Abstract
Keratoconus (KCN) is a leading cause for cornea grafting worldwide. Keratoconus is a multifactorial disease that causes progressive thinning of the cornea and whose etiology is poorly understood. Several studies have used proteomics on patient tear fluids to identify potential biomarkers. However, proteome of the cornea itself has not been investigated fully. We report here new findings from a case–control study using multiplexed mass spectrometry (MS) on individual (unpooled) corneas to gain deeper insights into proteins and biomarkers relevant to keratoconus. We employed a high-pressure approach to extract total protein from individual corneas from five cases and five controls, followed by trypsin digestion and tandem mass tag (TMT) labeling. The MS-derived data were searched using the Human NCBI RefSeq protein database v92, with peptides and proteins filtered at 1% false discovery rate. A total of 3132 proteins were detected, of which 627 were altered significantly (p ≤ 0.05) in keratoconus corneas. The increases were overwhelmingly in the mTOR/PI3/AKT signal-mediated regulations of cell survival and proliferation, nonsense-mediated decay of transcripts, and proteasomal pathways. The decreases were in several extracellular matrix proteins and in many members of the complement system. Importantly, this multiplexed proteomic study of keratoconus corneas identified, to our knowledge, the largest number of corneal proteins. The novel findings include changes in pathways that regulate transcript stability, proteasomal degradation, and the complement system in corneas with keratoconus. These observations offer new prospects toward future discovery of novel molecular targets for diagnostic and therapeutic innovations for patients with keratoconus.
Keywords: cornea, keratoconus, proteomics, ER stress, proteasomal degradation, complement
Introduction
Keratoconus is an ectatic thinning of the cornea that is often bilateral and progressive with early signs of the disease appearing in the first or the second decade of life. With increased steepening and thinning of the cornea and associated loss of vision, cornea grafting is often required, making keratoconus (KCN) the most common cause for cornea grafting worldwide.
Keratoconus is primarily asyndromic or sporadic, while the rare syndromic form(s) can be associated with Leber congenital amaurosis, Down syndrome, and other connective tissue syndromes such as arterial tortuosity and Marfan syndrome (Chang and Chodosh, 2013; Krachmer et al., 1984; Rabinowitz, 1998; Soiberman et al., 2017). The sporadic form of keratoconus is prevalent, ranging from 0.02% (Al-Amri, 2018) to 4.8% in some parts of the Middle East (Torres Netto et al., 2018). Keratoconus is also associated with eye rubbing, atopy, and contact lens wear (Macsai et al., 1990; Weed et al., 2008; Zadnik et al., 1998). There are no curative treatments for keratoconus, except management of the disease with corneal Intacs (Linebarger et al., 2000) and crosslinking of the corneal collagens with ultraviolet (UV) and riboflavin (Wollensak et al., 2004).
Attempts to understand the pathogenesis of keratoconus have examined the tear fluid of patients for cytokines and inflammatory dysregulations (Jun et al., 2011; Lema and Duran, 2005; Lema et al., 2008). In addition, whole proteomic profiling of the tear fluid has been performed for potential biomarkers that have identified prolactin-inducible protein as a potential biomarker for keratoconus (Priyadarsini et al., 2014). However, the major affected tissue in keratoconus is the cornea, which has not been studied fully by current proteomic approaches, the purpose of this study.
We report here new findings from a case–control study using multiplexed mass spectrometry (MS) on individual (unpooled) corneas to gain deeper insights into proteins and biomarkers relevant to keratoconus.
Materials and Methods
Ethics statement
All patient tissues were procured after informed consent following a Human Subject Research protocol approved by the KKESH Institutional Review Board. All study team members and analyses of the findings were approved by active Human Subject Research protocols in each of the institutions. Donor (DN) tissues deemed unsuitable for transplant usage were obtained from the Lions Eye Institute for Transplant and Research. This study adhered to the tenets of the Declaration of Helsinki.
Procurement of samples, protein extraction, trypsin digestion, and tandem mass tag labeling
Seven patient corneas (KCN) were procured during cornea transplantation from KKESH, Riyadh. Immediately after surgery, the central cornea was dissected and snap frozen in liquid nitrogen and stored in dry ice. Subsequently, all samples were shipped in dry ice to Baltimore for further processing. The keratoconus corneas included here were obtained after either penetrating keratoplasty or lamellar keratoplasty. Four patients were diagnosed without other associated conditions, while KCN12 had keratoconus with associated vernal keratoconjunctivitis (Table 1). All keratoconus samples had an intact or almost intact epithelium, some with reported breaks in the Bowman's layer (Table 2).
Table 1.
Patient and Control Donor Cornea Demography
| Sample ID | Group | Age | Sex | Other conditions |
|---|---|---|---|---|
| DN1 | Donor | 64 | M | N/A |
| DN3 | Donor | 58 | F | N/A |
| DN5 | Donor | 64 | M | N/A |
| DN8 | Donor | 71 | M | N/A |
| DN9 | Donor | 55 | F | N/A |
| KCN2 | Keratoconus | 25 | M | No |
| KCN8 | Keratoconus | 23 | F | No |
| KCN9 | Keratoconus | 33 | M | No |
| KCN10 | Keratoconus | 35 | F | No |
| KCN12 | Keratoconus | 22 | M | Vernal conjunctivitis |
DN, donor; KCN, keratoconus.
Table 2.
Patient Cornea Pathology
| Sample ID | Specimen description | Epithelium | Bowman's layer | Stroma |
|---|---|---|---|---|
| KCN2 | Partial thickness cornea section | Uniform | Intact | Unremarkable anterior stroma |
| KCN8 | Partial thickness cornea section | Irregular and variable in thickness | No definite breaks | Some scarring and fibrosis |
| KCN9 | Partial thickness cornea section | Centrally thickened | Multifocal breaks | Unremarkable anterior stroma |
| KCN10 | Partial thickness cornea section | Centrally thin | Multifocal breaks | Unremarkable anterior stroma |
| KCN12 | Full-thickness cornea section | Disrupted | Multifocal breaks | Unremarkable anterior stroma |
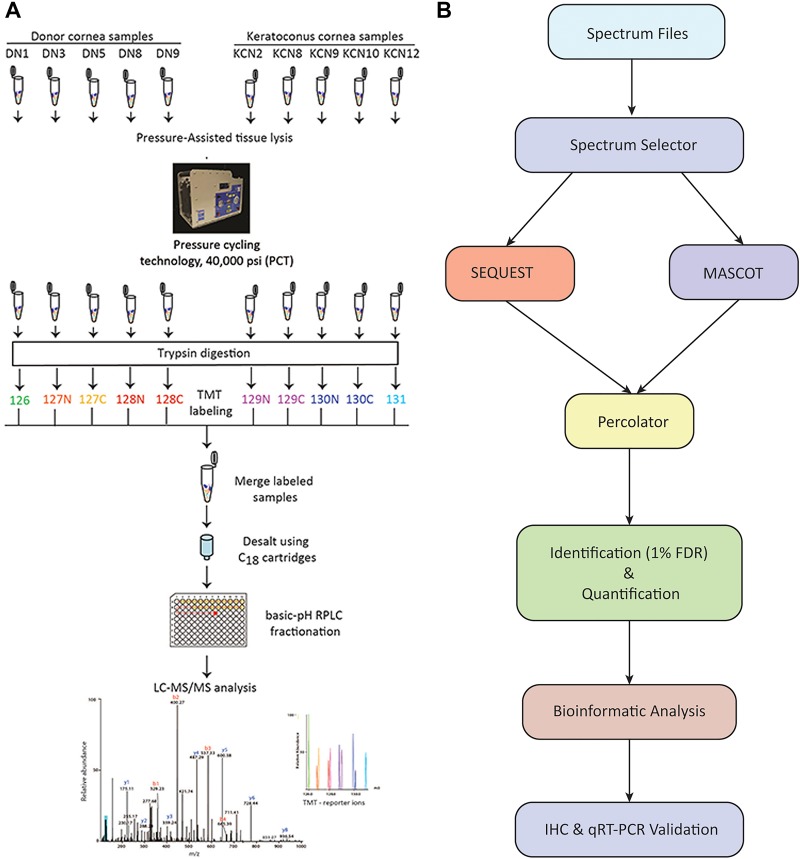
Five donor corneas were procured from the Lions Eye Institute for Transplant and Research, Florida. Donor corneas were placed in Optisol solution and shipped to Baltimore within 6–11 h after death. Upon receipt, the central cornea was dissected and snap frozen for further processing. Corneas were lysed in 8M urea using a pressure cycling technology containing 50 mM triethyl ammonium bicarbonate (TEAB), and transferred to 100 μL capacity PCT (pressure cycling technology) tubes and extracted using a barocycler at 95°C and 60 cycles of alternating pressure (1 cycle = 40,000 psi for 50 sec and 5000 psi for 10 sec). The lysates were transferred to microfuge tubes and centrifuged at 14,000 rpm for 10 min and the supernatant was collected (Fig. 1A).
