Abstract
Toxoplasma gondii is an important human and veterinary pathogen and the causative agent of toxoplasmosis, a potentially severe disease especially in immunocompromised or congenitally infected humans. Current therapeutic compounds are not well-tolerated, present increasing resistance, limited efficacy and require long periods of treatment. On this context, searching for new therapeutic targets is crucial to drug discovery. In this sense, recent works suggest that N-myristoyltransferase (NMT), the enzyme responsible for protein myristoylation that is essential in some parasites, could be the target of new anti-parasitic compounds. However, up to date there is no information on NMT and the extent of this modification in T. gondii. In this work, we decided to explore T. gondii genome in search of elements related with the N-myristoylation process. By a bioinformatics approach it was possible to identify a putative T. gondii NMT (TgNMT). This enzyme that is homologous to other parasitic NMTs, presents activity in vitro, is expressed in both intra- and extracellular parasites and interacts with predicted TgNMT substrates. Additionally, NMT activity seems to be important for the lytic cycle of Toxoplasma gondii. In parallel, an in silico myristoylome predicts 157 proteins to be affected by this modification. Myristoylated proteins would be affecting several metabolic functions with some of them being critical for the life cycle of this parasite. Together, these data indicate that TgNMT could be an interesting target of intervention for the treatment of toxoplasmosis.
Keywords: Toxoplasma gondii, protein myristoylation, N-myristoyltransferase, myristoylome, calcium homeostasis
Graphical Abstract
1. INTRODUCTION
N-myristoylation is a lipid modification that is estimated to affect between 1–4% of all proteins in higher eukaryotes (Martinez, et al., 2008). This modification is characterized by the covalent attachment of the fourteen-carbon fatty acid myristate to an essential N-terminal glycine residue via an amide bond after the removal of the initiator methionine. The N-myristoylation process is catalyzed by myristoyl-CoA:protein N-myristoyltransferase (NMT), a member of the GCN5-related N-acetyltransferases (GNAT) superfamily of proteins (Farazi, et al., 2001). The consensus sequence recognized by N-myristoyltransferase (NMT) is M-G-X-X-X-S/T/C where X represents most amino acids except for Proline (P), aromatic or charged residues in position X3 and preferentially a lysine or arginine residue at position 7 and/or 8 (Farazi, et al., 2001). This protein modification plays a direct role in cellular signaling and subcellular targeting by promoting membrane binding and modulating protein-protein interactions (Wright, et al., 2010). Currently, our knowledge of protein myristoylation has expanded to understand that the mechanism of protein myristoylation is well conserved in eukaryotic organisms including relevant human pathogens such as Plasmodium spp. and trypanosomatids like Trypanosma brucei, Trypanosoma cruzi and Leishmania spp. (reviewed in (Martin, et al., 2011)). Studies based on NMT inhibition in pathogens confirm that N-myristoylation is relevant for proliferation of these parasites since myristoylated proteins participate in cell replication, migration and host-cell invasion (Beck, et al., 2010, Frenal, et al., 2010). Furthermore, and contrary to what was observed for higher eukaryotes, parasites express only one NMT (Martin, et al., 2011, Wright, et al., 2014). In this manner, this acyltransferase is considered a good potential target for the development of new treatments for these and other pathogens (Wright, et al., 2014).
Toxoplasma gondii is a human and veterinary pathogen member of Apicomplexa phylum (Montoya and Liesenfeld, 2004, Tenter, et al., 2000). This protozoan is considered of great relevance for the human health since its infection produces toxoplasmosis, an infectious disease that affects about one third of the global population with high morbidity and mortality rates especially in South and Central America and Continental Europe (Bangoura, et al., 2011, Furtado, et al., 2011, Meerburg and Kijlstra, 2009). Currently, the most effective T. gondii treatment is a combination of pyrimethamine and sulfadiazine, although both drugs are not well-tolerated and present increasing resistance (Montoya and Liesenfeld, 2004, Petersen, 2007). Due to this, is that there is an urgent need of finding new targets of intervention. Given the fact that parasites express only one NMT that seems to be essential for their viability is that TgNMT appears as a good target of intervention. However, the scope and role of protein myristoylation in T. gondii has not been addressed yet.
Up to present, there are no reports on the existence of any N-myristoyltransferase in T. gondii or on the importance and extent of protein myristoylation on this parasite. As such, in this work we screened the Toxoplasma database (www.toxodb.org) and found a single putative gene product that could act as an N-myristoyltransferase (TgNMT). We confirmed that this gene product presents myristoyltransferase activity in vitro and is expressed in the cytoplasm of intra- and extracellular parasites. Furthermore, NMT activity would be required for a normal lytic cycle since inhibition of myristoylation with 2-hydroxymyristic acid results in a reduced total lysis area when compared to controls. Finally, an in silico screen of the database showed that T. gondii myristoylome would encompass a total of 157 potential myristoylable proteins with some of them found to interact with TgNMT. Interestingly, other putative myristoylated proteins are associated to calcium regulation, which is critical for T. gondii life-cycle and thus, supporting the effect observed on the lytic cycle. Together, these results position TgNMT as an attractive drug target of intervention for the treatment of toxoplasmosis and at the same time suggest a complex interplay between protein myristoylation and calcium homeostasis.
2. MATERIALS AND METHODS
2.1. Antibodies and reagents
Specialized and common reagents were from Sigma, unless specified. ECL Plus was from GE Biosciences. Alexa-conjugated secondary antibodies were from Molecular Probes. Tissue culture reagents were from Invitrogen.
2.2. Toxoplasma and host-cell cultures
T. gondii tachyzoites of the RH ∆hxgprt strain (Donald, et al., 1996) were used throughout the study. Parasites were maintained by serial passage on confluent monolayers of human foreskin fibroblasts (HFFs) in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% v/v bovine serum albumin (BSA), 100 i.u. (international units) /ml penicillin and 100 µg/ml streptomycin. When required, freshly lysed tachyzoites were purified using a 3 µm polycarbonate filter.
2.3. Toxoplasma gondii database analysis
The search for putative N-myristoyltransferase gene products in the T. gondii genome was performed using the TGME49 strain with the interpro domain tool (ToxoDB v28), using the pfam (www.pfam.org) database as default. The screening was performed using the pfam 01233 towards the N-terminal end of the protein and pfam 02799 towards the C-terminal end.
For the generation of the in silico myristoylome, we used the ToxoDB v28 web page (www.toxodb.org). The TGME49 strain was screened using the “similarity/pattern tool”, utilizing a combination of the conserved myristoylation pattern and its exceptions (Martin, et al., 2011). The results were analyzed with the gene ontology (GO) analysis tool for molecular function, biological process and cellular component with a p-value of 0.05. The p-value was calculated with the Fisher’s exact test. Proteins that were experimentally determined to be palmitoylated in T. gondii are marked with (+). Polybasic domains found in the first 25 amino acids were determined by the ExPASy Compute pI/Mw tool (http://web.expasy.org/compute_pi/) and lipidic modifications excluding myristoylation were determined by GPS-Lipid v1.0 program (Xie, et al., 2016).
2.4. Phylogenetic analysis of the predicted NMT
Reference sequences related to TGME49_209160 amino acid sequences were obtained by a BLASTp search on Apicomplexa (taxid:5794), Homo sapiens (taxid:9606), Trypanosomatidae (taxid:5654) and Fungi (taxid:4751) databases at NCBI domain (Madden, 2013). Reference amino acid sequences from Trypanosoma brucei, Mus musculus and Droshopila melanogaster were added manually; the final set of amino acid sequences used is listed in Table S1. A total of 36 sequences were aligned using MAFFT (Multiple Alignment using Fast Fourier Transform) under default settings (Katoh and Standley, 2013). A multiple alignment was subsequently analyzed and cleaned using the trimAl software in automated 1 mode (Capella-Gutierrez, et al., 2009). The trimmed alignments are shown in Figure S1. The tree was constructed by a maximum likelihood algorithm with RaxML software using the Gamma Blosum62 Protein model and rapid bootstrapping method (Stamatakis, 2014). Artwork and annotation was done by the interactive Tree Of Life (iTOL) program v 4.3 (Letunic and Bork, 2016).
