Abstract
Objective
Amid electronic health records, laboratory tests, and other technology, office-based patient and provider communication is still the heart of primary medical care. Patients typically present multiple complaints, requiring physicians to decide how to balance competing demands. How this time is allocated has implications for patient satisfaction, payments, and quality of care. We investigate the effectiveness of machine learning methods for automated annotation of medical topics in patient-provider dialog transcripts.
Materials and Methods
We used dialog transcripts from 279 primary care visits to predict talk-turn topic labels. Different machine learning models were trained to operate on single or multiple local talk-turns (logistic classifiers, support vector machines, gated recurrent units) as well as sequential models that integrate information across talk-turn sequences (conditional random fields, hidden Markov models, and hierarchical gated recurrent units).
Results
Evaluation was performed using cross-validation to measure 1) classification accuracy for talk-turns and 2) precision, recall, and F1 scores at the visit level. Experimental results showed that sequential models had higher classification accuracy at the talk-turn level and higher precision at the visit level. Independent models had higher recall scores at the visit level compared with sequential models.
Conclusions
Incorporating sequential information across talk-turns improves the accuracy of topic prediction in patient-provider dialog by smoothing out noisy information from talk-turns. Although the results are promising, more advanced prediction techniques and larger labeled datasets will likely be required to achieve prediction performance appropriate for real-world clinical applications.
Keywords: classification, supervised machine learning, patient care, communication
INTRODUCTION
Background
Appropriate documentation of the clinical visit is critical for communication among medical professionals,1,2 enabling quality assurance,3 and accurate billing and reimbursement.4 The traditional way of documenting a clinical visit in the electronic health record, namely physicians’ notes, provide a source of valuable information on what occurred during the interaction and what physicians consider to be important. Electronic health records have improved the accessibility of medical information,5 but patients are demanding access to information at a greater scale.6 Pressure to quickly document the medical visit may lead providers to type into the record during the medical visit, which can negatively impact patient-provider communication.7–9 Primary care providers spend about half of their time working on computers,10,11 which appears partly responsible for growing concerns of physician burnout across a wide range of physician specialties.12–14 In addition, physician generated notes do not always provide an accurate representation of what occurred during the visits.15,16
Technologies that could reduce the burden of documentation on providers, and increase convergence between documentation of a visit and the content of the clinical interaction are greatly needed. Natural language processing (NLP) technologies combined with advances in automatic speech recognition17,18 offer potentially promising solutions.19,20 Information extraction and summarization technologies built on top of resulting transcripts will be needed to take the next step in reducing the burden of documentation on physicians and in providing clinical decision support to both patients and physicians.18 If successful, machine learning–enabled automatic speech recognition and charting could free up valuable time for physicians to talk to their patients, rather than typing extensively during clinical encounters.21,22
Patient-provider conversations are complex, multidimensional, and multifunctional.23–27 Patients present multiple issues during an office visit requiring clinicians to divide time and effort during a visit to address competing demands,28,29 such as a patient could be concerned about blood pressure, knee pain, and blurry vision in a single appointment. Moreover, visit content does not solely focus on biomedical issues, but also on psychosocial matters, personal habits, mental health,30 patient-physician relationship,31 and small talk.28,32 Health communications researchers analyze the content of patient-provider communication by directly labeling the interaction using trained raters to label the topical content of clinical interactions. For example, during periodic health examinations with their primary care physicians, only one-third of patients with mental health needs had a discussion about mental health.30,33 These findings suggest that quality improvement efforts that evaluate the content of clinical interactions might help address gaps in service delivery.
However, labeling each talk-turn using coding systems designed to capture the content of medical visits, such as the Multi-Dimensional Interaction Analysis system,28,32,33 is labor intensive and costly. It requires training a team to label the text and establish and maintain reliability. Depending on the extensiveness of the labeling system, it can take several hours to label one patient-provider interaction.34,35 This level of effort means that even in research settings with resources for detailed evaluation, studies of patient-provider interaction are often limited in scale.28 As a result, the direct evaluation of clinical interactions is not feasible in routine care settings for quality improvement purposes.36,37 Automated methods capable of extracting the topical content of clinical encounters could support providers who overlook asking patients about critical issues (eg, suicide, blood pressure medication) when it is clinically indicated, reduce the burden of documentation currently placed on providers, and facilitate large-scale research on the quality of patient-provider communication.
Related work
The past decade has seen an explosion of interest in machine learning and NLP in medical contexts,38 targeting problems such as automated extraction of information from clinical notes and electronic health records.39–42 Of direct relevance to the present article is a growing body of work dedicated to applying methods from machine learning and NLP to the automatic annotation of conversations between providers and patients. The typical approach in such studies begins with labeling a corpus of transcript data (eg, providing human-generated labels for each utterance or talk-turn in each visit in the corpus). Machine learning techniques are then used to learn a classification model from a subset of the corpus, the training data, and the model’s predictions are evaluated by comparing its predictions with the known human labels on unseen test transcripts. For example, Mayfield et al43 analyzed patient-provider transcripts from the ECHO (Enhancing Communication and HIV Outcomes) study44 and employed a logistic regression model to classify utterances into the categories “information-giving” or “information-requesting.”45 More recently, Kotov et al46 analyzed transcripts from motivational interviewing related to pediatric obesity and developed probabilistic machine learning models for classifying patient utterances into classes of behavioral responses. In later work on the same dataset, Hasan et al47 compared probabilistic models with more recent recurrent neural network approaches and found the latter to be generally more accurate on that dataset.
There has been less prior work on the problem of automated classification of topical content in patient-provider dialog. Wallace et al48 developed machine learning models to classify dialog utterances into 1 of 6 high-level discussion topics: biomedical, logistics, psychosocial, antiretroviral (ARV), missing/other, and socializing. Using the same ECHO dataset44 as used in the Mayfield et al study, they evaluated the performance of conditional random fields (CRFs) for this prediction task and concluded that the results showed promise for automated classification of patient-provider interactions into clinically relevant topics. Gaut et al49 proposed the use of labeled topic models for classifying psychotherapy sessions with 161 possible topic labels, using a dataset published by Alexander Street Press, and again finding that these models showed promise in terms of predictive ability.
These earlier studies demonstrate that machine learning systems can generate plausible annotations of medical dialogues—our work in this article pursues this line of research further. We differ from earlier work on topic classification in a number of aspects. For example, we compare both probabilistic and neural network classification methods for topic classification of talk-turns, whereas Wallace et al48 and Gaut et al49 only focused on probabilistic approaches. We also evaluate performance for a more detailed set of 27 topics compared with the 6 high-level topics used in the Wallace et al study. The Gaut et al study also differs from our work in that it primarily focused on session-level labels and did not investigate the use of sequential information across talk-turns for talk-turn–level predictions as we do here. To our knowledge, this is the first study that systematically compares sequential and nonsequential classification methods, for both probabilistic and neural network models, on the problem of talk-turn topic classification from transcripts of patient-provider dialog.
