Abstract
The purpose of this study was to identify early circulating metabolite changes implicated in the mechanism of action of irinotecan, a DNA topoisomerase I inhibitor, in cancer patients. A liquid chromatography–tandem mass spectrometry–based targeted metabolomic platform capable of measuring 254 endogenous metabolites was applied to profile circulating metabolites in plasma samples collected pre- and post-irinotecan treatment from 13 cancer patients. To gain further mechanistic insights, metabolic profiling was also performed for the culture medium of human primary hepatocytes (HepatoCells) and 2 cancer cell lines on exposure to SN-38 (an active metabolite of irinotecan). Intracellular reactive oxygen species (ROS) was detected by dihydroethidium assay. Irinotecan induced a global metabolic change in patient plasma, as represented by elevations of circulating purine/pyrimidine nucleobases, acylcarnitines, and specific amino acid metabolites. The plasma metabolic signature was well replicated in HepatoCells medium on SN-38 exposure, whereas in cancer cell medium SN-38 induced accumulation of pyrimidine/purine nucleosides and nucleobases while having no impact on acylcarnitines and amino acid metabolites.SN-38 induced ROS in HepatoCells, but not in cancer cells. Distinct metabolite signatures of SN-38 exposure in HepatoCells medium and cancer cell medium revealed different mechanisms of drug action on hepatocytes and cancer cells. Elevations in circulating purine/pyrimidine nucleobases may stem from nucleotide degradation following irinotecan-induced DNA double-strand breaks. Accumulations of circulating acylcarnitines and specific amino acid metabolites may reflect, at least in part,irinotecan-induced mitochondrial dysfunction and oxidative stress in the liver. The plasma metabolic signature of irinotecan exposure provides early insights into irinotecan mechanism of action in patients.
Keywords: metabolomics, irinotecan, DNA double-strand break, oxidative stress, mitochondrial dysfunction, steatohepatitis
Irinotecan (CPT-11), a semisynthetic analogue of camptothecin, is used as the mainstay treatment for metastatic colorectal cancer and other solid tumors.1,2 It acts as a prodrug that is activated to 7-ethyl-10-hydroxycamptothecin (SN-38) by carboxylesterase 2 in the blood and liver, and SN-38 is subsequently detoxified in the liver by uridine diphosphate-glucuronosyltransferase 1A1 to form a β-glucuronide conjugate, SN-38G.3 SN-38 is a potent inhibitor of DNA topoisomerase I, an essential enzyme that controls DNA structural changes by relaxing DNA supercoil and religating cleaved DNA strands during DNA replication and transcription.3,4 SN-38 can bind to the DNA-topoisomerase I complex to form a stable ternary complex that prevents religation of DNA strands and interferes with the moving replication fork, thereby inducing replication arrest and lethal double-strand DNA breaks and ultimately cell death.3,4
Inhibition of DNA topoisomerase I is the central mechanism for the antitumor activity of irinotecan, whereas it may cause DNA damage and cell death in fast-proliferating normal cells such as bone marrow cells and intestinal basal cells. Thus, irinotecan treatment is commonly associated with hematologic and gastrointestinal toxicities, with neutropenia and severe delayed diarrhea as the major dose-limiting toxicities. In addition, a growing body of evidence indicates a link between irinotecan treatment and the development of steatohepatitis, which is pathologically characterized by fat accumulation (steatosis), inflammation, ballooning of hepatocytes, and fibrosis.5–7 It has been shown that steatohepatitis occurred in 20% of patients with metastatic colorectal cancer receiving irinotecan-based chemotherapy as adjuvant therapy before surgical resection of liver metastases, and the development of steatohepatitis increased by 10 times the 90-day postoperative mortality, specifically death from liver failure.5 Although the precise mechanism of irinotecan-induced steatohepatitis is not fully understood, mitochondrial dysfunction and oxidative stress appear to play a critical role.7,8 Given the significant negative clinical impact of steatohepatitis, early identification of individuals at risk is particular crucial. Preoperative detection of steatohepatitis, however, is challenging with current laboratory and clinical tools (eg, liver function test, imaging techniques, or preoperative biopsy). It is imperative to develop innovative approaches that could presage the development of steatohepatitis and therefore aid in the identification of at-risk individuals.
Novel metabolomic technologies allow high-throughput assessment of a large number of endogenous metabolites, which provide powerful tools for mapping biochemical pathways implicated in disease and in response to drug treatment.9,10 Pharmacometabolomics is an emerging field that applies metabolomics to define the metabolic signature of drug exposure, thereby enabling identification of biochemical pathways implicated in drug efficacy and adverse drug reactions.9 By defining metabolite profiles both at baseline (prior to) and after drug exposure, pharmacometabolomics has the potential to provide early insights into the mechanism of drug action and the molecular basis for variation in drug response. As such, pharmacometabolomics, as with other “omics” tools, holds the promise of identifying potential mechanistic biomarkers predictive of drug pharmacokinetics, pharmacodynamics, efficacy, or toxicity.11–16
We applied a liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based targeted metabolomics platform to define the plasma metabolic signature of irinotecan exposure in cancer patients. We further performed in vitro studies to better understand the mechanistic implications of observed plasma metabolic signature. Because circulating metabolites in plasma likely reflect the metabolites excreted or released from the liver (the major metabolism organ) and/or tumor in response to the treatment in patients, we profiled metabolite changes in the culture medium of primary human hepatocytes (Corning HepatoCells) and 2 cancer cell lines (MDA-MB-231 and T47D) following SN-38 exposure. Our goal was to identify early plasma metabolic changes implicated in the mechanism of irinotecan action in patients, which would serve as potential mechanistic circulating biomarkers for early prediction of irinotecan efficacy or toxicity, particularly irinotecan-induced steatohepatitis.
Methods
Patients and Plasma Samples
The protocol was approved by the institutional review board of Wayne State University, and written informed consent was obtained from each patient. All procedures were in accordance with the ethical standards of the responsible committee on human experimentation or with the Helsinki Declaration of 1975 (as revised in 1983). Plasma samples for metabolomics analysis were obtained from 13 cancer patients as part of a phase 1 clinical trial that evaluates the combination therapy of irinotecan and veliparib (ClinicalTrials.gov identifier ). Patients with metastatic and refractory solid malignancies and with adequate hematologic, renal, and liver functions were enrolled in the study. Details on the inclusion/exclusion criteria and study design have been published by us.17
Treatment cycles were 21 days. Irinotecan was administered as 1.5-hour intravenous infusion (100 mg/m2) on days 1 and 8. Twice-daily oral administration of veliparib began on day 3 of cycle 1 and continued until day 14, followed by 6 days of no treatment (days 15–20). Veliparib treatment continued in cycle 2 through days 1–14, followed by a 6-day rest. The plasma pharmacokinetics of irinotecan and its active metabolite, SN-38, were evaluated as described previously by us.17 Plasma samples for metabolite profiling were obtained at pre-dosing, 1.5, 5.5, 28, and 48 hours following a 1.5-hour intravenous infusion of irinotecan alone in cycle 1, day 1, as well as following coadministration of irinotecan and veliparib in cycle 2, day 8. All plasma samples were stored at −80°C until metabolomics analysis.
Cell Culture and Treatment
HepatoCells, which are single-use cryopreserved cells derived from primary human hepatocytes, were purchased from Corning Inc. (Corning, New York). Cells were thawed and seeded at 8 × 105 cells per well in a 12-well BioCoat Collagen I plate (Corning, New York), maintained daily in fresh Corning HepatoCells Culture Medium supplemented with 10% fetal bovine serum (FBS). On the third day after thawing, HepatoCells were cultured in fresh drug-free medium or medium containing SN-38 at clinically relevant concentration (50 or 500 nM) for 1, 6, and 24 hours. At the end of treatment, the culture medium was collected and centrifuged (at 3000g, 4°C, for 5 minutes), and the supernatant was collected and stored at −80°C until metabolomics analysis.
Human breast cancer cell lines, MDA-MB-231 and T47D, were purchased from Division of Cancer Treatment and Diagnosis of the National Cancer Institute (Frederick, Maryland) and maintained in RPMI-1640 medium, supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were seeded in 12-well plates at a density of 5 × 105 per well. After 24 hours, the cell culture medium was removed, and cells were cultured in fresh drug-free medium or medium containing SN-38 at 50 or 500 nM for 1, 6, and 24 hours. At the end of treatment, the culture medium was collected and centrifuged (at 3000g, 4°C, for 5 minutes), and the supernatant was collected and stored at −80°C until metabolomics analysis. Of note, because 24-hour SN-38 exposure caused significant cell death of MDA-MB-231 and T47D, metabolomics profiling was only performed for the 1- and 6-hour exposure samples.
