Abstract
Ultraconserved elements (UCEs) are among the most popular DNA markers for phylogenomic analysis. In at least three of five placental mammalian genomes (human, dog, cow, mouse, and rat), 2189 UCEs of at least 200 bp in length that are identical have been identified. Most of these regions have not yet been functionally annotated, and their associations with diseases remain largely unknown. This is an important knowledge gap in human genomics with regard to UCE roles in physiologically critical functions, and by extension, their relevance for shared susceptibilities to common complex diseases across several mammalian organisms in the event of their polymorphic variations. In the present study, we remapped the genomic locations of these UCEs to the latest human genome assembly, and examined them for documented polymorphisms in sequenced human genomes. We identified 29,983 polymorphisms within analyzed UCEs, but revealed that a vast majority exhibits very low minor allele frequencies. Notably, only 112 of the identified polymorphisms are associated with a phenotype in the Ensembl genome browser. Through literature analyses, we confirmed associations of 37 (i.e., out of the 112) polymorphisms within 23 UCEs with 25 diseases and phenotypic traits, including, muscular dystrophies, eye diseases, and cancers (e.g., familial adenomatous polyposis). Most reports of UCE polymorphism—disease associations appeared to be not cognizant that their candidate polymorphisms were actually within UCEs. The present study offers strategic directions and knowledge gaps for future computational and experimental work so as to better understand the thus far intriguing and puzzling role(s) of UCEs in mammalian genomes.
Keywords: genome, ultraconserved elements, UCEs, orthologous regions, polymorphism, complex diseases, phenotype
Introduction
Ultraconserved elements (UCEs) were first discovered by Bejerano et al. (2004) who reported the existence of 481 genomic segments over 200 bp in length that are absolutely conserved between orthologous regions of the human, rat, and mouse genomes. Stephen et al. (2008) subsequently expanded the set of known UCEs by comparing a wider set of mammalian genomes, identifying 2189 sequences ≥200 bp and 13,736 sequences ≥100 bp that are identical in at least 3 of 5 placental mammals (human, dog, cow, mouse, and rat). They also showed that these UCEs evolved relatively rapidly during tetrapod evolution, increasing in size and number, possibly under positive selection, but then became virtually frozen in the amniotes, with a mutation rate far lower than that of protein-coding sequences, suggesting fierce purifying selection upon fixation.
Such extraordinary conservation suggests that UCEs have acquired an indispensable function, although that function remains mysterious, particularly in view of the fact that each UCE has a distinct sequence. Initial deletion studies suggested that the absence of UCEs did not result in overt phenotypic changes in mice (Ahituv et al., 2007). More recently, however, clear phenotypic effects were detected when other UCEs were deleted from the mouse genome (Dickel et al., 2018; Nolte et al., 2014). A large fraction of UCEs have been found to be transcriptionally active and involved in multiple human cancers (Calin et al., 2007; reviewed in Fabris and Calin, 2017; Terracciano et al., 2017). The transcription of several UCEs is upregulated by hypoxia (Ferdin et al., 2013). A recent study suggested that UCEs might be important for genome organization and genome stability (McCole et al., 2018).
Only few studies have reported the frequency of polymorphisms within UCEs in the human population. At the time when UCEs were discovered, only 6 of 106,767 examined ultraconserved bases were validated single nucleotide polymorphisms (SNPs) (Bejerano et al., 2004). Three years later, Chen et al. (2007) identified 102 polymorphisms within UCEs, 24 of which were verified by 2 or more research groups. A study by Ovcharenko (2008) revealed that the number of UCE SNPs differs between populations, but overall, over 25% of UCEs harbor at least one SNP.
Wojcik et al. (2010) sequenced 9634 bp of UCEs and detected six SNPs, showing that UCEs are less conserved than initially suggested, but still several times less variable when compared with the rest of the human genome. Additionally, they observed a higher UCE mutations frequency in patients with chronic lymphocytic leukemia or colorectal cancer than in the general population.
Since the locations of the majority of UCEs are not yet included in main genomic browsers, most of subsequent studies of polymorphism/phenotype associations have overlooked the potential location of functional polymorphisms within UCEs. Partly as a consequence thereof, the role of these regions and the consequences of polymorphisms within them remain poorly understood.
We aimed in the present study to define genomic locations of UCEs according to the latest human genome release, to identify overlapping genes and to identify polymorphisms within UCEs, especially those that have been reported to be associated with a phenotype. The study attempts to address an important knowledge gap in human genomics with regard to UCE roles in critical, presumably indispensible functions, and their relevance for susceptibilities to common complex diseases across mammalian organisms in the event of their polymorphic variations.