MATERIALS AND METHODS
Dataset
The source data include transcripts of audio-recordings of primary care office visits from the MHD (Mental Health Discussion) study.33 Each transcript corresponds to a visit between a patient and a provider—a small fraction of the dialog corresponds to other participants in the conversation (such as a nurse and family member). Data collection occurred from 2007 to 2009 in a health system in Michigan with 26 ambulatory care clinics. Patients were 50-80 years of age, all had insurance, and were due for a colorectal cancer screening at the time of appointment. All aspects of the research protocol were approved by relevant organizations’ institutional review boards.
Each visit is comprised of a series of talk-turns, with 122 083 talk-turns in total across 279 visits (median and mean of 408 and 438 talk-turns per visit, respectively, with upper and lower quartiles of 312 and 522) from 59 providers. Each talk-turn was manually assigned by a human coder (labeler) to 1 of 39 different topic labels33 that were modified from the Multi-Dimensional Interaction Analysis coding system.32 A topic is defined as an issue raised in a conversation that required a response from the other member of the dyad and had at least 2 exchanges between the dyad. A small number of talk-turns were split into 2 if the turn straddled 2 topics. This resulted in a few of the original talk-turns being represented as 2 separate talk-turns after coding. Figure 1 illustrates how different topics were assigned to talk-turns during a short portion of a particular visit. Topic labels that occurred in less than 20 visits were merged into a single topic denoted as Other, resulting in a total of 27 unique topics in the corpus. Table 1 provides the names of the topic labels, a brief description of each, as well as the percentage of talk-turns assigned to each by the labelers across the corpus. The topic label distribution is skewed toward topics relevant to periodic health exams—the 3 most frequent topics (BiomedHistory, PreventiveCare, and MusSkePain) account for more than half of all talk-turns.
Figure 1.
A short excerpt from an annotated dialogue transcript. Topic labels are assigned to each talk-turn. MD and PT indicate the speaker for each talk-turn, where MD stands for “medical doctor” or “provider,” and PT stands for “patient.”
Table 1.
Name and brief description of each topic ordered by the percentage of each topic in talk-turns
| Short topic name | Brief description | Talk-turns assigned (%) |
|---|---|---|
| Biomed History | Biomedical history, symptoms, and medical condition | 29.85 |
| Preventive Care | Preventive medical measures | 14.67 |
| Mus Ske Pain | Musculoskeletal pain | 8.30 |
| Visit Flow Mgmt | Agenda setting, opening of visit, closing of visit | 6.38 |
| Gyn Genito Urinary | Gynecological and genitourinary problem | 4.72 |
| Physical Exam | Physical exam | 3.41 |
| Family | Family and significant other | 3.10 |
| Health Care System | Health care system | 2.89 |
| Work Leisure | Work and leisure activities | 2.73 |
| Tests | Tests and diagnostic procedures | 2.59 |
| Cigarette | Cigarette | 2.43 |
| Weight | Weight | 2.38 |
| Dizzy Dent Hear Vision | Dizzyness, vision, hearing, dental issues | 2.03 |
| Other | Other (various rare topics) | 1.94 |
| Exercise | Exercise | 1.89 |
| Depression | Depression | 1.86 |
| Medication | Medications | 1.84 |
| SmallTalk | Small talk | 1.72 |
| General Anxieties | General anxieties and worries | 1.38 |
| MDLife | Physician personal life | 1.04 |
| Diet | Diet, food (exclude supplements) | 0.69 |
| Alcohol | Alcohol | 0.57 |
| Sleep | Sleep | 0.53 |
| Therapeutic Intervention | Therapeutic intervention | 0.33 |
| Risky Behavior | Risky behaviors (eg, international travel, weapons at home) and risk avoidance preventive practices (eg, safe sex, wearing seatbelt or bike helmet) | 0.32 |
| OtherAddictions | Caffeine, or other addictions | 0.21 |
| Age | Age | 0.17 |
Text preprocessing
We applied a number of preprocessing steps to convert the dialog text into a set of tokenized words. We first replaced the patient names and numbers with -NAME- and -NUMBER- tokens to remove potentially identifiable information. After removing symbols, other than a set of punctuation symbols, such as “.”, “?”, “-”, the sentences in each talk-turn were tokenized into a list of words using the standard Python NLTK tokenizer.50
For the models that used bag-of-words encoding, the vocabulary included all unigrams and bigram noun phrases that occurred at least 5 times in the corpus, except for 2 sets of stopwords (see Supplementary Appendix A for more information), resulting in a vocabulary of size . For our neural network models, the vocabulary consisted of all unigrams, with neither set of stopwords removed (as is customary in neural network models) with a final vocabulary size of including an unknown token.
Representing talk-turns for classification models
The data for each visit , , is represented as a sequence of labeled talk-turns , with talk-turns in the th visit, . Let and represent the list of word tokens and the topic label, respectively, for the th talk-turn in the th visit. As mentioned earlier, can take values from 1 to 27, corresponding to each of the 27 unique topics. Each word in is encoded as a binary vector (“one-hot encoding”) of length , where is the vocabulary size. For example, if a word occurs in a talk-turn and the ID in the vocabulary for the word is 10, then the binary/one-hot-encoded vector becomes a vector of length , where the 10th entry of the binary word vector is set to 1 and all other entries are set to zero.
For the majority of the classification models we evaluated, the binary word vectors in each talk-turn were aggregated into a single talk-turn vector by adding the individual binary word vectors (also known as a bag-of-words encoding) and reweighting using tf-idf weights, a common text preprocessing step that downweights common and uninformative words. The vector represents each talk-turn and has dimensionality equal to the size of the vocabulary .
For our neural network models, we used a different representation as follows. We generated one vector per talk-turn using a network composed of an embedding layer and a bidirectional set of gated recurrent units (GRUs). The embedding layer, initialized with pretrained GloVe51 vectors, takes each binary word vector in the talk-turn and maps it to a dense embedded vector representation. The GRU component takes the sequence of embedded vectors within a talk-turn (1 embedded vector per word) and produces a single fixed-dimensional vector to represent the talk-turn. This approach has been shown to be useful in NLP applications for encoding variable-length sequential information from words in a sentence (talk-turn) into a fixed-dimensional vector that can be used as a feature vector for downstream classification.52 We used 128 for the GRU unit size, and the output talk-turn vector had size 256 because the GRU outputs in 2 directions are concatenated.