LC-MS/MS-Based Targeted Metabolomics
Metabolites in patient plasma and cell medium samples were quantitatively profiled using an LC-MS/MS-based targeted metabolomics platform, which quantitatively measures 254 metabolites involved in major human metabolic pathways. The list of 254 metabolites is presented in Supplementary Table 1. All LC-MS/MS analyses were performed on an AB SCIEX QTRAP 6500 LC-MS/MS system, which consists of a SHIMADZU Nexera ultra-high-performance liquid chromatography coupled with a triple quadrupole/linear ion trap mass spectrometer. Analyst 1.6 software was used for system control and data acquisition, and MultiQuant 3.0 software was used for data processing and quantitation.
Metabolites in plasma or cell medium samples were extracted as described previously with modifications.18 In brief, 1 mL methanol (cooled at −80°C) was added into 250 μL of plasma or cell medium followed by vortex-mixing and centrifugation (at 10 000 rpm, 4°C for 10 minutes). The supernatant was transferred to a new 2-mL microcentrifuge tube, and 0.8 mL 80% methanol (cooled to −80°C) was added to the precipitate followed by vortex-mixing and centrifugation. Supernatant from 2 extractions was combined and dried in a CentriVap Refrigerated Centrifugal Concentrator (Kansas City, Missouri) at 10°C. The residual was first reconstituted in 125 μL of acetonitrile-water (50/50, v/v) followed by vortex-mixing and centrifugation, and the supernatant was diluted and subjected to 3 runs of LC-MS/MS analyses based on different chromatographic separation mechanisms to cover all targeted metabolites with diverse physicochemical properties (Supplementary Table 1).
Chromatographic separation of small organic acids, amino acid metabolites, nucleobases, nucleosides, monophosphate nucleosides, sugar derivatives, and fatty acid derivatives was achieved based on reversed-phase liquid chromatography on a Synergi Polar-RP column (80A, 2.0 × 150 mm, 4 μm). The gradient elution consisted of mobile phase A (0.03% formic acid in water) and mobile phase B (0.03% formic acid in acetonitrile), at a flow rate of 0.25 mL/min. The gradient program was as follows: 0–0.3 minutes, 0% B; 0.3–25 minutes, 0%−95% B; 25–25.1 minutes, 95%−0%; 25.1–30 minutes, 0% B. Separation of highly polar metabolites such as amino acids was achieved based on hydrophilic-interaction liquid chromatography (HILIC) on an Atlantis HILIC Silica column (2.1 × 150 mm, 3 μm). The gradient elution consisted of mobile phase A (10 mM ammonium formate, pH 3.0, and 0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile) at a flow rate of 0.25 mL/min. The gradient program was as follows: 0–0.5 minutes, 95% B; 0.5–10.5 minutes, 95%−40% B; 10.5–15 minutes, 40% B; 15–17 minutes, 40% to 95% B; 17–24 minutes, 95% B. Chromatographic separation of coenzyme A and derivatives, mono-, di-, or triphosphate nucleotides, and sugar phosphates was achieved based on ion-pair liquid chromatography on an Atlantis T3 column (2.1 × 100 mm, 3.0 μm). The gradient elution consisted of mobile phase A (100 mM hexofluoro-2-propanol and 8.6 mM triethylammonium acetate in water; final pH, 8.3) and mobile phase B (10% acetonitrile in mobile phase A) at a flow rate of 0.5 mL/min. The gradient program was as follows: 0–0.3 minutes, 0% B; 0.3–5 minutes, 0% to 10% B; 5.0–20 minutes, 10% to 100% B; 20–21 minutes, 100% B; 21–22.1 minutes, 100%−0% B; 22–30 minutes, 0% B.
Column eluents were monitored under both positive and negative ionization modes using the multiple reaction monitoring (MRM) on a QTRAP 6500 mass spectrometer. Mass spectrometric parameters (including ionization polarity, product ion, collision energy, declustering potential, and cell exit potential) were optimized to obtain the most sensitive and specific mass transitions for individual metabolites by direct infusion of the standard solutions into the ion source with a syringe pump. Other optimized mass spectrometric parameters were as follows: ion spray potential was 5500 V for positive ionization mode and 4500 V for negative ionization mode; bebulizer gas and bath gas were 50 psi; curtain gas was 30 psi; collision gas was set at medium level; and source temperature was 475°C. The dwell time for each MRM transition was 3 milliseconds in the reversed-phase and HILIC methods and 10 milliseconds in the ion-pair method.
Calibration standards of individual metabolites were prepared in the appropriate mobile phase at the concentration range of 10 nM to 10 μM (Supplementary Table 1). Quality control samples were prepared in the appropriate mobile phase at a concentration of 200 nM. Because concentrations of individual metabolites in plasma or cell medium samples varied significantly, both original samples and 20-fold diluted samples were processed and subjected to LC-MS/MS analyses.
Detection of Reactive Oxygen Species in SN-38-Treated Cells
Dihydroethidium, an oxidative fluorescent dye, was used to detect intracellular reactive oxygen species (ROS) following SN-38 exposure.19 HepatoCells or cancer cells were incubated with the drug-free medium or medium containing SN-38 (50 or 500 nM) for 1–6 hours. At the end of treatment, cells were washed twice with phosphate buffer solution (pH 7.4) and stained by incubating with 5 μM of dihydroethidium in Hank’s buffer saline solution (HBSS) at 37°C for 30 minutes. Cells were washed once with HBSS and kept in HBSS. Intracellular ROS was determined from the conversion of nonfluorescent dihydroethidium to its fluorescent derivative, and fluorescent intensity was measured at the excitation wavelength of 488 nm and absorption wavelength of 585 nm. Live cell images were recorded at 4× and 10× magnification using Texas red filter by a Nikon Eclipse Ti microscope (Nikon Instruments, Inc., Melville, New York) and processed using software NIS Elements AR Analysis 4.20.02 (Nikon Instruments, Inc., Melville, New York).
Data Analysis
Metabolomics data analyses were performed using the MetaboAnalyst web-based statistical package (http://www.metaboanalyst.ca/). Features (ie, metabolites) with >50% missing values were removed from the analysis, and the remaining missing values were replaced by the minimum value of a feature. To meet the normality assumption, individual metabolite concentrations were log-transformed and then autoscaled (mean-centered and divided by the standard deviation of each metabolite). A 1-way analysis of variance (ANOVA), if significant, followed by Fisher’s least-significant differences post hoc analysis was performed to identify significantly changed metabolites among groups. The study of the metabolic signature of irinotecan exposure in 13 cancer patients was intended to be hypothesis-generating, and thus a false discovery rate (FDR) adjusted P < .2 along with unadjusted P < .05 was used to identify plasma metabolites significantly changed by irinotecan, whereas an FDR adjusted P < .05 was used for identifying metabolites significantly changed on SN-38 exposure in cell culture medium to validate the metabolic signature of irinotecan exposure detected in patients. A heat map was used to visualize metabolic signatures of irinotecan/SN-38 exposure, with significantly changed metabolites ranked in ascending order according to the FDR adjusted P-values from the ANOVA test.
Results
Plasma Metabolic Signature of Irinotecan Exposure in Cancer Patients
Plasma metabolic signature of irinotecan exposure was determined in 13 patients with solid tumors. The tumor types include breast cancer (5 patients), ovarian cancer (3), colon cancer (2), lung cancer (1), anus cancer (1), and esophagus cancer (1). Patient characteristics (shown as the median and range) were: age (53 years; 31–72 years), weight (71 kg; 51–107 kg), height (168 cm; 154–184 cm), body surface area (1.79 m2; 1.51–2.30 m2), total bilirubin (0.2 mg/dL; 0.1–0.8 mg/dL), aspartate aminotransferase (22 IU/L, 12–76 IU/L), and alanine aminotransferase (30 IU/L, 7–49 IU/L), and serum creatine (0.8 mg/dL; 0.4–1.2 mg/dL).