FIG. 1.
Workflow of sample preparation and TMT labeling (A), and data analysis (B). TMT, tandem mass tag.
Protein estimation was carried out using bicinchoninic acid (BCA) assay. An aliquot of 100 μg protein from each sample was reduced and alkylated and subjected to in-solution trypsin digestion as described previously (Zahari et al., 2015). Briefly, the samples were reduced using 10 mM dithiothreitol (DTT) at 37°C for 1 h followed by alkylation using 30 mM iodoacetamide at room temperature for 20 min, protected from light. The protein samples were digested using sequencing grade trypsin at 1:20 enzyme to protein ratio at 37°C overnight. An aliquot of each digested sample was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to assess uniformity of extraction and digestion of the samples.
The samples were then acidified to 1% trifluoroacetic acid followed by C18-based desalting. The desalted peptides were vacuum dried and labeled using tandem mass tag (TMT) reagents. Peptides from each sample were labeled using TMT-10-plex labeling method as per manufacturer's instructions (Catalog No. 90110; Thermo Fisher Scientific). Peptides derived from DN1 were labeled with TMT tag 126, DN3 with TMT tag 127N, DN5 with TMT tag 127C, DN8 with TMT tag 128N, DN9 with TMT tag 128C, and keratoconus samples (KCN), KCN2 with TMT tag 129N, KCN8 with TMT tag 129C, KCN9 with TMT tag 130N, KCN10 with TMT tag 130C, and KCN12 with TMT tag 131 (Fig. 1A). The TMT-labeled peptides were pooled and freeze dried.
Basic-pH reversed-phase liquid chromatography
TMT-labeled peptides were subjected to basic-pH reversed-phase liquid chromatography fractionation (Guan et al., 2014) as follows: the vacuum-dried labeled peptides were reconstituted in solvent A (10 mM triethylammonium bicarbonate, pH 8.5) and loaded onto Zorbax Extend C18, 3.5 μm 250 × 4.6 mm column (Agilent Technologies, Santa Clara, CA). The peptide separation was carried out using a gradient of 0 to 50% solvent B (10 mM triethylammonium bicarbonate in 90% acetonitrile, pH 8.5) in 70 min. A total of 96 fractions were collected, which were then concatenated into 24 fractions, vacuum dried, and stored at −80°C until further LC-MS (liquid chromatography-mass spectrometry) analysis.
LC-MS/MS analysis
Fractionated peptide samples were loaded onto an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) interfaced with Ultimate 3000 RSLCnano ultra-high-pressure liquid chromatography system (Thermo Fisher Scientific, San Jose, CA). Peptide fractions were reconstituted in 0.1% formic acid and loaded on a precolumn (100 μm id × 5 mm, C18 PepMap100, 5 μm; Thermo Fisher Scientific, San Jose, CA) at 8 μL/min for 6 min using 0.1% formic acid in water. After 6 min, the peptides were separated on an EasySpray column (75 μm id × 50 cm, C18 PepMap, 2 μm; Thermo Fisher Scientific, San Jose, CA) at 300 nL/min using 0.1% formic acid in 95% acetonitrile.
The gradient consisted of an initial step of 8–32% in B over 100 min followed by 32–50% over 7 min, 32–90% in B over 3 min, held at 90% of B for 5 min, and then equilibrated for 20 min at 5% B, where the mobile phase A consisted of water containing 0.1% formic acid and mobile phase B consisted of 95% acetonitrile in 0.1% formic acid in water with a total run time of 140 min.
The mass spectrometer was fitted with an EasySpray source (Thermo Fisher Scientific, San Jose, CA) and operated in a data-dependent acquisition (DDA) manner. Each DDA cycle consisted of one OT (Orbitrap) MS survey scan acquired at 120,000 resolution at m/z 200 and precursor ions meeting and user-defined criteria, such as, charge state, monoisotopic precursor selection, intensity, and dynamic exclusion, were selected for MS2 based on “cycle time” of 3 sec. The AGC (automatic gain control) target was set at 5e5 for MS1 scan, while 5e4 for MS2 scans. The precursor ions were fragmented using higher-energy collisional dissociation with normalized collision energy of 35%. Maximum injection times were set at 50 msec for MS1 scans, while 100 ms for MS2 scans. An internal calibration was carried out using lock-mass from ambient air (m/z 445.120025) (Olsen et al., 2005).
Data analysis
The MS-derived data were searched against Human NCBI RefSeq protein database (version 92, containing 81498 protein entries with common contaminants added) using SEQUEST and MASCOT search algorithms through Proteome Discoverer software suite (version 2.2; Thermo Fisher Scientific, Bremen, Germany). For both algorithms, search parameters included maximum of two missed cleavages, carbamidomethylation at cysteine, TMT 6-plex (+229.163) modification at N-terminus of peptide and lysine was set as fixed modifications. Oxidation of methionine and proline, phosphorylation at serine, threonine, and tyrosine, sulfation of tyrosine, and acetylation of protein N-termini were set as variable modifications. For MS data, monoisotopic peptide mass tolerance was set to 10 ppm and MS/MS tolerance to 0.1 Da. Peptides and proteins were filtered at 1% false discovery rate using Percolator (Fig. 1B).
Statistical analysis
We performed two-tailed Student's t-tests (independent samples) to calculate probabilities (p-value) with the null hypothesis (H0) defined as no difference in means between the sample groups compared. Then Benjamini-Hochberg method of multiple hypothesis correction was employed to calculate adjusted p-values. Ratio of sample means from replicate measurements in each study group was then taken to derive protein fold change values.
Immunohistochemistry on corneal sections
For donor corneas, the peripheral scleral tissue was removed from the corneoscleral ring with a razor blade under a dissecting microscope, and the central corneal buttons were then cut into two equal segments. Patient samples consisted of central corneal button halves. All tissues were fixed in 4% paraformaldehyde and paraffin-embedded blocks were sectioned (6 micron thick). Sections were collected on clean glass slides and dried for 2 h at room temperature. For immunohistochemistry (IHC), sections were blocked with blocking buffer (10% animal serum, bovine serum albumin, and phosphate-buffered saline), followed by overnight incubation at 4°C in primary antibody.
On the next day, the sections were washed with TBS thrice and incubated with the secondary antibody for 2 h and finally stained with DAPI and processed for imaging. DN and KCN corneal sections were stained with antibodies against apolipoprotein E (APOE; Cat. No. ab1906, 1:500; Abcam), C4BP (Cat. No. HPA000926, 1:50; Sigma-Aldrich) and ADRIP/APM2 (Cat. No. NBP2–30736, 1:200; Novus Biologicals). Fluorescence images of immunostained corneas were acquired with Zeiss 700 microscope and Zen software.
Results
Sample information and SDS-PAGE of extracted proteins
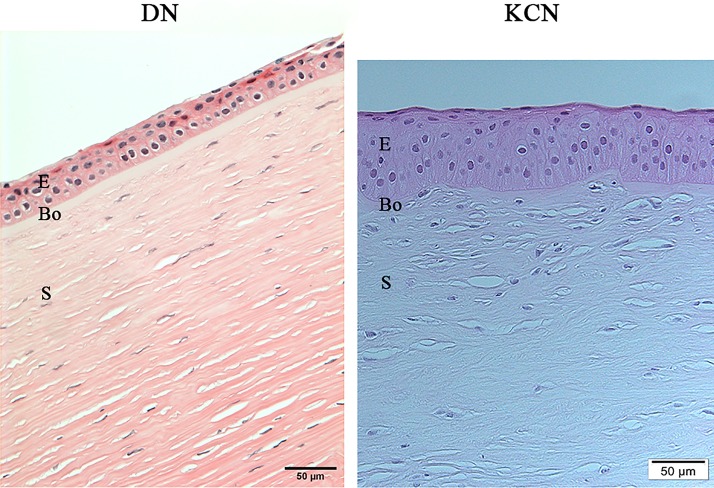
Keratoconus diagnoses of patients were performed by trained cornea specialists from the KKESH. Disease severity was assessed by clinical cornea specialists using the Amsler-Krumeich classification system (Krumeich et al., 1998) and all patients for this study were in stage 3–4. Hematoxylin and eosin staining of representative sections of KCN corneas showed somewhat thickened epithelia with slightly enlarged basal cells, while the stroma appeared unremarkable (Fig. 2).