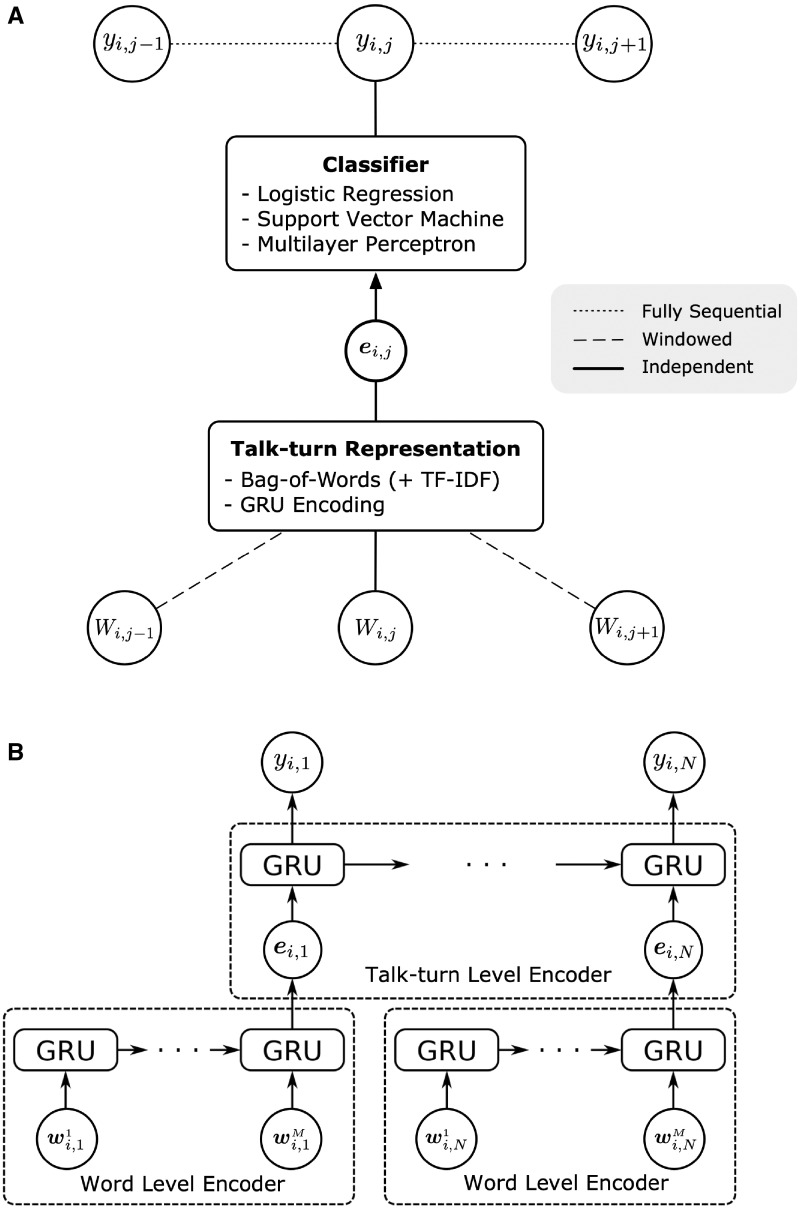
Given the talk-turn vectors , we classify each talk-turn either independently or by using sequential information across talk-turns. Figure 2A provides a high-level overview of the 3 primary different types of models we explored: 1) independent models that classify each talk-turn only using the words in talk-turn , 2) window-based models that also use words from a window of talk-turns both before and after talk-turn , and 3) fully sequential models that also consider the topic labels (or predictions) of talk-turns before and after when predicting a topic for talk-turn . In addition, we consider another type of sequential model that uses the talk-turn–level GRUs on top of word level GRU outputs, which is depicted in Figure 2B. We describe each of the 3 approaches in more detail below. Additional implementation details can be found in Supplementary Appendix C.
Figure 2.
(A) A high-level diagram of the various models discussed in the article. i, j stands for talk-turn j in session i. Wi, j is the list of tokenized words in talk-turn j. For each talk-turn j, we first generate the vectorized talk-turn representation ei, j, and the talk-turn representation ei, j is used as an input to different classifiers to predict the topic label yi, j, which is the topic label for talk-turn j. Windowed models use the adjacent talk-turns to create the talk-turn–level representation, and the fully sequential models make use of the sequential dependencies between the topic labels. (B) Simplified diagram of the hierarchical gated recurrent units (Hier-GRU). Each one-hot-encoded word vector in talk-turn j in visit i (shown as wi, j k for the k-th word in talk-turn j) is fed into the word level encoder to get a talk-turn representation ei, j, which becomes the input to the talk-turn–level encoder. The model has dependencies at the hidden state of talk-turn–level gated recurrent units (GRUs). In our experiments, both encoders were bidirectional.
Independent models
Independent models classify each talk-turn by using only the words in that talk-turn, , independently of the other talk-turns. The independent models used in our study were the logistic regression (LR) classifiers, support vector machines (SVMs), and feedforward neural networks with a single hidden layer. The LR and SVM classifiers used the bag-of-words vectors with tf-idf weights as input to predict the topic label . For the feedforward neural network, the output talk-turn vectors of the bidirectional GRU units were used as inputs, and the softmax function was used for the activation function in the perceptron. The parameters (weights) of the embedding layer, the GRU, and the feedforward neural network were all trained together as a single network, and we refer to this model as independent GRU.
Window-based models
Instead of using a single talk-turn vector as an input (as in the independent models), for the window-based models, we concatenated the adjacent vectors before and after each talk-turn. The input vector was defined as , with dimension , where is the size of the vocabulary for bag-of-words representations. This windowed input vectors were then used as input to either the LR or SVM classifier. In Figure 2, the window-based approach is indicated with dashed lines at the input level.
The window-based representation captures potentially useful sequential information across talk-turns in a manner not available to the independent models—but at the cost of having on the order of times as many parameters. The approach is particularly useful when making topic predictions for short talk-turns with very little information—information from neighboring talk-turns can be used to help to make predictions in such cases. We used in our experiments based on the knowledge that the transcripts were labeled with the convention that each topic label must span at least 2 exchanges (4 talk-turns).
Fully sequential models
To model the sequential dependencies between the topic labels, we used linear-chain CRFs and hidden Markov models (HMMs). The linear-chain CRF is widely used for sequence labeling tasks such as named-entity recognition or part-of-speech tagging48,53—here, we applied it to the problem of predicting the topic label of a talk-turn, given a sequence of talk-turns. The HMMs are constructed by using the output class label probabilities from each of the independent models discussed above. We converted the class probabilities from the classifiers, , to emission probabilities, (needed by the HMM), by using the fact that , given that the talk-turn probabilities do not depend on a particular topic . The emission probabilities are combined with the Markov transition probabilities (which can be directly estimated from label sequences in the training data) via the Viterbi algorithm to compute the sequence of topic labels that has the highest joint probability across talk-turns, conditioned on the observed talk-turn words.
We also found that using speaker-specific transition matrices improved the accuracy of our sequential models. Not surprisingly, providers tend to start new topics during a conversation more than the patients do. The figure in Supplementary Appendix B shows the percentage of time that a particular speaker (provider, patient, or other) starts each topic. To incorporate speaker information in the HMM approach, we augmented the standard HMM to use 2 topic transition matrices, 1 for the provider and 1 for the patient or other speakers. Each transition matrix corresponds to the speaker of the state that the HMM is transitioning to (eg, one transition matrix for transitioning to provider and the other for transitioning to all others). The decoding process in the Viterbi algorithm is modified so that it uses the appropriate matrix depending on the speaker.