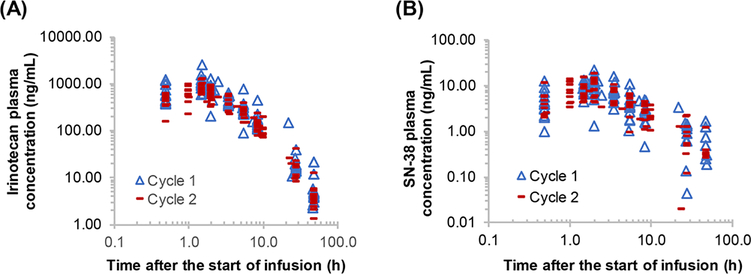
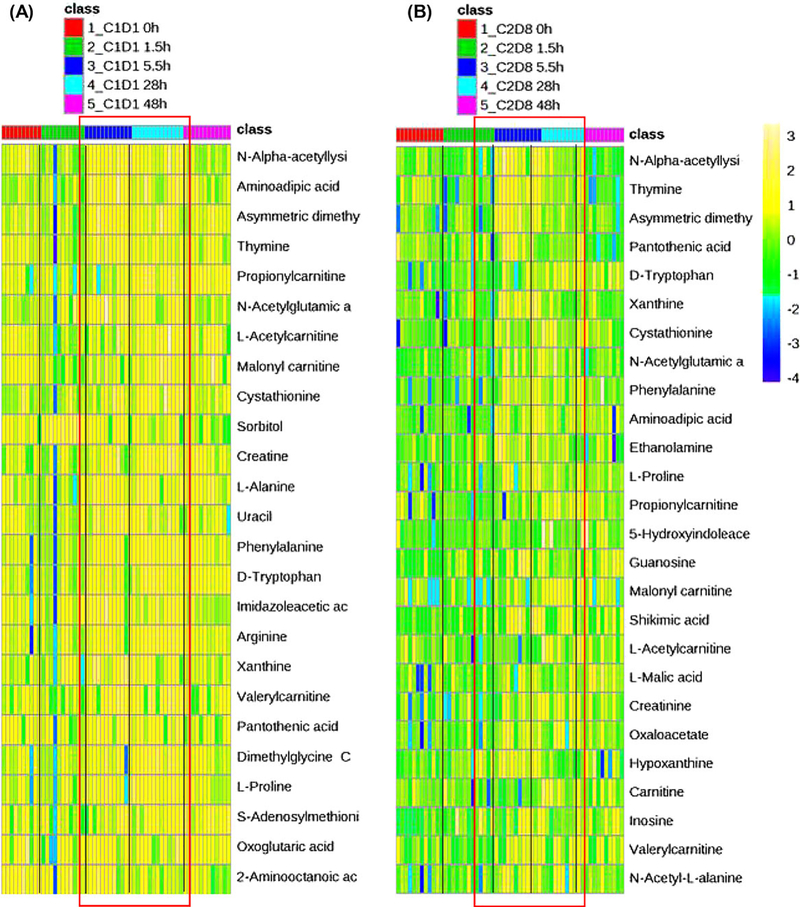
Figure 1A,B shows the plasma concentration-time profiles of irinotecan and its active metabolite SN-38 in 13 individual patients following a 1.5-hour intravenous infusion of irinotecan alone (in cycle 1, day 1) and in combination with veliparib (in cycle 2, day 8). Of 199 quantifiable metabolites (ie, those above the lower limit of quantitation), the plasma levels of 26 and 33 metabolites were significantly changed in a time-dependent manner (ANOVA, FDR adjusted P < .2) following the administration of irinotecan alone and in combination with veliparib, respectively (Figure 2 and Supplementary Table 2 and 3). The metabolite changes were generally delayed relative to the increase of plasma concentrations of irinotecan and SN-38. Specifically, although the maximum plasma concentrations of irinotecan and SN-38 were achieved at the end of infusion (1.5 hours), the most apparent elevations in plasma levels of metabolites (including amino acid metabolites, acylcarnitine derivatives, and pyrimidine/purine nucleobases) occurred at 5.5 and/or 28 hours (Figures 1 and 2). The metabolite changes following coadministration of irinotecan and veliparib were similar to those after administration of irinotecan alone, suggesting these metabolite changes were induced by irinotecan exposure and not influenced by concomitant veliparib (Figure 2B and Supplementary Table 3). The Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis indicated that the circulating metabolites significantly altered by irinotecan exposure were enriched for the pathways of amino acid metabolism, oxidation of fatty acids, and pyrimidine/purine metabolism.
Figure 1.
Plasma concentration-time profiles of irinotecan (A) and its active metabolite, SN-38 (B), in 13 individual patients following 1.5-hour intravenous infusion of irinotecan alone on cycle 1, day 1 and in combination with veliparib on cycle 2, day 8.
Figure 2.
Plasma metabolic signature of irinotecan exposure in cancer patients. The most apparent elevations in plasma levels of metabolites, including amino acid metabolites (eg, N-α-acetyllysine, 2-aminoadipic acid, asymmetric dimethylarginine, and cystathionine), acylcarnitine derivatives (eg, propionylcarnitine, L-acetylcarnitine, malonylcarnitine, and valerylcarnitine), and pyrimidine/purine nucleobases (eg, thymine, uracil, and xanthine) occurred at 5.5 and/or 28 hours. (A) Heat map of the significantly changed circulating metabolites (ANOVA, FDR-adjusted P < .2) following 1.5-hour intravenous infusion of irinotecan (100 mg/m2) on cycle 1, day 1 in patients. (B) Heat map of the top 25 most significantly changed circulating metabolites (ANOVA, FDR-adjusted P < .2) following the coadministration of irinotecan (100 mg/m2) and veliparib on cycle 2, day 8 in patients. The order of metabolites is based on the FDR-adjusted P values (from the most significant). The scales from −4 to 3 (ie, from blue to yellow) represent the normalized metabolite levels from low to high. The 5.5- and 28-hour data are boxed in red for visualization of the most apparent metabolite changes, which generally occurred 5.5 and 28 hours following irinotecan administration.
Metabolic Signature of SN-38 Exposure in the Culture Medium of HepatoCells and Cancer Cells
Circulating metabolites in plasma might stem from metabolites released from tissues such as the liver and tumor. To gain insight into the source of circulating metabolites altered by irinotecan exposure in patients, primary human hepatocytes (HepatoCells) and 2 cancer cell lines (MDA-MB-231 and T47D) were exposed to drug-free medium or SN-38 (50 or 500 nM) for 1, 6, or 24 hours, and the metabolites excreted into the culture medium were profiled using the targeted metabolomics platform. The maximum plasma concentration of SN-38 in individual patients ranged from 10 to 20 nM following irinotecan infusion (100 mg/m2); see Figure 1. Although SN-38 concentrations in patient liver and tumors were not measured, it is generally believed that SN-38 liver concentrations are higher than its plasma concentration. Therefore, we treated the cells with SN-38 at 50 and 500 nM, which were considered as the clinically relevant concentrations.
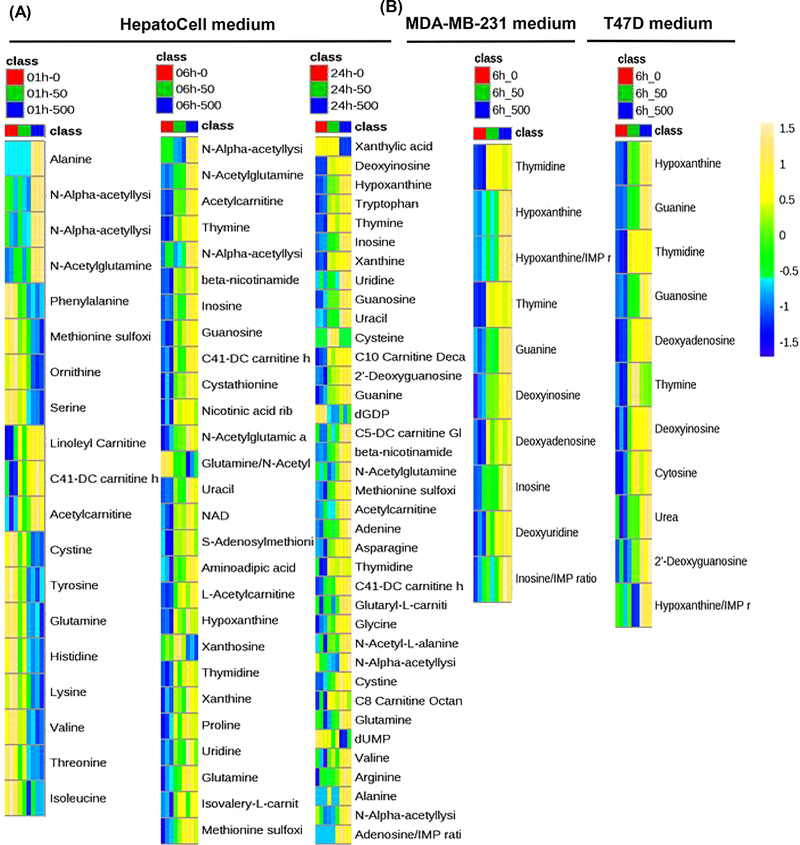
The observed plasma metabolic signature in patients was well replicated in the culture medium of HepatoCells, whereby SN-38 exposure induced concentration- and time-dependent accumulation of amino acid metabolites, acylcarnitines, as well as pyrimidine/purine nucleosides and nucleobases (Figure 3). Of 207 quantifiable metabolites in the medium, 19, 27, and 37 metabolites were significantly changed (ANOVA, FDR-adjusted P < .05) following 1-, 6-, and 24-hour SN-38 exposure, respectively (Figure 3). Specifically, early metabolite changes (at 1 hour) included the accumulation of amino acid metabolites (eg, N-α-acetyllysine) and acylcarnitines (including linoleylcarnitine, hexanoylcarnitine, and acetylcarnitine) and the decrease of amino acids (Figure 3). With longer exposure times (6 and 24 hours), the accumulation of amino acid metabolites (eg, N-α-acetyllysine, aminoadipic acid, and cystathionine) and acylcarnitines became more pronounced, and in addition, a number of pyrimidine/purine nucleosides (eg, uridine, thymidine, inosine, guanosine, deoxyinosine, and deoxyguanosine) and nucleobases (eg, thymine, uracil, hypoxanthine, xanthine, adenine, and guanine) were significantly accumulated in the medium, particularly at 24 hours (Figure 3).
