Abstract
Objective
Clinical prediction models require updating as performance deteriorates over time. We developed a testing procedure to select updating methods that minimizes overfitting, incorporates uncertainty associated with updating sample sizes, and is applicable to both parametric and nonparametric models.
Materials and Methods
We describe a procedure to select an updating method for dichotomous outcome models by balancing simplicity against accuracy. We illustrate the test’s properties on simulated scenarios of population shift and 2 models based on Department of Veterans Affairs inpatient admissions.
Results
In simulations, the test generally recommended no update under no population shift, no update or modest recalibration under case mix shifts, intercept correction under changing outcome rates, and refitting under shifted predictor-outcome associations. The recommended updates provided superior or similar calibration to that achieved with more complex updating. In the case study, however, small update sets lead the test to recommend simpler updates than may have been ideal based on subsequent performance.
Discussion
Our test’s recommendations highlighted the benefits of simple updating as opposed to systematic refitting in response to performance drift. The complexity of recommended updating methods reflected sample size and magnitude of performance drift, as anticipated. The case study highlights the conservative nature of our test.
Conclusions
This new test supports data-driven updating of models developed with both biostatistical and machine learning approaches, promoting the transportability and maintenance of a wide array of clinical prediction models and, in turn, a variety of applications relying on modern prediction tools.
Keywords: predictive analytics, calibration, model updating
INTRODUCTION
Clinical risk prediction models are developed to support patient and provider decision making,1,2 assist in resource allocation,3 and adjust quality metrics for acuity4–6 across an array of clinical specialties and settings.4,6–11 Opportunities for the deployment of prediction models to support patient-level decision making are arising as the adoption of electronic health records accelerates.4,12–16 At the same time, our understanding of the challenges of incorporating predictive analytics into clinical care is rapidly evolving, requiring new methods and evidence-based recommendations.
One challenge central to the long-term, prospective application of prediction tools is the tendency of model performance, particularly in terms of calibration, to deteriorate over time.10,11,17–22 Performance drift results from applying models in nonstationary environments with changing outcome rates, shifting patient populations, and evolving clinical practice.6,10,11,21–23 Patient mix may change gradually or quickly as populations age, new facilities bring new populations to a health system, or models are transported across clinical settings.23–26 Predictor-outcome associations may shift along with practice patterns or the healthcare process model, such as changes in clinical guidelines, provider experience, coding practices, measurement accuracy, electronic health record interfaces, data entry workflows, and data definitions.10,11,26,27 Such changes impact model calibration in ways that vary in magnitude and form depending on the model’s underlying learning algorithm.17,18,28
As clinical use of patient-level predictions requires highly accurate models that consistently perform well, methods to update and correct models in response to deteriorating performance are critical. A range of updating approaches are available to correct performance drift, from simple recalibration to full model revision (ie, refitting) and even model extension with the incorporation of new predictors.10,11,25,26,29 However, simple updating methods are often overlooked in favor of training entirely new models.11,21 The challenge with full retraining is the question of how to prioritize new data without losing relevant information in old data. If the old data are completely discarded, the new (and often smaller) dataset is used alone, and overfitting becomes a real risk.10,11,21 Recalibration, however, can correct accuracy while preserving and extending prior knowledge by incorporating new data into an existing model.11,21 When similar or more stable performance can be achieved through recalibration,25,30,31 model refitting may not be the best or most appropriate choice for models in clinical use.
Updating methods vary in complexity, data requirements, and analytical resource demands10,11,25,26,29; thus, guidance is needed to select among updating methods while balancing both the amount of available evidence in new patient data and the desire to avoid overfitting. In addition, as the volume and complexity of prediction models implemented into production electronic health record and ancillary clinical systems explodes, automated surveillance procedures that can be deployed on active models are needed. Vergouwe et al30 recently proposed such a procedure to select between updating methods. However, their approach is limited to use with parametric models and may exhibit too strong a preference for refitting due to overfitting and the assumption that a refit model is always the leading choice.30,31
In this study, we develop a testing procedure to recommend updating methods to maintain performance over time that minimizes overfitting, accounts for uncertainty associated with the updating sample size, and is widely applicable to both parametric and nonparametric models. Our procedure selects between a set of updating methods based on a scoring rule, with both elements defined by the user. Updating approaches may include commonly applicable methods such as recalibration, as well as model-specific methods such as reweighting the leaf nodes of each tree in a random forest model. We illustrate the properties of our procedure on both simulated scenarios of population shifts that impact clinical use cases and 2 models developed and applied over time to Department of Veterans Affairs inpatient admissions.