FIG. 2.
Histology of donor and keratoconus corneas from Saudi Arabian patients. KCN corneal section show thickened epithelia (E), enlarged basal cells, irregular Bowman's layer (Bo), and unremarkable stroma (S). DN, donor; KCN, keratoconus.
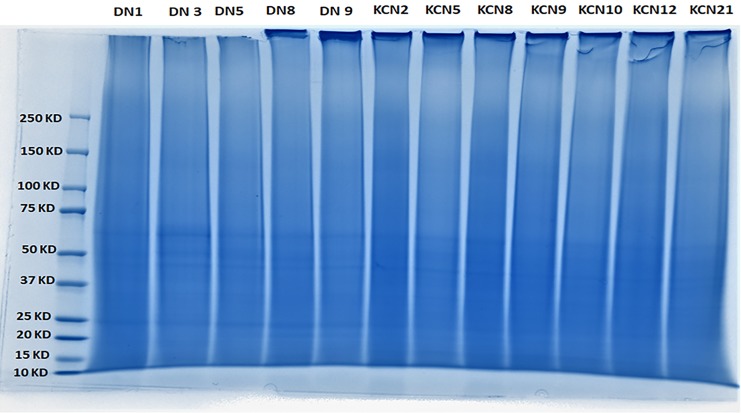
Before performing MS, we ensured that the high-pressure extraction approach was performed uniformly across all samples. Therefore, we first examined the extracted corneal samples by conventional SDS-PAGE to assess their overall extraction and digestion. The DN samples and five of the seven KCN samples appeared similar in their staining and banding pattern and were used for MS (Fig. 3).
FIG. 3.
SDS polyacrylamide gel electrophoresis of digested proteins extracted from five donor and seven keratoconus corneas. SDS, sodium dodecyl sulfate.
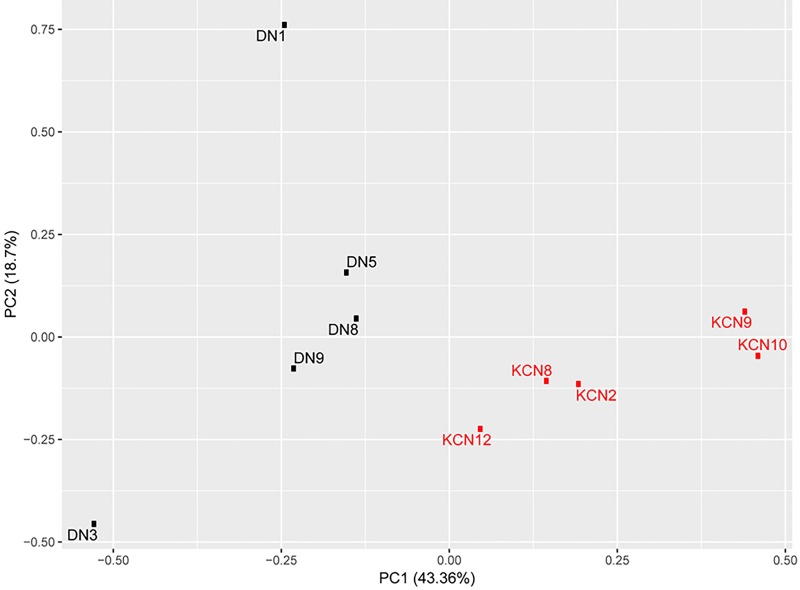
Principal component analysis of KCN and DN corneas
A total of 3132 proteins were detected as differentially present in donor and KCN samples at expressed values >0 in any one sample. Principal Component Analysis was conducted using log2 transformation of 1+ the expressed values of 3132 proteins (Fig. 4). The first principal component (PC1) accounted for the majority of differences comprising 43.36% of the total variability and showed separation between diseased and control tissues along the primary axis. The second principal component accounted for 18.7% of the total variability and reflected minor sample differences within patient and control groups, with the control samples having more variance than the patient group along the minor PC2 axis.
FIG. 4.
PCA of five keratoconus case and five donor control samples. The 3132 differentially expressed proteins were used for the PCA and show separation between diseased and control DN samples in the primary PC1 axis. PC, principal component; PCA, principal component analysis.
Overview of significant changes
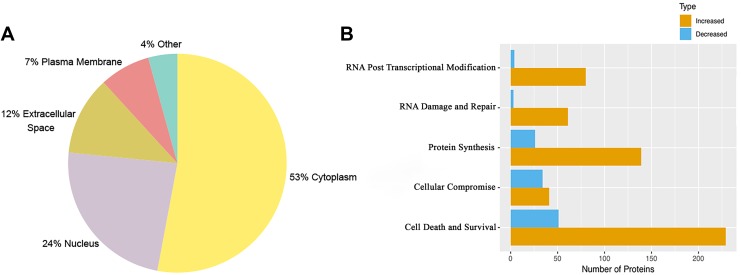
Out of 3132 proteins, 205 were modified with proline oxidation and all of the detected collagens fell in this group. In addition, 48 out of 3132 were phosphorylated at serine and threonine residues (Supplementary Table S1). A total of 627 were changed at a weighted p-value of ≤0.05 (Supplementary Table S2). A larger set of 1159 proteins was flagged as changed when we used a less stringent weighted p-value of ≤0.1 (Supplementary Table S3). All further analyses in this study used the 627 significantly changed proteins (p value of ≤0.05). Of the 627 proteins, 106 were decreased and 521 elevated in KCN. Cytoplasmic proteins form the largest group (53%), with nucleus (24%), extracellular space (12%), and plasma membrane (7%) trailing behind (Fig. 5A).
FIG. 5.
Classification of the 627 significantly altered proteins. (A) Protein classification based on subcellular localization. (B) Protein classification based on molecular functions using the IPA software. IPA, Ingenuity pathway analysis.
The significantly altered proteins were further classified based on molecular functions using the Ingenuity pathway analysis (IPA) software. The largest functional group was cell death and survival; other significant categories were RNA posttranscription modification, protein synthesis, and RNA damage and repair (Fig. 5B). A detailed breakdown into networks based on molecular functions is shown in Supplementary Table S4. Major canonical pathways that were increased in the keratoconus samples included EIF2, EIF4, and mTOR signaling related to cell death and survival, while most significant decreases were in the complement system and acute-phase response proteins (Table 3).
Table 3.
Most Significant Canonical Pathways
| Pathway | p | Overlapa | |
|---|---|---|---|
| 1 | EIF2 signaling | 6.89E-25 | 19.4% (45/232) |
| 2 | Complement system | 9.80E-16 | 43.2% (16/37) |
| 3 | Regulation of eIF4 and p70S6K signaling | 4.52E-15 | 17.3% (29/168) |
| 4 | Acute-phase response signaling | 2.51E-14 | 16.2% (29/179) |
| 5 | mTOR signaling | 1.07E-12 | 13.6% (30/221) |
Number of proteins detected in our samples out of the total number of proteins included in the pathway by the Ingenuity pathway analysis.