2.5. Enzymatic activity determination
NMT activity determination was carried out as described by Goncalves and coworkers (Goncalves, et al., 2012). Briefly, 6.8 nM of purified TgNMT (E. coli expressed recombinant protein obtained as described in supplementary material) was incubated with the reaction mixture in a final volume of 220 ml. This mixture contained 20 mM Na2HPO4/NaH2PO4 pH 7.4, 0.5 mM EDTA, 0.5 mM EGTA and 0.1 % Triton X-100 (v/v), 15 mM myristoyl-CoA and 8 mM CPM (7-diethylamino-3-(4-maleimido-phenyl)-4-methylcoumarin; a fluorescent probe). The reaction was initiated by the addition of 12 ml of a peptide containing-solution (final concentration 15 mM). The peptide substrate used corresponded to the N-terminal end of either pp60src (amino acid sequence: NH2-GSNKSKPK-CO-NH2 (King and Sharma, 1994)) or the putative T. gondii substrate TGME49_268960 (amino acid sequence: NH2-GSQTSNSR-CO-NH2). Both peptides were synthesized at GenScript, USA. The reaction was incubated at 25°C for 30 minutes a nd the fluorescence emission at 470 nm was determined (excitation at 384). The NMT activity is expressed as percentage of the relative fluorescence units obtained when pp60src is used as substrate. Determinations were carried out in 96 well black bottom microplates (FLUOTRACTM 200, Greiner) using Multi modal Synergy H1 (BioTek).
2.6. Indirect immunofluorescence studies
All the steps were carried out at room temperature. For intracellular parasites, 8×105 freshly lyzed tachyzoytes were allowed to invade a confluent monolayer of HFF cells. After 1-hour incubation at 37°C, media was discarded and the cells were washed twice with PBS. Cells were fixed with formaldehyde 4% v/v in PBS for 30 minutes followed by a 1 minute wash in PBS. Then, cells were permeabilized with 0.3% v/v Triton X-100 in PBS for 20 minutes and blocked with 3% w/v BSA in PBS for 30 minutes. After this, primary antibodies were diluted in 3% w/v BSA in PBS: mouse anti-SAG1:1/200 (kindly provided by Dr Clemente, INTECH, Argentina; (Albarracin, et al., 2015)) and rabbit anti-TgNMT: 1/1,000 (Supplemental material) and then incubated for 60 minutes followed by extensive washes. Secondary antibodies used were Alexa Fluor 488-conjugated goat anti-mouse IgG (1/1,000) and Alexa Fluor 594- conjugated goat anti-rabbit (1/1,000; Invitrogen). Finally, samples were mounted with Fluoromont (Abcam). When extracellular parasites were used (2×106), they were fixed on microscopy slides with formaldehyde 4% v/v in PBS for 30 minutes followed by a 1 minute wash in PBS. Then, the same protocol mentioned above was followed. Parasites were imaged using an Eclipse E600 microscope (Nikon).
2.7. Plaque assay
Freshly lysed purified parasites (200 parasites) were used to infect HFF cell monolayers seeded in 24-well plates in presence of 0 (vehicle), 6.25, 12.5, 25, 50 or 100 mM 2-hydroxy myristate (2HMA) in DMSO (final concentration 0.1% v/v). Then, infected cells were incubated for 7 days at 37°C, 5 % CO2 without movement. Cells were then fixed with 70% ethanol and stained with 0.1% crystal violet. Plaques were scanned using an Epson scanner. For plaque count and size, plaque images were opened with the ImageJ software, converted to grayscale and a threshold adjustment was performed to highlight plaque limits. The plaques area was approximated in pixels by the measure tool. Experiments were repeated twice independently with two replicates each time.
2.8. Cell proliferation assay
Monolayers of hTERT cells were grown in 96-well flat-bottom tissue culture microtiter plates at 37°C 5% CO2 and incubated with different concentrations of 2-hydroxy myristic acid (2HMA), supplied at a final concentration of 0.5 % v/v DMSO in complete DMEM. After 6 days, cells were washed and 0.5 mg/ml of MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide] added. Plates were kept 4 hours at 37°C, then cells disrupted and formazan product solubilized in DMSO 100 % and absorbance at 540 nm recorded using a 96-well plate reader (Synergy Mx BioTek). Values are presented as percentages relative to cells incubated without 2HMA (100% survival).
2.9. Generation of TgNMT interactome
Immunoprecipitation was performed as previously described by Adams et al. (Adams, 2002) with minor modifications. Briefly, freshly lysed tachyzoites (2×108) were resupended in 2 ml lysis buffer (Tris-HCl 50 mM, NaCl 120 mM and 0.5% v/v Nonidet P-40, pH8). Lysates were obtained by sonication and cellular debris was discarded by centrifugation for 15 minutes at 10,000 xg, 4°C. Then, the sample was divided in 2 halves adding in each half either 10 ml anti-TgNMT or 10 ml pre-immune sera. After 16–18 hour incubation at 4°C, protein A/G sepharose (previously equilibrated in lysis buffer) was added to both samples and incubation continued for another hour at 4°C. Then, immunocomplexes were was hed 5 times in lysis buffer and finally resuspended in 50 ml lysis buffer. Bound proteins were eluted by heating samples for 5 minutes at 80°C. An aliquot of the eluates (10% of total volume) were analyzed by SDS-PAGE and the rest acetone-precipitated to be analyzed by MudPIT.
2.10. MudPIT analysis of immunoprecipitates
Protein pellets were washed 2 times with 350 ml ice-cold acetone. Air-dried pellets were dissolved in 8 M urea/ 100 mM Tris-HCl pH 8.5. Proteins were reduced with 1 M Tris (2-carboxyethyl) phosphine hydrochloride and alkylated with 500 mM 2-chloroacetamide. Proteins were digested for 18 hr at 37 °C in 2 M urea, 100 mM Tris-HCl pH 8.5, 1 mM CaCl2 with 2 mg trypsin (Promega, Madison, WI). Digest was stopped with formic acid, 5% final concentration. Debris was removed by centrifugation, 30 minutes 18,000 xg.
A MudPIT microcolumn (Wolters, et al., 2001) was prepared by first creating a Kasil frit at one end of an undeactivated 250 mm ID/360 mm OD capillary (Agilent Technologies, Inc., Santa Clara, CA). The Kasil frit was prepared by briefly dipping a 20 – 30 cm capillary in well-mixed 300 ml Kasil 1624 (PQ Corporation, Malvern, PA) and 100 ml formamide, curing at 100°C for 4 hrs, and cutting the frit to ~2 mm in length. Strong cation exchange particles (SCX Partisphere, 5 mm dia., 125 Å pores, Phenomenex, Torrance, CA) were packed in-house from particle slurries in methanol 2.5 cm. An additional 2.5 cm reversed phase particles (C18 Aqua, 3 µm dia., 125 Å pores, Phenomenex) were then similarly packed into the capillary using the same method as SCX loading, to create a biphasic column. An analytical RPLC column was generated by pulling a 100 mm ID/360 mm OD capillary (Polymicro Technologies, Inc, Phoenix, AZ) to 5 mm ID tip. Reversed phase particles (Aqua C18, 3 mm dia., 125 Å pores, Phenomenex, Torrance, CA) were packed directly into the pulled column at 800 psi until 12 cm long. The MudPIT microcolumn was connected to an analytical column using a zero-dead volume union (Upchurch Scientific (IDEX Health & Science), P-720–01, Oak Harbor, WA).