Materials and METHODS
Testing procedure
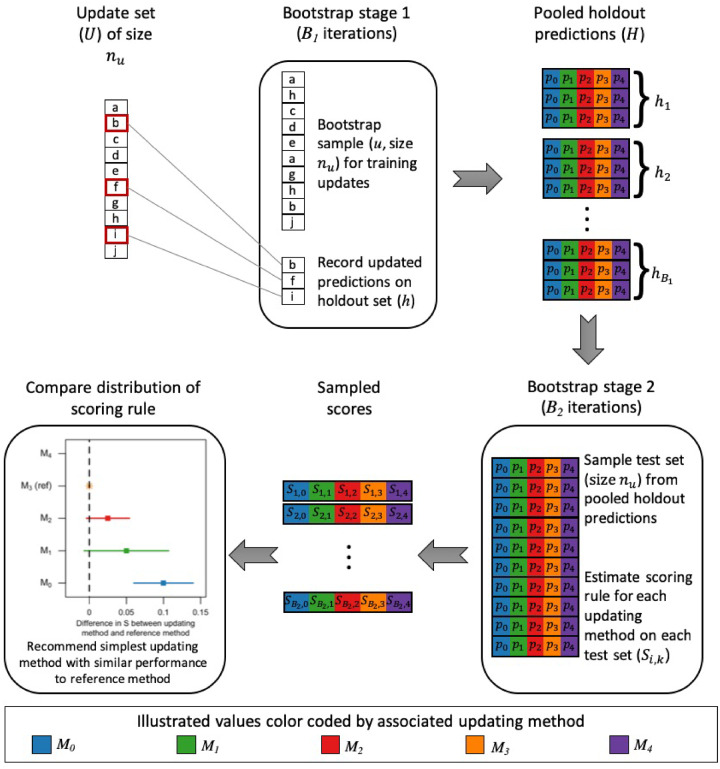
Our testing procedure recommends the simplest updating method that maximizes model performance in terms of accuracy, discrimination, or calibration. The procedure is designed for situations involving an existing prediction model and a set of new observations available for updating. The updating set may comprise observations from a new clinical setting in which the model will be applied or observations accruing since the model was trained. Figure 1 provides an overview of the procedure, which is based on a 2-stage bootstrapping approach. The first bootstrapping stage minimizes the influence of overfitting on the procedure’s recommendations by providing an out-of-bag set of updated predictions. The second bootstrapping stage utilizes these predictions to evaluate the updating methods on samples of equal size to the updating set, incorporating uncertainty associated with the updating sample size into decision making. We selected bootstrapping as the resampling method for both stages to maintain the sample size of the updating set for all assessments.
Figure 1.
Steps of testing procedure.
Given an update set () of size and a current model (), we define a set of updating methods as , where methods are sorted by increasing statistical complexity or user preference. For the purposes of this article, we define the set of updating methods as intercept correction (), linear logistic recalibration (), flexible logistic recalibration (), and model refitting (). Intercept correction and linear logistic recalibration are common approaches correcting systematic over- or underprediction and overfitting, respectively.25,29 Flexible logistic recalibration extends the linear logistic recalibration approach to allow nonlinearity in the association between outcomes and baseline predictions, potentially correcting more complex forms of miscalibration.32
We develop a pooled set of holdout predictions () via the first bootstrapping stage. For each of iterations, we randomly sample with replacement observations from , defining this sample as , and identify the holdout set () as those observations from not included in . Predicted probabilities from are estimated for all observations in and . Based on , we calculate the adjustments required for each updating methods. We apply these adjustments to , resulting in a set of predicted probabilities for each observation in based on the current model and each updating method. Holdout set predictions are pooled across iterations to construct . Here we set to be 100; however, fewer may be permissible as long as is large enough to capture variability in predictions and each observation is included in with similar probability.
The performance of each updating method is evaluated on via the second bootstrapping stage. For each of iterations, we randomly sample with replacement observations from and measure performance of each updating method in this sample using the user-defined scoring rule (S). For the purposes of this article, we use the Brier score, which incorporates both discrimination and calibration.33,34 See Supplementary Appendix S3 for sensitivity analyses using the logarithmic score. To enable stable quantile estimates for the scoring rule for each updating method, we set at 1000. This process results in a set of where indexes the iteration and indexes the updating method.
We define as the updating method for which the median S is closest in terms of absolute value to the scoring rule’s ideal value. No other method will have significantly better performance than as their accuracy cannot be significantly closer to the scoring rule’s ideal value; however, other methods may exhibit similar performance with a score that is not significantly different than that of . In our case, is the updating method with the minimum median Brier score as this measure tends toward 0 with increasing accuracy. As our procedure aims to recommend the simplest updating method that does not compromise performance, we need only consider whether any methods simpler than are comparably accurate. If is the current model (), then no comparisons are needed and the procedure recommends retaining the current model. Otherwise, starting with , we estimate the percentile-based 100 confidence interval for the paired difference in S between and . If this interval contains 0, indicating no significant difference, the procedure recommends . Otherwise, we increment and repeat until a recommendation is made or , in which case is recommended.
We do not correct for multiple comparisons in this final step of the procedure because we do not seek to control the familywise error rate of rejecting 1 or more null hypotheses of no difference between models. Rather, we seek to identify significant differences with a standard correction for uncertainty that does not depend on the number of comparisons being made. Operationally, users can control the stringency of this correction uniformly through setting in the 100 confidence interval. We are also not comparing all methods to simultaneously. Instead, we are filtering options and defining pairwise comparisons based on predefined preferences. For similar reasons, we encourage using the 100 confidence interval framework over a hypothesis testing framework. However, that approach is equally easy to execute. Rather than forming an interval from the and quantiles of the distribution of differences, the user estimates , the quantile represented by 0. A “P value” equal to is compared with , where being less than is equivalent to 0 being excluded from the interval.
Simulations
We conducted a simulation study to characterize the performance of our procedure under population shifts that may impact model performance.17,18,35,36 Such shifts involve changes in outcome prevalence, distributions of risk factors (ie, case mix), and predictor-outcome associations. We constructed separate populations for model development, updating, and evaluation for scenarios in which (1) no population shifts occurred, (2) the outcome became more prevalent, (3) the case mix became more homogenous, (4) the case mix became more heterogenous, and (5) predictor-outcome associations changed. Our simulated scenarios illustrate situations in which the testing procedure is applied to a fully shifted population, rather than a gradually shifting population in which observations are a mixture of the pre- and postshift patterns. This may reflect updating after transporting a model to a new clinical setting or after a long delay.
