1 drives similar changes to peroxisomes while the nonenveloped adenovirus (ADV5) does not, in conjunction with the knowledge that plasmalogens are enriched in HCMV virions [41], the authors proposed that peroxisomal lipid metabolism is a general feature of enveloped virus infections. In support of this, a previous study of influenza virus, an RNA virus, showed that ether lipid metabolism was induced and required upon infection, and that peroxisome-derived lipids were enriched in influenza virions [43]. Along a different vein, Sychev et al. discovered that peroxisomal lipid metabolism, notably synthesis of DHA, was required for maintenance of KSHV latency, protecting infected cells from premature death [44].
A range of changes in peroxisome numbers, composition, and shape were shown to accompany the above mentioned infection-induced metabolic rewiring (Figure 2B). HCMV, HSV-1, and KSHV infections increase the number of peroxisomes per cell – specifically by modulation of peroxisome fission in the case of HCMV [37,44]. HCMV and KSHV were further shown to alter peroxisome composition, increasing proteins involved in biogenesis (HCMV [37,45]) and lipid metabolism (HCMV [37,45], KSHV [44]). Additionally, HCMV and HSV-1 perturbed peroxisome morphology by inducing flattened and irregularly shaped peroxisomes that caused enhanced plasmalogen synthesis during infection [37]. While these peroxisome phenotypes have not yet been investigated in influenza infection, the conserved functional changes suggest that influenza is altering peroxisome plasticity in similar ways. Alternatively, other viruses are known to drive these peroxisome phenotypes, but their functions remain unclear. Human immunodeficiency virus (HIV) infection, for example, drives the expression of brain miRNAs (e.g., miR-500a-5p, miR-34c-3p, miR-93-3p, miR-381-3p) that change peroxisome numbers, cause elongated peroxisome morphology, and alter peroxisome composition [46]. As HIV is also an enveloped virus, it is possible that these peroxisome features are being rewired to enhance lipid synthesis for virion assembly.
Peroxisomes and Plant Viruses
In addition to this growing body of knowledge in human infections, peroxisomes have long been recognized as key players in the replication cycles of plant viruses. While plant pathogens engage peroxisome biology in ways that often contrast with viral mechanisms in humans, underlying themes can be found between these strategies. For example, tombusviruses, a family of nonenveloped RNA plant viruses that includes cucumber necrosis virus (CNV), tomato bushy stunt virus (TBSV), and Cymbidium ringspot virus (CymRSV), use peroxisomes as scaffolds for virus replication factories (Figure 2B). Upon infection, the viral auxiliary replicase (p33) and polymerase (p92) are targeted to peroxisome membranes to generate peroxisome-derived multivesicular bodies (pMVBs), which serve as the location for virus genome replication [47-49]. The formation of pMVBs requires the careful orchestration of host and viral components within peroxisomes, involving ESCRT, SNARE, and PEX proteins, as well as a specific p33–Pex19 interaction required for pMVB initiation [50-52]. In CNV infection this has been further linked to increased de novo peroxisome biogenesis, facilitating high viral output [48]. As discussed above, Pex19-mediated virus–host interactions and the modulation of peroxisome biogenesis have both been shown in human infections, but peroxisomes have not been specifically implicated in viral genome replication. However, similar viral mechanisms exist for other organelles, such as the ER (e.g., DENV, vaccinia virus) and Golgi (e.g., poliovirus) [11].