Figure 3.
Metabolite signatures of SN-38 exposure in the culture medium of human primary hepatocytes (HepatoCells) and cancer cells. SN-38 exposure induced concentration- and time-dependent accumulation of amino acid metabolites, acylcarnitines, and pyrimidine/purine nucleosides and nucleobases in HepatoCells, whereas, in the culture medium of 2 cancer cell lines, SN-38 exposure only led to significant accumulations of pyrimidine/purine nucleosides and nucleobases, while having no apparent impact on amino acid metabolites and acylcarnitines. (A) Heat maps of the significantly changed metabolites (ANOVA, FDR-adjusted P < .05) in the culture medium of human primary hepatocytes (HepatoCells) and (B) the culture medium of cancer cells (MDA-MB-231 and T47D) in response to exposure to SN-38 at 50 and 500 nM for 1, 6, or 24 hours. The order of metabolites is based on the FDR-adjusted P values (from the most significant). The scales from −4 to 3 (ie, from blue to yellow) represent the normalized metabolite levels from low to high.
Different from the metabolic signature in HepatoCells medium, SN-38 exposure induced significant accumulation of pyrimidine/purine nucleosides and nucleobases (ANOVA, FDR-adjusted P < .05) in cancer cell medium, while having no apparent impact on the levels of acylcarnitines or amino acid metabolites (Figure 3). Specifically, at early exposure times (1 hour), only a few pyrimidine/purine nucleosides and nucleobases were significantly accumulated (ie, guanine and thymine in MDA-MB-231 medium; guanine, cytosine, and deoxyadenosine in T47D medium). With longer exposure time (eg, 6 hours), a number of pyrimidine/purine nucleosides and nucleobases were significantly accumulated in cancer cell medium (ANOVA, FDR-adjusted P < .05; Figure 3). Because a large proportion of cells died after 24-hour SN-38 exposure, the 24-hour samples were not profiled.
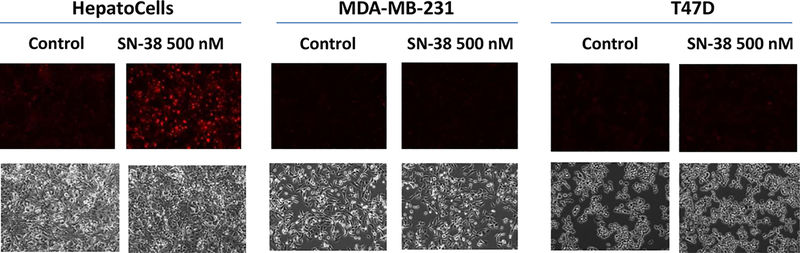
SN-38-Induced Oxidative Stress in HepatoCells, But Not in Cancer Cells
To gain further insights into the mechanism of irinotecan action on hepatocytes and cancer cells, HepatoCells and cancer cells (MDA-MB-231 and T47D) were exposed to drug-free medium or SN-38 (50 or 500 nM) for 1–6 hours, and the formation of reactive oxygen species (ROS), as the indicator of oxidative stress, was detected using dihydroethidium assay.19 On SN-38 exposure at either 50 or 500 nM, the formation of ROS, as measured by the red fluorescence resulting from the oxidation of dihydroethidium by superoxide, was detected in HepatoCells after 1 to 6 hours of SN-38 exposure, whereas ROS was not detected in MDA-MB-231 or T47D cells (Figure 4). These data suggested that hepatocytes, but not cancer cells (at least MDA-MB-231 or T47D), were susceptible to SN-38-induced oxidative stress. On the contrary, cancer cells were more sensitive to the antiproliferative effect of SN-38, as indicated by the observation of significant cell death in MDA-MB-231 or T47D, but not HepatoCells, after 24-hour SN-38 exposure.
Figure 4.
Detection of intracellular reactive oxygen species (ROS) by dihydroethidium (DHE) assay in human primary hepatocytes (HepatoCells) and cancer cells (MDA-MB-231 and T47D) on exposure to SN-38 (500 nM) or drug-free medium (control) for 2 hours. The formation of ROS, as measured by the red fluorescence resulting from the oxidation of dihydroethidium by superoxide,was detected in HepatoCells but not in cancer cells. Live cell images in the upper panel were recorded at 10× magnification using Texas red filter (excitation, 540–580 nm; emission, 600–660 nm), and those in the lower panel were recorded at 10× magnification using white light.
Discussion
Using an LC-MS/MS-based targeted metabolomics platform, we identified that plasma levels of a panel of amino acid metabolites, acylcarnitine derivatives, and pyrimidine/purine nucleobases were elevated following intravenous infusion of irinotecan in cancer patients. The observed plasma metabolic signature in patients was well replicated in the culture medium of HepatoCells, whereby SN-38 exposure induced concentration- and time-dependent accumulation of amino acid metabolites, acylcarnitines, as well as pyrimidine/purine nucleosides and nucleobases. Differently, in the culture medium of 2 cancer cell lines, SN-38 exposure only led to significant accumulations of pyrimidine/purine nucleosides and nucleobases, while having no apparent impact on amino acid metabolites and acylcarnitines. Further in vitro studies demonstrated that SN-38, at clinically relevant concentrations, induced oxidative stress in HepatoCells, but not in cancer cells. Collectively, these data imply that distinct metabolite signatures of SN-38 exposure in HepatoCells medium and cancer-cell medium reflect different mechanisms of drug action on hepatocytes and cancer cells. As cancer cells are rapidly proliferative, DNA double-strand breaks from DNA topoisomerase I inhibition is the central mechanism for cancer-cell killing activity of irinotecan/SN-38. In contrast, hepatocytes are relatively quiescent in terms of cell proliferation, but they are rich in mitochondria, and therefore hepatocytes are susceptible to drug-induced mitochondria dysfunction and oxidative stress.7,20,21 Compared with hepatocytes, many cancer types exhibit depletion of mitochondrial DNA (mtDNA) copy number, decreases in mtDNA expression (mtRNA) levels, and reduced mitochondrial respiratory activity,22,23 which collectively may render cancer cells resistant to irinotecan/SN-38-induced mitochondrial dysfunction.