Reflecting a variety of predictor types that may be observed in clinical datasets, the simulated data covariates generated from multivariate normal, gamma, binary, Poisson, and multinomial distributions. We defined coefficients for 2 reference logistic regression models, one with 10 predictors and another with 40, referred to subsequently as the simple and complex models, respectively. We generated binary outcomes under each model by comparing the probabilities calculated from each model with randomly generated values from a uniform [0, 1] distribution. For the model development population, we simulated 100 000 observations with baseline distributions and coefficients. For each population drift scenario, we simulated 200 000 observations with adjusted parameters, assigning half to the updating population and half to the evaluation population. Under the scenario of no population shift, these data were simulated with the same settings as the development population. To simulate event rate shift, we adjusted the intercept of the logistic models to increase the event rate from 25% to 30%. Observations for the more homogenous and heterogenous case mix scenarios were generated by decreasing and increasing the variability of predictor distributions, respectively. For the predictor-outcome association shift scenario, we adjusted half the logistic models’ coefficients by 20%, with some increasing and others decreasing in strength of association. See Supplementary Appendix S1 for additional details.
We explored the impact of population shifts on updating recommendations under varying training (= 1000, 5000, and 10 000) and updating (= 1000, 5000, and 10 000) sample sizes. We expect larger may lead to more robust, generalizable models that are more amenable to recalibration rather than requiring refitting under some scenarios. As larger provide more information to support updating, we expect them to lead to more complex updating recommendations than smaller under all population shift scenarios. We trained either the simple or complex logistic regression model on observations sampled from the development population. We sampled observations from the updating and evaluation populations of each population shift scenario. To determine the recommended updating method, we applied our testing procedure to the updating sample. To document the impact of updating recommendations, we assessed the performance of each available updating method on the evaluation sample. This process was repeated 1000 times for each combination of model complexity, , and .
Case study
As illustrative examples on clinical data, we explored the decisions of our procedure on 2 logistic regression models, one for 30-day all-cause mortality and another for hospital-acquired acute kidney injury (AKI). Both models were developed and updated on a national set of inpatient admissions to Department of Veterans Affairs facilities.17,18 These models experienced documented calibration drift across several years.17,18 Drift of the AKI model accelerated 3 years after development due to a complex mix of event rate, case mix, and predictor-outcome association changes.18 Performance of the mortality model drifted more consistently over 7 years as a result of steady event rate and case mix shifts.17 We applied our procedure at multiple points after development of each model to assess the need for updating. This study was approved by the Institutional Review Board and the Research and Development committee of the VA Tennessee Valley Healthcare System.
The mortality model was trained on admissions from 2006 (n = 235 548) and the AKI model on admissions from 2003 (n = 170 675). We updated both models 1, 3, and 5 years after development, defining updating points at the end of 2007, 2009, and 2011 for the mortality model and at the end of 2004, 2006, and 2008 for the AKI model. Calibration of the mortality model steadily declined across this period, whereas calibration drift of the AKI model accelerated 4 years after development. We applied the testing procedure with updating sets defined as admissions in the prior 1, 3, 6, and 12 months. For simplicity, we refer to the 12-month update set as a large update cohort, the 1-month update set as a small update cohort, and the 3- and 6-month update sets as moderate update cohorts. We documented performance of the original and updated models on a prospective evaluation set of admissions in the 3 months after each updating point, reflecting the notion that an updated model would ideally perform well immediately after updating.
Baseline comparison
For baseline comparison with the most applicable method in the literature, we evaluated a closed testing procedure recently proposed by Vergouwe et al.30 Using sequential likelihood ratio tests, this procedure selects the simplest updating method providing a fit similar to model refitting. We extended Vergouwe et al’s procedure to incorporate flexible logistic recalibration32 in addition to the methods originally included. We applied Vergouwe et al’s procedure to both the simulated and case study data, comparing the recommendations with those of our test and the impact of these decisions on subsequent model performance. Results are provided in Supplementary Appendix S2.
RESULTS
Simulations
The updating recommendations of our testing procedure by population shift scenario, training sample size, and updating sample size for the complex (=40) model are detailed in Table 1. See Supplementary Appendix S4 for additional results.
Table 1.