LC-MS/MS analysis was performed using an Agilent Technologies 1200 HPLC pump and a Thermo LTQ-Orbitrap XL using an in-house built electrospray stage. MudPIT experiments were performed with steps of 0% buffer C, 20% buffer C, 50% buffer C, 100% C, and 90/10 % buffer C/B, being run for 4 minutes at the beginning of each gradient of buffer B. Electrospray was performed directly from the analytical column by applying the ESI voltage at a tee (150 mm ID, Upchurch Scientific; (Wolters, et al., 2001)). Electrospray directly from the LC column was done at 2.5 kV with an inlet capillary temperature of 275 OC. Data-dependent acquisition of MS/MS spectra with the LTQ –Orbitrap XL were performed with the following settings: MS/MS on the 10 most intense ions per precursor scan, 1 microscan, reject charge state 1; dynamic exclusion repeat count, 1, repeat duration, 30 second; exclusion list size 500; and exclusion duration, 60 second.
Protein and peptide identification and protein quantitation were done with Integrated Proteomics Pipeline - IP2 (Integrated Proteomics Applications, Inc., San Diego, CA. http://www.integratedproteomics.com/). Tandem mass spectra were extracted from raw files using RawConverter and were searched against Uniprot Toxoplasma protein database with reversed sequences using ProLuCID (He, et al., 2015, Peng, et al., 2003). The search space included all fully-tryptic peptide candidates. Carbamidomethylation (+57.02146) of cysteine was considered as a static modification. Peptide candidates were filtered using DTASelect (Tabb, et al., 2002).
3. RESULTS
3.1. Toxoplasma gondii encodes for a single putative N-myristoyltransferase
In order to identify sequences coding for proteins with N-myristoyltransferase activity in Toxoplasma gondii, a search using the TOXODB database was performed (www.toxodb.org; (Gajria, et al., 2008)). The screening was carried out using the PFAM consensus sequences typical of the NMT family of proteins (the PF01233, myristoyl-CoA binding region and the PF02799 substrate protein-binding region; (Weston, et al., 1998)). Only one protein, product of the TGME49_209160 gene was retrieved (E value = 9.0E−75). This suggests that only one copy of NMT is present in T. gondii (paralog count = 0), as is observed in many lower eukaryotes.
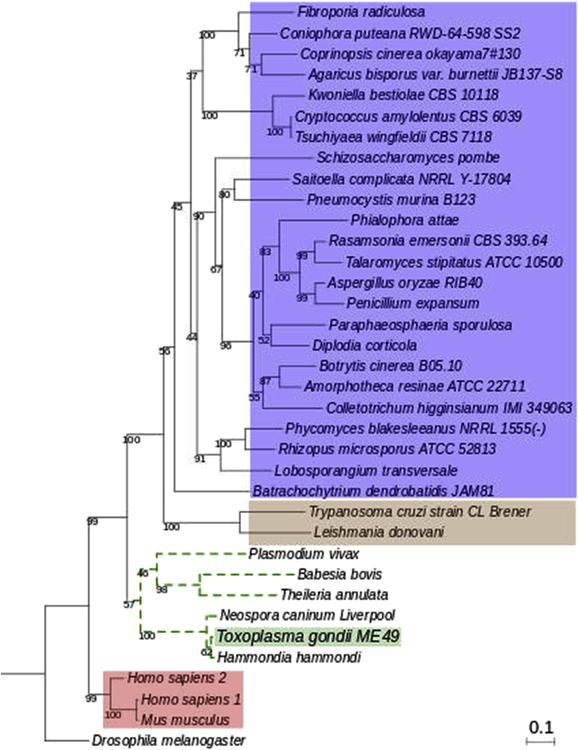
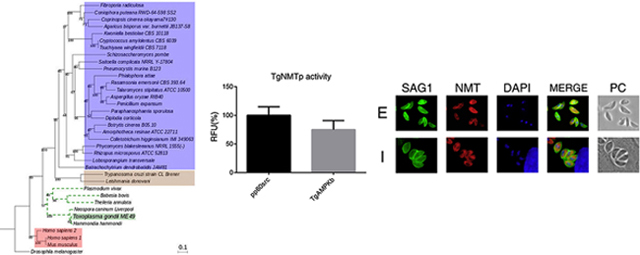
A phylogenetic analysis was generated with the deduced amino acid sequence of TGME49_209160 and myristoyl-CoA:protein N-myristoyltransferases (NMTs) from different eukaryotic organisms including parasites, fungi and mammals. Figure 1 shows that the predicted T. gondii NMT (TgNMTp) is in the same evolutionary branch as other members of the phylum Apicomplexa while sequences related to trypanosomatid parasites are grouped in a distant clade where sequences of fungi organisms are the most represented. This analysis highlights that humans NMTs (HsNMTs) and Mus musculus sequences are grouped in a distant clade from TgNMTp. This result suggests that there are significant evolutionary differences between these NMTs and that this difference could be of importance in attempts to design specific inhibitors.
Figure 1. Phylogenetic analysis of TGME49_209160 amino acid sequence.
Phylogenetic tree representing evolutionary relationships between the TGME49_209160 amino acid sequence and reference sequences of known and predicted NMTs of eukaryotic organisms such as other apicomplexans (dotted green lines), trypanosomatids (brown box), mammals (red box) and fungi (blue box). Sequence of Drosophila melanogaster was included as outgroup.
Multiple amino acid sequence alignments shows highly conserved regions between putative T. gondii NMT and NMTs described in other species (Figure S2). Notably, the residues that integrate the two PROSITE signature sequences of NMT enzymes (PS00975 at the N-terminal domain and PS0076 at the C-terminal domain), together with the presence of key amino acids for the catalysis, strongly suggest that TGME49_209160 codes for a functional myristoyl-CoA: protein N-myristoyltransferase.
3.2. TgNMTp is a functional N-myristoyltransferase that expresses throughout the lytic cycle
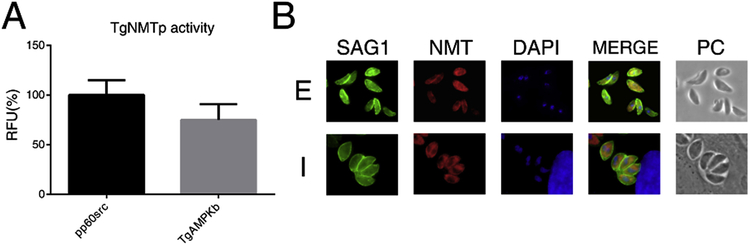
In order to determine if TgNMTp presents N-myristoyltransferase activity, TGME49_209160 coding sequence was successfully expressed in E. coli and the protein purified (Supplemental Material). TgNMTp, obtained with high degree of purity and homogeneity, was used for the determination of enzymatic activity and to generate specific antibodies (Supplemental material). The N-myristoyltransferase activity was assayed according to Goncalves et al. using two octapeptides as a substrates (Goncalves, et al., 2012). The first one, G-S-N-K-S-K-P-K-NH2, is a derivative of the N-terminal sequence of H. sapiens proto-oncogene tyrosine kinase pp60src. This peptide was used as a standard substrate since humans and S. cerevisiae NMTs were able to transfer the myristate moiety to their amino terminal Gly residue (Rocque, et al., 1993). The second octapeptide (G-S-Q-T-S-N-S-R-NH2) was synthesized based on the results obtained in our in silico myristoylome (Table S3). This putative myristoylable peptide corresponds to the N-terminal end of T. gondii TgAMPK β subunit protein (accession number ToxoDB: TGME49_268960). Figure 2A shows that TgNMTp presented myristoyl transferase activity towards both substrates.
Figure 2. Enzymatic activity and cellular localization of TgNMT.
(A) TgNMTp was incubated with either with a standard substrate (pp60src) or a putative T. gondii substrate (TgAMPKb; access number Toxodb: TGME49_268960) as described in the Material and Methods section. Activity is expressed as a percentage of the relative fluorescence units (RFU) obtained with pp60src. Appropriate blanks (without Myristoyl-CoA or substrate) are already subtracted. (B) Indirect Immunofluorescence assay using the anti-TgNMT serum (NMT, red) on intra- (I) or extracellular (E) parasites. Mouse anti-SAG1 (SAG1, green) was used to label the plasma membrane and DAPI was used to mark nuclei. Parasites were observed by phase-contrast microscopy (PC).