Another type of sequential model that is entirely neural-network-based does not have direct dependencies between the topic labels, but has bidirectional connections between the hidden states of the talk-turn–level GRUs. The model, which we also refer to as Hier-GRU, has a hierarchical structure having 2 different GRUs, one at the word level to generate talk-turn–level representation, and the other which takes the talk-turn vectors as inputs to predict the output label for each talk-turn . Similar to independent GRU, the talk-turn–level GRU output is connected to a fully-connected layer and then a softmax to make prediction.
RESULTS
Experimental methods
We evaluated all models using 10-fold cross-validation and computed evaluation metrics both at the talk-turn level and at the visit level. At the talk-turn level, we computed the classification accuracy by comparing 1) the predicted topic from a model with 2) the human-generated topic for the talk-turn. To obtain results at the visit level, we aggregated the predictions from the individual talk-turns within each visit to generate a visit-level binary-valued prediction vector of dimension 27 (the number of topic labels) with 1 for topic label if the model predicted topic for 1 or more talk-turns in the visit, and 0 otherwise. Using such a vector for each visit, we calculated the accuracy, precision, recall, and F1 scores. The metrics were micro-averaged by globally counting the true positives, false positives, and so on, for all the topic labels.
To evaluate the performance of the classifiers, we compared the results with those of simple baseline models that predict the most common topics. The baseline at the talk-turn level just predicts the most common topic in the corpus, BiomedHistory (as shown in Table 1). At the visit level, the baseline always predicts the set of topic labels that occur in 50% or more of all visits, irrespective of the words within each visit.
Summary of experimental results
Table 2 shows the classification accuracies at the talk-turn level for independent models, windowed models, and sequential models. The most accurate independent model is the GRU and the most accurate windowed model is the windowed LR. The models with sequential information clearly outperform independent models with the Hier-GRU yielding the highest accuracy of 61.77% over all the models. The improvement in accuracy of Hier-GRU and HMM-GRU are both statistically significant, with , relative to each of the independent GRU and windowed LR models (using dependent t tests for paired samples across the 10 folds of cross-validation).
Table 2.
Accuracies for topic prediction at the level of talk-turns for different prediction models
| Model | Talk-turn level accuracy (%) | |
|---|---|---|
| Baseline | 29.85 | |
| Independent models | LR | 37.00 |
| SVM | 36.64 | |
| GRU | 38.85a | |
| Window-based models | Windowed LR | 51.12a |
| Windowed SVM | 50.46 | |
| Fully sequential models | CRF | 48.37 |
| HMM-LR | 56.89 | |
| HMM-SVM | 51.52 | |
| HMM-GRU | 57.60b | |
| Hier-GRU | 61.77ab | |
Micro-averaged precision and recall scores are the same as accuracy.
CRF: conditional random field; Hier-GRU: hierarchical gated recurrent units; HMM-GRU: hidden Markov model gated recurrent units; LR: logistic regression; SVM: support vector machine;
Highest talk-turn level accuracy in each model type.
Scores from the two best models.
The visit-level evaluation scores are shown in Table 3. Interestingly, the gap in performance (as measured by the micro-averaged F1 score) between independent, windowed, and sequential models is much smaller in the visit-level scores. The primary reason for this is that the independent models tend to have relatively high recall scores, whereas sequential models have relatively high precision scores.
Table 3.
Micro-averaged accuracy, precision, recall, and F1 scores, at the visit level, for different prediction models
| Visit level (%) |
|||||
|---|---|---|---|---|---|
| Model | Accuracy | Precision | Recall | F1 | |
| Baseline | 72.29 | 73.79 | 62.22 | 67.42 | |
| Independent models | LR | 75.19 | 67.91 | 84.25 | 75.15a |
| SVM | 72.45 | 63.40 | 90.40 | 74.50 | |
| GRU | 72.47 | 64.19 | 86.59 | 73.68 | |
| Window-based models | Windowed LR | 77.28 | 69.82 | 86.47 | 77.21 |
| Windowed SVM | 79.81 | 75.06 | 82.13 | 78.37a | |
| Fully sequential models | CRF | 74.19 | 80.43 | 58.42 | 67.64 |
| HMM-LR | 80.00 | 80.16 | 73.31 | 76.55 | |
| HMM-SVM | 75.21 | 78.70 | 60.90 | 68.63 | |
| HMM-GRU | 79.00 | 74.98 | 79.35 | 77.06 | |
| Hier-GRU | 78.96 | 73.69 | 82.43 | 77.78a | |
CRF: conditional random field; Hier-GRU: hierarchical gated recurrent units; HMM-GRU: hidden Markov model gated recurrent units; LR: logistic regression; SVM: support vector machine;
Highest F1 score in each model type.
The prediction models also can be evaluated at the level of individual topics to understand variability in prediction accuracy across topics. Precision, recall, and F1 scores were calculated by treating each topic label separately at the talk-turn level. Scores from the 2 best-performing models (Hier-GRU and HMM-GRU) are shown in Table 4, sorted by the percentage of each topic. The more common topics (that occur for example in at least 5% of the talk-turns) generally have higher F1 scores. However, there are some less common but highly specific topics, such as the cigarette topic (in 2.43% of talk-turns), that also have relatively high F1 scores.
Table 4.
Precision, recall, and F1 scores of each topic, calculated at the talk-turn level using Hier-GRU and HMM-GRU prediction results.