Percent of iterations for which each updating methods was recommended by our testing procedure under each simulated scenario, training sample size (), and updating sample size () for the complex model
|
= 1000 |
= 5000 |
= 10000 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Scenario | Updating method | = 1000 | = 5000 | = 10 000 | = 1000 | nu= 5000 | nu= 10 000 | = 1000 | = 5000 | = 10 000 |
| No population shift | Not updating | 94 | 7.7 | 0 | 100 | 99.7 | 87 | 100 | 100 | 100 |
| Intercept correction | 1.2 | 1.3 | 0 | 0 | 0.2 | 0 | 0 | 0 | 0 | |
| Linear recalibration | 4.8 | 7.1 | 0 | 0 | 0.1 | 0 | 0 | 0 | 0 | |
| Flexible recalibration | 0 | 0.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Model refitting | 0 | 83.8 | 100 | 0 | 0 | 13 | 0 | 0 | 0 | |
| Increased event rate | Not updating | 43.6 | 0.1 | 0 | 43.4 | 0 | 0 | 48.1 | 0 | 0 |
| Intercept correction | 48 | 5.2 | 0 | 56.6 | 99.2 | 81.7 | 51.9 | 100 | 100 | |
| Linear recalibration | 8.4 | 4.4 | 0 | 0 | 0.8 | 5.2 | 0 | 0 | 0 | |
| Flexible recalibration | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Model refitting | 0 | 90.3 | 100 | 0 | 0 | 13.1 | 0 | 0 | 0 | |
| Less variable case mix | Not updating | 79.9 | 22.5 | 1 | 100 | 88.5 | 68.4 | 100 | 96.3 | 89.4 |
| Intercept correction | 14.1 | 14.6 | 0.5 | 0 | 10.3 | 21.1 | 0 | 3.6 | 7 | |
| Linear recalibration | 6 | 20.7 | 1.6 | 0 | 1.2 | 5.2 | 0 | 0.1 | 3.6 | |
| Flexible recalibration | 0 | 0.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Model refitting | 0 | 42.1 | 96.9 | 0 | 0 | 5.3 | 0 | 0 | 0 | |
| More variable case mix | Not updating | 59 | 0 | 0 | 95.5 | 60.1 | 15.8 | 96.4 | 74.7 | 74.9 |
| Intercept correction | 30.4 | 0 | 0 | 4.5 | 28.4 | 13.3 | 3.6 | 24.7 | 18 | |
| Linear recalibration | 8.2 | 0.2 | 0 | 0 | 1.7 | 7.8 | 0 | 0.4 | 3.5 | |
| Flexible recalibration | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Model refitting | 2.4 | 99.8 | 100 | 0 | 9.8 | 63.1 | 0 | 0.2 | 3.6 | |
| Association changes | Not updating | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Intercept correction | 0 | 0 | 0 | 3.6 | 0 | 0 | 2.4 | 0 | 0 | |
| Linear recalibration | 0 | 0 | 0 | 0 | 0 | 0 | 1.1 | 0 | 0 | |
| Flexible recalibration | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Model refitting | 100 | 100 | 100 | 96.4 | 100 | 100 | 96.5 | 100 | 100 | |
When no population shifts occurred, our test generally recommended retaining the original model. As the updating samples increasingly outweighed training samples, model refitting became the primary recommendation. A similar pattern emerged when the event rate increased. In this case, intercept correction was recommended; however, a shift toward model refitting was apparent as the updating sample dominated the training sample. With small updating samples, test recommendations were split between not updating and intercept correction. Under both population shifts, the recommended updates provided superior or similar calibration to that achieved with more complex updates. Recommendations for more complex updating when training samples were very small compared with updating samples improved performance compared with the original model.
In response to changes in predictor-outcome associations, our test recommended model refitting, regardless of the relative sizes of the training and updating samples. Refitting under predictor-outcome association shift improved accuracy compared with simpler updates, even when updating samples were smaller than training samples.
Case mix shifts resulted in the most variable recommendations. When variability in case mix decreased between the training and updating populations, recommendations varied across the spectrum of updating methods. However, the overall trend was toward retaining the original model, particularly when updating samples included similar or smaller volumes of data than training samples. When >1000, no significant improvement in performance was observed with updating, supporting the recommendation to retain the original model.
With increasing variability in case mix, for the simple model, refitting was recommended for updating samples of similar or larger size as training samples; however, not updating was the dominant recommendation for smaller update samples. Recommendations for the complex model were primarily split between not updating and intercept correction, although refitting the model was recommended as updating samples grew larger than training samples. Calibration under the procedure’s recommendations was generally less variable, but not significantly different, than that of the original model. More complex updates than those recommended did not provide additional improvement in performance. For the smallest training samples, however, refitting with larger updating samples, as recommended, improved discrimination but not calibration compared with recalibration methods.