To determine TgNMT subcellular localization and expression throughout the lytic cycle, indirect immunofluorescence study (IFI) on intra- and extracellular tachyzoites was carried out. As other NMTs, T. gondii N-myristoyltransferase is localized to the cytosol of both on intra- and extracellular parasites (Figure 2B).
3.3. TgNMT activity is important for in T. gondii lytic cycle
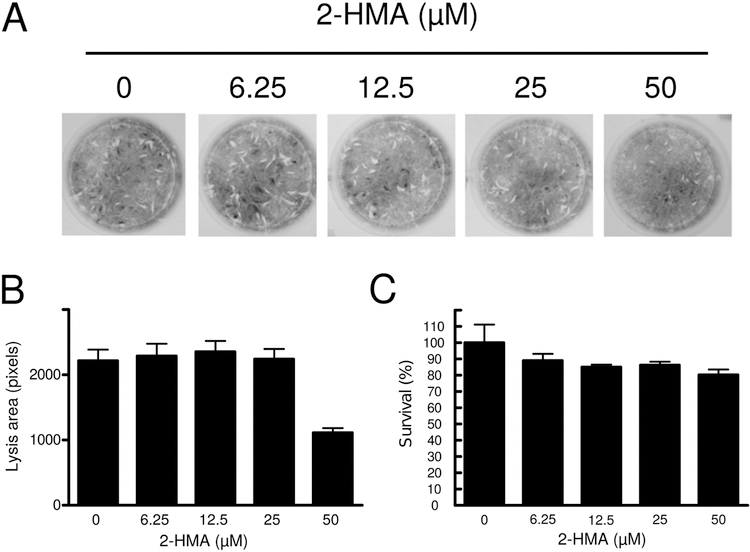
Based on our previous observations, we decided to assess whether TgNMT activity is required during tachyzoite lytic cycle. In order to do this, we incubated infected HFF monolayers with increasing concentrations of 2-hydroxymyristic acid (2HMA). This analog of myristic acid becomes metabolically activated in cells to form 2-hydroxymyristoyl-CoA, a potent inhibitor of NMT (Paige, et al., 1990). Figure 3 shows that when infected cells are treated with 2HMA at low concentrations no effect was observed. However, at the highest concentration tested (50 mM) a decrease of approximately 50% of the total area lysed by the tachyzoites was observed (Figure 3). On the other hand, no effect in host-cell viability was observed under our experimental conditions. These data suggest that NMT activity is important for optimal lytic cycle under tissue-culture conditions.
Figure 3. NMT activity is necessary for T. gondii lytic cycle.
(A) Plaque lysis assay performed on HFF monolayers infected with tachyzoites and incubated in presence of 0 (vehicle) or increasing concentrations of 2HMA. (B) Quantitation of lysis area. (C) Cellular cytotoxicity and of increasing concentrations of 2HMA was evaluated on HFF monolayers through the reduction of MTT. The experiments were carried out in duplicate in three independent experiments. Asterisk indicates significant differences with p < 0.05.
3.4. TgNMT interacts with various proteins
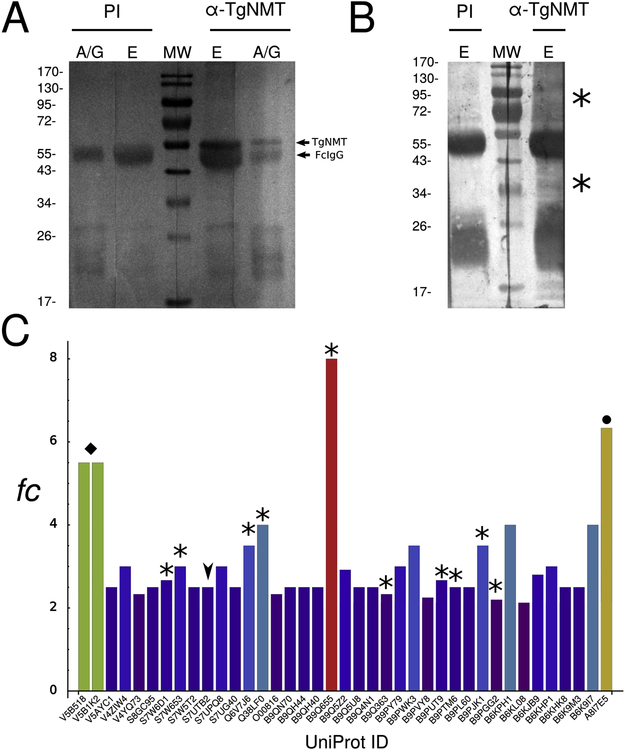
In order to obtain information on proteins interacting with TgNMT, we performed immunoprecipitations using the anti-TgNMT serum generated in our laboratory (Supplemental material) followed by MudPIT analysis for subsequent identification by spectral count analysis. Spectral Counting (SpC) is a viable strategy for free labeling proteomics, where the number of spectra corresponding to the peptides of a protein is used as a measure of its abundance (Liu, et al., 2004). Our crude results show a total of 433 putative interacting proteins and we calculated the enrichment of every protein based on fold change in SpC detection. Thus, proteins having a fold change value equal or greater than two were considered as enriched. In this manner we obtained a total of 66 putative interacting proteins (Figure 4 and Table S2, highlighted in red).
Figure 4. TgNMT interactome.
(A) Western blot analysis of T. gondii NMT immunoprecipitation with TgNMT antiserum (α-TgNMT). The pre-immune fraction (PI) was used as negative control. A/G: proteins bound to A/G sepharose beads even after elution; E: eluted proteins from with A/G sepharose; MW: molecular weight pattern. TgNMT migration is indicated with an arrowhead. (B) Ponceau red stain of Western blot described above. Asterisks indicate bands corresponding to potential TgNMT partners. (C) Bar chart of spectral count (SpC) for every protein detected in the MudPIT analysis. Only those proteins whose SpC fold change (fc) exceeded the value of 2 were plotted. Asterisks indicate proteins that participate in the translation process. Arrowheads indicate a predicted TgNMT substrate. A black diamond indicates proteins that participate in vesicular transport. A black circle indicates GRA7 antigen, a well-represented antigen in the total proteome.
Since protein myristoylation is in general a co-translational modification, it is not surprising to find ribosomal proteins in our analysis or relevant proteins for the translation process (Figure 4C, bars with asterisk). Ribosomal proteins detected in our analysis allow inferring an interaction with TgNMT as it was observed in other eukaryotes (Farazi, et al., 2001).
3.5. T. gondii myristoylome would encompass several proteins with varied functions
For a given protein to be recognized and modified by NMT, it should contain at its N-terminus the following amino acid consensus sequence: M1−G2−X3−X4−X5−(S/T/C)6, where X represents most amino acids except for Proline (P), aromatic or charged residues in position X3. It is important to highlight that other amino acids can be tolerated at position X6 (Alanine,A; Glycine,G) and even a Lysine(L) residue at position 7 can reduce the stringency for certain amino acids at position 2 (revised in (Martin, et al., 2011)). With the aim of determining the extent of myristoylation in T. gondii, an in silico screening over the TGME49 strain gene products was performed. Using the tool for protein motif pattern identification, and the consensus amino acid sequence with the exceptions described above, we found 181 gene products that could be myristoylated. Since protein myristoylation occurs mainly on cytoplasmic proteins, we discarded sequences containing predicted signal peptides, thus obtaining a total of 157 gene products (Table 1 and Table S3 for the complete list of proteins). This represents 1.77% of the total T. gondii ME49 strain proteome which is in accordance with other reports stating that protein myristoylation covers between 1–4% of all proteins on a given proteome (Martinez, et al., 2008).
Table 1.