Materials and Methods
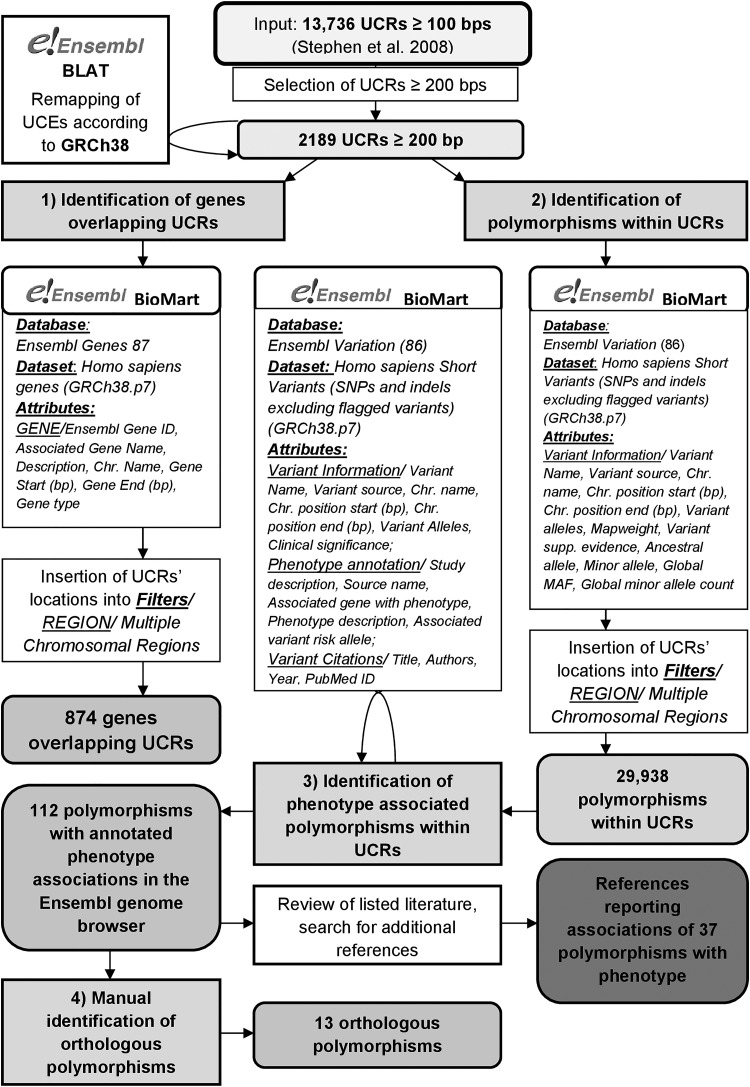
The workflow of the study is presented in Figure 1. UCEs ≥100 bp, which are identical in at least three of five placental mammals (human, dog, cow, mouse, and rat) were obtained from Stephen et al. (2008). Among 13,736 reported UCEs, a subset of 2189 UCEs, which are ≥200 bp in length was selected for further analysis. Selected UCEs were remapped according to the latest human genome release (GRCh38) using the Ensembl BLAT (BLAST-like alignment tool) tool (Kent, 2002).
FIG. 1.
Overview of the study.
Using the Ensembl BioMart data mining tool (Smedley et al., 2015), we collected genes, within which 2189 ≥ 200 bp UCEs are located. Ensembl Genes 87 was set as database and Homo sapiens genes (GRCh38.p7) was chosen as dataset. In the attributes tab, the following features were selected: GENE/Ensembl Gene ID, Associated Gene Name, Description, Chromosome Name, Gene Start (bp), Gene End (bp), and Gene type. UCEs' locations were then inserted into Filters/REGION/Multiple Chromosomal Regions. A code in Python was used to allocate UCEs to their corresponding genes according to their locations.
In the next step, we collected names and locations of all known polymorphisms within ≥200 bp long UCEs in the human genome using the BioMart data mining tool (Smedley et al., 2015). Ensembl Variation, release 86, was set as database and Homo sapiens Short Variants (SNPs and indels, excluding flagged variants; GRCh38.p7) was chosen as dataset.
In the Attributes tab, the following variant-associated information was ticked: Variant Information/Variant Name, Variant source, Chromosome name, Chromosome position start (bp), Chromosome position end (bp), Variant alleles, Mapweight, Variant supporting evidence, Ancestral allele, Minor allele (ALL), Global minor allele frequency (all individuals), Global minor allele count (all individuals). UCEs' locations were inserted into Filters/REGION/Multiple Chromosomal Regions. A code in Python was used to allocate polymorphisms to their corresponding UCEs according to their locations.
Using BioMart we also screened the UCEs for phenotype-associated polymorphisms and retrieved scientific literature describing these associations. We adjusted the settings used in the previous step—in the Attributes tab, the following variant-associated information was ticked: Variant Information/Variant Name, Variant source, Chromosome name, Chromosome position start (bp), Chromosome position end (bp), Clinical significance; Phenotype annotation/Study description, Source name, Associated gene with phenotype, Phenotype description, Associated variant risk allele; Variant Citations/Title, Authors, Year, PubMed ID. UCEs' locations were inserted into Filters/REGION/Multiple Chromosomal Regions. Scientific articles obtained with BioMart and with additional manual search of the listed databases were manually reviewed to verify given polymorphism/phenotype associations.
Orthologous polymorphisms were considered as polymorphisms in other species' genome located within the same nucleotide triplet within exonic regions. For all 112 phenotype-associated polymorphisms we manually checked whether orthologous polymorphisms in other species exist. This was done by performing multiple text alignments in Ensembl.
We chose 18 eutherian mammals EPO alignment, in which DNA sequences from 18 species are aligned, including Human (Homo sapiens), assembly GRCh38; Gorilla (Gorilla gorilla gorilla), assembly gorGor3.1; Chimpanzee (Pan troglodytes), assembly CHIMP2.1.4; Orangutan (Pongo abelii) assembly PPYG2; Macaque (Macaca mulatta), assembly Mmul_8.0.1; Olive baboon (Papio anubis), assembly PapAnu2.0; Vervet-AGM (Chlorocebus sabaeus), assembly ChlSab1.1; Marmoset (Callithrix jacchus), assembly C_jacchus3.2.1; Mouse (Mus musculus), assembly GRCm38; Mouse SPRETEiJ (Mus spretus), assembly SPRET_EiJ_v1; Rat (Rattus norvegicus), assembly Rnor_6.0; Rabbit (Oryctolagus cuniculus), assembly OryCun2.0; Horse (Equus caballus), assembly EquCab2; Cat (Felis catus), assembly Felis_catus_6.2; Dog (Canis lupus familiaris), assembly CanFam3.1; Pig (Sus scrofa), assembly Sscrofa10.2; Cow (Bos taurus), assembly UMD3.1, and Sheep (Ovis aries), assembly Oar_v3.1.