| Hier-GRU |
HMM-GRU |
||||||
|---|---|---|---|---|---|---|---|
| Label | Precision | Recall | F1 | Precision | Recall | F1 | Assigned topic (%) |
| BiomedHistory | 65.80 | 76.61 | 70.79a | 74.24 | 56.26 | 64.01 | 29.85 |
| PreventiveCare | 73.34 | 83.41 | 78.05a | 77.98 | 72.49 | 75.13a | 14.67 |
| MusSkePain | 67.48 | 69.14 | 68.30a | 67.68 | 64.27 | 65.93a | 8.30 |
| VisitFlowMgmt | 63.64 | 63.58 | 63.61a | 64.97 | 64.54 | 64.76a | 6.38 |
| GynGenitoUrinary | 67.62 | 54.76 | 60.51 | 72.15 | 57.66 | 64.09a | 4.72 |
| PhysicalExam | 48.91 | 52.33 | 50.56 | 50.21 | 63.20 | 55.96 | 3.41 |
| Family | 49.31 | 47.56 | 48.42 | 42.79 | 54.21 | 47.83 | 3.10 |
| HealthCareSystem | 37.81 | 28.85 | 32.73 | 46.10 | 39.49 | 42.54 | 2.89 |
| WorkLeisureActivity | 47.26 | 46.20 | 46.72 | 52.90 | 53.14 | 53.02 | 2.73 |
| TestDiagnostics | 49.67 | 32.43 | 39.24 | 37.24 | 52.88 | 43.70 | 2.59 |
| Cigarette | 71.38 | 85.84 | 77.94a | 81.38 | 72.73 | 76.81a | 2.43 |
| Weight | 49.54 | 52.60 | 51.02 | 47.20 | 46.77 | 46.98 | 2.38 |
| Other | 21.42 | 9.59 | 13.25 | 14.45 | 15.20 | 14.81 | 2.03 |
| Depression | 50.82 | 64.27 | 56.76 | 45.71 | 44.24 | 44.96 | 1.94 |
| DizzyDentHearVision | 23.08 | 14.01 | 17.44 | 46.96 | 50.83 | 48.82 | 1.89 |
| Medication | 38.97 | 27.09 | 31.96 | 22.46 | 64.29 | 33.29 | 1.86 |
| Exercise | 56.94 | 59.23 | 58.06 | 45.42 | 67.01 | 54.14 | 1.84 |
| SmallTalk | 20.61 | 18.70 | 19.61 | 32.79 | 31.09 | 31.92 | 1.72 |
| GeneralAnxieties | 32.51 | 18.63 | 23.68 | 16.36 | 32.86 | 21.84 | 1.38 |
| MDLife | 33.81 | 14.19 | 19.99 | 12.79 | 11.69 | 12.22 | 1.04 |
| Diet | 27.15 | 19.62 | 22.78 | 27.52 | 57.10 | 37.14 | 0.69 |
| Alcohol | 56.08 | 36.12 | 43.94 | 52.19 | 64.91 | 57.86 | 0.57 |
| Sleep | 13.40 | 2.33 | 3.97 | 29.80 | 40.32 | 34.27 | 0.53 |
| TherapeuticIntervention | 0.00 | 0.00 | 0.00 | 2.04 | 5.30 | 2.95 | 0.33 |
| RiskyBehavior | 33.90 | 5.87 | 10.00 | 44.87 | 41.06 | 42.88 | 0.32 |
| OtherAddictions | 0.00 | 0.00 | 0.00 | 23.55 | 41.40 | 30.02 | 0.21 |
| Age | 0.00 | 0.00 | 0.00 | 13.23 | 20.00 | 15.93 | 0.17 |
The rows are sorted by the percentage of talk-turns of each topic. In general, the more frequently discussed topics have higher F1 scores.
Hier-GRU: hierarchical gated recurrent units; HMM-GRU: hidden Markov model gated recurrent units.
Highest F1 scores.
DISCUSSION
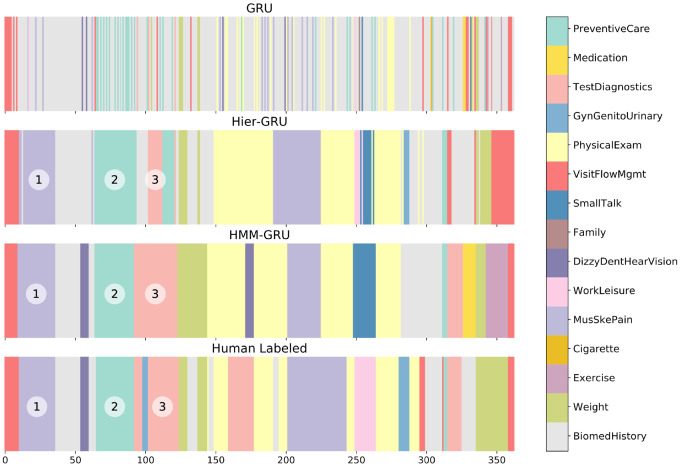
Using sequential information across talk-turns in predictive models systematically leads to more accurate predictions, particularly when predicting topic labels for talk-turns. To illustrate this point in more detail, Figure 3 shows sequences of predicted and human-assigned topic labels, for one particular visit, where different colors represent different topic labels. The top plot is from the independent GRU model. The lack of sequential information in the model leads to predictions that are noisy and lack the sequential smoothness of the human-assigned labels (bottom plot). The second plot is from the Hier-GRU model, and the third plot shows the predicted sequence of topics from the Viterbi parse of the HMM-GRU model (ie, the probabilistic predictions from the same GRU model in the top plot, but which are now sequentially smoothed by the HMM transition matrices). It is visually apparent (not only for this visit but for all visits) that the sequential models (second and third) are much more similar to the human labeling (bottom) than the independent model (top).
Figure 3.
Sequences of color-coded topic labels for one of the visits in our dataset. The upper plot shows the predicted topic labels from an independent model, and the center 2 plots show those from fully sequential models. The lower plot corresponds to the human-coded labels. The segments for the MusSkePain topic (1) had lengths of 23 talk-turns for hierarchical gated recurrent units (Hier-GRU), 27 for hidden Markov model gated recurrent units (HMM-GRU), and 26 for human labeled. Similarly, (2) the PreventiveCare segments had lengths 30, 28, and 27, and (3) the TestDiagnostics segment had lengths 10, 31, and 22 in talk-turns, respectively, for Hier-GRU, HMM-GRU, and human labeled. See Supplementary Appendix E for the boxplots of topic segment lengths for the 4 sequences of labels.
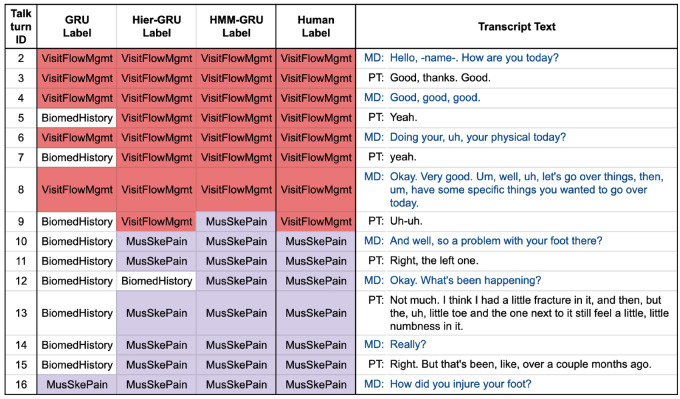
Figures 4 and 5 provide a more detailed look at portions of the transcript corresponding to Figure 3. We see for example that there are quite a few short talk-turns that have no words with topic-relevant information, such as “Yeah” (talk-turns 5 and 7 in Figure 4 and 222 in Figure 5) and “Okay” (talk-turn 224 in Figure 5). The sequential models are able to use the context information to assign these talk-turns to the same topic as the human labeler. The independent GRU model, however, does not have any context and assigns these talk-turns by default to the topic with the highest marginal probability (BiomedHistory).
Figure 4.
The beginning part of the visit shown in Figure 3. Each talk-turn is presented with predicted labels from 3 different models (independent gated recurrent units [GRU], hierarchical gated recurrent units [Hier-GRU], and hidden Markov model gated recurrent units [HMM-GRU]) and the human-coded labels. For the short talk-turns the BiomedHistory topic label is predicted quite often by the independent GRU, while the 2 other models produce label sequences that are more similar to human-coded labels. MD: medical doctor or provider; PT: patient.