Selected putative myristoylated proteins
| Accession ID* | Description | Protein Function | Fitness score^ | Report on experimental myristoylation |
|---|---|---|---|---|
| TGME49_207460 | Rab 5b | Intracellular membrane trafficking | −1.35 | (Ebine, et al., 2016) Ψ |
| TGME49_223940 | GAP45 | Gliding motility | −4.14 | (Frenal, et al., 2010) |
| TGME49_237820 | ISP2 | Parasite division | −2.04 | (Beck, et al., 2010) |
| TGME49_245710 | phosphatidylinositol-4-phosphate 5-kinase, putative | Calcium signaling | 1.15 | (Wright, et al., 2015) # |
| TGME49_249970 | acylated pleckstrin-homology domain-containing protein (APH) | Microneme exocytosis | −5.53 | (Bullen, et al., 2016) |
| TGME49_260820 | ISP1 | Parasite division | −2.04 | (Beck, et al., 2010) |
| TGME49_262860 | ADP-ribosylation factor family protein 1, putative | Dense granule proteins release | −0.52 | (Wright, et al., 2015)# |
| TGME49_268960 | 5’-AMP-activated protein kinase subunit beta-1 family protein, putative | Energy sensor | −3.95 | (Wright, et al., 2016, Wright, et al., 2015) #,& |
| TGME49_269780 | ADP-ribosylation factor, putative | Vesicle biogenesis 32 | −1.28 | (Wright, et al., 2015) # |
| TGME49_281580 | PP2C-like, putative | Protein phosphatase | −1.06 | (Wright, et al., 2016, Wright, et al., 2015) #,& |
| TGME49_305860 | CDPK3 | Parasite egress | −0.09 | (Garrison, et al., 2012) |
| TGME49_316540 | ISP3 | Parasite division | −0.63 | (Beck, et al., 2010) |
Toxodb release 40 (15 Oct 2018).
Data represent mean CRISPR phenotype scores reported by Sidik et al. (Sidik, et al., 2016)
Reported to be myristoylated in L. donovani
Reported to be myristoylated in P. falciparum
Reported to be myristoylated in T. brucei
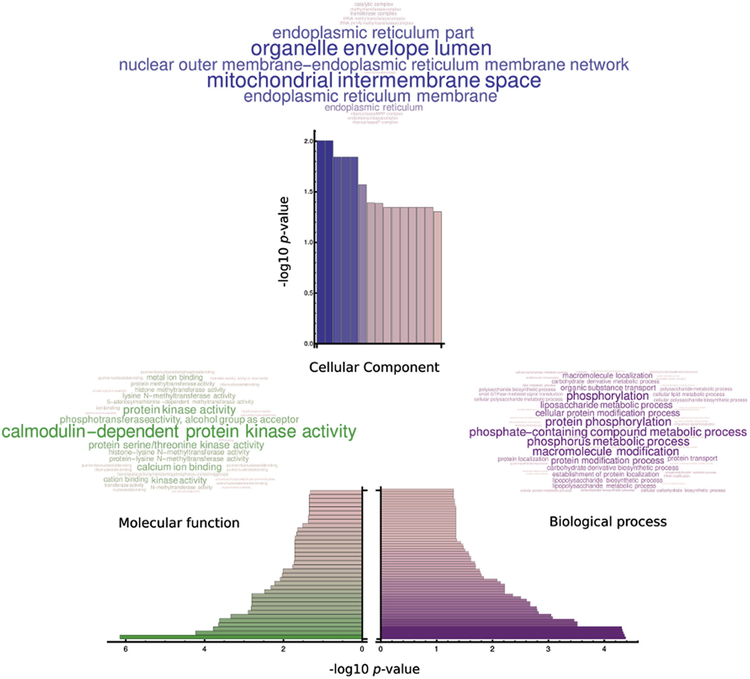
A Gene Ontology analysis (GO) of the theoretical myristoylome suggests a possible role of the gene products in the life cycle of the parasite. The most enriched products participate in processes such as protein phosphorylation (GO:0006468; Table S4) and lipopolysaccharide biosynthetic process (GO:0009103). Twenty three gene products share the five most enriched biological processes in our in silico myristoylome (-log10= 4.37; Figure 5, Biological Process), many of which are kinases, principally calcium dependent protein kinases (CDPKs). Additionally our molecular function analysis show that these twenty three products are associated with kinase activity being calmodulin-dependent protein kinase activity the most enriched function (GO:0004683,-log10= 6.15; Figure 5, Molecular Function). These gene products are annotated to contain at least one kinase domain, where CDPKs are again the kinase family most represented among other gene products with confirmed or predicted kinase activity (Table S5). CDPKs are proposed as good drug target candidates in Plasmodium spp. and Toxoplasma spp (Tate, et al., 2014), are predicted to be myristoylated and could function as calcium –myristoyl switches. Furthermore, CDPKs and some of these twenty three proteins showed a phenotype when the corresponding genes were deleted (Sidik, et al., 2016).
Figure 5. Gene Ontology (GO) analysis of the in silico myristoylome.
Possible biological process affected (A); predicted molecular function involved (B) and putative cellular components affected (C) by protein myristoylation.
A cellular component GO analysis revels that myristoylated proteins are widely distributed within the cell. This is attributed mainly to gene products that present kinase activity (the most represented in the myristoylome) which are not associated to any subcellular compartment (Figure 5, Cellular component). The vast representation of kinases in our in silico analysis could open a new study in TgNMT activity and localization regulation by phosphorylation, as TgNMT is confirmed to be phosphorylated in vivo by the T. gondii phosphoproteome (Treeck, et al., 2011). This analysis suggests a relevant role of protein myristoylation in T. gondii, as our results reveal gene products that were confirmed to be essential for the pathogen survival, with an important percentage of these proteins confirmed to be palmitoylated as well. As already shown in previous studies protein myristoylation is essential for subsequent palmitoylation (Resh, 2016). In this manner, myristoylation can directly and indirectly affect a vast diversity of proteins in T. gondii that are confirmed to influence its life cycle.
3.6. Human and T. gondii NMT substrates differ in their N-terminal amino acid occurrence
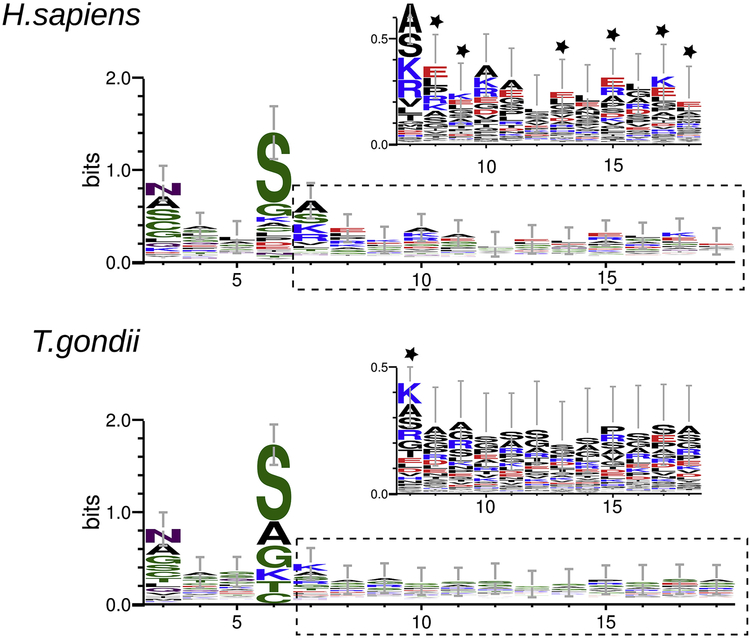
Analyzing NMT substrates amino acid sequence could help to understand the specificity for this acyl-transferase. With this in mind, we compared the amino acid composition of HsNMT and putative TgNMT peptide substrates focusing in the amino terminal region. Initial methionine and the conserved glycine at the second position in analyzed sequences were deleted with the aim of highlighting differences between myristoylated sequences. A WebLogo analysis shows that the first six residues are similar in both species being asparagine (N) in position three and serine (S) in position six the most characteristic. A clear difference is shown at position seven were the most representative amino acid in human sequences is alanine (A) while lysine (K) is preferred at this position in T. gondii sequences. Amino acids at positions 7 to 18 show high variability, but in-depth analysis over the logo sequences show relevant differences: charged amino acids are preferred in H. sapiens sequences (positions 8, 9, 13, 15, 17 and 18), while in T. gondii neutral and small amino acids are more frequent (Figure 6).