Plant viruses are also known to engage peroxisome functions that are currently a black box in human infections, such as ROS homeostasis (Figure 2B). CNV infection, in addition to forming pMVBs, causes peroxide accumulation in infected cells, likely contributing to the virus-induced increase in peroxisome biogenesis [48]. Alternatively, barley stripe mosaic virus (BSMV) dysregulates peroxisome ROS processing in order to bypass the host immune response [53]. In plants, the balance between hydrogen ions, radical oxygens, and peroxide dictates immune signaling, and elevated peroxide causes hypersensitivity to pathogen invasion [48,54,55]. BSMV has coevolved a mechanism to combat ROS-induced host defense via the multifunctional viral protein γb, which localizes to peroxisomes and suppresses the function of glycolate oxidase (GOX), a peroxisome protein involved in redox. This γb–GOX interaction serves a proviral role by decreasing peroxide production, therefore inhibiting host defense and promoting virus replication. Intriguingly, ROS dysregulation and oxidative stress is a known feature of many human virus infections and has even been linked to immune evasion in some, such as influenza and poxvirus infections, but how the peroxisome regulates these processes remains unclear.
Beyond ROS processing, the major antiviral response mechanism in plants is the generation and cell–cell spread of small silencing RNAs (siRNAs) that target viral genomes [56,57]. Although long thought to be a peroxisome-independent process, Incarbone et al. recently demonstrated that viruses can bypass this defense by shuttling host siRNAs into the peroxisome matrix, sequestering them from the effector molecules necessary to induce a response (Figure 2B) [58]. Peanut clump virus (PCV), specifically, uses its viral suppressor of RNA silencing (VSR) protein P15 to bind host siRNAs and translocate them from the cytoplasm into the peroxisome, facilitating productive infection and systemic spread [58]. While it remains to be seen if other plant or animal viruses participate in analogous modes of organelle confinement, this study highlighted the diverse strategies viruses have evolved to hijack peroxisome plasticity. Moreover, it has become increasingly evident that human viruses modulate protein functions by inducing translocations between organelles [59], and it is likely that peroxisomes are involved in similar processes.
Concluding Remarks
Peroxisomes are critical cellular organelles that play dual roles during the progression of human virus infections. Either the host or pathogen can leverage peroxisome functions to achieve antiviral defense or virus replication, a subcellular battle that plays out across infection time. Upon virus entry, the host engages peroxisomal MAVS to quickly respond to invasion and limit the spread of infection. However, reflective of the constant coevolution between viruses and hosts, the discovery of this antiviral mechanism was followed by the discovery of viral mechanisms that subvert or bypass this host defense. By controlling peroxisome biogenesis, morphology, and composition at the later stages of infection, viruses have coevolved strategies to turn peroxisome functions to the benefit of virus production and spread. Peroxisomes are thus poised at the virus–host interface, contributing to the outcome of infections across both viral and host species.
Due to the highly plastic character of peroxisomes and overlapping biology with other host organelles (e.g., mitochondria), defining the molecular pathways that regulate peroxisome dynamics has remained an unresolved challenge for researchers. We now know that studies of viral infections provide the appropriate context and the opportunity for addressing questions at the core of peroxisome biology. For example, the observation that peroxisome numbers and protein composition are manipulated during a variety of virus infections suggests that there may be a common transcriptional switch that is induced upon infection. One possibility is the PPAR family of transcription factors (PPARα, PPARγ, and PPARβ/δ). However, PPARα is the only member known to induce peroxisome biogenesis, while the roles of PPARγ and PPARβ/δ in peroxisome biogenesis remain controversial [16]. Other factors, such as the transcriptional coactivator PGC-1α [60] and several peroxisome proliferator drugs [61], are known to act independently of PPARα, indicating additional yet-to-be-discovered peroxisome regulators. Interestingly, multiple viruses are known to modulate these noncanonical transcriptional activators, including PPARγ [46,62-64] and PGC-1α [65], but how this plays into the regulation of peroxisome dynamics during infection is not understood. As the upregulation of peroxisome genes and numbers is essential for the progression of multiple human virus infections, identifying a common transcriptional switch would have major implications for the development of antiviral therapeutics. In addition, changes in peroxisome numbers are often linked to virus-induced immune modulation, so examining how virus infection modulates peroxisome biogenesis may be the next key step in understanding the cell signaling capacities of peroxisomes.