Case study
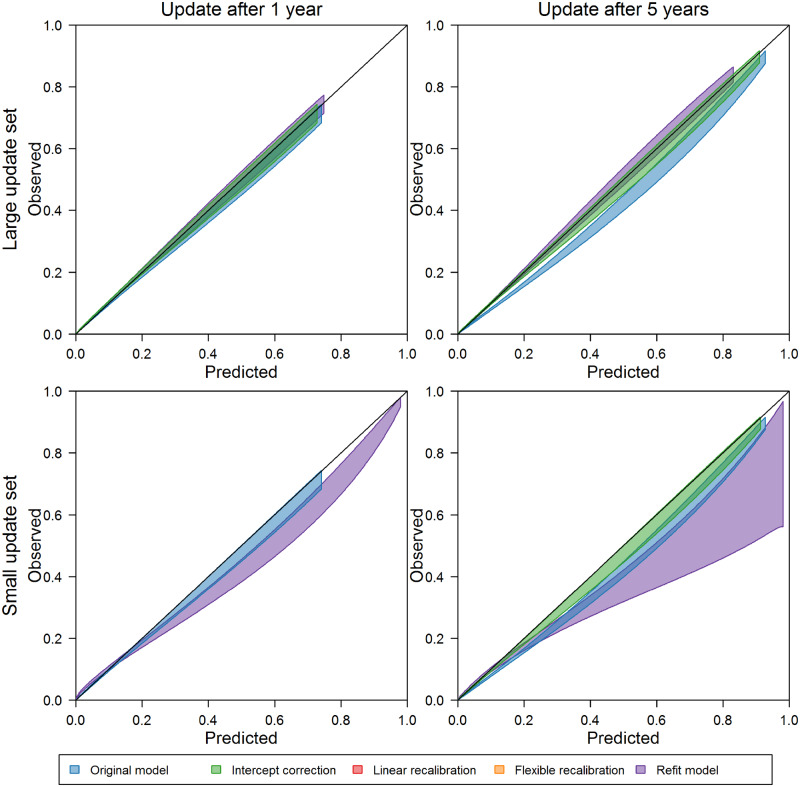
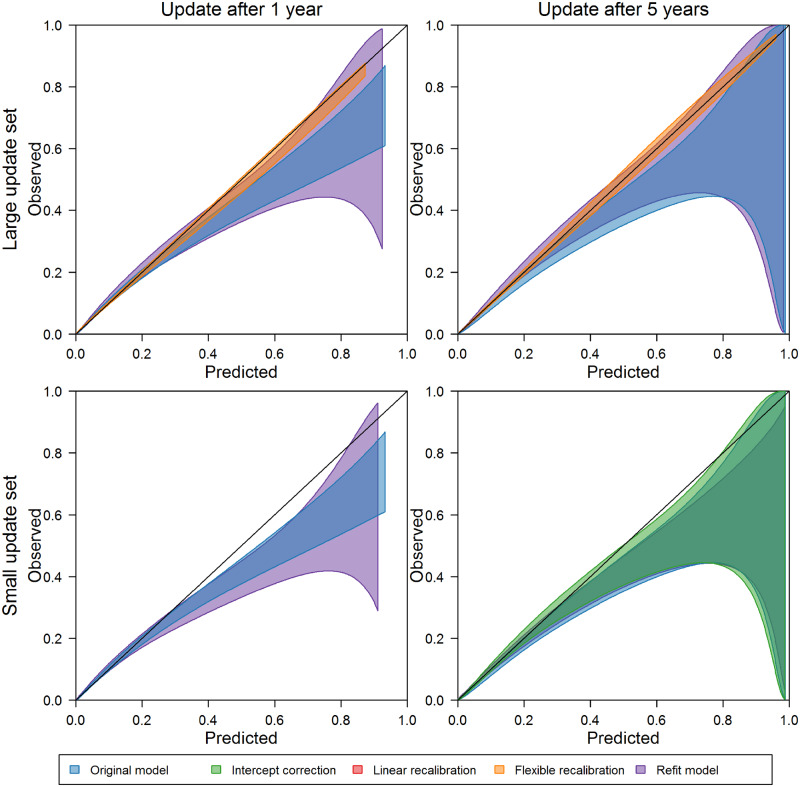
The updating recommendations of our testing procedure at 1, 3, and 5 years after model development are documented in Table 2. Corresponding calibration belts37 for the original model, the refitted model, and our procedure’s recommended update are presented in Figures 2 and 3. See Supplementary Appendix S5 for full results.
Table 2.
Updating method recommended by our testing procedure by time since model development and size of updating set based on number of months of admissions used for updating
| Time from development to updating | Update set size |
|||
|---|---|---|---|---|
| Large(12 mo) | Moderate(6 mo) | Moderate(3 mo) | Small(1 mo) | |
| Acute kidney injury | ||||
| 1 y | Intercept correction | Intercept correction | No update | No update |
| 3 y | Intercept correction | Intercept correction | Intercept correction | No update |
| 5 y | Intercept correction | Intercept correction | Intercept correction | Intercept correction |
| 30-d mortality | ||||
| 1 y | Flexible logistic recalibration | Flexible logistic recalibration | Flexible logistic recalibration | No update |
| 3 y | Flexible logistic recalibration | Flexible logistic recalibration | Intercept correction | No update |
| 5 y | Flexible logistic recalibration | Flexible logistic recalibration | Flexible logistic recalibration | Intercept correction |
Figure 2.
Calibration belts in the 3 months after updating based on large (12 months) and small (1 month) update sets for the original acute kidney injury model, the refit model, and the recommended update (if different [eg, no update to original model recommended for bottom left panel]).
Figure 3.
Calibration belts in the 3 months after updating based on large (12 months) and small (1 month) update sets for the original 30-day mortality model, the refit model, and the recommended update (if different [eg, no update to original model recommended for bottom left panel]).
Intercept correction was the most complex updating method recommended for the AKI model. This change was recommended with large update sets at all time points; however, calibration among inpatient admissions in the 3 months after updating was not significantly improved by intercept correction 1 year after model development (see upper left panel of Figure 2). Five years after model development, intercept correction with the large update set, as recommended by our procedure, improved calibration over the original model and provided in similar calibration to that of the refitted model. With small update samples, the calibration curves reflect lower calibration after refitting compared with both the original and intercept-corrected models. At the 5-year update point, intercept correction, as recommended, with the small updating set improved upon the calibration of the original model. In each case, more complex recalibration did not provide additional performance improvements.
For the 30-day mortality model, large and moderate update sets prompted flexible logistic recalibration both soon after model development and as time passed. The calibration curves associated with flexible logistic recalibration highlight well-calibrated predictions over a wide range of probabilities, while the curves for the original and refitted models highlight uncertainty and overprediction as predicted probabilities increase. Simpler recalibration approaches did not provide as much improvement in performance as the recommended flexible logistic recalibration. With small update sets and more time since model development, intercept correction was recommended. Although calibration was somewhat improved with this update compared with the original and refitted models, the calibration curves reflect uncertainty in performance after updating, which could have been further improved on by more complex recalibration in this case (see Supplementary Appendix Figure 5.4).