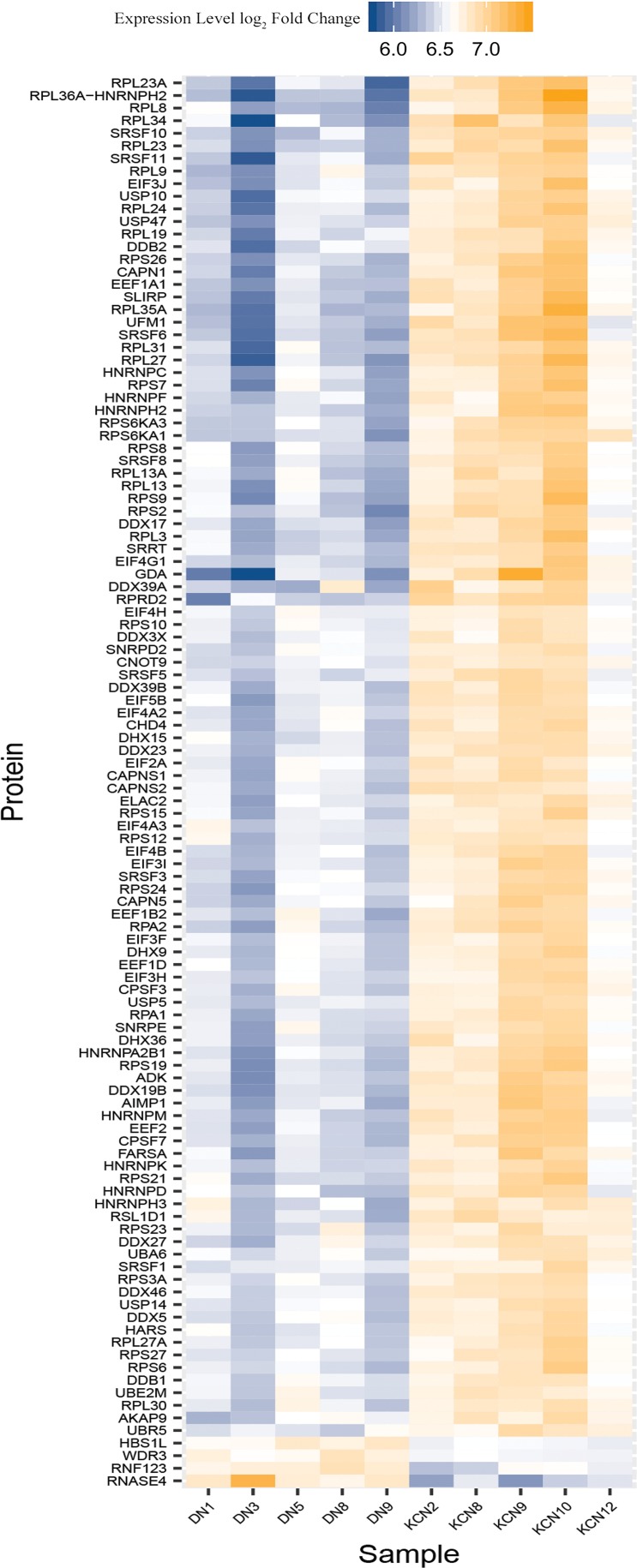
Translation, nonsense-mediated decay, ubiquitin proteasomal degradation pathway members elevated in KCN
These proteins regulate translation initiation, transcript stability, transcript splicing, or RNA metabolism (Fig. 6). A subset constitutes the EIF2 signaling pathway. These increases include exon junction complex proteins, UPF2, MAGOHB, and PABPC1, translation initiation and elongation factors, EIF1, EIF2A, EIF3H, EIF3I, EIF3J, and EIF4A2, EIF4A3, and EIF4H, which form complexes with the 40S ribosome to form the preinitiation complex that will eventually complex with the 60S ribosome for translation initiation. These proteins are involved in quality check of gene expression and nonsense-mediated decay (Kurosaki et al., 2019). EIF4G1 that binds EIFAs (RNA helicases) and increases their activities was also elevated in the KCN corneas.
FIG. 6.
Significantly altered proteins from translation, nonsense-mediated decay, and protein degradation pathways. This heat map shows proteins that regulate translation initiation, transcript stability, splicing, RNA metabolism, protein ubiquitination, and degradation elevated in the keratoconus samples.
Consistent with these increases, we also detected increases in ribosomal proteins that associate with the 40S subunit and the 60S subunit; a total of 31 ribosomal proteins were elevated that regulate transcript modification and NMD (nonsense-mediated decay) of translation-stalled transcripts or those with premature stop codons.
We further detected elevations in proteins that regulate unwinding, processing of transcripts and translation (DDX17, DDX19b, DDX23, and DHX15), and multiple splicing factors (SRSFs). Several proteins that are induced by oxidative stress and form part of the cullin-RING ligase-mediated proteasomal degradation system were elevated (Wang and Martin, 2015). These include CAND1, COPS4, and COPS8 (of the COP9 signalosome), and RNF123, CUL2, CUL3, and CUL4B (cullin-RING ligases). Additional proteins involved in ubiquitination and degradation of proteins include SUMO3, UBA6, UBE2M, UFM1, UBR5, several USPs, and calpains and form a large fraction of the increased proteins in KCN.
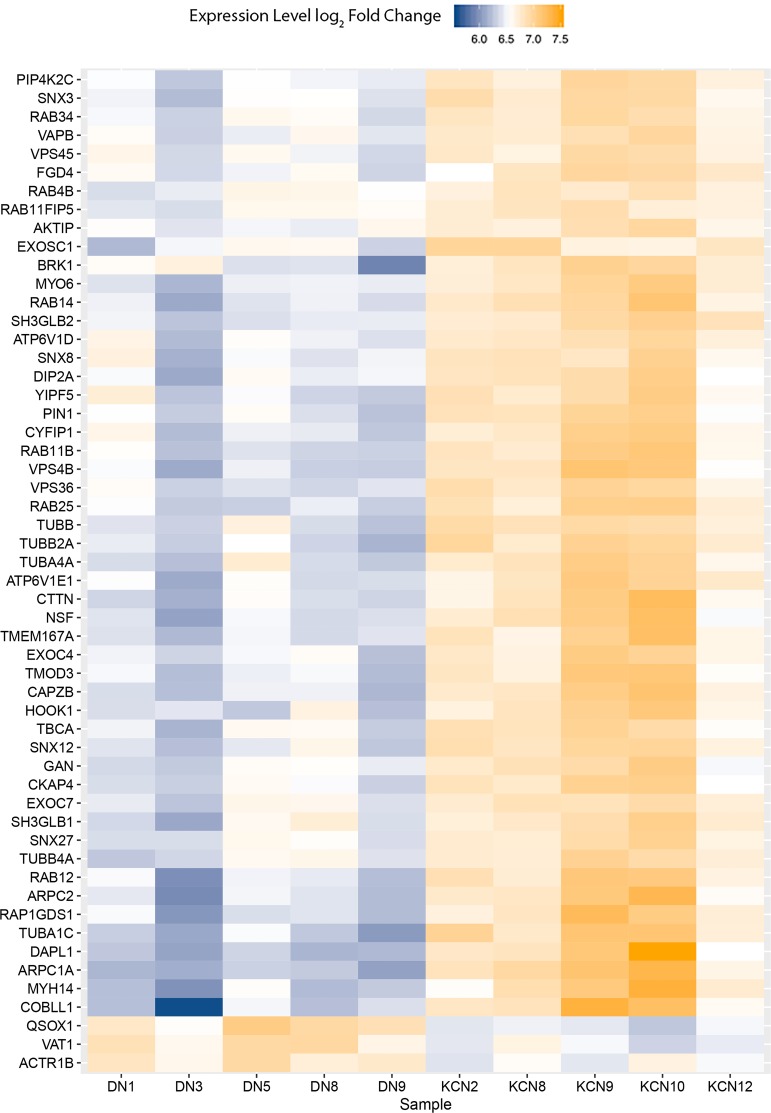
Increases in cytoskeletal and vesicular transport-related proteins
Multiple proteins that regulate cytoskeletal structure, vesicular transport, and endosomal trafficking were elevated in the KCN samples (Fig. 7). These include actin-related proteins (ARPC1A, ARPC2, and COBLL1), tubulins (TUBA1C, TUBA4A, TUBB2A, and TUBB4A), tubulin-folding cofactor (TBCA), and those involved in vesicular transport, endosome, ER, and Golgi functions (multiple RABs, VPS36, VPS4B, and VPS45). Several of these are also involved in the regulation of autophagy (ATP6V1D, ATP6V1E1, and RAB11FIP5) and cell death (DAPL1). Only three proteins from this group were decreased in KCN, which were QSOX1 regulating cell proliferation, VAT1 regulating mitochondrial oxidation reduction functions, and ACTR1B involved in antigen presentation by MHC class II.
FIG. 7.
Altered cytoskeletal and vesicular transport proteins. Heat map of increased and decreased cytoskeletal and vesicular transport proteins. Several proteins that regulate cytoskeletal structure, vesicular transport, endosomal trafficking, and autophagy were elevated in keratoconus corneas.