Results
In the present work, we adhered to the definition of polymorphisms suggested by Karki et al. (2015). We manually curated the genomic locations of the 2189 UCEs ≥200 bp reported by Stephen et al. (2008), identified overlapping genes, screened the UCEs for polymorphisms and identified those previously associated with a phenotype in the Ensembl database (Zerbino et al., 2018). We also checked whether any colocated polymorphisms occur in other species. The workflow of the study and the main results are shown in Figure 1.
Genes overlapping UCEs
Among 2189 UCEs, 541 (24.7%) are intergenic, whereas 1648 (75.3%) are located within 874 genes (Supplementary Table S1). Most of these genes are protein coding (629), but a substantial fraction express long intergenic noncoding RNA (lincRNAs; 119) or antisense RNA (90) (Supplementary Table S2).
Among the 874 identified genes, 337 genes include more than 1 UCE and among these, 24 extend over at least 10 UCEs. Genes that overlap with the most UCEs are forkhead box P2 (FOXP2; 28 UCEs), leucine-rich melanocyte differentiation associated (LRMDA; 21 UCEs), neuronal PAS domain protein 3 (NPAS3; 19 UCEs), LINC01122 (18 UCEs), zinc finger E-box-binding homeobox 2 (ZEB2; 18 UCEs), and activator of transcription and developmental regulator AUTS2 (AUTS2; 18 UCEs).
Polymorphisms within UCEs are abundant, but rare
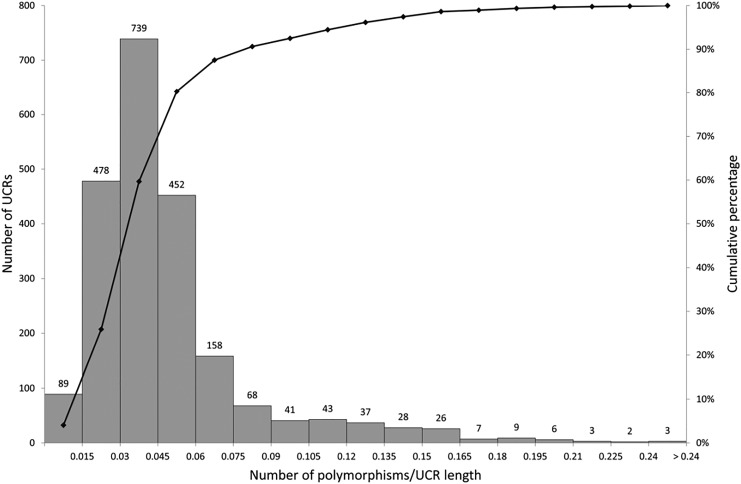
Within the 2189 screened UCEs, 29,983 polymorphisms were found using the BioMart tool (Smedley et al., 2015) (Supplementary Table S3). Each of the UCEs harbors at least one polymorphism, but most (90%; 1974 out of 2189) contain more than 5 polymorphisms. Among these, 19 UCEs include more than 50 polymorphisms (Fig. 2). The most polymorphisms (n = 70) are located within UCE 5114, which is 554 bp in length. The density of polymorphisms is the highest for UCE 5138, which is 206 bp in length and contains 64 polymorphisms (i.e., 0.311 polymorphism/bp) (Fig. 3 and Supplementary Table S3).
FIG. 2.
Graphic representation of UCEs according to the number of polymorphisms within them. UCEs, ultraconserved elements.
FIG. 3.
Graphic representation of UCEs according to polymorphism density within them.
Since finding so many polymorphisms within UCEs was unexpected, we examined their population frequencies. Using the BioMart tool, we extracted the polymorphisms' global minor allele frequencies (MAFs) and global minor allele counts (MACs), where available. Collectively, we retrieved MAFs and MACs for 12,458 among 29,983 polymorphisms within UCEs (Supplementary Table S3). The median MAF is 0.0002, whereas the average MAF equals 0.006224. Around 94.25% of polymorphisms have MAFs ≤0.01. However, a substantial number of polymorphisms exhibit high values of MAFs, 37 SNPs having MAFs of 0.4–0.5.
Most phenotype-associated polymorphisms are in coding regions
One hundred and twelve out of 29,938 polymorphisms within UCEs, including 85 SNPs, 18 deletions, 7 insertions, and 2 indels, have annotated phenotype associations in the Ensembl genome browser (Supplementary Table S4). Eighty-three out of the 112 phenotype-associated polymorphisms are located within coding regions; they include 47 missense variants, 18 frameshift variants, 8 stop codon gains, 5 synonymous variants, 2 lost stop codons, 1 protein-altering variant, 1 inframe insertion, and 1 inframe deletion. The rest of the phenotype-associated polymorphisms reside within 3′ untranslated regions (13 polymorphisms) or introns of coding genes (10 intron variants, 1 splice region variant, 1 splice acceptor variant, and 1 splice donor variant), whereas 3 polymorphisms are intergenic (Fig. 4).
FIG. 4.
Number of phenotype-associated UCE polymorphisms according to the most severe consequence of the polymorphism.
Phenotype-associated polymorphisms are concentrated within particular UCEs and genes
One hundred and twelve phenotype-associated polymorphisms are located within 39 UCEs, with 13 UCEs containing more than 1 phenotype-associated polymorphism (Supplementary Table S4), and 3 UCEs containing at least 10 phenotype-associated polymorphisms. The most (19) occur in UCE 10358 within the APC gene (APC regulator of WNT-signaling pathway) on chromosome 5. UCE 13736 within the MECP2 (methyl-CpG-binding protein 2) on chromosome X and UCE 7376 within the MBD5 (methyl-CpG-binding domain protein 5) on chromosome 2 contain 12 and 10 phenotype-associated polymorphisms, respectively.
Some of the genes contain multiple UCEs with phenotype-associated polymorphisms (Supplementary Table S4). Two UCEs (7376 and 7380) within the MBD5 gene on chromosome 2 contain 10 and 5 functionally annotated polymorphisms, respectively. Two UCEs (4568 and 4577) within the MAP2K5 (mitogen-activated protein kinase kinase 5) on chromosome 15 each contain one phenotype-associated polymorphism. Within the FHL1 (four and a half LIM domains 1) on chromosome X UCE 13666 contains three and UCE 13667 one phenotype-associated polymorphism.