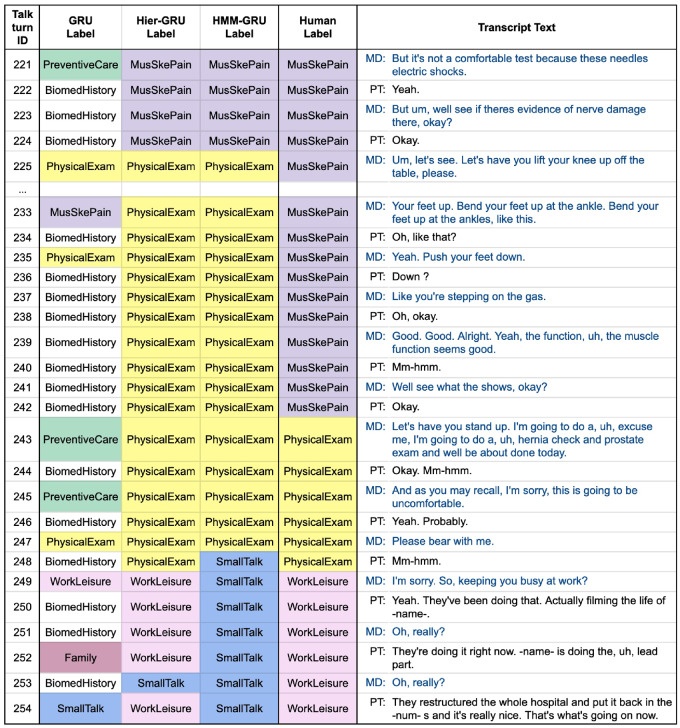
Figure 5.
Another excerpt from the same visit in Figure 3. Topics that are semantically similar are confusable (PhysicalExam and MusSkePain in talk-turns 233-242, and SmallTalk and WorkLeisure in talk-turns 249-254). GRU: gated recurrent units; Hier-GRU: hierarchical gated recurrent units; HMM-GRU: hidden Markov model gated recurrent units; MD: medical doctor or provider; PT: patient.
While the smoothing in sequential models helps to improve prediction accuracy, it can also produce errors due to oversmoothing. In particular, we found that the HMM-GRU model tends to predict longer topic segments relative to the human-labeled results, as can be seen in talk-turns 100 to 200 of the visit in Figure 3. The human labels contain short bursts of topics GynGenitoUrinary and BiomedHistory that are not detected by the HMM-GRU model. This is further quantified by the visit-level results in Table 3, where the recall scores of the fully sequential models are systematically lower than the independent models, and the reverse for the precision scores.
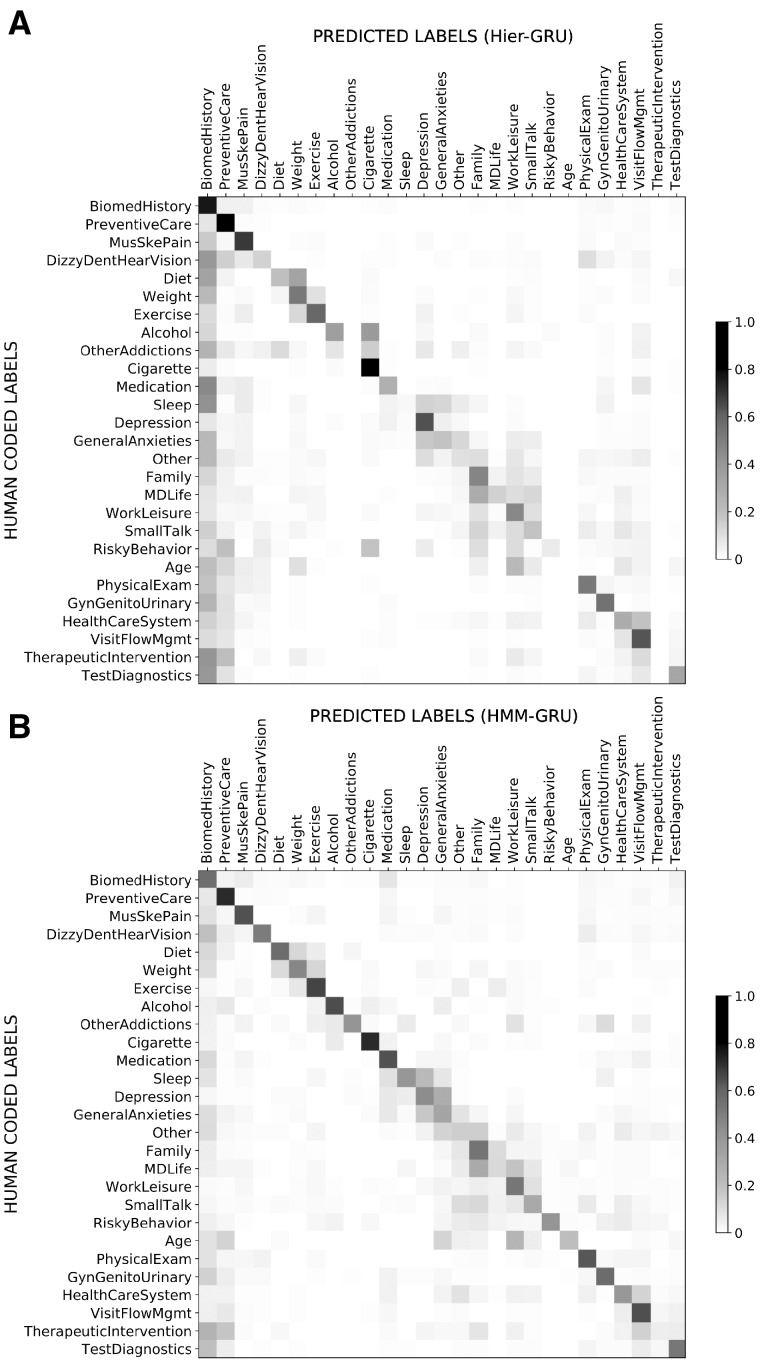
We also observed that some topics are semantically similar and easily confusable. For example, in Figure 5, from talk-turn 233 to 242, the 2 sequential models predict the topic PhysicalExam, while the human labeled MusSkePain—from the corresponding transcript text either prediction seems reasonable. Similarly, from talk-turn 249 to 254 the HMM-GRU predicts SmallTalk, while the human labeled WorkLeisure—from the text the corresponding talk-turns appear to be a mixture of both. We also found other examples across the corpus where the model frequently gets confused among small groups of related topics (eg, GeneralAnxieties and Depression; Weight and Diet). The full confusion matrices are shown in Figure 6. There is inevitably a subjective aspect to the human labeling, suggesting that there is likely to be a performance ceiling in terms of the accuracy of any algorithm relative to human labels on this data.
Figure 6.
Confusion matrices generated by (A) hierarchical gated recurrent units (Hier-GRU) and (B) hidden Markov model gated recurrent units (HMM-GRU), where the intensity of each cell shows the conditional probabilities of p(predicted label | human-coded label) and each row sums to 1. A number of subsets of topics have high confusion probabilities, including Diet/Weight/Exercise, Depression/GeneralAnxieties, and Family/MDLife/WorkLeisure.