DISCUSSION
We presented a new testing procedure to recommend prediction model updating methods that minimizes overfitting, accounts for uncertainty associated with updating sample size, incorporates a preference for simpler updating, and is applicable regardless of the modeling technique generating predicted probabilities. This procedure supports clinical prediction models underlying informatics applications for decision support, population management, and quality benchmarking, both when transporting such models in a new setting or applying them over time in evolving clinical environments.
As is desirable based on statistical theory, the test displayed a preference for recommending more complex updates as the updating sample size increased or as the training and updating populations became increasingly disparate (See Table 3). Our findings reflect the concept that when more information is available to support updating (ie, update sets are large), more complex updating may be appropriate. For example, when our simulated update set was 10 times larger than the training set, our test most often recommended refitting the model, even in scenarios for which there were no differences between training and updating populations. This pattern is reassuring, as we intuitively would want to refit a model when substantially more information is available for learning associations, even if we do not expect associations to have changed. For models trained on small datasets, updating recommendations varied when update sets were equally small, highlighting uncertainty in both the original and adjusted models.
Table 3.
General patterns of our testing procedure’s updating recommendations
| Situation | Most common recommendation |
|---|---|
| Changes in outcome prevalence, similar volumes of training and updating data | Intercept correction |
| Changes in case mix, similar volumes of training and updating data | No updating or recalibration |
| Changes in predictor-outcome associations, regardless of sample sizes | Refit the model |
| Updating set substantially larger than training set | Refit the model |
Generally, in those situations in which the updating and training sets provided the same volume of data, no population shift resulted in recommendations to retain the original model, case mix shift resulted in recommendations of retain the original model or conduct modest recalibration, and outcome rate shifts resulted in recommendations of intercept correction. Reassuringly, when predictor-outcome associations shifted between simulated training and updating populations, our test predominantly recommended refitting regardless of the training or updating sample sizes.
Our case study results similarly highlighted recommendations for more complex updating as population shift increased and updating sample sizes grew. With large updating samples, our test recommended updating at each time points considered and suggested more complex recalibration compared with recommendations based on smaller updating samples. As calibration of the original models deteriorated over time, our test responded by recommending recalibration even when updating samples were limited. By recommending recalibration to varying degrees rather than refitting the models, the test allowed us to avoid refitting in cases where this more data-intensive updating approach would have provided no benefit to or even harmed prospective performance.
Our testing procedure’s recommendations were frequently different from those provided by Vergouwe et al’s closed testing procedure. Vergouwe et al’s procedure commonly recommended model refitting, even for simulations involving no population shifts and update sets substantially smaller than training sets. These refitted models resulted in either similar or inferior performance to that which was achieved through recalibration. This highlights the importance of controlling for overfitting and avoiding the assumption that model refitting is the ideal updating methods against which other methods should compete.
We have described our testing procedure previously as filtering based on any statistically significant difference in performance between updating methods. However, the procedure can be adjusted to filter based on clinically significant differences in performance between updating methods. This would be achieved by adjusting the final step of the procedure to consider a user-specified minimum difference in S between and simpler updating methods. Although we have used 0 to find any difference in accuracy, small differences in the scoring metric may not be associated with clinically meaningful differences in performance and may result in users questioning the value of updating. Methods for defining clinically meaningful differences in performance as measured by various scoring rules remain an open area of research.
While our analyses focus on logistic regression models, the testing procedure is applicable to any model, as the method makes no assumptions regarding the learning approach and relies only on observed and predicted values. In addition to applications with other dichotomous outcome models, extension to multiclass models is straightforward by providing an appropriate scoring rule, such as the multiclass definition of the Brier score. Although the updating methods implemented in these analyses are generalizable across learning algorithms, users may tailor the set of updating methods to be considered based on use case-specific needs and preferences. The only requirement for defining a custom set of updating methods is that users provide an order of complexity or preference among the included methods, which may require careful consideration for cases lacking a natural ordering. Some users may also prefer to optimize a different scoring metric than implemented in these analyses. Such an adjustment is easily made by replacing the Brier score in the second bootstrapping stage. In sensitivity analyses using the logarithmic scoring rule, we observed strong agreement with the test recommendations based on the Brier score (see Supplementary Appendix S3).
Our testing procedure supports periodic updating of static models. Alternatively, online learning algorithms continuously update as new observations become available, incorporating changes in the environment as they occur.27,38,39 Such models have been applied to health outcomes, but have yet to be incorporated into clinical tools.26,27,40 The shift to an online paradigm is not straightforward for clinical use cases, as new validation methods are required27,38 and the regulatory framework for implementing dynamic models is evolving.41
There are several key limitations of our test and the evaluations presented here. The case study highlights the conservative nature of our test, which may be a limitation for certain use cases. When only a small updating set was available to update the 30-day mortality model, our test recommended not updating 1 year after model development and only minimal recalibration after 5 years. Calibration assessment based on admissions in the 3 months following these update points indicated that flexible recalibration, as recommended with larger update sets, could have provided additional improvement in calibration. While we view the decision to recommend less complex updating as a benefit, given the relatively small size of the updating set in this example, the requirements of some use cases may view any improvement in calibration to be desirable and the test’s recommendation as a missed opportunity. As a second limitation, the first bootstrapping stage may be computationally expensive, particularly for complex models. Although advancements in computational resources continue to reduce computation times, a refinement to the number of iterations in the first bootstrap stage may be warranted. Finally, updating per the test’s recommendation, or any of the considered methods, may not result in sufficient improvement in model performance to warrant continued clinical application of the model. Users should evaluate performance of the updated model to determine if clinically acceptable performance is achieved or whether model extension or alternative models may be required.