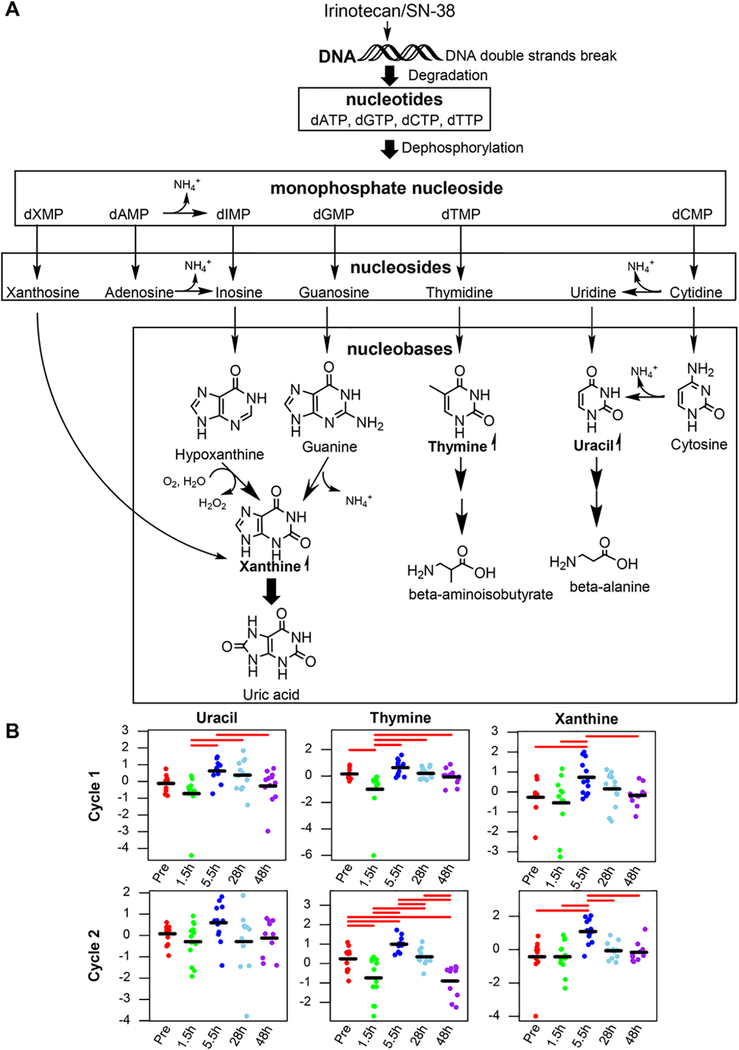
Following cellular DNA double-strand breaks, damaged DNA would break down to purine and pyrimidine nucleotides, which subsequently undergo rapid degradation.24 Purine nucleotide degradation involves dephosphorylation, deamination, and cleavage of glycosidic bonds to liberate purine nucleobases that are further converted to xanthine and then uric acid (Figure 5A). Specifically, purine nucleotides undergo dephosphorylation via 5′-nucleotidase to form nucleosides (adenosine and guanosine). Adenosine is then deaminated via adenosine deaminase and hydrolyzed via nucleosidase to form hypoxanthine, which is oxidized to form xanthine and then uric acid through the action of xanthine oxidase. The other purine nucleoside, guanosine, is cleaved to form guanine that is subsequently deaminated via guanine deaminase to form xanthine and then uric acid. Uric acid is the final product of purine degradation, which can be excreted into urine. Likewise, pyrimidine nucleotides undergo similar degradation reactions (dephosphorylation, deamination, and cleavage of glycosidic bonds) to liberate pyrimidine nucleobases including cytosine, uracil, and thymine (Figure 5A). Unlike the purine ring that is not cleaved in humans, the pyrimidine ring can be opened and finally degraded to highly water-soluble products, including β-alanine (from degradation of cytosine and uracil) and β-aminoisobutyrate (from degradation of thymine), which are excreted into urine. Although the final purine/pyrimidine degraded products (ie, uric acid, β-alanine, and β-aminoisobutyrate) were not measured by our targeted metabolomics platform, we observed significant accumulations of purine/pyrimidine degradation intermediates including nucleosides and nucleobases in the culture medium of both HepatoCells and cancer cells on SN-38 exposure, with more significant accumulation in cancer-cell medium (Figure 3). This observation fits with the notion that purine and pyrimidine nucleotide degradation occurs following cellular DNA double-strand breaks induced by irinotecan/SN-38. In agreement with the purine/pyrimidine metabolite signature of SN-38 exposure in cell culture medium, we observed time-dependent change in circulating purine and pyrimidine nucleobases (specifically xanthine, thymine, and uracil) in patient plasma following irinotecan treatment (Figure 5B). Because highly proliferating cells, particularly tumor cells, are more susceptible to irinotecan/SN-38-induced DNA damage than relatively quiescent cells such as hepatocytes, elevations of circulating purine and pyrimidine nucleobases in patients might reflect, in large part, the extent of DNA double-strand breaks induced by irinotecan/SN-38 in tumor cells and possibly highly proliferative normal cells (eg, bone marrow and intestinal cells). As such, the circulating levels of nucleobases (xanthine, thymine, and uracil) on irinotecan exposure could be potential early indicators for the antitumor effect and/or side effects (eg, hematological and intestinal toxicity) in patients. Further larger studies are needed to test this hypothesis.
Figure 5.
Elevation of circulating purine/pyrimidine nucleobases (eg, uracil, thymine, and xanthine) in cancer patients following irinotecan administration indicates purine and pyrimidine nucleotide degradation following irinotecan/SN-38-induced DNA double-strand breaks. (A) Schematic illustration of purine and pyrimidine nucleotide degradation pathways following DNA double-strand breaks. (B) Observed time course of circulating purine nucleobases (xanthine) and pyrimidine nucleobases (uracil and thymine) following 1.5-hour intravenous infusion of irinotecan alone on cycle 1, day 1 (upper panel) and in combination with veliparib in cycle 2, day 8 (lower panel) in 13 cancer patients. Data points represent log-transformed and autoscaled (mean-centered and divided by the standard deviation) metabolite concentrations in individual patients, and the black bar represents the mean of normalized data at each point. The significantly different pairs are indicated with red bars using pairwise Fisher’s LSD post hoc analyses at a 5% level.
In addition, we observed time-dependent elevations in a panel of circulating acylcarnitines (including propionylcarnitine, L-acetylcarnitine, malonylcarnitine, and valerylcarnitine) and amino acid metabolites (including N-α-acetyllysine, aminoadipic acid, asymmetric dimethylarginine, and cystathionine) in patient plasma following irinotecan administration (Figure 2 and Supplementary Table 2). This plasma metabolic signature was in line with the metabolic signature of SN-38 exposure in HepatoCells medium but not cancer-cell medium (Figure 3). Taken together with the observation that SN-38, at clinically relevant concentrations, induced ROS in HepatoCells but not cancer cells (Figure 4), these findings imply that elevated circulating levels of acylcarnitines and specific amino acid metabolites in patient plasma might reflect, at least in part, irinotecan-induced mitochondrial dysfunction and oxidative stress in the liver. This speculation is supported by the existing knowledge regarding the role of mitochondrial dysfunction in drug hepatotoxicity as well as functional implications of acylcarnitines and specific amino acid metabolites in hepatic mitochondrial dysfunction and oxidative stress.7,21,25–30
Mitochondria are essential organelles for fat oxidation and energy production and are also a primary source for cellular ROS. Numerous investigations suggest that many drugs (including irinotecan) or their reactive metabolites can induce hepatic mitochondrial dysfunction via various mechanisms such as enhancement of mitochondrial membrane permeabilization, impairment of the oxidative phosphorylation process, inhibition of mitochondrial fatty acid β-oxidation, and deletion or damage of mitochondrial DNA.21,25,31,32 Mitochondrial dysfunction further causes increased production of ROS through the damaged respiratory chain, increased lipid peroxidation, and impairment of fatty acid β-oxidation. High levels of ROS, in turn, aggravate mitochondrial dysfunction, thus leading to a vicious circle.31 Although the precise mechanisms underlying irinotecan-induced steatohepatitis are yet to be defined, emerging evidence reveals that mitochondrial dysfunction, fatty acid β-oxidation impairment, and oxidative stress play a crucial role in the development of nonalcoholic or drug-induced steatohepatitis.21,33,34
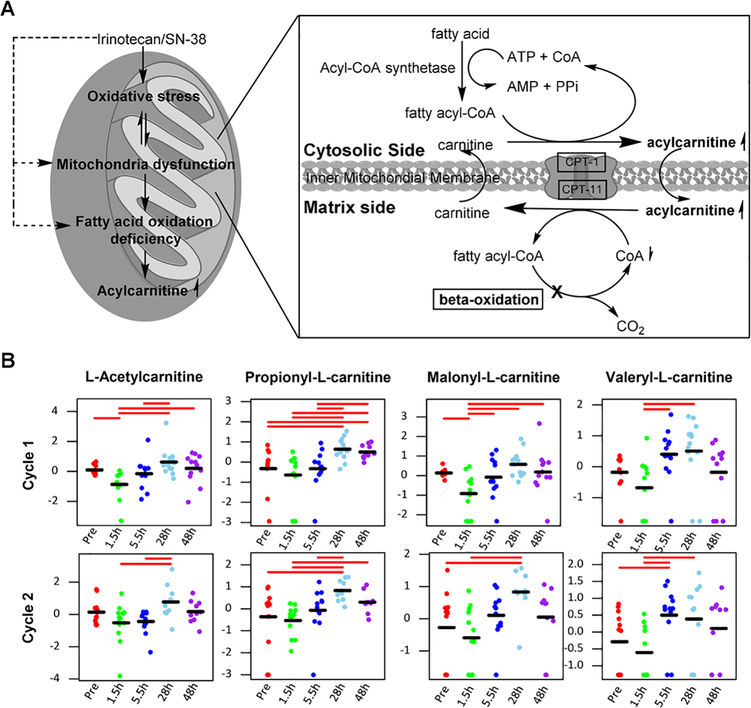
Impairment of mitochondrial fatty acid β-oxidation has been associated with elevated circulating fatty acid derivatives such as acylcarnitines and reduced ketone bodies in plasma.25,32 Fatty acid β-oxidation is a multistep process that involves entry of the fatty acid into the cytosol, activation to acyl-CoA by conjugation with coenzyme A in the cytosol, conversion to acylcarnitine by the enzyme carnitine palmitoyltransferase-1, transport across the inner mitochondrial membrane by the transporter carnitine translocase, and then conversion back to acyl-CoA by carnitine palmitoyltransferase-2 inside the mitochondrion, where fatty acid oxidation (β-oxidation) takes place (Figure 6A).35 Although short- and medium-chain fatty acids can enter mitochondria unassisted, the formation of fatty acylcarnitines is necessary to facilitate the entry of long-chain fatty acids into the mitochondria matrix for β-oxidation. When fatty acid β-oxidation is impaired, acylcarnitines would be accumulated in the liver and subsequently released into circulation. Indeed, circulating acylcarnitines have been postulated as specific biomarkers of mitochondrial dysfunction.26,36 Serum acylcarnitines are routinely measured in the screening of neonates for mitochondrial fatty acid β-oxidation deficiency disorders caused by defects in either carnitine/acylcarnitine transporters or enzymes involved in β-oxidation.37 Of 10 acylcarnitines measured by our targeted metabolomics, the circulating levels of 4 acylcarnitines (including propionylcarnitine, acetylcarnitine, malonylcarnitine, and valerylcarnitine) were significantly elevated in patient plasma, generally 5.5 and 28 hours after irinotecan administration (Figure 6B). Our finding fits with published data showing that these particular acylcarnitines are accumulated in the plasma of individuals with fatty acid β-oxidation deficiency from a variety of mechanisms.38–40 The acylcarnitine profiles of irinotecan/SN-38 exposure observed in patients as well as in HepatoCells medium suggest that irinotecan/SN-38 might inhibit mitochondrial fatty acid β-oxidation through a direct mechanism or secondary to mitochondrial dysfunction caused by inhibiting mitochondrial DNA replication. This raises the possibility that the plasma acylcarnitine profile could presage irinotecan hepatotoxicity and thus aid in the identification of “at-risk” patients. Further, larger studies are needed to test this hypothesis.