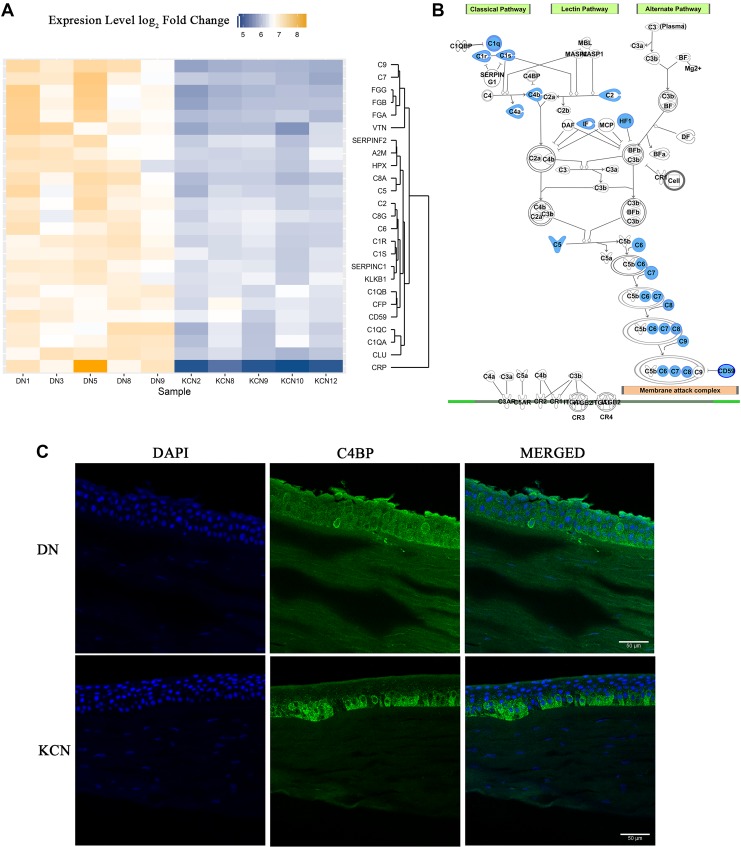
Decreased proteins of acute-phase and tissue injury responses
Several members of the classical complement pathway and the common downstream pathway that mediate removal of infected and damaged cells and cellular debris (Noris and Remuzzi, 2013) were reduced in the KCN samples (Fig. 8A). Induction of the classical pathway is marked by the activation of the pattern recognition molecule C1q, activation of the C1r and C1s proteases, and cleavage of C4 into C4a and C4b. These molecules, C1q, C1r, C1s, C4, C4a, C4b, and C2a, were all reduced in KCN. Of the downstream steps common to all three pathways, C5, C6, C7, C8, and C9, which form the membrane attack complex were all reduced (Fig. 8B).
FIG. 8.
Complement system and acute-phase response protein changes. (A) Heat map of altered complement system and acute-phase response-associated proteins. Several proteins from the classical complement pathway and the common downstream pathway were reduced in the Keratoconus samples. (B) Complement system and acute-phase response-related protein pathway. Proteins highlighted in blue denote decreases in KCN proteomes. (C) Immunostaining of C4BP in donor and KCN corneal sections.
Since multiple complement system members were decreased, we wondered whether C4BP, which is a negative regulator of the classical pathway (Fujita et al., 1978; Nagasawa et al., 1980; Scharfstein et al., 1978; Suankratay et al., 1999), was altered in keratoconus corneas. By IHC, we detected a preferential staining of C4BP in the basal epithelial cell layer of KCN corneas, whereas it was uniformly distributed in the epithelial cell layers of the donor cornea (Fig. 8C).
Decreases in extracellular matrix and extracellular matrix regulating proteins in KCN
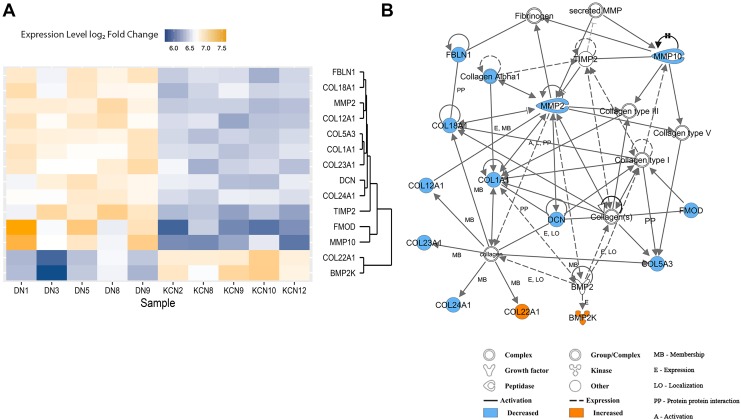
The major phenotype in keratoconus is thinning and weakening of the corneal stroma, which largely comprised structural connective tissue proteins, collagens, other extracellular matrix (ECM) proteins, and proteoglycans (Hassell and Birk, 2010). Therefore, we searched for changes in ECM-related proteins, expecting decreases mostly in ECM proteins and increases in ECM-degrading enzymes. Concordantly, we found a select few collagens that were significantly decreased in the KCN group—stromal collagens, COL1A1, COL5A3, fibrillar collagen-associated COL12A1, COL18A1, and transmembrane COL23A1 (Fig. 9A, B).
FIG. 9.
ECM protein changes. (A) Heat map of altered ECM proteins. Several proteins responsible for regulating corneal thickness such as collagens and proteoglycans were decreased in keratoconus cornea. (B) ECM protein network showing interactions between ECM proteins. ECM, extracellular matrix.
Fibrillar collagen binding proteoglycans, fibromodulin (FMOD) and decorin (DCN), were also decreased. Fibulin 1 (FBLN1) that reportedly promotes fibrosis and deposition of fibronectin and HSPG2 matrices (Ge et al., 2015) was also reduced in our KCN samples. Two MMPs were reduced, while TIMP2 (Arpino et al., 2015) that suppresses several MMP activities was also reduced. COL22A1 was the only collagen that was elevated, and unlike collagen types I and V, it resides in myotendinous junctions (Koch et al., 2004). BMP2K, a bone morphogenetic kinase, was also elevated in KCN, and interestingly, one study has reported a variant in this gene in high myopia (Liu et al., 2009).
Validation of proteomic findings by immunostaining of cornea sections
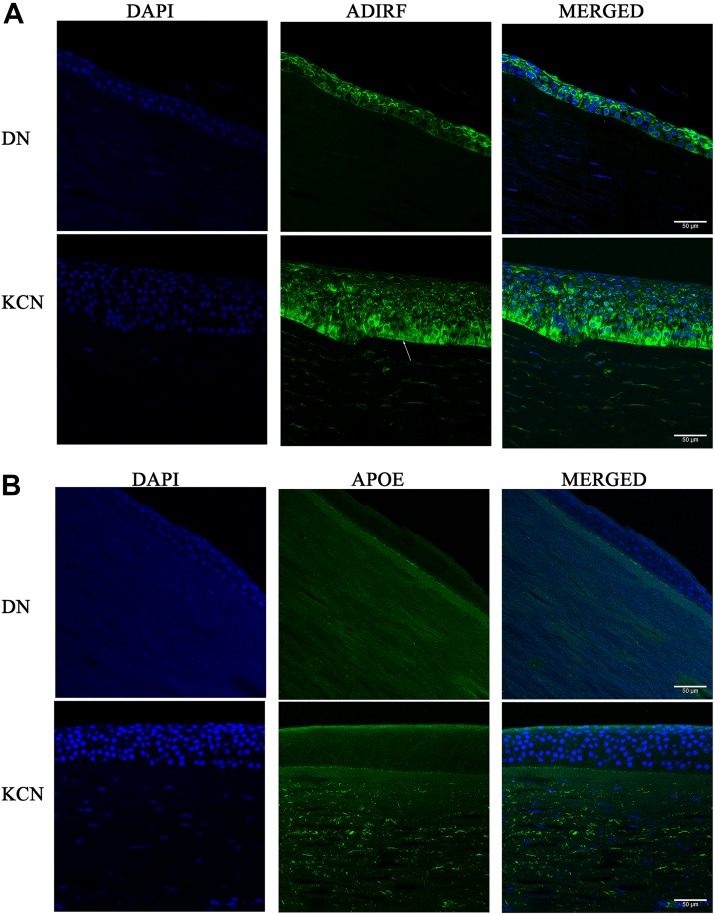
Some of our current findings were validated by similar observations in our earlier proteomic study. For example, ALDHA1, PRDX1, TXN1, SOD1, COL1A1, COL3A1, C7, and C9 were also detected as decreased in our earlier proteomics on pooled keratoconus corneas (Chaerkady et al., 2013). In this study, we selected to test the presence of one of the most increased proteins in both studies, ADIRF (Adipogenesis Regulatory Factor) also known as APM2, by immunofluorescence staining of KCN and DN corneal sections. There are no reports of the ADIRF protein in the cornea, or the eye for that matter; it is expressed in adipose tissues, may have a role in membrane trafficking, and has been linked to cisplatin resistance in tumors (Scott et al., 2009).