CONCLUSION
As clinical prediction models continue to be developed and deployed in complex, ever-changing environments, maintenance of these models will become increasingly crucial to their utility. Models underlying population health management, quality assessment, and clinical decision support applications require a high degree of accuracy and developers must be responsive to any degradation in performance. We describe a new testing procedure to support data-driven updating of categorical prediction models, with the intent to increase the long-term sustainability of those models in a continuously evolving clinical environment. Our procedure encourages small corrections when only a small amount of new data are available, and graduates to recommending full model retraining when the new dataset is large enough to support it. The procedure is applicable to models developed with either biostatistical or machine learning approaches, and is customizable to user needs and preferences.
Supplementary Material
ACKNOWLEDGMENTS
Initial results of this study were presented at the American Medical Informatics Association 2018 Annual Symposium, with an associated abstract published in the conference proceedings.
FUNDING
This work was supported by funding from the National Library of Medicine grant number 5T15LM007450 (Davis); the Veterans Health Administration grant numbers VA HSR&D IIR 11-292 and 13-052 (Matheny); and the National Institutes of Health grant number BCHI-R01-130828 (Lasko).
CONTRIBUTIONS
SED, MEM, and RAG designed the methodological framework. SED and MEM acquired the case study data and implemented the study plan. TAL, CF, and CGW provided critical methodological feedback and contributed to the design and interpretation of the simulation and case studies. SED conducted all data analysis and drafted the initial manuscript. All authors contributed to interpretation of the results and critical revision of the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Conflict of interest statement
None declared.
REFERENCES
- 1. Hall LM, Jung RT, Leese GP.. Controlled trial of effect of documented cardiovascular risk scores on prescribing. BMJ 2003; 3267383: 251–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Feldman M, Stanford R, Catcheside A, Stotter A.. The use of a prognostic table to aid decision making on adjuvant therapy for women with early breast cancer. European Journal of Surgical Oncology 2002; 286: 615–9. [DOI] [PubMed] [Google Scholar]
- 3. Amarasingham R, Patel PC, Toto K, et al. Allocating scarce resources in real-time to reduce heart failure readmissions: a prospective, controlled study. BMJ Qual Saf 2013; 2212: 998–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Amarasingham R, Patzer RE, Huesch M, Nguyen NQ, Xie B.. Implementing electronic health care predictive analytics: considerations and challenges. Health Aff (Millwood) 2014; 337: 1148–54. [DOI] [PubMed] [Google Scholar]
- 5. Jarman B, Pieter D, van der Veen AA, et al. The hospital standardised mortality ratio: a powerful tool for Dutch hospitals to assess their quality of care? Qual Saf Health Care 2010; 191: 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Steyerberg EW, Moons KG, van der Windt DA, et al. Prognosis research strategy (PROGRESS) 3: prognostic model research. PLoS Med 2013; 102: e1001381.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ohno-Machado L, Resnic FS, Matheny ME.. Prognosis in critical care. Annu Rev Biomed Eng 2006; 81: 567–99. [DOI] [PubMed] [Google Scholar]
- 8. Matheny ME, Miller RA, Ikizler TA, et al. Development of inpatient risk stratification models of acute kidney injury for use in electronic health records. Med Decis Making 2010; 306: 639–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kansagara D, Englander H, Salanitro A, et al. Risk prediction models for hospital readmission: a systematic review. JAMA 2011; 30615: 1688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Toll DB, Janssen KJ, Vergouwe Y, Moons KG.. Validation, updating and impact of clinical prediction rules: a review. J Clin Epidemiol 2008; 6111: 1085–94. [DOI] [PubMed] [Google Scholar]
- 11. Moons KG, Kengne AP, Grobbee DE, et al. Risk prediction models: II. External validation, model updating, and impact assessment. Heart 2012; 989: 691–8. [DOI] [PubMed] [Google Scholar]
- 12. Sajda P. Machine learning for detection and diagnosis of disease. Annu Rev Biomed Eng 2006; 81: 537–65. [DOI] [PubMed] [Google Scholar]
- 13. Steyerberg EW, van der Ploeg T, Van Calster B.. Risk prediction with machine learning and regression methods. Biom J 2014; 564: 601–6. [DOI] [PubMed] [Google Scholar]
- 14. Pencina MJ, Peterson ED.. Moving from clinical trials to precision medicine: the role for predictive modeling. JAMA 2016; 31516: 1713–4. [DOI] [PubMed] [Google Scholar]
- 15. Parikh RB, Kakad M, Bates DW.. Integrating predictive analytics into high-value care: the dawn of precision delivery. JAMA 2016; 3157: 651–2. [DOI] [PubMed] [Google Scholar]
- 16. Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI.. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J 2015; 13: 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davis SE, Lasko TA, Chen G, Matheny ME.. Calibration drift among regression and machine learning models for hospital mortality. AMIA Annu Symp Proc 2018; 2017: 625–34 [PMC free article] [PubMed] [Google Scholar]