From the topic-specific results in Table 4 we can see that while the predictions are relatively accurate for some topics, for others (eg, Age, TherapeuticIntervention, OtherAddictions, Other, MDLife), the scores are quite low. The broad nature of these topics is a likely contributor to the low accuracies, but the relative lack of training data per topic may also be another contributing factor. These topics account for roughly 1% (or less) of talk-turns in the corpus and many of these talk-turns are relatively short with little topical content, leading to relatively less signal, particularly for training neural network models. One possible approach to improve accuracy would be to incorporate additional external information relevant to these topics, such as incorporating lists of relevant words from ontological sources such as Unified Medical Language System into the training of prediction models, or adding relevant information from sources such as physician or specialist notes.
CONCLUSION
Patient-provider communication is an essential component of health care. In this context, prediction models that annotate patient-provider transcripts can in principle provide both useful information about the nature of topics discussed in specific conversations as well as contribute to a broader understanding of patient-provider communication. We have demonstrated that machine learning methods show promise for building models that can automatically predict discussion topics in dialog at the talk-turn and visit level. In particular, using a large real-world patient-provider dialog corpus, we investigated the performance of a variety of classification models including LR, SVMs, feedforward neural network model with GRUs, hierarchical GRUs, CRFs, and HMMs. We found that sequential models (eg, Hier-GRU and HMM-GRU) are more accurate compared with nonsequential models for predicting topic labels for talk-turns. In addition, we found that semantic similarity of discussion topics can be a significant contributor to prediction error.
While additional research and model improvement is needed, our results show promise for a number of medical topics that are critical quality indicators in primary care (eg, cigarette smoking, pain). Potential applications might include exploring systems that incorporate prior information from a list of problem areas or prior diagnoses found in the medical record. For example, the presence or absence of smoking cessation counseling during primary care encounters may inform population health management programs aimed at helping patients quit smoking. Deployment of systems such as this in real-world primary care may also be useful for obtaining the scale of data needed to improve model performance.
FUNDING
This work was supported by Patient-Centered Outcomes Research Institute number ME-1602-34167. Zac E Imel is the one who received the PCORI grant as an overall PI of the project.
AUTHOR CONTRIBUTIONS
All authors assisted in the writing of the manuscript. JP carried out the analyses and was responsible for writing the Materials and Methods and Results sections, with assistance from DK, RLL, SS, and EK-T. PS supervised the analyses. MT and PK were responsible for data management and cleaning along with JP. JEL and MT-S were responsible for collection of source data and description of data and human labeling. ZI, MT-S, PS, and DCA jointly conceived of the project, and ZEI, KM, and PK wrote the introduction.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Mary Ann Cook for helpful comments and feedback, Keith Gunnerson and Taylor Shuman for assistance in formatting of the data, and Abhishek Jindal for useful discussions on sequential models.
Conflict of interest statement
ZEI, DCA, and MT are co-founders with equity stakes in a technology company, Lyssn.io, focused on tools to support training, supervision, and quality assurance of psychotherapy and counseling. Also, within the past five years, PS has received research funding from SAP, Qualcomm, eBay, Google, Adobe, and Cylance, and has served as a consultant for Toshiba, Samsung, General Motors, and First American.
REFERENCES
- 1. Simon HB. The write stuff: how good writing can enhance patient care and professional growth. Am J Med 2013; 1266: 467–71. [DOI] [PubMed] [Google Scholar]
- 2. Hewett DG, Watson BM, Gallois C, et al. Communication in medical records: intergroup. Language and patient care . J Lang Soc Psychol 2009; 282: 119–38. [Google Scholar]
- 3. Curtis JR, Sathitratanacheewin S, Starks H, et al. Using electronic health records for quality measurement and accountability in care of the seriously ill: opportunities and challenges. J Palliat Med 2018; 21: S52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hsiao WC, Yntema DB, Braun P, et al. Measurement and analysis of intraservice work. JAMA 1988; 26016: 2361–70. [PubMed] [Google Scholar]
- 5. White A, Danis M.. Enhancing patient-centered communication and collaboration by using the electronic health record in the examination room. JAMA 2013; 30922: 2327–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singh K, Meyer SR, Westfall JM.. Consumer-facing data, information, and tools: self-management of health in the digital age. Health Aff (Millwood) 2019; 383: 352–8. [DOI] [PubMed] [Google Scholar]
- 7. Shachak A, Reis S.. The impact of electronic medical records on patient-doctor communication during consultation: a narrative literature review. J Eval Clin Pract 2009; 154: 641–9. [DOI] [PubMed] [Google Scholar]
- 8. Ventres W, Kooienga S, Vuckovic N, et al. Physicians, patients, and the electronic health record: an ethnographic analysis. Ann Fam Med 2006; 42: 124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sinsky CA. On presence: a tale of two visits. December 29, 2016. https://catalyst.nejm.org/electronic-health-record-tale-two-visits/ Accessed June 27, 2019.