Polymorphisms in UCEs are associated with various diseases
Among the 112 phenotype-associated polymorphisms, 76 (67.9%) had available phenotype descriptions in Ensembl, whereas the associated phenotype was unspecified for the remaining 36 polymorphisms. In total, we found articles regarding 37 polymorphism/phenotype associations (Table 1).
Table 1.
An Overview of Phenotype-Associated Polymorphisms Within Ultraconserved Elements for Which Literature References Have Been Found
| Polymorphism name | Chr. | UCE ID | Gene | Associated phenotype description (comment) | Source |
|---|---|---|---|---|---|
| rs17105335 | 1 | 371 | AGBL4 | Amyotrophic lateral sclerosis (in Irish cohort) | Cronin et al. (2008) |
| rs2020906 | 2 | 6629 | FBXO11, MSH6 | Lynch syndrome (likely neutral variant) | Hansen et al. (2014) |
| rs10496382 | 2 | 7038 | / | Height (among top highly constrained SNPs associated with height detected in 23,764 European American samples from the National Heart, Lung, and Blood Institute Candidate Gene Association Resource, but after adding the data from the GIANT consortium, significance was lost) | Chiang et al. (2012) |
| rs13382811 | 2 | 7246 | ZEB2 | Severe myopia | Khor et al. (2013) |
| rs104893634 | 2 | 7789 | HOXD10, HOXD9, HOXD-AS2 (AS) | Vertical talus congenital | Dobbs et al. (2006); Shrimpton et al. (2004) |
| rs2307121 | 5 | 10019 | ADAMTS6 | Central corneal thickness | Lu et al. (2013) |
| rs587777277 | 5 | 10277 | NR2F1, NR2F1-AS1 (AS) | Bosch-Boonstra-Schaaf optic atrophy syndrome | Bosch et al. (2014) |
| rs587777275 | 5 | 10277 | NR2F1, NR2F1-AS1 (AS) | Bosch-Boonstra-Schaaf optic atrophy syndrome | Bosch et al. (2014) |
| rs587777274 | 5 | 10277 | NR2F1, NR2F1-AS1 (AS) | Bosch-Boonstra-Schaaf optic atrophy syndrome | Bosch et al. (2014) |
| rs387906239 | 5 | 10358 | APC | Familial adenomatous polyposis 1 attenuated | Soravia et al. (1999) |
| rs3797704 | 5 | 10358 | APC | No association with breast cancer | Chang et al. (2016) |
| rs387906232 | 5 | 10358 | APC | Familial adenomatous polyposis 1 | Fodde et al. (1992) |
| rs387906237 | 5 | 10358 | APC | Familial adenomatous polyposis 1 attenuated | Curia et al. (1998) |
| rs121434591 | 5 | 10453 | MATR3 (ENSG00000280987, ENSG00000015479) | Distal myopathy | Senderek et al. (2009) |
| rs587777300 | 5 | 10453 | MATR3 (ENSG00000280987, ENSG00000015479) | Amyotrophic lateral sclerosis 21 | Johnson et al. (2014) |
| rs863223403 | 9 | 12957 | HNRNPK | Au-Kline syndrome | Au et al. (2015) |
| rs121917900 | 10 | 1446 | ERCC6 | Cockayne syndrome B | Mallery et al. (1998) |
| rs75462234 | 10 | 1766 | PAX2 | Papillorenal syndrome | Schimmenti et al. (1999) |
| rs77453353 | 10 | 1766 | PAX2 | Renal coloboma syndrome | Amiel et al. (2000) |
| rs76675173 | 10 | 1766 | PAX2 | Papillorenal syndrome | Schimmenti et al. (1997) |
| rs587777708 | 10 | 1766 | PAX2 | Focal segmental glomerulosclerosis 7 | Barua et al. (2014) |
| rs11190870 | 10 | 1798 | / | Adolescent idiopathic scoliosis (severe), no association with breast cancer | Chettier et al. (2015); Gao et al. (2013); Grauers et al. (2015); Jiang et al. (2013); Londono et al. (2014); Miyake et al. (2013); Shen et al. (2011); Takahashi et al. (2011) |
| rs724159963 | 11 | 2195 | FAR1 | Peroxisomal fatty acyl-CoA reductase 1 disorder | Buchert et al. (2014) |
| rs16932455 | 11 | 2242 | SOX6 | Capecitabine sensitivity | O'Donnell et al. (2012) |
| rs997295 | 15 | 4568 | MAP2K5 | Motion sickness; BMI | De et al. (2015); Guo et al. (2013); Hromatka et al. (2015) |
| rs587777373 | 15 | 4731 | NR2F2 | Congenital heart defects multiple types 4 | Al Turki et al. (2014) |
| rs398123839 | X | 13372 | DMD | Duchenne muscular dystrophy | Hofstra et al. (2004); Roberts et al. (1992) |
| rs863224976 | X | 13372 | DMD | Becker muscular dystrophy | Tuffery-Giraud et al. (2005) |
| rs132630295 | X | 13568 | PLP1 | Spastic paraplegia 2 X-linked | Gorman et al. (2007) |
| rs132630287 | X | 13568 | PLP1 | Spastic paraplegia 2 X-linked | Saugier-Veber et al. (1994) |
| rs132630292 | X | 13568 | PLP1 | Pelizaeus/Merzbacher disease atypical | Hodes et al. (1997) |
| rs137852350 | X | 13607 | GRIA3 | Mental retardation X-linked 94 | Wu et al. (2007) |
| rs122459149 | X | 13666 | FHL1 | Emery-Dreifuss muscular dystrophy 6 X-linked | Gueneau et al. (2009); Knoblauch et al. (2010) |
| rs122458141 | X | 13666 | FHL1 | Myopathy X-linked with postural muscle atrophy | Schoser et al. (2009); Windpassinger et al. (2008) |
| rs786200914 | X | 13666 | FHL1 | Myopathy X-linked with postural muscle atrophy | Schoser et al. (2009) |
| rs267606811 | X | 13667 | FHL1 | Myopathy X-linked with postural muscle atrophy | Windpassinger et al. (2008) |
| rs62621672 | X | 13736 | MECP2 | Rett syndrome (nonpathogenic variant) | Zahorakova et al. (2007) |
ADAMTS6, ADAM metallopeptidase with thrombospondin type 1 motif 6 gene; BMI, body mass index; FHL1, four and a half LIM domains 1; MAP2K5, mitogen-activated protein kinase kinase 5; MECP2, methyl-CpG-binding protein 2; SNPs, single nucleotide polymorphisms; SOX6, SRY-box 6 gene; UCE, ultraconserved element; ZEB2, zinc finger E-box-binding homeobox 2.