We detected cytoplasmic staining of this protein in both DN and KCN corneal epithelial layers, suggesting a function for ADIRF/APM2 of unknown significance at the ocular surface. In addition, the staining appeared to be more in the superficial epithelial cells in the DN, but stained basal epithelial cells in the KCN cornea (Fig. 10A). We also tested APOE in DN and KCN corneas by immunostaining (Fig. 10B). It is one of the most significantly decreased proteins in our current study. It plays a key role in lipid homeostasis by facilitating lipid transport between tissues or cells. There are 3 major alleles of APOE (Mahley and Rall, 2000), and the encoded protein regulates cholesterol transport, platelet aggregation, immune response, and oxidative processes (Bhattacharjee et al., 2008). By IHC, we detected preferential staining of APOE in the keratoconus corneal stroma only (Fig. 10B); primary controls did not show staining (Supplementary Fig. S1).
FIG. 10.
IHC of selected proteins in normal donor and KCN corneal sections. Paraffin-embedded corneas were sectioned and immunostained for (A) ADIRF and (B) apolipoprotein E. ADIRF, adipogenesis regulatory factor; IHC, immunohistochemistry.
Discussion
Using multiplexed high-resolution MS, we examined the corneal proteomes of five individual cornea samples with keratoconus from Saudi Arabia. The most notable changes in the current proteomic data relate to regulations of RNA metabolism, protein synthesis, protein degradation, cell death, and complement pathways. Within these broad categories, the majority of the increases were in translation and RNA metabolism, proteins regulating ubiquitination, neddylation, and degradation of proteins.
We detected increases in several nuclear ribosomal proteins, RNA-helicase, exon junction spliceosomal proteins (Ejp), translation initiation, and elongation factors. Although the increases were not large in magnitude, they were all highly significant and are beginning to show a more complete picture of the dysregulations that occur in the corneal tissue itself. These collectively speak of ER stress, integrated stress response (ISR) (Pakos-Zebrucka et al., 2016), and protein folding disorders that have been repeatedly associated with degenerative diseases such as Alzheimer's and Parkinson's diseases (Forloni et al., 2002; Sweeney et al., 2017).
Acute ER stress, due to cell intrinsic issues or external stress-causing factors such as hypoxia, nutrient paucity, viral infections, or UV exposure and DNA damage lead to ISR. This enables PERK kinase-mediated translation reprogramming (Pakos-Zebrucka et al., 2016; Ron, 2002) and reduction of global Cap-dependent messenger RNA (mRNA) translation, while increasing expression of selected transcription factors like ATF4 to induce apoptosis and autophagy-related genes (Goetz and Wilkinson, 2017).
Chronic ER stress, on the other hand, engages an amino acid transporter network to mTORC1 pathway activation, inhibition of autophagy, and increased translation (Wang and Kaufman, 2016). While we did not detect elevations in PERK, or GCN2 (EIF2AK4) or downstream ATF4, as hallmarks of ISR in the KCN proteome, we detected increases in multiple translation initiation and elongation factors (EEF1A1, EEF1B2, EEF1D, EIF3F, EIF3H, and others) downstream of ATF4, which do speak of increased translational activities consistent with chronic ER stress and chronic ISR (Guan et al., 2014).
Another response that is intricately linked to ER stress and ISR is NMD (Goetz and Wilkinson, 2017; Kurosaki et al., 2019), which involves detection of pre-mRNA with premature termination codons or those tagged by the NMD machinery and to launch their degradation. NMD is usually suppressed by acute ER stress and ISR; however, its hyperactivation has been reported in neurodegenerative conditions (Kamelgarn et al., 2018). Increases in key NMD proteins in our data suggest increased efforts to remove aberrant transcripts in the diseased cornea.
Our 2013 tissue proteomic study of the corneal stroma had also shown elevations in proteins involved in ER stress, proteins of the translation machinery, and NMD (Chaerkady et al., 2013). In addition, in a previous proteomic study of cultured stromal cells from patient corneas placed in a low-nutrient environment, we also detected signs of ER stress and ISR, and consistently decreased ECM synthesis (Foster et al., 2018). However, many of the ER stress NMD-related changes seen in the corneal tissue proteomics data were not seen in the cell culture proteomics. A possible explanation is that in culture, many of the stress-causing factors are lifted or reduced and the cells recover.
The complement system comprised the classical (antibody mediated), alternative, and lectin-mediated pathways, is a network of signaling hubs, produced largely by circulating white blood cells, and orchestrate immune surveillance, removal of cellular debris, and inflammatory responses (Noris and Remuzzi, 2013). The cornea, although avascular, has low, but detectable levels of complement proteins, and these are certainly present at high levels in the vasculature-populated limbal area (Mondino and Hoffman, 1980; Mondino et al., 1980).
The widespread decrease in members of the complement system, lipid and lipoprotein catabolism, and acute-phase and tissue injury responses seen here as well as in the 2013 study is remarkable. This raises the intriguing possibility that there may be some structural or functional impairments in the transport of circulating factors from the limbal vasculature or the aqueous humor. Decreased complement functions most likely mean poor clearance of protein and cellular debris resulting from ISR and ER stress to further contribute to degenerative conditions in the cornea.
Oxidative stress has long been recognized as centrally important in the pathogenesis of keratoconus (Kenney et al., 2000, 2005; Mondino et al., 1980). As the outermost barrier to the eye, the cornea is constantly exposed to external stresses such as UV light, hypoxia, and mechanical and microbial insults. The healthy cornea has multiple protective mechanisms to counteract reactive oxygen species (ROS) and lipid peroxidation products that accumulate as a result of such external stresses (Chen et al., 2009). These protective mechanisms include superoxide dismutases that convert ROS to less damaging H2O2, thioredoxins and peroxiredoxins that regulate cellular redox homeostasis, and aldehyde dehydrogenases that remove damaging aldehydes produced during lipid peroxidation (Chen et al., 2009; Lassen et al., 2008).
Landmark studies on keratoconus showed decreases in these protective mechanisms and increases in tissue degradations (Arnal et al., 2011; Behndig et al., 2001; Cristina Kenney and Brown, 2003; Olofsson et al., 2007). Unfolded protein response, ER stress, NMD, and perturbed global protein synthesis identified in our proteomic studies provide context to these downstream events. Our studies have identified interactive signaling pathways and genes as potential candidates in keratoconus susceptibility. A limitation of this study is the relatively small number of individual patient samples profiled. Thus, as we increase the patient sample size over time, the proteins and pathways we identify will have greater significance.
Contextualizing this study with the broader literature
The cornea has been reported to develop increased reflectivity (Hollingsworth et al., 2005; Wittig-Silva et al., 2013) haze, epithelial alterations (Sykakis et al., 2012), break in Bowman's layer-reduced stromal thickness, stromal cell alterations (Efron and Hollingsworth, 2008), and altered collagen fibril structure (Hayes et al., 2012; Mikula et al., 2018). Earlier biochemical studies of the cornea reported elevated level of degenerative enzymes, acid esterases, acid phosphatases, acid lipases, cathepsin B and G, and reduced level of protease inhibitors as factors contributing to the thinning of the cornea (Lema and Duran, 2005; Lema et al., 2008; Sutton et al., 2010).
Unlike the tear fluid studies, the corneal proteome from keratoconus patients has not been characterized extensively. The few corneal proteomic studies that have been performed have considerable technical differences between studies as these span a larger period of time. The first such study examined the epithelium and stroma individually from four keratoconus and four control corneas using a nonlabel shot gun proteomic and two-dimensional gel electrophoresis approaches that detected 104 epithelial and 44 stromal proteins (Joseph et al., 2011). In 2013, we used a more sensitive 4plex iTRAQ (isobaric tag for relative and absolute quantification) labeling and MS on pooled stromal and epithelial tissue extracts from five keratoconus and donor corneas. This identified a much larger set of 957 epithelial and 1157 stromal proteins (Chaerkady et al., 2013).