Another intriguing avenue for discovery is the virus-induced regulation of peroxisome morphology. Altered peroxisome shape, particularly elongation, has been observed during proliferation or external stress (UV radiation and ROS) [4,24,25]. However, how peroxisome shape influences peroxisome functions remains an intriguing question. It has been speculated that tubular or other complex morphologies enhance processes associated with peroxisome membranes [7], as the coupling of membrane expansion to irregular structure would increase interactions between luminal and membrane-associated proteins. This, in turn, could increase the efficiency of pathways that share functional components between the peroxisome membrane and lumen, such as transport of fatty acids for oxidation and lipid synthesis [40,66]. Evidence for this lies in the recent discovery that herpesvirus infections cause the formation of enlarged, flattened peroxisomes that lead to increased plasmalogen synthesis [37]. As this was found to be important for herpesvirus assembly, it is likely that other viruses known to modulate peroxisome morphology (e.g., elongation in HIV infection [46]) are similarly using peroxisome shape to enhance function, and – conversely – viruses known to alter specific peroxisome functions (e.g., lipid metabolism in influenza virus [43], WNV [36], and KSHV [45] infections, among others [67]) are also regulating peroxisome shape. Therefore, elucidating the processes underlying peroxisome shape may provide strategies to selectively inhibit peroxisome-mediated functions required for infection.
Due to the fundamental nature of peroxisomes as multifunctional and dynamic organelles, the aspects of peroxisome biology required for virus infections will be broadly applicable to human health and disease. This is already apparent in the themes present across biological conditions, as discussed above: changes to peroxisome biogenesis are found in metabolic disorders [27,30], development [2,4,6], cancer [9], and stress responses [23], while enhanced peroxisome-mediated ROS processing occurs in tumors [68] and plant infections [29,55], and peroxisomal immune signaling has been implicated in a variety of processes [3,9,31], among others. Additionally, in a strikingly similar vein to virus infections, the gut microbiota is known to widely regulate PPARs in a tissue-specific manner [69], and pathological microbial infections require peroxisome metabolism, both of ROS and lipids [70]. Although the relationships between peroxisome dynamics and functions are becoming increasingly apparent, how these processes are linked remains largely unknown. Further investigations geared towards defining the mechanisms that regulate peroxisome plasticity in tandem with peroxisome functions promise to provide a critical next step forward (see Outstanding Questions), impacting our understanding of both the biology of virus infections and basic cellular processes.
Outstanding Questions.
What virus- or host-driven mechanisms underlie the dynamic regulation of the structure and function of peroxisomes during infection?
Is there a common transcriptional switch that is induced upon infection to control peroxisome numbers and composition?
How does peroxisome shape influence metabolic regulation in the context of infection?
What is the role of peroxisome-mediated ROS processing during infection with human viruses?
Highlights.
The peroxisome has emerged as a multifunctional organelle with the capacity to act at the interface between host defense and virus replication.
Nearly a decade ago, the antiviral protein MAVS was found to localize to the peroxisome, marking peroxisome-mediated immune signaling as a critical component of host defense. In response, viruses have acquired mechanisms for suppressing peroxisome signaling.
Virus-induced rewiring of peroxisome function revealed that peroxisomes also have proviral functions via lipid synthesis necessary for enveloped viruses.
The opposing peroxisome functions are temporally regulated, acting in immune response early in infection and facilitating virus assembly at later stages.
Peroxisome numbers and morphology are finely tuned in tandem with peroxisomal functions during infection, providing a powerful example of a link between shape and function.
Acknowledgments
We are grateful for funding from the National Institutes for Health (NIH) (R01 GM114141) and Mallinckrodt Scholar Award to I.M.C., as well as an NIH training grant from National Institute of General Medical Sciences (NIGMS) (T32GM007388).