Figure 6.
Elevation of circulating acylcarnitines (eg, L-acetylcarnitine, propionylcarnitine, malonylcarnitine, and valerylcarnitine) in cancer patients following irinotecan administration is likely a result of fatty acid β-oxidation deficiency because of irinotecan-induced hepatic mitochondrial dysfunction. (A) Schematic illustration of the transport of fatty acids across the mitochondrial inner membrane and fatty acid β-oxidation process in the mitochondrial matrix. CPTI, carnitine palmitoyltransferase-1; CPTII, carnitine palmitoyltransferase-2. (B) Observed time course of circulating acylcarnitines following 1.5-hour intravenous infusion of irinotecan alone on cycle 1, day 1 (upper panel) and in combination with veliparib on cycle 2, day 8 (lower panel). Data points represent log-transformed and autoscaled (mean-centered and divided by the standard deviation) metabolite concentrations in individual patients, and the black bar represents the mean of normalized data at each point. The significantly different pairs are indicated with red bars using pairwise Fisher’s LSD post hoc analyses at a 5% level.
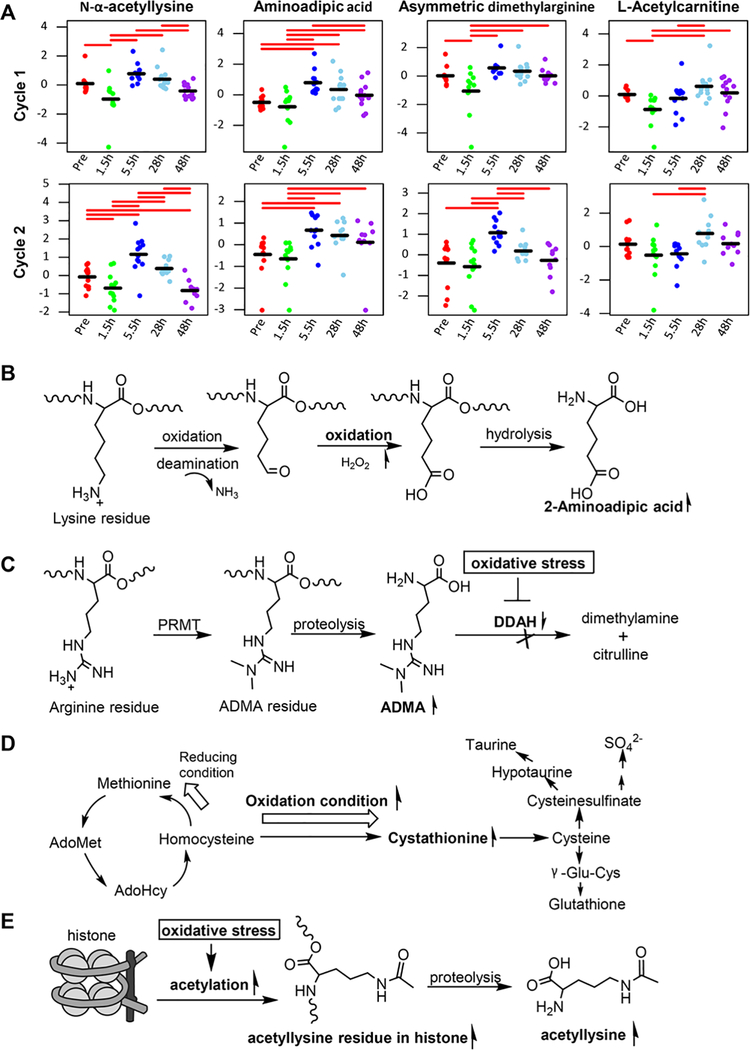
In addition to acylcarnitines, we also observed a time-dependent change in circulating specific amino acid metabolites (including N-α-acetyllysine, 2-aminoadipic acid, asymmetric dimethylarginine, and cystathionine) following irinotecan exposure in patients (Figures 2 and 7A). Although the functional relevance of this finding is yet to be determined, emerging evidence indicates an association of these amino acid metabolites with oxidative stress. For example, 2-aminoadipic acid is a product of lysine degradation. Under oxidation conditions, the β-amino group of lysine residues in proteins can undergo deamination by metal-catalyzed oxidation to form the intermediate allysine, which further undergoes oxidation to form 2-aminoadipic acid (Figure 7B).41 It has been shown that in conditions of aging, diabetes, sepsis, and renal failure, increased oxidation of lysyl residues to 2-aminoadipic acid occurs in human skin collagen and potentially other tissues, leading to the release of free 2-aminoadipic acid into circulation.27,28 It has also been shown that the elevation of 2-aminoadipic acid after long-term hyperglycemia of endothelial cells is a sign of lysine breakdown through oxidative stress and reactive oxygen species.42 Thus, 2-aminoadipate has been proposed as a potential small-molecule marker of oxidative stress.43 Likewise, there is evidence that circulating asymmetric dimethylarginine levels increase in conditions of oxidative stress.44,45 Asymmetric dimethylarginine is a product of methylation of arginine residues in proteins by the action of protein-arginine methyltransferases.46 Because more than 90% of asymmetric dimethylarginine generated is metabolized by dimethylargine dimethylaminohydrolase (DDAH),47 hepatic DDAH activity is an important determinant of circulating asymmetric dimethylarginine levels.48,49 DDAH is known to be highly sensitive to oxidative stress because of the presence of sulfhydryl groups at the active site, and it has been shown that oxidative stress inactivates this enzyme by modifying critical cysteine residues in its active site.50,51 As such, inactivation of DDAH because of irinotecan-induced oxidative stress may lead to the accumulation of asymmetric dimethylarginine in the liver, which would be subsequently released into circulation (Figure 7C). In fact, elevation of circulating asymmetric dimethylarginine has been observed in various pathological conditions associated with liver dysfunction.30,44,52,53 Circulating levels of cystathionine may be augmented as an autocorrective response to oxidative stress.54 The conversion of methionine to cysteine is rate-limited by cystathionine β-synthase, whose activity is regulated by the redox balance.55,56 Under conditions of oxidative stress, activation of cystathionine β-synthase would enhance conversion of methionine to cystathionine and then cysteine, leading to enhanced synthesis of glutathione, which serves as an autocorrective response to an oxidative insult (Figure 7D).29 Interestingly, N-α-acetyllysine was identified as the most significantly changed amino acid metabolite on irinotecan/SN-38 exposure in both patient plasma and HepatoCells medium. The acetylation of lysine residues in proteins has been known as an important mechanism of epigenetics, whereby histone acetyltransferases catalyze the addition of acetyl groups from acetyl-CoA onto the lysine residues of histones and nonhistone proteins. Emerging evidence suggests that oxidative stress can induce histone acetylation.57–59 Although the functional relevance of N-α-acetyllysine is as yet unknown, it is possible that elevated circulating N-α-acetyllysine is a consequence of enhanced histone acetylation in response to oxidative stress induced by irinotecan/SN-38 (Figure 7E). Collectively, taken together with the published and our data, the plasma amino acid metabolite profile in patient plasma might reflects, at least in part, irinotecan/SN-38-induced oxidative stress in the liver.
Figure 7.