Khor et al. (2013) found highly suggestive evidence of association of rs13382811 (a SNP within ZEB2) on chromosome 2 with severe myopia. Lu et al. (2013) found a connection between rs2307121 (a SNP located in the ADAM metallopeptidase with thrombospondin type 1 motif 6 gene [ADAMTS6]) and corneal structure. Sequencing of positional candidate genes of a large family of Bulgarian descent revealed a missense mutation rs121434591 as a cause for vocal cord and pharyngeal weakness with distal myopathy (Senderek et al., 2009). rs11190870, an intergenic SNP located 6745 bp downstream of the Ladybird homeobox 1 gene (LBX1), is associated with adolescent idiopathic scoliosis (Chettier et al., 2015; Gao et al., 2013; Grauers et al., 2015; Jiang et al., 2013; Londono et al., 2014; Miyake et al., 2013; Takahashi et al., 2011).
Sequence variant rs16932455 within SRY-box 6 gene (SOX6) is among the top SNPs from the multipopulation meta-analysis of genome-wide association findings for capecitabine susceptibility (O'Donnell et al., 2012). rs997295, a SNP located in the MAP2K5 gene, is among lead SNPs for motion sickness (Hromatka et al., 2015). This polymorphism is also among SNPs associated with body mass index or binary obesity (De et al., 2015; Guo et al., 2013; Yazdi et al., 2015). Among UCE polymorphisms with cited references rs997295 is the only polymorphism with more than one phenotype association.
Polymorphisms within orthologous regions
We manually checked if 112 phenotype-associated polymorphisms located within UCEs have orthologous polymorphisms located within the same triplet codon in exonic regions in other species. We aligned human UCEs containing phenotype-associated polymorphisms with 17 other mammalian species and found 10 orthologous polymorphisms for 8 human phenotype-associated polymorphisms within UCEs (Supplementary Table S5). Among the 10 orthologous polymorphisms, 9 were found in cow and 1 in pig, respectively. According to the Ensembl genome browser these orthologous polymorphisms have not yet been associated with phenotype in other species. One example of an orthologous polymorphism is shown in Supplementary Figure S1.
Discussion
At present, UCEs are among the most popular DNA markers for phylogenomic analysis (Tagliacollo and Lanfear, 2018). They might also be good candidates for targeted sequencing projects and association studies (Silla et al., 2014) and might present valuable and easily detectable disease biomarkers. Bao et al. (2016) for example found association between rs8004379 and prostate cancer-specific mortality. SNPs within UCEs may also be valuable prognostic biomarkers for patients with locally advanced colorectal cancer who receive 5-fluorouracil-based chemotherapy (Lin et al., 2012).
Although UCEs are absolutely conserved between orthologous regions of several species genomes, our study revealed as many as 29,938 polymorphisms within the 2189 analyzed UCEs that comprise 628,364 bp of the human genome (Supplementary Table S3). The number of polymorphisms recorded in Ensembl release 86, genome-wide, is one per 22.7 bp. Our data suggest that the polymorphism density within UCEs is slightly higher than the genomic average: one polymorphism per 21.0 bp is present within these regions. How can UCEs be so conserved among species but simultaneously harbor so many polymorphisms? One possible explanation would be that polymorphisms in UCEs might be less stable or rarer within populations than polymorphisms outside of UCEs.
Remarkably, our results indeed show that a vast majority of UCE polymorphisms exhibit extremely low MAFs—less than 6% occur at a frequency of >1%. Depletion of prevalent polymorphisms from UCEs has also been confirmed by Silla et al. (2014). It may be assumed that recurrent polymorphisms are less likely associated with fitness, while those occurring at lower MAFs more likely have deleterious consequences.
Our results show that the density of polymorphisms is not uniform among all UCEs—a substantial proportion of UCEs contains many polymorphisms (Figs. 2 and 3). These data suggest that these regions have not all been under the same purifying selection. Considering the fact that analyzed UCEs are defined as identical in at least three of five placental mammals (human, dog, cow, mouse, and rat) (Stephen et al., 2008), this may reflect a decrease of conservation throughout primate evolution (Ovcharenko, 2008). Some UCEs might have lost their crucial functions recently and therefore, neutral polymorphisms might have started to accumulate within these regions.
A vast majority of the UCE polymorphisms has not (yet) been associated with diseases or phenotypic traits, but 112 are annotated as phenotype associated in the Ensembl genome browser. Interestingly, they are concentrated within certain UCEs: in 13 UCEs more than 1 phenotype-associated polymorphism is present, wherein 3 UCEs include 10 or more phenotype-associated polymorphisms although this may reflect an ascertainment bias towards protein-coding regions.