A recent study compared the affected cone area with the noncore area in four unpooled keratoconus corneas and detected two dozen proteins and a larger set of 400–500 proteins when comparing keratoconus versus donor samples (Yam et al., 2019). We were interested in carrying out multiplex proteomic studies on individual corneas to determine consistent changes that speak of common underlying biological processes that distinguish keratoconus from healthy donor corneas. The granularity and robustness of the findings will only increase with increasing numbers of such studies. To that end, here, we used an improved high-pressure method to extract corneal proteins and investigated the proteome profile of corneas from keratoconus patients in Saudi Arabia.
Compared to our earlier MS study on pooled keratoconus corneas, this study used a more efficient extraction technique, and the detection and analysis methods were also more sensitive. However, several key observations from this study were consistent with those in our previous study of pooled patient samples (Chaerkady et al., 2013).
For example, collagen type XII detected as decreased in the current set of patients is in agreement with our earlier MS study (Chaerkady et al., 2013), and an early IHC of study of keratoconus corneas (Cheng et al., 2001). ECM proteins such as COL1A1 and FMOD, and complement proteins, C1Q, C2, C5, C6, and CFH, as well as APOE, decreased in this study, were also noted as decreased in our previous proteomic study. Overall, 222 proteins were elevated and 98 proteins decreased commonly in this and our previous proteomic study (Supplementary Table S5).
Another technical difference between this and our previous MS study is that, here, the epithelial and the stromal tissue were extracted together, whereas in the earlier study, we had attempted to separate the epithelium and the stroma and conducted MS on these layers separately (Chaerkady et al., 2013). We opted not to separate the epithelium from the stroma in this MS as in the previous study, this did not completely rid the stroma of epithelial proteins. Despite these and other technical differences, as well as population-based differences in the samples, the similarities in the findings of the two studies underscore biological dysregulations common to the pathogenesis of keratoconus.
We note that this study had a modest sample size that requires validation in studies with larger sample sizes from independent populations, and control donor samples that are closely matched with the cases. Because access to corneal samples is often a limiting constraint in visual health-related biomarker research, we call for future comparative studies in different world populations as well.
Conclusions
To our knowledge, we have identified the largest set of differentially regulated proteins in individual keratoconic corneas by MS, making this study the most comprehensive thus far. We identified novel changes in the complement system, nonsense-mediated mRNA decay pathway, ubiquitination, protein degradation, and transport mechanisms in keratoconus that emphasize cellular changes possibly dictating the ECM compromise seen in keratoconus. These findings provide novel proteins and pathways as potential candidates for genetic and therapeutic targets.
Supplementary Material
Acknowledgments
The authors thank Mrs. Muneera AlFutais, Mr. Ches Souru, and Ms. Khitam Alhati, Technical Research Associates (Research Department, KKESH), for their technical assistance. The authors also thank NYU School of Medicine's Center for Biospecimen Research and Development (CBRD) for sectioning donor corneas.
Abbreviations Used
- ADIRF
adipogenesis regulatory factor
- APOE
apolipoprotein E
- DDA
data-dependent acquisition
- DN
donor
- ECM
extracellular matrix
- FMOD
fibromodulin
- IHC
immunohistochemistry
- ISR
integrated stress response
- KCN
keratoconus
- mRNA
messenger RNA
- MS
mass spectrometry
- PC
principal component
- ROS
reactive oxygen species
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- TMT
tandem mass tag
- UV
ultraviolet
Author Disclosure Statement
The authors declare they have no financial conflicts of interest.
Funding Information
This study was supported by funds from King Khaled Eye Specialist Hospital to S.C. and S.A.-S. and NIH/NEI funding EY026104 to S.C.
Supplementary Material
References
- Al-Amri AM. (2018). Prevalence of keratoconus in a refractive surgery population. J Ophthalmol 2018, 5983530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnal E, Peris-Martinez C, Menezo JL, Johnsen-Soriano S, and Romero FJ. (2011). Oxidative stress in keratoconus? Invest Ophthalmol Vis Sci 52, 8592–8597 [DOI] [PubMed] [Google Scholar]
- Arpino V, Brock M, and Gill SE. (2015). The role of TIMPs in regulation of extracellular matrix proteolysis. Matrix Biol 44–46, 247–254 [DOI] [PubMed] [Google Scholar]
- Behndig A, Karlsson K, Johansson BO, Brannstrom T, and Marklund SL. (2001). Superoxide dismutase isoenzymes in the normal and diseased human cornea. Invest Ophthalmol Vis Sci 42, 2293–2296 [PubMed] [Google Scholar]
- Bhattacharjee PS, Neumann DM, Foster TP, et al. (2008). Effect of human apolipoprotein E genotype on the pathogenesis of experimental ocular HSV-1. Exp Eye Res 87, 122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaerkady R, Shao H, Scott SG, Pandey A, Jun AS, and Chakravarti S. (2013). The keratoconus corneal proteome: Loss of epithelial integrity and stromal degeneration. J Proteomics 87, 122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HY, and Chodosh J. (2013). The genetics of keratoconus. Semin Ophthalmol 28, 275–280 [DOI] [PubMed] [Google Scholar]
- Chen Y, Mehta G, and Vasiliou V. (2009). Antioxidant defenses in the ocular surface. Ocul Surf 7, 176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng EL, Maruyama I, Sundarraj N, Sugar J, Feder RS, and Yue BY. (2001). Expression of type XII collagen and hemidesmosome-associated proteins in keratoconus corneas. Curr Eye Res 22, 333–340 [DOI] [PubMed] [Google Scholar]
- Cristina Kenney M, and Brown DJ. (2003). The cascade hypothesis of keratoconus. Cont Lens Anterior Eye 26, 139–146 [DOI] [PubMed] [Google Scholar]
- Efron N, and Hollingsworth JG. (2008). New perspectives on keratoconus as revealed by corneal confocal microscopy. Clin Exp Optom 91, 34–55 [DOI] [PubMed] [Google Scholar]
- Forloni G, Terreni L, Bertani I, et al. (2002). Protein misfolding in Alzheimer's and Parkinson's disease: Genetics and molecular mechanisms. Neurobiol Aging 23, 957–976 [DOI] [PubMed] [Google Scholar]
- Foster JW, Shinde V, Soiberman US, et al. (2018). Integrated stress response and decreased ECM in cultured stromal cells from keratoconus corneas. Invest Ophthalmol Vis Sci 59, 2977–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Gigli I, and Nussenzweig V. (1978). Human C4-binding protein. II. Role in proteolysis of C4b by C3b-inactivator. J Exp Med 148, 1044–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Q, Chen L, Jaffar J, et al. (2015). Fibulin1C peptide induces cell attachment and extracellular matrix deposition in lung fibroblasts. Sci Rep 5, 9496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz AE, and Wilkinson M. (2017). Stress and the nonsense-mediated RNA decay pathway. Cell Mol Life Sci 74, 3509–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan BJ, Krokowski D, Majumder M, et al. (2014). Translational control during endoplasmic reticulum stress beyond phosphorylation of the translation initiation factor eIF2alpha. J Biol Chem 289, 12593–12611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassell JR, and Birk DE. (2010). The molecular basis of corneal transparency. Exp Eye Res 91, 326–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S, Khan S, Boote C, et al. (2012). Depth profile study of abnormal collagen orientation in keratoconus corneas. Arch Ophthalmol 130, 251–252 [DOI] [PubMed] [Google Scholar]
- Hollingsworth JG, Efron N, and Tullo AB. (2005). In vivo corneal confocal microscopy in keratoconus. Ophthalmic Physiol Opt 25, 254–260 [DOI] [PubMed] [Google Scholar]
- Joseph R, Srivastava OP, and Pfister RR. (2011). Differential epithelial and stromal protein profiles in keratoconus and normal human corneas. Exp Eye Res 92, 282–298 [DOI] [PubMed] [Google Scholar]