Figure 6. Web logo of HsNMT and TgNMT peptides substrates.
Amino acid alignment of HsNMT and TgNMT peptide substrates from positions 3 to 18. Residues are highlighted according to their chemistry (polar: green; neutral: purple; basic: blue; acidic: red; hydrophobic: black). For both enzymes, a zoomed view over positions 6 to 18 (dotted lines) is shown; residues are highlighted by charge (neutral: black; positive: blue; negative: red). Stars indicate differences.
4. DISCUSSION
Protein N-myristoylation refers to the irreversible covalent attachment of 14-carbon myristic acid onto an N-terminal glycine by N-myristoyltransferases (NMTs, (Farazi, et al., 2001)). This important protein modification plays roles in cellular signaling pathways, mediating subcellular targeting and protein-protein interactions (thoroughly reviewed in (Martin, et al., 2011)). However, there is scarce information on this protein modification in Toxoplasma gondii. With this in mind, we decided to explore the extent and roles of protein myristoylation in this parasite.
First we decided to determine whether T. gondii expressed a NMT. In order to do this, we performed an in silico screening across the T. gondii database and found only one gene product that could be T. gondii NMT (TgNMTp). The finding of a single putative TgNMT is not unusual, since contrary to what was observed in higher eukaryotes where two NMT isoforms are expressed, parasites express only one NMT which is essential. In fact, single NMTs have been described for Leishmania major, Leishmania donovani, Trypanosoma brucei and P. falciparum (Brannigan, et al., 2010, Price, et al., 2003, Wright, et al., 2014). Besides, our phylogenetic analysis showed that this protein presents homology to other apicomplexan NMTs and at the same time highlights that humans NMTs (HsNMTs) and Mus musculus sequences are grouped in a distant clade from TgNMTp. This difference between parasitic and their host NMTs could be exploited for the design of specific inhibitors. In fact, Plasmodium falciparum NMT is being the target of small molecule inhibitors to treat malaria (Schlott, et al., 2018, Wright, et al., 2014). The presence of conserved amino acids that have been shown to be critical for NMT activity on TgNMTp led us to think that this protein could present enzymatic activity. In line with this, here we showed that TgNMTp presents NMT activity in vitro and the typical cytosolic localization (McIlhinney and McGlone, 1996) in intra and extracellular tachyzoites, suggesting that its expression is needed through the lytic cycle. This was further confirmed when incubation of intracellular parasites with the NMT inhibitor 2HMA showed a decreased total lysed area, suggesting that the lytic cycle was altered. Since protein myristoylation can direct a protein to its site of action influencing its correct functioning, a decrease in host-cell lysis by the parasite can be attributed to myristoylation inhibition of proteins that are important for motility (TgGAP45/70, TgCDPK1), early tachyzoite endodyogeny (TgCDPK7, TgISP2), egress (TgCDPK3) and/or invasion (TgCDPK1, TgGAP45). In this manner the lytic cycle would be deregulated, leading to lower performance of host-cell lysis by T. gondii. The effect of 2HMA could be observed at concentrations that did not affect host-cell viability, suggesting a differential affinity between TgNMT and its host counterparts. The lack of effect observed on host-cells could be due to the concentrations of 2HMA used. It is well documented that the highest concentration of 2HMA that produces alteration in myristoylation without having a cytotoxic effect in mammalian cells is 1 mM (Galbiati, et al., 1996, Martin, et al., 2008, Paige, et al., 1990).
Immunoprecipitation assays provided a glimpse of TgNMT interactors. As expected of a co-translational modification, several ribosomal proteins were detected (Figure 4 and Table S2). It was reported that the N-terminal region of all described NMTs interacts with the ribosome during protein translation (Farazi, et al., 2001). Interestingly 12 proteins found to interact with TgNMT were also predicted to be myristoylated by our in silico myristoylome and thus, were considered putative TgNMT substrates (Table S6). Only one of the twelve possible substrates was significantly enriched ((fold change: 2.5; ID: TGME49_310420; Figure 4C, bar with arrowhead), while the others present low fold change values (Table S6). Despite this, we detected TgGAP45 (a reported myristoylated protein; (Frenal, et al., 2010)) and three MORN repeat-containing protein, a microneme protein (TgMIC7), a SRS (SAG related sequence) protein, a protein phosphatase 2C-like (reported to be myristoylated in L. donovani; (Wright, et al., 2015)) and three uncharacterized proteins. The low enrichment of these gene products could be due either to technical limitations, since immunoprecipitation assays detect proteins that interact with the protein of interest only at the time of the experiment or to the dynamics and distribution of the process, which is linked to translation. In this manner, possible myristoylable substrates are detected as a function of their abundance at this particular stage. As such, future studies of T. gondii myristoylome should include other techniques such as metabolic tagging followed by MudPIT for protein identification as already described for Leishmania donovani (Wright, et al., 2015).
In order to understand the full extent and role of myristoylation in Toxoplasma gondii, we used a bioinformatics approach to generate an in silico myristoylome. Our results show that 1.77% of T. gondii total proteome could be myristoylated. This is in agreement with repvious reports stating that myristoylation covers 0.5– 3% of all the proteins in a given proteome (Martinez, et al., 2008, Maurer-Stroh, et al., 2002).
Gene ontology analyses show that the most represented protein-function is the calmodulin protein kinase activity; this molecular function is related to our myristoylome by gene products like CDPKs (calcium-dependent protein kinases), ULK kinases and CAM Kinase family. Particularly, CDPKs have been thoroughly studied and documented to play a key role in T. gondii life cycle. Thus, CDPKs are being proposed as drug targets for antiparasitic design (Billker, et al., 2009). These kinases contain consensus motifs for N-myristoylation or palmitoylation, a feature also observed in plant CDPKs, many of which show membrane localization (Dammann, et al., 2003). In fact, mutation of the N-terminal glycine of TgCDPK3 prevents myristoylation which alters the membrane location that is required to carry out its function (Garrison, et al., 2012). Finally, a comparative analysis with the experimental data published by Sidik and coworkers revealed that 66% of the proteins present in our analysis are relevant to the progression of the lytic cycle (Sidik, et al., 2016). Hence, studies on the influence of myristoylation on these proteins would be of great importance. Myristoylomes have been described for parasites including trypanosomatids, Leishmania donovani and Plasmodium falciparum (Herrera, et al., 2016, Ritzefeld, et al., 2018, Wright, et al., 2016, Wright, et al., 2015). A comparative analysis of these myristoylomes with our in silico study reveals some similarities between substrates of P. falciparum and T. gondii NMTs. Proteins participating in traffic and secretion (ADP-ribosylation factor family proteins, Rab5b), motility (GAP45, CDPK1) and IMC integrity (ISP1–3, GAP45) are found in both myristoylomes (Wright, et al., 2014). Besides, T. gondii myristoylome include homologs of proteins described to be myristoylated in L. donovani such as protein phosphatase 2C-like, phosphatidylinositol-4-phosphate 5-kinase and the ADP-ribosylation factor family of proteins (Wright, et al., 2015). These comparative results support the strategy used in this work to obtain T. gondii myristoylome. It is important to highlight that this myristoylome includes proteins that have already been reported to be myristoylated such as TgISP1–3, TgGAP45 and TgCDPK3 (Beck, et al., 2010, Frenal, et al., 2010, Lourido, et al., 2012). TgGAP45, TgCDPK7 and TgCDPK1 are essential proteins required for the parasite’s life-cycle (Frenal, et al., 2010, Lourido, et al., 2010, Morlon-Guyot, et al., 2014). Another essential protein that is included in our list is the enzyme carbamoyl phosphate synthase (CPS), documented to be essential for T. gondii virulence (Fox and Bzik, 2002). Additionally 28 genes products detected in this work are confirmed to be palmitoylated as well (Table S3; (Caballero, et al., 2016, Foe, et al., 2015)), including three morn-repeat containing proteins and the membrane occupation and recognition nexus protein 2 (TgMORN2). MORN motifs play critical roles in several proteins with roles in the organization of membranous and cytoskeletal structures while TgMORN2 is a redundant member of the cell assembly process during the replication of the parasite (Gubbels, et al., 2006). Additionally, 63 gene products are listed as hypothetical proteins in the T. gondii database and described to be essential (Sidik, et al., 2016), which suggest that the role and extent of protein myristoylation in this parasite is an unexplored field and that deserves to be studied in more detail.