Associated phenotypes include different types of muscular dystrophies (e.g., distal myopathy, X-linked myopathy with postural muscle atrophy, Duchene-, Becker-, and Emery-Dreifuss muscular dystrophy), adolescent idiopathic scoliosis, amyotrophic lateral sclerosis, as well as a number of eye-related diseases (e.g., myopia, Bosch-Boonstra-Schaaf optic atrophy syndrome, papillorenal syndrome), cancer (e.g., familial adenomatous polyposis) etc. (Table 1). Our results imply that conservation of at least some UCEs still is of high importance for normal phenotype, which is in accordance with published UCEs knockout studies (Dickel et al., 2018; Nolte et al., 2014).
Only four among all reviewed articles regarding polymorphism/phenotype associations have mentioned that locations of analyzed polymorphisms are within conserved genomic regions (Chen et al., 2007; Chiang et al., 2012; Senderek et al., 2009; Shen et al., 2011), which strongly indicates that UCEs mostly remain unnoticed by the authors. Simultaneously, our identification of the 37 polymorphisms within UCEs with phenotype annotations highlights the fact that a wealth of data regarding functional annotation of UCEs is readily available in established databases. Tools that would enable integration of functional data regarding polymorphisms with their possible location within UCEs would be a big step toward better understanding of the UCEs' roles. Apart from additional computational analyses, also much more experimental study will be needed.
There are some limitations worthy of consideration in the interpretation of the current study. It is very likely that our method using the BioMart tool (Smedley et al., 2015) missed some polymorphism/phenotype associations, since not all of them are annotated in the Ensembl genome browser (Zerbino et al., 2018) and, therefore, could not be found in the BioMart analysis. Additionally, it is possible that some of the reported phenotype-associated polymorphisms do not play a causal role, but are, instead, false positives.
Results presented in this study would greatly benefit from additional studies utilizing other high-performance bioinformatics tools for retrieving phenotype-associated polymorphisms located within UCEs and published literature on these polymorphisms. However, due to the novel releases of genome assemblies, there is also a need to update and perform new genome alignments and to define UCEs on an ongoing basis.
Conclusions
In the present study, we remapped UCEs according to the latest human genome release, identified genes overlapping UCEs, and uncovered a large number of polymorphisms within these regions. Majority of the identified polymorphisms exhibit extremely low MAFs, which implies vital importance of UCEs' conservation. In accordance, we found a number of polymorphisms within UCEs, which have already been associated with diseases or phenotypic traits in the literature. Our study serves as a basis for further computational and experimental work that is crucially needed for a better understanding of the puzzling role(s) of UCEs in mammalian genomes.
Supplementary Material
Abbreviations Used
- MAC
minor allele count
- MAF
minor allele frequency
- MAP2K5
mitogen-activated protein kinase kinase 5
- MBD5
methyl-CpG-binding domain protein 5
- NIH
National Institutes of Health
- SNP
single nucleotide polymorphism
- UCE
ultraconserved element
- ZEB2
zinc finger E-box-binding homeobox 2
Author Disclosure Statement
The authors declare they have no competing financial interests.
Funding Information
This work was supported by the Slovenian Research Agency (ARRS) through the Research program Comparative genomics and genome biodiversity (grant No. P4-0220). Dr. Calin is the Felix L. Haas Endowed Professor in Basic Science. Work in Dr. Calin's laboratory is supported by National Institutes of Health (NIH/NCATS) grant UH3TR00943-01 through the NIH Common Fund, Office of Strategic Coordination (OSC), the NIH/NCI grant 1 R01 CA182905-01, a U54 grant No. CA096297/CA096300–UPR/MDACC Partnership for Excellence in Cancer Research 2016 Pilot Project, a Team DOD (CA160445P1) grant, a Ladies Leukemia League grant, a CLL Moonshot Flagship project, a SINF 2017 grant, and the Estate of C.G. Johnson, Jr.
Supplementary Material
References
- Ahituv N, Zhu Y, Visel A, et al. (2007). Deletion of ultraconserved elements yields viable mice. PLoS Biol 5, e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Turki S, Manickaraj AK, Mercer CL, et al. (2014). Rare variants in NR2F2 cause congenital heart defects in humans. Am J Hum Genet 94, 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiel J, Audollent S, Joly D, et al. (2000). PAX2 mutations in renal–coloboma syndrome: Mutational hotspot and germline mosaicism. Eur J Hum Genet 8, 820–826 [DOI] [PubMed] [Google Scholar]
- Au PYB, You J, Caluseriu O, et al. (2015). GeneMatcher aids in the identification of a new malformation syndrome with intellectual disability, unique facial dysmorphisms, and skeletal and connective tissue abnormalities caused by de novo variants in HNRNPK. Hum Mutat 36, 1009–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao BY, Lin VC, Yu CC, et al. (2016). Genetic variants in ultraconserved regions associate with prostate cancer recurrence and survival. Sci Rep 6, 22124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua M, Stellacci E, Stella L, et al. (2014). Mutations in PAX2 associate with adult-onset FSGS. J Am Soc Nephrol 25, 1942–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, et al. (2004). Ultraconserved elements in the human genome. Science 304, 1321–1325 [DOI] [PubMed] [Google Scholar]
- Bosch DGM, Boonstra FN, Gonzaga-Jauregui C, et al. (2014). NR2F1 mutations cause optic atrophy with intellectual disability. Am J Hum Genet 94, 303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchert R, Tawamie H, Smith C, et al. (2014). A peroxisomal disorder of severe intellectual disability, epilepsy, and cataracts due to fatty acyl-CoA reductase 1 deficiency. Am J Hum Genet 95, 602–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Ferracin M, et al. (2007). Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 12, 215–229 [DOI] [PubMed] [Google Scholar]