- 18. Davis SE, Lasko TA, Chen G, Siew ED, Matheny ME.. Calibration drift in regression and machine learning models for acute kidney injury. J Am Med Inform Assoc 2017; 246: 1052–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Minne L, Eslami S, De Keizer N, De Jonge E, De Rooij SE, Abu-Hanna A.. Effect of changes over time in the performance of a customized SAPS-II model on the quality of care assessment. Intensive Care Med 2012; 381: 40–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Steyerberg EW. Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating. New York: NY: Springer; 2009. [Google Scholar]
- 21. Moons KG, Altman DG, Vergouwe Y, Royston P.. Prognosis and prognostic research: application and impact of prognostic models in clinical practice. BMJ 2009; 338: b606.. [DOI] [PubMed] [Google Scholar]
- 22. Hickey GL, Grant SW, Murphy GJ, et al. Dynamic trends in cardiac surgery: why the logistic EuroSCORE is no longer suitable for contemporary cardiac surgery and implications for future risk models. Eur J Cardiothorac Surg 2013; 436: 1146–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Debray TP, Vergouwe Y, Koffijberg H, Nieboer D, Steyerberg EW, Moons KG.. A new framework to enhance the interpretation of external validation studies of clinical prediction models. J Clin Epidemiol 2015; 683: 279–89. [DOI] [PubMed] [Google Scholar]
- 24. Kappen TH, Vergouwe Y, van Klei WA, van Wolfswinkel L, Kalkman CJ, Moons KG.. Adaptation of clinical prediction models for application in local settings. Med Decis Making 2012; 323: E1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Janssen KJ, Moons KG, Kalkman CJ, Grobbee DE, Vergouwe Y.. Updating methods improved the performance of a clinical prediction model in new patients. J Clin Epidemiol 2008; 611: 76–86. [DOI] [PubMed] [Google Scholar]
- 26. Siregar S, Nieboer D, Vergouwe Y, et al. Improved prediction by dynamic modelling: an exploratory study in the adult cardiac surgery database of the Netherlands association for cardio-thoracic surgery. Interact Cardiovasc Thorac Surg 2014; 19(suppl 1): S8. [DOI] [PubMed] [Google Scholar]
- 27. Jenkins DA, Sperrin M, Martin GP, Peek N.. Dynamic models to predict health outcomes: current status and methodological challenges. Diagn Progn Res 2018; 21: 23.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Minne L, Eslami S, de Keizer N, de Jonge E, de Rooij SE, Abu-Hanna A.. Statistical process control for monitoring standardized mortality ratios of a classification tree model. Methods Inf Med 2012; 514: 353–8.. [DOI] [PubMed] [Google Scholar]
- 29. Steyerberg EW, Borsboom GJ, van Houwelingen HC, Eijkemans MJ, Habbema JD.. Validation and updating of predictive logistic regression models: a study on sample size and shrinkage. Stat Med 2004; 2316: 2567–86. [DOI] [PubMed] [Google Scholar]
- 30. Vergouwe Y, Nieboer D, Oostenbrink R, et al. A closed testing procedure to select an appropriate method for updating prediction models. Stat Med 2017; 3628: 4529–39. [DOI] [PubMed] [Google Scholar]
- 31. Van Calster B, Van Hoorde K, Vergouwe Y, et al. Validation and updating of risk models based on multinomial logistic regression. Diagn Progn Res 2017; 11: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dalton JE. Flexible recalibration of binary clinical prediction models. Stat Med 2013; 322: 282–9. [DOI] [PubMed] [Google Scholar]
- 33. Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 2010; 211: 128–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brier GW. Verification of forecasts expressed in terms of probability. Mon Weather Rev 1950; 751: 1–3. [Google Scholar]
- 35. Tsymbal A. The problem of concept drift: definitions and related work. Comput Sci Dep Trinity College Dublin 2004; 1062: 58. [Google Scholar]
- 36. Murphy-Filkins R, Teres D, Lemeshow S, Hosmer DW.. Effect of changing patient mix on the performance of an intensive care unit severity-of-illness model: how to distinguish a general from a specialty intensive care unit. Crit Care Med 1996; 2412: 1968–73. [DOI] [PubMed] [Google Scholar]
- 37. Nattino G, Finazzi S, Bertolini G.. A new calibration test and a reappraisal of the calibration belt for the assessment of prediction models based on dichotomous outcomes. Stat Med 2014; 3314: 2390–407. [DOI] [PubMed] [Google Scholar]
- 38. Su TL, Jaki T, Hickey GL, Buchan I, Sperrin M.. A review of statistical updating methods for clinical prediction models. Stat Methods Med Res 2018; 271: 185–97. [DOI] [PubMed] [Google Scholar]
- 39. Gama J, Žliobaitė I, Bifet A, Pechenizkiy M, Bouchachia A.. A survey on concept drift adaptation. ACM Comput Surv 2014; 464: 44. [Google Scholar]
- 40. Hickey GL, Grant SW, Caiado C, et al. Dynamic prediction modeling approaches for cardiac surgery. Circ Cardiovasc Qual Outcomes 2013; 66: 649–58. [DOI] [PubMed] [Google Scholar]
- 41. U.S. Food & Drug Administration. Proposed Regulatory Framework for Modifications to Artificial Intelligence/Machine Learning (AI/ML)-Based Software as a Medican Device (SaMD). Discussion paper and request for feedback. 2019. https://www.fda.gov/media/122535/download. Accessed April 15, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.