References
- 1.Dixit E et al. (2010) Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141, 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Odendall C et al. (2014) Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol 15, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vazquez C and Horner SM (2015) MAVS coordination of antiviral innate immunity. J. Virol 89, 6974–6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qian G et al. (2015) Peroxisomes in different skeletal cell types during intramembranous and endochondral ossification and their regulation during osteoblast differentiation by distinct peroxisome proliferator-activated receptors. PLoS One 10, e0143439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wanders RJA and Poll-The BT (2017) Role of peroxisomes in human lipid metabolism and its importance for neurological development. Neurosci. Lett 637, 11–17 [DOI] [PubMed] [Google Scholar]
- 6.Jauregui M and Kim PK (2014) Probing peroxisome dynamics and biogenesis by fluorescence imaging. Curr. Protoc. Cell Biol 62, 21.9.1–21.9.20 [DOI] [PubMed] [Google Scholar]
- 7.Schrader M and Fahimi HD (2006) Peroxisomes and oxidative stress. Biochim. Biophys. Acta 1763, 1755–1766 [DOI] [PubMed] [Google Scholar]
- 8.Bonekamp NA et al. (2009) Reactive oxygen species and peroxisomes: struggling for balance. BioFactors 35, 346–355 [DOI] [PubMed] [Google Scholar]
- 9.Tripathi DN and Walker CL (2016) The peroxisome as a cell signaling organelle. Curr. Opin. Cell Biol 39, 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verchot J (2011) Wrapping membranes around plant virus infection. Curr. Opin. Virol 1, 388–395 [DOI] [PubMed] [Google Scholar]
- 11.Jean Beltran PM et al. (2017) Exploring and exploiting proteome organization during viral infection. J. Virol 91, JVI.00268–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith JJ and Aitchison JD (2013) Peroxisomes take shape. Nat. Rev. Mol. Cell Biol 14, 803–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costello JL and Schrader M (2018) Unloosing the Gordian knot of peroxisome formation. Curr. Opin. Cell Biol 50, 50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geuze HJ et al. (2003) Involvement of the endoplasmic reticulum in peroxisome formation. Mol. Biol. Cell 14, 2900–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koch J et al. (2012) PEX11 proteins attract Mff and human Fis1 to coordinate peroxisomal fission. J. Cell Sci 125, 3813–3826 [DOI] [PubMed] [Google Scholar]
- 16.Schrader M et al. (2016) Proliferation and fission of peroxisomes – an update. Biochim. Biophys. Acta 1863, 971–983 [DOI] [PubMed] [Google Scholar]
- 17.Sugiura A et al. (2017) Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature 542, 251–254 [DOI] [PubMed] [Google Scholar]
- 18.Fransen M et al. (2017) The peroxisome-mitochondria connection: how and why? Int. J. Mol. Sci 18,1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua R and Kim PK (2016) Multiple paths to peroxisomes: mechanism of peroxisome maintenance in mammals. Biochim. Biophys. Acta 1863, 881–891 [DOI] [PubMed] [Google Scholar]
- 20.Castro IG et al. (2018) A role for MIRO1 in motility and membrane dynamics of peroxisomes. Traffic 19, 229–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua R et al. (2017) VAPs and ACBD5 tether peroxisomes to the ER for peroxisome maintenance and lipid homeostasis. J. Cell Biol 216, 367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costello JL et al. (2017) ACBD5 and VAPB mediate membrane associations between peroxisomes and the ER. J. Cell Biol 216, 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez-Huertas E et al. (2000) Stress induces peroxisome biogenesis genes. EMBO J. 19, 6770–6777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schrader M et al. (1999) Induction of tubular peroxisomes by UV irradiation and reactive oxygen species in HepG2 cells. J. Histochem. Cytochem 47, 1141–1148 [DOI] [PubMed] [Google Scholar]
- 25.Sinclair AM et al. (2009) Peroxule extension over ER-defined paths constitutes a rapid subcellular response to hydroxyl stress. Plant J. 59, 231–242 [DOI] [PubMed] [Google Scholar]
- 26.Novikoff AB and Novikoff PM (1982) Microperoxisomes and peroxisomes in relation to lipid metabolism. Annu. New York Acad. Sci 386, 138–152 [DOI] [PubMed] [Google Scholar]