- Chang YS, Lin CY, Yang SF, Ho CM, and Chang JG. (2016). Analysing the mutational status of adenomatous polyposis coli (APC) gene in breast cancer. Cancer Cell Int 16, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CTL, Wang JC, and Cohen BA. (2007). The strength of selection on ultraconserved elements in the human genome. Am J Hum Genet 80, 692–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chettier R, Nelson L, Ogilvie JW, Albertsen HM, and Ward K. (2015). Haplotypes at LBX1 have distinct inheritance patterns with opposite effects in adolescent idiopathic scoliosis. PLoS One 10, e0117708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CWK, Liu CT, Lettre G, et al. (2012). Ultraconserved elements in the human genome: Association and transmission analyses of highly constrained single-nucleotide polymorphisms. Genetics 192, 253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin S, Berger S, Ding J, et al. (2008). A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum Mol Genet 17, 768–774 [DOI] [PubMed] [Google Scholar]
- Curia MC, Esposito DL, Aceto G, et al. (1998). Transcript dosage effect in familial adenomatous polyposis: Model offered by two kindreds with exon 9 APC gene mutations. Hum Mutat 11, 197–201 [DOI] [PubMed] [Google Scholar]
- De R, Verma SS, Drenos F, et al. (2015). Identifying gene-gene interactions that are highly associated with body mass index using quantitative multifactor dimensionality reduction (QMDR). BioData Min 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickel DE, Ypsilanti AR, Pla R, et al. (2018). Ultraconserved enhancers are required for normal development. Cell 172, 491.e15–499.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs MB, Gurnett CA, Pierce B, et al. (2006). HOXD10 M319K mutation in a family with isolated congenital vertical talus. J Orthop Res 24, 448–453 [DOI] [PubMed] [Google Scholar]
- Fabris L, and Calin GA. (2017). Understanding the genomic ultraconservations: T-UCRs and cancer. Int Rev Cell Mol Biol 333, 159–172 [DOI] [PubMed] [Google Scholar]
- Ferdin J, Nishida N, Wu X, et al. (2013). HINCUTs in cancer: Hypoxia-induced noncoding ultraconserved transcripts. Cell Death Differ 20, 1675–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodde R, van der Luijt R, Wijnen J, et al. (1992). Eight novel inactivating germ line mutations at the APC gene identified by denaturing gradient gel electrophoresis. Genomics 13, 1162–1168 [DOI] [PubMed] [Google Scholar]
- Gao W, Peng Y, Liang G, et al. (2013). Association between common variants near LBX1 and adolescent idiopathic scoliosis replicated in the Chinese Han population. PLoS One 8, e53234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman MP, Golomb MR, Walsh LE, et al. (2007). Steroid-responsive neurologic relapses in a child with a proteolipid protein-1 mutation. Neurology 68, 1305–1307 [DOI] [PubMed] [Google Scholar]
- Grauers A, Wang J, Einarsdottir E, et al. (2015). Candidate gene analysis and exome sequencing confirm LBX1 as a susceptibility gene for idiopathic scoliosis. Spine J 15, 2239–2246 [DOI] [PubMed] [Google Scholar]
- Gueneau L, Bertrand AT, Jais JP, et al. (2009). Mutations of the FHL1 gene cause Emery-Dreifuss muscular dystrophy. Am J Hum Genet 85, 338–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Lanktree MB, Taylor KC, Hakonarson H, Lange LA, and Keating BJ. (2013). Gene-centric meta-analyses of 108 912 individuals confirm known body mass index loci and reveal three novel signals. Hum Mol Genet 22, 184–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen MF, Neckmann U, Lavik LAS, et al. (2014). A massive parallel sequencing workflow for diagnostic genetic testing of mismatch repair genes. Mol Genet Genomic Med 2, 186–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodes ME, Blank CA, Pratt VM, Morales J, Napier J, and Dlouhy SR. (1997). Nonsense mutation in exon 3 of the proteolipid protein gene (PLP) in a family with an unusual form of Pelizaeus-Merzbacher disease. Am J Med Genet 69, 121–125 [PubMed] [Google Scholar]
- Hofstra RMW, Mulder IM, Vossen R, et al. (2004). DGGE-based whole-gene mutation scanning of the dystrophin gene in Duchenne and Becker muscular dystrophy patients. Hum Mutat 23, 57–66 [DOI] [PubMed] [Google Scholar]
- Hromatka BS, Tung JY, Kiefer AK, Do CB, Hinds DA, and Eriksson N. (2015). Genetic variants associated with motion sickness point to roles for inner ear development, neurological processes and glucose homeostasis. Hum Mol Genet 24, 2700–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Qiu X, Dai J, et al. (2013). Association of rs11190870 near LBX1 with adolescent idiopathic scoliosis susceptibility in a Han Chinese population. Eur Spine J 22, 282–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Pioro EP, Boehringer A, et al. (2014). Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci 17, 664–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R, Pandya D, Elston RC, and Ferlini C. (2015). Defining “mutation” and “polymorphism” in the era of personal genomics. BMC Med Genomics 8, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. (2002). BLAT—The BLAST-like alignment tool. Genome Res 12, 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor CC, Miyake M, Chen LJ, et al. (2013). Genome-wide association study identifies ZFHX1B as a susceptibility locus for severe myopia. Hum Mol Genet 22, 5288–5294 [DOI] [PubMed] [Google Scholar]
- Knoblauch H, Geier C, Adams S, et al. (2010). Contractures and hypertrophic cardiomyopathy in a novel FHL1 mutation. Ann Neurol 67, 136–140 [DOI] [PubMed] [Google Scholar]
- Lin M, Eng C, Hawk ET, et al. (2012). Identification of polymorphisms in ultraconserved elements associated with clinical outcomes in locally advanced colorectal adenocarcinoma. Cancer 118, 6188–6198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londono D, Kou I, Johnson TA, et al. (2014). A meta-analysis identifies adolescent idiopathic scoliosis association with LBX1 locus in multiple ethnic groups. J Med Genet 51, 401–406 [DOI] [PubMed] [Google Scholar]