- 10. Tai-Seale M, Olson CW, Li J, et al. Electronic health record logs indicate that physicians split time evenly between seeing patients and desktop medicine. Health Aff (Millwood) 2017; 364: 655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arndt BG, Beasley JW, Watkinson MD, et al. Tethered to the EHR: primary care physician workload assessment using EHR event log data and time-motion observations. Ann Fam Med 2017; 155: 419–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shanafelt TD, Hasan O, Dyrbye LN, et al. Changes in burnout and satisfaction with work-life balance in physicians and the general US working population between 2011 and 2014. Mayo Clin Proc 2015; 9012: 1600–13. [DOI] [PubMed] [Google Scholar]
- 13. Tai-Seale M, Dillon E, Yang Y, et al. Physicians’ well-being linked to in-basket messages generated by algorithms in electronic health records. Health Aff (Millwood) 2019; 38:1073–78.. [DOI] [PubMed] [Google Scholar]
- 14. Friedberg MW, Chen PG, Van Busum KR, et al. Factors affecting physician professional satisfaction and their implications for patient care, health systems, and health policy. Rand Health Q 2014; 3: 1.. [PMC free article] [PubMed] [Google Scholar]
- 15. Thielke S, Hammond K, Helbig S.. Copying and pasting of examinations within the electronic medical record. Int J Med Inform 2007; 76 Suppl 1: S122–8. [DOI] [PubMed] [Google Scholar]
- 16. Hammond KW, Helbig ST, Benson CC, et al. Are electronic medical records trustworthy? Observations on copying, pasting and duplication. AMIA Annu Sympos Proc 2003; 2003: 269–73. [PMC free article] [PubMed] [Google Scholar]
- 17. Chiu C-C, Tripathi A, Chou K, et al. Speech recognition for medical conversations. Proc Interspeech 2018; 2018: 2972–6. [Google Scholar]
- 18. Rajkomar A, Kannan A, Chen K, et al. Automatically charting symptoms from patient-physician conversations using machine learning. JAMA Intern Med 2019; 1796: 836–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Elwyn G, Barr PJ, Grande SW.. Patients recording clinical encounters: a path to empowerment? Assessment by mixed methods. BMJ Open 2015; 58: e008566.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barr PJ, Dannenberg MD, Ganoe CH, et al. Sharing annotated audio recordings of clinic visits with patients-development of the open recording automated logging system (ORALS): study protocol. JMIR Res Protoc 2017; 67: e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hill RG Jr, Sears LM, Melanson SW.. 4000 clicks: a productivity analysis of electronic medical records in a community hospital ED. Am J Emerg Med 2013; 3111: 1591–4. [DOI] [PubMed] [Google Scholar]
- 22. Verghese A, Shah NH, Harrington RA.. What this computer needs is a physician: humanism and artificial intelligence. JAMA 2018; 3191: 19–20. [DOI] [PubMed] [Google Scholar]
- 23. Hall JA, Roter DL, Katz NR.. Meta-analysis of correlates of provider behavior in medical encounters. Med Care 1988; 267: 657–75. [DOI] [PubMed] [Google Scholar]
- 24. Beck RS, Daughtridge R, Sloane PD.. Physician-patient communication in the primary care office: a systematic review. J Am Board Fam Pract 2002; 15: 25–38. [PubMed] [Google Scholar]
- 25. Mishler EG. The Discourse of Medicine: Dialectics of Medical Interviews. Westport, CT: Greenwood Publishing Group; 1984. [Google Scholar]
- 26. van Osch M, van Dulmen S, van Vliet L, et al. Specifying the effects of physician’s communication on patients' outcomes: a randomised controlled trial. Patient Educ Couns 2017; 1008: 1482–9. [DOI] [PubMed] [Google Scholar]
- 27. Hojat M. The interpersonal dynamics in clinician–patient relationships In: Hojat M, ed. Empathy in Health Professions Education and Patient Care. Cham, Switzerland: Springer International Publishing; 2016: 129–50. [Google Scholar]
- 28. Tai-Seale M, McGuire TG, Zhang W.. Time allocation in primary care office visits. Health Serv Res 2007; 425: 1871–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Foo PK, Frankel RM, McGuire TG, et al. Patient and physician race and the allocation of time and patient engagement efforts to mental health discussions in primary care. J Ambul Care Manage 2017; 403: 246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tai-Seale M, McGuire T, Colenda C, et al. Two-minute mental health care for elderly patients: inside primary care visits. J Am Geriatr Soc 2007; 5512: 1903–11. [DOI] [PubMed] [Google Scholar]
- 31. Eton DT, Ridgeway JL, Linzer M, et al. Healthcare provider relational quality is associated with better self-management and less treatment burden in people with multiple chronic conditions. Patient Prefer Adherence 2017; 11: 1635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Charon R, Greene MG, Adelman RD.. Multi-dimensional interaction analysis: a collaborative approach to the study of medical discourse. Soc Sci Med 1994; 397: 955–65. [DOI] [PubMed] [Google Scholar]
- 33. Tai-Seale M, Hatfield LA, Wilson CJ, et al. Periodic health examinations and missed opportunities among patients likely needing mental health care. Am J Manag Care 2016; 22: e350–7. [PMC free article] [PubMed] [Google Scholar]
- 34. Moyers TB, Martin T, Manuel JK, et al. Assessing competence in the use of motivational interviewing. J Subst Abuse Treat 2005; 281: 19–26. [DOI] [PubMed] [Google Scholar]
- 35. Caperton DD, Atkins DC, Imel ZE.. Rating motivational interviewing fidelity from thin slices. Psychol Addict Behav 2018; 324: 434–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Levinson W, Lesser CS, Epstein RM.. Developing physician communication skills for patient-centered care. Health Aff (Millwood) 2010; 297: 1310–8. [DOI] [PubMed] [Google Scholar]
- 37. Hoerger M, Epstein RM, Winters PC, et al. Values and options in cancer care (VOICE): study design and rationale for a patient-centered communication and decision-making intervention for physicians, patients with advanced cancer, and their caregivers. BMC Cancer 2013; 131: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nadkarni PM, Ohno-Machado L, Chapman WW.. Natural language processing: an introduction. J Am Med Inform Assoc 2011; 185: 544–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deleger L, Molnar K, Savova G, et al. Large-scale evaluation of automated clinical note de-identification and its impact on information extraction. J Am Med Inform Assoc 2013; 201: 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roberts K, Harabagiu SM.. A flexible framework for deriving assertions from electronic medical records. J Am Med Inform Assoc 2011; 185: 568–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mork JG, Bodenreider O, Demner-Fushman D, et al. Extracting Rx information from clinical narrative. J Am Med Inform Assoc 2010; 175: 536–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dernoncourt F, Lee JY, Uzuner O, et al. De-identification of patient notes with recurrent neural networks. J Am Med Inform Assoc 2017; 243: 596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mayfield E, Laws MB, Wilson IB, et al. Automating annotation of information-giving for analysis of clinical conversation. J Am Med Inform Assoc 2014; 21 (e1): e122–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Beach MC, Saha S, Korthuis PT, et al. Patient-provider communication differs for black compared with white HIV-infected patients. AIDS Behav 2011; 154: 805–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Laws MB, Beach MC, Lee Y, et al. Provider-patient adherence dialogue in HIV care: results of a multisite study. AIDS Behav 2013; 171: 148–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kotov A, Hasan M, Carcone A, et al. Interpretable probabilistic latent variable models for automatic annotation of clinical text. AMIA Annu Symp Proc 2015; 2015: 785–94. [PMC free article] [PubMed] [Google Scholar]
- 47. Hasan M, Kotov A, Carcone A, et al. A study of the effectiveness of machine learning methods for classification of clinical interview fragments into a large number of categories. J Biomed Inform 2016; 62: 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wallace BC, Laws MB, Small K, et al. Automatically annotating topics in transcripts of patient-provider interactions via machine learning. Med Decis Mak 2014; 344: 503–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gaut G, Steyvers M, Imel ZE, et al. Content coding of psychotherapy transcripts using labeled topic models. IEEE J Biomed Health Inform 2017; 212: 476–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bird S, Klein E, Loper E.. Natural Language Processing with Python: Analyzing Text with the Natural Language Toolkit. Newton, MA: O’Reilly Media, Inc; 2009. [Google Scholar]
- 51. Pennington J, Socher R, Manning CD.. GloVe: global vectors for word representation. In: Proceedings of Empirical Methods in Natural Language Processing (EMNLP); 2014: 1532–43. [Google Scholar]
- 52. Goldberg Y. Neural Network Methods in Natural Language Processing (Synthesis Lectures on Human Language Technologies). San Rafael, CA: Morgan & Claypool; 2017. [Google Scholar]
- 53. Lafferty JD, McCallum A, Pereira F.. Conditional random fields: probabilistic models for segmenting and labeling sequence data In: Proceedings of the Eighteenth International Conference on Machine Learning. Burlington, MA: Morgan Kaufmann; 2001: 282–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.