- 27.Lodhi IJ and Semenkovich CF (2014) Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 19, 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller JM et al. (1993) Peroxisome through cell differentiation and neoplasia. Biol. Cell 77, 77–88 [DOI] [PubMed] [Google Scholar]
- 29.Hu J et al. (2012) Plant peroxisomes: biogenesis and function. Plant Cell 24, 2279–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Argyriou C et al. (2016) Peroxisome biogenesis disorders. Transl. Sci. Rare Dis 1, 111–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bender S et al. (2015) Activation of type I and III interferon response by mitochondrial and peroxisomal MAVS and inhibition by hepatitis C virus. PLoS Pathog. 11, e1005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreira AR et al. (2016) Hepatitis C virus NS3-4A inhibits the peroxisomal MAVS-dependent antiviral signalling response. J. Cell. Mol. Med 20, 750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magalhães AC et al. (2016) Peroxisomes are platforms for cytomegalovirus’ evasion from the cellular immune response. Sci. Rep 6, 26028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi YYB et al. (2018) Peroxisomes support human herpesvirus 8 latency by stabilizing the viral oncogenic protein vFLIP via the MAVS-TRAF complex. PLoS Pathog. 14, e1007058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.You J et al. (2015) Flavivirus infection impairs peroxisome biogenesis and early antiviral signaling. J. Virol 89, 12349–12361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coyaud E et al. (2018) Global interactomics uncovers extensive organellar targeting by Zika virus. Mol. Cell. Proteomics 17, 2242–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jean Beltran PM et al. (2018) Infection-induced peroxisome biogenesis is a metabolic strategy for herpesvirus replication. Cell Host Microbe 24, 526–541.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun GY et al. (2018) Docosahexaenoic acid (DHA): an essential nutrient and a nutraceutical for brain health and diseases. Prostagland. Leukot. Essent. Fat. Acids 136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dorninger F et al. (2017) From peroxisomal disorders to common neurodegenerative diseases - the role of ether phospholipids in the nervous system. FEBS Lett. 591, 2761–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braverman NE and Moser AB (2012) Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta 1822, 1442–1452 [DOI] [PubMed] [Google Scholar]
- 41.Liu STH et al. (2011) Synaptic vesicle-like lipidome of human cytomegalovirus virions reveals a role for SNARE machinery in virion egress. Proc. Natl. Acad. Sci. U. S. A 108, 12869–12874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martín-Acebes MA et al. (2014) The composition of West Nile virus lipid envelope unveils a role of sphingolipid metabolism in flavivirus biogenesis. J. Virol 88, 12041–12054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanner LB et al. (2014) Lipidomics identifies a requirement for peroxisomal function during influenza virus replication. J. Lipid Res 55, 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sychev ZE et al. (2017) Integrated systems biology analysis of KSHV latent infection reveals viral induction and reliance on peroxisome mediated lipid metabolism. PLoS Pathog. 13, e1006256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jean Beltran PM et al. (2016) A portrait of the human organelle proteome in space and time during cytomegalovirus infection. Cell Syst. 3, 361–373.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Z et al. (2017) MicroRNAs upregulated during HIV infection target peroxisome biogenesis factors: implications for virus biology, disease mechanisms and neuropathology. PLoS Pathog. 13, e1006360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panavas T et al. (2005) The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of cucumber necrosis tombusvirus. Virology 338, 81–95 [DOI] [PubMed] [Google Scholar]
- 48.Rochon D et al. (2014) The p33 auxiliary replicase protein of cucumber necrosis virus targets peroxisomes and infection induces de novo peroxisome formation from the endoplasmic reticulum. Virology 452–453, 133–142 [DOI] [PubMed] [Google Scholar]