Regarding amino acid occurrence on NMT substrates, it has been described that the first eight N-terminal amino acids are determinant for NMT specificity and selectivity (Castrec, et al., 2018). However, it was also reported that positions in substrate sequence beyond position 8 could have an impact on NMT specificity (Maurer-Stroh, et al., 2002). Here we analyze the amino acid sequences of the putative TgNMT substrates predicted by our in silico approach and compare them with host myristoylated substrates. We found that human myristoylated substrates present charged amino acids in positions 7 to 18 more frequently whereas in T. gondii neutral amino acids are preferred on these positions. These differences in the amino acidic composition of NMTs substrates in positions beyond the classical sequence could be useful for the design of specific inhibitors to target parasitic NMTs. This observation could be related with the NMT structure, since it has been documented that a region distal to the active site of the enzyme might accommodate myristoylable peptides (Maurer-Stroh, et al., 2002). Is important to highlight that no crystal structure for TgNMT is available and a structural study of TgNMT could be valuable to understand the differences observed in our study between human and T. gondii NMT substrates. Furthermore, a structural analysis of TgNMT could be of great relevance for inhibitor designing.
In summary, our results strongly suggest that myristoylation is a mechanism operating in T. gondii with impact on its lytic cycle, particularly in calcium-dependent processes which are essential for growth. As such, is that protein myristoylation could have a central role in T. gondii pathogenesis. Furthermore, as observed in other parasites, T. gondii expresses only one NMT with apparent differences to its host counterparts. These two observations suggest that TgNMT could be an interesting target for anti-toxoplasmic compunds. Finally, here we provide the first evidence of possible TgNMT substrates, being necessary confirm myristoylation targets by specific protocols such as click-chemistry assays (Yap, et al., 2010).
Supplementary Material
Highlights.
Toxoplasma gondii encodes for a single putative N-myristoyltransferase
A phylogenetic analysis suggests significant differences between TgNMT and HsNMT
T. gondii myristoylome include 157 predicted proteins
TgNMT activity is important for the lytic cycle
5. ACKNOWLEDGMENTS
BDO, JJM, and JRY were supported by the National Institute of General Medical Sciences (8 P41 GM103533). This work was partially supported by ANPCyT grant PICT 2014–1917 (MMC) and from Bunge & Born Foundation to infectious diseases research (MMC). AMA is a PhD fellow from National Council of Research (CONICET). VRT and DMR are researchers from CONICET. MMC is a researcher from CONICET and UNSAM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. REFERENCES
- Adams PD, Seeholzer S & Ohh M, 2002. Identification of Associated Proteins by Coimmunoprecipitation In: Argentine J, (Ed., Molecular Cloning: a Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., pp. 59–74. [Google Scholar]
- Albarracin RM, Becher ML, Farran I, Sander VA, Corigliano MG, Yacono ML, Pariani S, Lopez ES, Veramendi J, and Clemente M, 2015. The fusion of Toxoplasma gondii SAG1 vaccine candidate to Leishmania infantum heat shock protein 83-kDa improves expression levels in tobacco chloroplasts. Biotechnol J 10, 748–759. [DOI] [PubMed] [Google Scholar]
- Bangoura B, Zoller B, and Daugschies A, 2011. [Prevalence and relevance of avian Toxoplasma gondii infections in Europe]. Berl Munch Tierarztl Wochenschr 124, 485–496. [PubMed] [Google Scholar]
- Beck JR, Rodriguez-Fernandez IA, de Leon JC, Huynh MH, Carruthers VB, Morrissette NS, and Bradley PJ, 2010. A novel family of Toxoplasma IMC proteins displays a hierarchical organization and functions in coordinating parasite division. PLoS Pathog 6, e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billker O, Lourido S, and Sibley LD, 2009. Calcium-dependent signaling and kinases in apicomplexan parasites. Cell Host Microbe 5, 612–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannigan JA, Smith BA, Yu Z, Brzozowski AM, Hodgkinson MR, Maroof A, Price HP, Meier F, Leatherbarrow RJ, Tate EW, Smith DF, and Wilkinson AJ, 2010. N-myristoyltransferase from Leishmania donovani: structural and functional characterisation of a potential drug target for visceral leishmaniasis. J Mol Biol 396, 985–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen HE, Jia Y, Yamaryo-Botte Y, Bisio H, Zhang O, Jemelin NK, Marq JB, Carruthers V, Botte CY, and Soldati-Favre D, 2016. Phosphatidic Acid-Mediated Signaling Regulates Microneme Secretion in Toxoplasma. Cell Host Microbe 19, 349–360. [DOI] [PubMed] [Google Scholar]
- Caballero MC, Alonso AM, Deng B, Attias M, de Souza W, and Corvi MM, 2016. Identification of new palmitoylated proteins in Toxoplasma gondii. Biochim Biophys Acta 1864, 400–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, and Gabaldon T, 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrec B, Dian C, Ciccone S, Ebert CL, Bienvenut WV, Le Caer JP, Steyaert JM, Giglione C, and Meinnel T, 2018. Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern. Nat Chem Biol. [DOI] [PubMed]
- Dammann C, Ichida A, Hong B, Romanowsky SM, Hrabak EM, Harmon AC, Pickard BG, and Harper JF, 2003. Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol 132, 1840–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald RG, Carter D, Ullman B, and Roos DS, 1996. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J Biol Chem 271, 14010–14019. [DOI] [PubMed] [Google Scholar]
- Ebine K, Hirai M, Sakaguchi M, Yahata K, Kaneko O, and Saito-Nakano Y, 2016. Plasmodium Rab5b is secreted to the cytoplasmic face of the tubovesicular network in infected red blood cells together with N-acylated adenylate kinase 2. Malar J 15, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Waksman G, and Gordon JI, 2001. The biology and enzymology of protein N-myristoylation. J Biol Chem 276, 39501–39504. [DOI] [PubMed] [Google Scholar]
- Foe IT, Child MA, Majmudar JD, Krishnamurthy S, van der Linden WA, Ward GE, Martin BR, and Bogyo M, 2015. Global Analysis of Palmitoylated Proteins in Toxoplasma gondii. Cell Host Microbe 18, 501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox BA and Bzik DJ, 2002. De novo pyrimidine biosynthesis is required for virulence of Toxoplasma gondii. Nature 415, 926–929. [DOI] [PubMed] [Google Scholar]
- Frenal K, Polonais V, Marq JB, Stratmann R, Limenitakis J, and Soldati-Favre D, 2010. Functional dissection of the apicomplexan glideosome molecular architecture. Cell Host Microbe 8, 343–357. [DOI] [PubMed] [Google Scholar]
- Furtado JM, Smith JR, Belfort R Jr., Gattey D, and Winthrop KL, 2011. Toxoplasmosis: a global threat. J Glob Infect Dis 3, 281–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajria B, Bahl A, Brestelli J, Dommer J, Fischer S, Gao X, Heiges M, Iodice J, Kissinger JC, Mackey AJ, Pinney DF, Roos DS, Stoeckert CJ Jr., Wang H, and Brunk BP, 2008. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res 36, D553–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati F, Guzzi F, Magee AI, Milligan G, and Parenti M, 1996. Chemical inhibition of myristoylation of the G-protein Gi1 alpha by 2-hydroxymyristate does not interfere with its palmitoylation or membrane association. Evidence that palmitoylation, but not myristoylation, regulates membrane attachment. Biochem J 313 ( Pt 3), 717–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Treeck M, Ehret E, Butz H, Garbuz T, Oswald BP, Settles M, Boothroyd J, and Arrizabalaga G, 2012. A forward genetic screen reveals that calcium-dependent protein kinase 3 regulates egress in Toxoplasma. PLoS Pathog 8, e1003049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves V, Brannigan JA, Thinon E, Olaleye TO, Serwa R, Lanzarone S, Wilkinson AJ, Tate EW, and Leatherbarrow RJ, 2012. A fluorescence-based assay for N-myristoyltransferase activity. Anal Biochem 421, 342–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbels MJ, Vaishnava S, Boot N, Dubremetz JF, and Striepen B, 2006. A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J Cell Sci 119, 2236–2245. [DOI] [PubMed] [Google Scholar]