Elevation of circulating specific amino acid metabolites (eg, N-α-acetyllysine, 2-aminoadipic acid, asymmetric dimethylarginine, and cystathionine) in cancer patients following irinotecan administration is implicated in irinotecan-induced oxidative stress. (A) Observed time course of circulating levels of specific amino acid metabolites following 1.5-hour intravenous infusion of irinotecan alone in cycle 1, day 1 (upper panel) and in combination with veliparib in cycle 2, day 8 (lower panel) in cancer patients. Data points represent log-transformed and autoscaled (mean-centered and divided by the standard deviation) metabolite concentrations in individual patients, and the black bar represents the mean of normalized data at each point. The significantly different pairs are indicated with red bars using pairwise Fisher’s LSD post hoc analyses at a 5% level. (B) Schematic illustration of oxidation of lysyl residues to 2-aminoadipic acid under oxidized conditions. (C) Schematic illustration of accumulation of asymmetric dimethylarginine (ADMA) because of inactivation of dimethylargine dimethylaminohydrolases (DDAH), a major enzyme responsible for the metabolism of ADMA, under oxidative stress. PRMT, protein-arginine methyltransferases. (D) Schematic illustration of increased cystathionine circulating levels as an autocorrective response to oxidative stress. (E) Schematic illustration of elevated circulating N-α-acetyllysine as a consequence of enhanced histone acetylation in response to oxidative stress.
Two limitations of our study need comment. First, a targeted metabolomics approach was used, which focuses on water-soluble small molecules such as amino acid metabolites, acylcarnitines, and nucleotide metabolites. Although targeted metabolomics provides highly specific, sensitive, and reliable identification and quantification of metabolites, this approach may miss some metabolites implicated in irinotecan exposure. Future study using untargeted metabolomics may provide comprehensive screening of metabolites and identify new metabolites relevant to irinotecan exposure. Second, our study is limited by the small patient sample size, and it is thus considered to be hypothesis-generating to provide a basis for further investigation in a larger patient population.
In summary, irinotecan exposure induces a global metabolic change in patient plasma, as represented by elevated circulating levels of purine and pyrimidine nucleobases, acylcarnitines, and specific amino acid metabolites. Accumulations of purine and pyrimidine nucleobases in plasma may stem, in large part, from purine/pyrimidine nucleotide degradation following irinotecan-induced DNA double-strand breaks in tumor cells and possibly highly proliferative normal cells such as bone marrow cells and intestinal epithelial cells. Elevated circulating levels of acylcarnitines and specific amino acid metabolites may reflect, at least in part, irinotecan-induced mitochondrial dysfunction and oxidative stress in the liver. The plasma metabolic signature of irinotecan exposure sheds light on the mechanisms of irinotecan action in patients. This study provides a foundation for future larger studies to investigate associations between plasma metabolite changes and the antitumor effect and toxicity of irinotecan, which may identify mechanistic circulating biomarkers for early prediction of irinotecan efficacy or toxicity, in particular irinotecan-induced steatohepatitis.
Supplementary Material
Acknowledgments
We thank the patients participating in the study.
Funding
This study was supported by the United States Public Health Service Cancer Center Support Grant P30 CA022453.
Footnotes
Declaration of Conflicting Interests
The authors declare that there are no conflicts of interest.
Data Sharing Statement
The authors are willing to share the data that are presented in this study. Please contact Dr. Jing Li (lijin@karmanos.org) to obtain the data.
Supporting Information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
References
- 1.Saltz LB, Cox JV, Blanke C, et al. Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. Irinotecan Study Group. N Engl J Med. 2000;343(13):905–914. [DOI] [PubMed] [Google Scholar]
- 2.Punt CJ, Koopman M. Capecitabine and irinotecan as first-line treatment of advanced colorectal cancer. J Clin Oncol. 2008;26(11):1907–1908; author reply 1908–1909. [DOI] [PubMed] [Google Scholar]
- 3.Mathijssen RH, Loos WJ, Verweij J, Sparreboom A. Pharmacology of topoisomerase I inhibitors irinotecan (CPT-11) and topotecan. Curr Cancer Drug Targets. 2002;2(2):103–123. [DOI] [PubMed] [Google Scholar]
- 4.Liu LF, Desai SD, Li TK, Mao Y, Sun M, Sim SP. Mechanism of action of camptothecin. Ann N Y Acad Sci. 2000;922:1–10. [DOI] [PubMed] [Google Scholar]
- 5.Vauthey JN, Pawlik TM, Ribero D, et al. Chemotherapy regimen predicts steatohepatitis and an increase in 90-day mortality after surgery for hepatic colorectal metastases. J Clin Oncol. 2006;24(13):2065–2072. [DOI] [PubMed] [Google Scholar]
- 6.Cleary JM, Tanabe KT, Lauwers GY, Zhu AX. Hepatic toxicities associated with the use of preoperative systemic therapy in patients with metastatic colorectal adenocarcinoma to the liver. Oncologist. 2009;14(11):1095–1105. [DOI] [PubMed] [Google Scholar]
- 7.McWhirter D, Kitteringham N, Jones RP, Malik H, Park K, Palmer D. Chemotherapy induced hepatotoxicity in metastatic colorectal cancer: a review of mechanisms and outcomes. Crit Rev Oncol Hematol. 2013;88(2):404–415. [DOI] [PubMed] [Google Scholar]
- 8.Schumacher JD, Guo GL. Mechanistic review of drug-induced steatohepatitis. Toxicol Appl Pharmacol. 2015;289(1):40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaddurah-Daouk R, Weinshilboum RM, Pharmacometabolomics Research N. Pharmacometabolomics: implications for clinical pharmacology and systems pharmacology. Clin Pharmacol Ther. 2014;95(2):154–167. [DOI] [PubMed] [Google Scholar]
- 10.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–683. [DOI] [PubMed] [Google Scholar]
- 11.Phapale PB, Kim SD, Lee HW, et al. An integrative approach for identifying a metabolic phenotype predictive of individualized pharmacokinetics of tacrolimus. Clin Pharmacol Ther. 2010;87(4):426–436. [DOI] [PubMed] [Google Scholar]
- 12.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A. 2009;106(34):14728–14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trupp M, Zhu H, Wikoff WR, et al. Metabolomics reveals amino acids contribute to variation in response to simvastatin treatment. PLoS One. 2012;7(7):e38386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yerges-Armstrong LM, Ellero-Simatos S, Georgiades A, et al. Purine pathway implicated in mechanism of resistance to aspirin therapy: pharmacometabolomics-informed pharmacogenomics. Clin Pharmacol Ther. 2013;94(4):525–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Backshall A, Sharma R, Clarke SJ, Keun HC. Pharmacometabonomic profiling as a predictor of toxicity in patients with inoperable colorectal cancer treated with capecitabine. Clin Cancer Res. 2011;17(9):3019–3028. [DOI] [PubMed] [Google Scholar]
- 16.Winnike JH, Li Z, Wright FA, Macdonald JM, O’Connell TM, Watkins PB. Use of pharmaco-metabonomics for early prediction of acetaminophen-induced hepatotoxicity in humans. Clin Pharmacol Ther. 2010;88(1):45–51. [DOI] [PubMed] [Google Scholar]
- 17.LoRusso PM, Li J, Burger A, et al. Phase I safety, pharmacokinetic, and pharmacodynamic study of the poly(ADP-ribose) polymerase (PARP) inhibitor veliparib (ABT-888) in combination with irinotecan in patients with advanced solid tumors. Clin Cancer Res. 2016;22(13):3227–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7(5):872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dikalov SI, Harrison DG. Methods for detection of mitochondrial and cellular reactive oxygen species. Antioxid Redox Signal. 2014;20(2):372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Degli Esposti D, Hamelin J, Bosselut N, et al. Mitochondrial roles and cytoprotection in chronic liver injury. Biochem Res Int. 2012;2012:387626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Begriche K, Massart J, Robin MA, Borgne-Sanchez A, Fromenty B. Drug-induced toxicity on mitochondria and lipid metabolism: mechanistic diversity and deleterious consequences for the liver. J Hepatol. 2011;54(4):773–794. [DOI] [PubMed] [Google Scholar]