- 49.Sasvari Z et al. (2018) Assembly-hub function of ER-localized SNARE proteins in biogenesis of tombusvirus replication compartment. PLoS Pathog. 14, e1007028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pathak KB et al. (2008) The host Pex19p plays a role in peroxisomal localization of tombusvirus replication proteins. Virology 379, 294–305 [DOI] [PubMed] [Google Scholar]
- 51.Nagy PD (2016) Tombusvirus-host interactions: co-opted evolutionarily conserved host factors take center court. Annu. Rev. Virol 3, 491–515 [DOI] [PubMed] [Google Scholar]
- 52.Kovalev N et al. (2017) The role of co-opted ESCRT proteins and lipid factors in protection of tombusviral double-stranded RNA replication intermediate against reconstituted RNAi in yeast. PLoS Pathog. 13, e1006520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang M et al. (2018) Barley stripe mosaic virus γb interacts with glycolate oxidase and inhibits peroxisomal ROS production to facilitate virus infection. Mol. Plant 11, 338–341 [DOI] [PubMed] [Google Scholar]
- 54.Chamnongpol S et al. (1998) Defense activation and enhanced pathogen tolerance induced by H2O2 in transgenic tobacco. Proc. Natl. Acad. Sci. U. S. A 95, 5818–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mhamdi A et al. (2012) Plant catalases: peroxisomal redox guardians. Arch. Biochem. Biophys 525, 181–194 [DOI] [PubMed] [Google Scholar]
- 56.Hamilton A et al. (2002) Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deleris A et al. (2006) Hierarchical action and inhibition of plant dicer-like proteins in antiviral defense. Science 313, 68–71 [DOI] [PubMed] [Google Scholar]
- 58.Incarbone M et al. (2017) Neutralization of mobile antiviral small RNA through peroxisomal import. Nat. Plants 3, 17094. [DOI] [PubMed] [Google Scholar]
- 59.Cook KC and Cristea IM (2019) Location is everything: protein translocations as a viral infection strategy. Curr. Opin. Chem. Biol 48, 34–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bagattin A et al. (2010) Transcriptional coactivator PGC-1 promotes peroxisomal remodeling and biogenesis. Proc. Natl. Acad. Sci. U. S. A 107, 20376–20381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sexton JZ et al. (2010) High content screening for non-classical peroxisome proliferators. Int. J. High Throughput Screen 2010, 127–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rauwel B et al. (2010) Activation of peroxisome proliferator-activated receptor gamma by human cytomegalovirus for de novo replication impairs migration and invasiveness of cytotrophoblasts from early placentas. J. Virol 84, 2946–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leghmar K et al. (2015) Cytomegalovirus infection triggers the secretion of the PPARg agonists 15-hydroxyeicosatetraenoic acid (15-HETE) and 13-hydroxyoctadecadienoic acid (13-HODE) in human cytotrophoblasts and placental cultures. PLoS One 10, e0132627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dharap A et al. (2015) Mutual induction of transcription factor PPARγ and microRNAs miR-145 and miR-329. J. Neurochem 135, 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaarbø M et al. (2011) Human cytomegalovirus infection increases mitochondrial biogenesis. Mitochondrion 11, 935–945 [DOI] [PubMed] [Google Scholar]
- 66.Smith BT et al. (2000) Intraperoxisomal localization of very-long-chain fatty acyl-CoA synthetase: implication in X-adrenoleukodystrophy. Exp. Cell Res 254, 309–320 [DOI] [PubMed] [Google Scholar]
- 67.Lange P et al. (2019) Chewing the fat: the conserved ability of DNA viruses to hijack cellular lipid metabolism. Viruses 11, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang J et al. (2015) ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol 17, 1259–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hasan A et al. (2019) Interactions between host PPARs and gut microbiota in health and disease. Int. J. Mol. Sci 20, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Di Cara F et al. (2017) Peroxisome-mediated metabolism is required for immune response to microbial infection. Immunity 47, 93–106.e7 [DOI] [PubMed] [Google Scholar]