- Lu Y, Vitart V, Burdon KP, et al. (2013). Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat Genet 45, 155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallery DL, Tanganelli B, Colella S, et al. (1998). Molecular analysis of mutations in the CSB(ERCC6) gene in patients with Cockayne syndrome. Am J Hum Genet 62, 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCole RB, Erceg J, Saylor W, and Wu CT. (2018). Ultraconserved elements occupy specific arenas of three-dimensional mammalian genome organization. Cell Rep 24, 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake A, Kou I, Takahashi Y, et al. (2013). Identification of a susceptibility locus for severe adolescent idiopathic scoliosis on chromosome 17q24.3. PLoS One 8, e72802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte MJ, Wang Y, Deng JM, et al. (2014). Functional analysis of limb transcriptional enhancers in the mouse. Evol Dev 16, 207–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell PH, Stark AL, Gamazon ER, et al. (2012). Identification of novel germline polymorphisms governing capecitabine sensitivity. Cancer 118, 4063–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I. (2008). Widespread ultraconservation divergence in primates. Mol Biol Evol 25, 1668–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RG, Bobrow M, and Bentley DR. (1992). Point mutations in the dystrophin gene. Proc Natl Acad Sci U S A 89, 2331–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saugier-Veber P, Munnich A, Bonneau D, et al. (1994). X-linked spastic paraplegia and Pelizaeus–Merzbacher disease are allelic disorders at the proteolipid protein locus. Nat Genet 6, 257–262 [DOI] [PubMed] [Google Scholar]
- Schimmenti LA, Cunliffe HE, McNoe LA, et al. (1997). Further delineation of renal-coloboma syndrome in patients with extreme variability of phenotype and identical PAX2 mutations. Am J Hum Genet 60, 869–878 [PMC free article] [PubMed] [Google Scholar]
- Schimmenti LA, Shim HH, Wirtschafter JD, et al. (1999). Homonucleotide expansion and contraction mutations of PAX2 and inclusion of Chiari 1 malformation as part of renal-coloboma syndrome. Hum Mutat 14, 369–376 [DOI] [PubMed] [Google Scholar]
- Schoser B, Goebel HH, Janisch I, et al. (2009). Consequences of mutations within the C terminus of the FHL1 gene. Neurology 73, 543–551 [DOI] [PubMed] [Google Scholar]
- Senderek J, Garvey SM, Krieger M, et al. (2009). Autosomal-dominant distal myopathy associated with a recurrent missense mutation in the gene encoding the nuclear matrix protein, matrin 3. Am J Hum Genet 84, 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Lu C, Jiang Y, Tang J, et al. (2011). Genetic variants in ultraconserved elements and risk of breast cancer in Chinese population. Breast Cancer Res Treat 128, 855–861 [DOI] [PubMed] [Google Scholar]
- Shrimpton AE, Levinsohn EM, Yozawitz JM, et al. (2004). A HOX gene mutation in a family with isolated congenital vertical talus and Charcot-Marie-Tooth disease. Am J Hum Genet 75, 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silla T, Kepp K, Tai ES, et al. (2014). Allele frequencies of variants in ultra conserved elements identify selective pressure on transcription factor binding. PLoS One 9, e110692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Haider S, Durinck S, et al. (2015). The BioMart community portal: An innovative alternative to large, centralized data repositories. Nucleic Acids Res 43, W589–W598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soravia C, Sugg SL, Berk T, et al. (1999). Familial adenomatous polyposis-associated thyroid cancer: A clinical, pathological, and molecular genetics study. Am J Pathol 154, 127–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen S, Pheasant M, Makunin IV, and Mattick JS. (2008). Large-scale appearance of ultraconserved elements in tetrapod genomes and slowdown of the molecular clock. Mol Biol Evol 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Tagliacollo VA, and Lanfear R. (2018). Estimating improved partitioning schemes for ultraconserved elements. Mol Biol Evol 35, 1798–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Kou I, Takahashi A, et al. (2011). A genome-wide association study identifies common variants near LBX1 associated with adolescent idiopathic scoliosis. Nat Genet 43, 1237–1240 [DOI] [PubMed] [Google Scholar]
- Terracciano D, Terreri S, de Nigris F, Costa V, Calin GA, and Cimmino A. (2017). The role of a new class of long noncoding RNAs transcribed from ultraconserved regions in cancer. Biochim Biophys Acta Rev Cancer 1868, 449–455 [DOI] [PubMed] [Google Scholar]
- Tuffery-Giraud S, Saquet C, Thorel D, et al. (2005). Mutation spectrum leading to an attenuated phenotype in dystrophinopathies. Eur J Hum Genet 13, 1254–1260 [DOI] [PubMed] [Google Scholar]
- Windpassinger C, Schoser B, Straub V, et al. (2008). An X-linked myopathy with postural muscle atrophy and generalized hypertrophy, termed XMPMA, is caused by mutations in FHL1. Am J Hum Genet 82, 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik SE, Rossi S, Shimizu M, et al. (2010). Non-codingRNA sequence variations in human chronic lymphocytic leukemia and colorectal cancer. Carcinogenesis 31, 208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Arai AC, Rumbaugh G, et al. (2007). Mutations in ionotropic AMPA receptor 3 alter channel properties and are associated with moderate cognitive impairment in humans. Proc Natl Acad Sci U S A 104, 18163–18168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdi FT, Clee SM, and Meyre D. (2015). Obesity genetics in mouse and human: Back and forth, and back again. PeerJ 3, e856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahorakova D, Rosipal R, Hadac J, et al. (2007). Mutation analysis of the MECP2 gene in patients of Slavic origin with Rett syndrome: Novel mutations and polymorphisms. J Hum Genet 52, 342–348 [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, et al. (2018). Ensembl 2018. Nucleic Acids Res 46, D754–D761 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.