- 22.Reznik E, Wang Q, La K, Schultz N, Sander C. Mitochondrial respiratory gene expression is suppressed in many cancers. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reznik E, Miller ML, Senbabaoglu Y, et al. Mitochondrial DNA copy number variation across human cancers. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen A, Barankiewicz J. Metabolic consequences of DNA damage: alteration in purine metabolism following poly(ADP ribosyl)ation in human T-lymphoblasts. Arch Biochem Biophys. 1987;258(2):498–503. [DOI] [PubMed] [Google Scholar]
- 25.Fromenty B, Pessayre D. Inhibition of mitochondrial beta-oxidation as a mechanism of hepatotoxicity. Pharmacol Ther. 1995;67(1):101–154. [DOI] [PubMed] [Google Scholar]
- 26.McGill MR, Li F, Sharpe MR, et al. Circulating acylcarnitines as biomarkers of mitochondrial dysfunction after acetaminophen overdose in mice and humans. Arch Toxicol. 2014;88(2):391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sell DR, Strauch CM, Shen W, Monnier VM. 2-aminoadipic acid is a marker of protein carbonyl oxidation in the aging human skin: effects of diabetes, renal failure and sepsis. Biochem J. 2007;404(2):269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sell DR, Strauch CM, Shen W, Monnier VM. Aging, diabetes, and renal failure catalyze the oxidation of lysyl residues to 2-aminoadipic acid in human skin collagen: evidence for metal-catalyzed oxidation mediated by alpha-dicarbonyls. Ann N Y Acad Sci. 2008;1126:205–209. [DOI] [PubMed] [Google Scholar]
- 29.Taoka S, Ohja S, Shan X, Kruger WD, Banerjee R. Evidence for heme-mediated redox regulation of human cystathionine beta-synthase activity. J Biol Chem. 1998;273(39):25179–25184. [DOI] [PubMed] [Google Scholar]
- 30.Richir MC, Ellger B, Teerlink T, et al. The effect of rosiglitazone on asymmetric dimethylarginine (ADMA) in critically ill patients. Pharmacol Res. 2009;60(6):519–524. [DOI] [PubMed] [Google Scholar]
- 31.Fromenty B, Robin MA, Igoudjil A, Mansouri A, Pessayre D. The ins and outs of mitochondrial dysfunction in NASH. Diabetes Metab. 2004;30(2):121–138. [DOI] [PubMed] [Google Scholar]
- 32.Labbe G, Pessayre D, Fromenty B. Drug-induced liver injury through mitochondrial dysfunction: mechanisms and detection during preclinical safety studies. Fundam Clin Pharmacol. 2008;22(4):335–353. [DOI] [PubMed] [Google Scholar]
- 33.Begriche K, Igoudjil A, Pessayre D, Fromenty B. Mitochondrial dysfunction in NASH: causes, consequences and possible means to prevent it. Mitochondrion. 2006;6(1):1–28. [DOI] [PubMed] [Google Scholar]
- 34.Serviddio G, Sastre J, Bellanti F, Vina J, Vendemiale G, Altomare E. Mitochondrial involvement in non-alcoholic steatohepatitis. Mol Aspects Med. 2008;29(1–2):22–35. [DOI] [PubMed] [Google Scholar]
- 35.Berg J, Tymoczko J, Stryer L. Fatty acid metabolism In: Berg J, Tymoczko J, eds. Biochemistry. 5th ed New York: WH Freeman; 2002. [Google Scholar]
- 36.Bhattacharyya S, Pence L, Beger R, et al. Acylcarnitine profiles in acetaminophen toxicity in the mouse: comparison to toxicity, metabolism and hepatocyte regeneration. Metabolites. 2013;3(3):606–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinaldo P, Matern D, Bennett MJ. Fatty acid oxidation disorders. Annu Rev Physiol. 2002;64:477–502. [DOI] [PubMed] [Google Scholar]
- 38.Wolf A, Weir P, Segar P, Stone J, Shield J. Impaired fatty acid oxidation in propofol infusion syndrome. Lancet. 2001;357(9256):606–607. [DOI] [PubMed] [Google Scholar]
- 39.Santer R, Fingerhut R, Lassker U, et al. Tandem mass spectrometric determination of malonylcarnitine: diagnosis and neonatal screening of malonyl-CoA decarboxylase deficiency. Clin Chem. 2003;49(4):660z–662. [DOI] [PubMed] [Google Scholar]
- 40.Vollmer JP, Haen S, Wolburg H, et al. Propofol related infusion syndrome: ultrastructural evidence for a mitochondrial disorder. Crit Care Med. 2018;46(1):e91–e94. [DOI] [PubMed] [Google Scholar]
- 41.Requena JR, Chao CC, Levine RL, Stadtman ER. Glutamic and aminoadipic semialdehydes are the main carbonyl products of metal-catalyzed oxidation of proteins. Proc Natl Acad Sci U S A. 2001;98(1):69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan W, Zhang J, Li S, Edwards JL. Amine metabolomics of hyperglycemic endothelial cells using capillary LC-MS with isobaric tagging. J Proteome Res. 2011;10(11):5242–5250. [DOI] [PubMed] [Google Scholar]
- 43.Zeitoun-Ghandour S, Leszczyszyn OI, Blindauer CA, Geier FM, Bundy JG, Sturzenbaum SR. C. elegans metallothioneins: response to and defence against ROS toxicity. Mol Biosyst. 2011;7(8):2397–2406. [DOI] [PubMed] [Google Scholar]
- 44.Bekpinar S, Vardagli D, Unlucerci Y, Can A, Uysal M, Gurdol F. Effect of rosiglitazone on asymmetric dimethylarginine metabolism in thioacetamide-induced acute liver injury. Pathophysiology. 2015;22(3):153–157. [DOI] [PubMed] [Google Scholar]
- 45.Leiper J, Murray-Rust J, McDonald N, Vallance P. S-nitrosylation of dimethylarginine dimethylaminohydrolase regulates enzyme activity: further interactions between nitric oxide synthase and dimethylarginine dimethylaminohydrolase. Proc Natl Acad Sci U S A. 2002;99(21):13527–13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McBride AE, Silver PA. State of the arg: protein methylation at arginine comes of age. Cell. 2001;106(1):5–8. [DOI] [PubMed] [Google Scholar]
- 47.Ogawa T, Kimoto M, Watanabe H, Sasaoka K. Metabolism of NG,NG-and NG,N’G-dimethylarginine in rats. Arch Biochem Biophys. 1987;252(2):526–537. [DOI] [PubMed] [Google Scholar]
- 48.Nijveldt RJ, Teerlink T, Siroen MP, van Lambalgen AA, Rauwerda JA, van Leeuwen PA. The liver is an important organ in the metabolism of asymmetrical dimethylarginine (ADMA). Clin Nutr. 2003;22(1):17–22. [DOI] [PubMed] [Google Scholar]
- 49.Vallance P, Leiper J. Cardiovascular biology of the asymmetric dimethylarginine:dimethylarginine dimethylaminohydrolase pathway. Arterioscler Thromb Vasc Biol. 2004;24(6):1023–1030. [DOI] [PubMed] [Google Scholar]
- 50.Jia SJ, Jiang DJ, Hu CP, Zhang XH, Deng HW, Li YJ. Lysophosphatidylcholine-induced elevation of asymmetric dimethylarginine level by the NADPH oxidase pathway in endothelial cells. Vascul Pharmacol. 2006;44(3):143–148. [DOI] [PubMed] [Google Scholar]
- 51.Giriwono PE, Shirakawa H, Hokazono H, Goto T, Komai M. Fermented barley extract supplementation maintained antioxidative defense suppressing lipopolysaccharide-induced inflammatory liver injury in rats. Biosci Biotechnol Biochem. 2011;75(10):1971–1976. [DOI] [PubMed] [Google Scholar]
- 52.Mookerjee RP, Dalton RN, Davies NA, et al. Inflammation is an important determinant of levels of the endogenous nitric oxide synthase inhibitor asymmetric dimethylarginine (ADMA) in acute liver failure. Liver Transpl. 2007;13(3):400–405. [DOI] [PubMed] [Google Scholar]
- 53.Mookerjee RP, Malaki M, Davies NA, et al. Increasing dimethylarginine levels are associated with adverse clinical outcome in severe alcoholic hepatitis. Hepatology. 2007;45(1):62–71. [DOI] [PubMed] [Google Scholar]
- 54.Banerjee R, Zou CG. Redox regulation and reaction mechanism of human cystathionine-beta-synthase: a PLP-dependent hemesensor protein. Arch Biochem Biophys. 2005;433(1):144–156. [DOI] [PubMed] [Google Scholar]
- 55.Taoka S, Lepore BW, Kabil O, Ojha S, Ringe D, Banerjee R. Human cystathionine beta-synthase is a heme sensor protein. Evidence that the redox sensor is heme and not the vicinal cysteines in the CXXC motif seen in the crystal structure of the truncated enzyme. Biochemistry. 2002;41(33):10454–10461. [DOI] [PubMed] [Google Scholar]
- 56.Mosharov E, Cranford MR, Banerjee R. The quantitatively important relationship between homocysteine metabolism and glutathione synthesis by the transsulfuration pathway and its regulation by redox changes. Biochemistry. 2000;39(42):13005–13011. [DOI] [PubMed] [Google Scholar]
- 57.Tomita K, Barnes PJ, Adcock IM. The effect of oxidative stress on histone acetylation and IL-8 release. Biochem Biophys Res Commun. 2003;301(2):572–577. [DOI] [PubMed] [Google Scholar]
- 58.Gu X, Sun J, Li S, Wu X, Li L. Oxidative stress induces DNA demethylation and histone acetylation in SH-SY5Y cells: potential epigenetic mechanisms in gene transcription in Abeta production. Neurobiol Aging. 2013;34(4):1069–1079. [DOI] [PubMed] [Google Scholar]
- 59.Gilmour PS, Rahman I, Donaldson K, MacNee W. Histone acetylation regulates epithelial IL-8 release mediated by oxidative stress from environmental particles. Am J Physiol Lung Cell Mol Physiol. 2003;284(3):L533–L540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.