- Jun AS, Cope L, Speck C, et al. (2011). Subnormal cytokine profile in the tear fluid of keratoconus patients. PLoS One 6, e16437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamelgarn M, Chen J, Kuang L, Jin H, Kasarskis EJ, and Zhu H. (2018). ALS mutations of FUS suppress protein translation and disrupt the regulation of nonsense-mediated decay. Proc Natl Acad Sci U S A 115, E11904–E11913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney MC, Brown DJ, and Rajeev B. (2000). Everett Kinsey lecture. The elusive causes of keratoconus: A working hypothesis. CLAO J 26, 10–13 [PubMed] [Google Scholar]
- Kenney MC, Chwa M, Atilano SR, et al. (2005). Increased levels of catalase and cathepsin V/L2 but decreased TIMP-1 in keratoconus corneas: Evidence that oxidative stress plays a role in this disorder. Invest Ophthalmol Vis Sci 46, 823–832 [DOI] [PubMed] [Google Scholar]
- Koch M, Schulze J, Hansen U, et al. (2004). A novel marker of tissue junctions, collagen XXII. J Biol Chem 279, 22514–22521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmer JH, Feder RS, and Belin MW. (1984). Keratoconus and related noninflammatory corneal thinning disorders. Surv Ophthalmol 28, 293–322 [DOI] [PubMed] [Google Scholar]
- Krumeich JH, Daniel J, and Knulle A. (1998). Live-epikeratophakia for keratoconus. J Cataract Refract Surg 24, 456–463 [DOI] [PubMed] [Google Scholar]
- Kurosaki T, Popp MW, and Maquat LE. (2019). Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat Rev Mol Cell Biol 20, 406–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen N, Black WJ, Estey T, and Vasiliou V. (2008). The role of corneal crystallins in the cellular defense mechanisms against oxidative stress. Semin Cell Dev Biol 19, 100–112 [DOI] [PubMed] [Google Scholar]
- Lema I, and Duran JA. (2005). Inflammatory molecules in the tears of patients with keratoconus. Ophthalmology 112, 654–659 [DOI] [PubMed] [Google Scholar]
- Lema I, Duran JA, Ruiz C, Diez-Feijoo E, Acera A, and Merayo J. (2008). Inflammatory response to contact lenses in patients with keratoconus compared with myopic subjects. Cornea 27, 758–763 [DOI] [PubMed] [Google Scholar]
- Linebarger EJ, Song D, Ruckhofer J, and Schanzlin DJ. (2000). Intacs: The intrastromal corneal ring. Int Ophthalmol Clin 40, 199–208 [DOI] [PubMed] [Google Scholar]
- Liu HP, Lin YJ, Lin WY, et al. (2009). A novel genetic variant of BMP2K contributes to high myopia. J Clin Lab Anal 23, 362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macsai MS, Varley GA, and Krachmer JH. (1990). Development of keratoconus after contact lens wear. Patient characteristics. Arch Ophthalmol 108, 534–538 [DOI] [PubMed] [Google Scholar]
- Mahley RW, and Rall SC., Jr. (2000). Apolipoprotein E: Far more than a lipid transport protein. Annu Rev Genomics Hum Genet 1, 507–537 [DOI] [PubMed] [Google Scholar]
- Mikula E, Winkler M, Juhasz T, et al. (2018). Axial mechanical and structural characterization of keratoconus corneas. Exp Eye Res 175, 14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondino BJ, and Hoffman DB. (1980). Hemolytic complement activity in normal human donor corneas. Arch Ophthalmol 98, 2041–2044 [DOI] [PubMed] [Google Scholar]
- Mondino BJ, Ratajczak HV, Goldberg DB, Schanzlin DJ, and Brown SI. (1980). Alternate and classical pathway components of complement in the normal cornea. Arch Ophthalmol 98, 346–349 [DOI] [PubMed] [Google Scholar]
- Nagasawa S, Ichihara C, and Stroud RM. (1980). Cleavage of C4b by C3b inactivator: Production of a nicked form of C4b, C4b’, as an intermediate cleavage product of C4b by C3b inactivator. J Immunol 125, 578–582 [PubMed] [Google Scholar]
- Noris M, and Remuzzi G. (2013). Overview of complement activation and regulation. Semin Nephrol 33, 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson EM, Marklund SL, Pedrosa-Domellof F, and Behndig A. (2007). Interleukin-1alpha downregulates extracellular-superoxide dismutase in human corneal keratoconus stromal cells. Mol Vis 13, 1285–1290 [PubMed] [Google Scholar]
- Olsen JV, De Godoy LM, Li G, et al. (2005). Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, and Gorman AM. (2016). The integrated stress response. EMBO Rep 17, 1374–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarsini S, Hjortdal J, Sarker-Nag A, Sejersen H, Asara JM, and Karamichos D. (2014). Gross cystic disease fluid protein-15/prolactin-inducible protein as a biomarker for keratoconus disease. PLoS One 9, e113310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz YS. (1998). Keratoconus. Surv Ophthalmol 42, 297–319 [DOI] [PubMed] [Google Scholar]
- Ron D. (2002). Translational control in the endoplasmic reticulum stress response. J Clin Invest 110, 1383–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfstein J, Ferreira A, Gigli I, and Nussenzweig V. (1978). Human C4-binding protein. I. Isolation and characterization. J Exp Med 148, 207–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott BJ, Qutob S, Liu QY, and Ng CE. (2009). APM2 is a novel mediator of cisplatin resistance in a variety of cancer cell types regardless of p53 or MMR status. Int J Cancer 125, 1193–1204 [DOI] [PubMed] [Google Scholar]
- Soiberman U, Foster JW, Jun AS, and Chakravarti S. (2017). Pathophysiology of keratoconus: What do we know today. Open Ophthalmol J 11, 252–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suankratay C, Mold C, Zhang Y, Lint TF, and Gewurz H. (1999). Mechanism of complement-dependent haemolysis via the lectin pathway: Role of the complement regulatory proteins. Clin Exp Immunol 117, 442–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G, Madigan M, Roufas A, and Mcavoy J. (2010). Secreted frizzled-related protein 1 (SFRP1) is highly upregulated in keratoconus epithelium: A novel finding highlighting a new potential focus for keratoconus research and treatment. Clin Exp Ophthalmol 38, 43–48 [DOI] [PubMed] [Google Scholar]
- Sweeney P, Park H, Baumann M, et al. (2017). Protein misfolding in neurodegenerative diseases: Implications and strategies. Transl Neurodegener 6, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykakis E, Carley F, Irion L, Denton J, and Hillarby MC. (2012). An in depth analysis of histopathological characteristics found in keratoconus. Pathology 44, 234–239 [DOI] [PubMed] [Google Scholar]
- Torres Netto EA, Al-Otaibi WM, Hafezi NL, et al. (2018). Prevalence of keratoconus in paediatric patients in Riyadh, Saudi Arabia. Br J Ophthalmol 102, 1436–1441 [DOI] [PubMed] [Google Scholar]
- Wang M, and Kaufman RJ. (2016). Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529, 326–335 [DOI] [PubMed] [Google Scholar]
- Wang X, and Martin DS. (2015). The COP9 signalosome and cullin-RING ligases in the heart. Am J Cardiovasc Dis 5, 1–18 [PMC free article] [PubMed] [Google Scholar]
- Weed KH, Macewen CJ, Giles T, Low J, and Mcghee CN. (2008). The Dundee University Scottish Keratoconus study: Demographics, corneal signs, associated diseases, and eye rubbing. Eye (Lond) 22, 534–541 [DOI] [PubMed] [Google Scholar]
- Wittig-Silva C, Chan E, Pollock G, and Snibson GR. (2013). Localized changes in stromal reflectivity after corneal collagen cross-linking observed with different imaging techniques. J Refract Surg 29, 410–416 [DOI] [PubMed] [Google Scholar]
- Wollensak G, Spoerl E, and Seiler T. (2003). Riboflavin/ultraviolet-a-induced collagen crosslinking for the treatment of keratoconus. Am J Ophthalmol 135, 620–627 [DOI] [PubMed] [Google Scholar]
- Yam GH, Fuest M, Zhou L. et al. (2019). Differential epithelial and stromal protein profiles in cone and non-cone regions of keratoconus corneas. Sci Rep 9, 2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadnik K, Barr JT, Edrington TB, et al. (1998). Baseline findings in the Collaborative Longitudinal Evaluation of Keratoconus (CLEK) Study. Invest Ophthalmol Vis Sci 39, 2537–2546 [PubMed] [Google Scholar]
- Zahari MS, Wu X, Blair BG, et al. (2015). Activating mutations in PIK3CA lead to widespread modulation of the tyrosine phosphoproteome. J Proteome Res 14, 3882–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.