- He L, Diedrich J, Chu YY, and Yates JR 3rd, 2015. Extracting Accurate Precursor Information for Tandem Mass Spectra by RawConverter. Anal Chem 87, 11361–11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera LJ, Brand S, Santos A, Nohara LL, Harrison J, Norcross NR, Thompson S, Smith V, Lema C, Varela-Ramirez A, Gilbert IH, Almeida IC, and Maldonado RA, 2016. Validation of N-myristoyltransferase as Potential Chemotherapeutic Target in Mammal-Dwelling Stages of Trypanosoma cruzi. PLoS Negl Trop Dis 10, e0004540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K and Standley DM, 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MJ and Sharma RK, 1994. Mechanisms of action of NIP71 on N-myristoyltransferase activity. Mol Cell Biochem 141, 79–86. [DOI] [PubMed] [Google Scholar]
- Letunic I and Bork P, 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44, W242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, and Yates JR 3rd, 2004. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76, 4193–4201. [DOI] [PubMed] [Google Scholar]
- Lourido S, Shuman J, Zhang C, Shokat KM, Hui R, and Sibley LD, 2010. Calcium-dependent protein kinase 1 is an essential regulator of exocytosis in Toxoplasma. Nature 465, 359–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourido S, Tang K, and Sibley LD, 2012. Distinct signalling pathways control Toxoplasma egress and host-cell invasion. EMBO J 31, 4524–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden T, 2013. The BLAST Sequence Analysis Tool. National Center for Biotechnology Information (US), Bethesda (MD). [Google Scholar]
- Martin DD, Beauchamp E, and Berthiaume LG, 2011. Post-translational myristoylation: Fat matters in cellular life and death. Biochimie 93, 18–31. [DOI] [PubMed] [Google Scholar]
- Martin DD, Vilas GL, Prescher JA, Rajaiah G, Falck JR, Bertozzi CR, and Berthiaume LG, 2008. Rapid detection, discovery, and identification of post-translationally myristoylated proteins during apoptosis using a bio-orthogonal azidomyristate analog. FASEB J 22, 797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Traverso JA, Valot B, Ferro M, Espagne C, Ephritikhine G, Zivy M, Giglione C, and Meinnel T, 2008. Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics 8, 2809–2831. [DOI] [PubMed] [Google Scholar]
- Maurer-Stroh S, Eisenhaber B, and Eisenhaber F, 2002. N-terminal N-myristoylation of proteins: refinement of the sequence motif and its taxon-specific differences. J Mol Biol 317, 523–540. [DOI] [PubMed] [Google Scholar]
- McIlhinney RA and McGlone K, 1996. Immunocytochemical characterization and subcellular localization of human myristoyl-CoA: protein N-myristoyltransferase in HeLa cells. Exp Cell Res 223, 348–356. [DOI] [PubMed] [Google Scholar]
- Meerburg BG and Kijlstra A, 2009. Changing climate-changing pathogens: Toxoplasma gondii in North-Western Europe. Parasitol Res 105, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya JG and Liesenfeld O, 2004. Toxoplasmosis. Lancet 363, 1965–1976. [DOI] [PubMed] [Google Scholar]
- Morlon-Guyot J, Berry L, Chen CT, Gubbels MJ, Lebrun M, and Daher W, 2014. The Toxoplasma gondii calcium-dependent protein kinase 7 is involved in early steps of parasite division and is crucial for parasite survival. Cell Microbiol 16, 95–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige LA, Zheng GQ, DeFrees SA, Cassady JM, and Geahlen RL, 1990. Metabolic activation of 2-substituted derivatives of myristic acid to form potent inhibitors of myristoyl CoA:protein N-myristoyltransferase. Biochemistry 29, 10566–10573. [DOI] [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, and Gygi SP, 2003. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2, 43–50. [DOI] [PubMed] [Google Scholar]
- Petersen E, 2007. Toxoplasmosis. Semin Fetal Neonatal Med 12, 214–223. [DOI] [PubMed] [Google Scholar]
- Price HP, Menon MR, Panethymitaki C, Goulding D, McKean PG, and Smith DF, 2003. Myristoyl-CoA:protein N-myristoyltransferase, an essential enzyme and potential drug target in kinetoplastid parasites. J Biol Chem 278, 7206–7214. [DOI] [PubMed] [Google Scholar]
- Resh MD, 2016. Fatty acylation of proteins: The long and the short of it. Prog Lipid Res 63, 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzefeld M, Wright MH, and Tate EW, 2018. New developments in probing and targeting protein acylation in malaria, leishmaniasis and African sleeping sickness. Parasitology 145, 157–174. [DOI] [PubMed] [Google Scholar]
- Rocque WJ, McWherter CA, Wood DC, and Gordon JI, 1993. A comparative analysis of the kinetic mechanism and peptide substrate specificity of human and Saccharomyces cerevisiae myristoyl-CoA:protein N-myristoyltransferase. J Biol Chem 268, 9964–9971. [PubMed] [Google Scholar]
- Schlott AC, Holder AA, and Tate EW, 2018. N-Myristoylation as a Drug Target in Malaria: Exploring the Role of N-Myristoyltransferase Substrates in the Inhibitor Mode of Action. ACS Infect Dis 4, 449–457. [DOI] [PubMed] [Google Scholar]
- Sidik SM, Huet D, Ganesan SM, Huynh MH, Wang T, Nasamu AS, Thiru P, Saeij JPJ, Carruthers VB, Niles JC, and Lourido S, 2016. A Genome-wide CRISPR Screen in Toxoplasma Identifies Essential Apicomplexan Genes. Cell 166, 1423–1435 e1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, and Yates JR 3rd, 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate EW, Bell AS, Rackham MD, and Wright MH, 2014. N-Myristoyltransferase as a potential drug target in malaria and leishmaniasis. Parasitology 141, 37–49. [DOI] [PubMed] [Google Scholar]
- Tenter AM, Heckeroth AR, and Weiss LM, 2000. Toxoplasma gondii: from animals to humans. Int J Parasitol 30, 1217–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treeck M, Sanders JL, Elias JE, and Boothroyd JC, 2011. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites’ boundaries. Cell Host Microbe 10, 410–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston SA, Camble R, Colls J, Rosenbrock G, Taylor I, Egerton M, Tucker AD, Tunnicliffe A, Mistry A, Mancia F, de la Fortelle E, Irwin J, Bricogne G, and Pauptit RA, 1998. Crystal structure of the anti-fungal target N-myristoyl transferase. Nat Struct Biol 5, 213–221. [DOI] [PubMed] [Google Scholar]
- Wolters DA, Washburn MP, and Yates JR 3rd, 2001. An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem 73, 5683–5690. [DOI] [PubMed] [Google Scholar]
- Wright MH, Clough B, Rackham MD, Rangachari K, Brannigan JA, Grainger M, Moss DK, Bottrill AR, Heal WP, Broncel M, Serwa RA, Brady D, Mann DJ, Leatherbarrow RJ, Tewari R, Wilkinson AJ, Holder AA, and Tate EW, 2014. Validation of N-myristoyltransferase as an antimalarial drug target using an integrated chemical biology approach. Nat Chem 6, 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MH, Heal WP, Mann DJ, and Tate EW, 2010. Protein myristoylation in health and disease. J Chem Biol 3, 19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MH, Paape D, Price HP, Smith DF, and Tate EW, 2016. Global Profiling and Inhibition of Protein Lipidation in Vector and Host Stages of the Sleeping Sickness Parasite Trypanosoma brucei. ACS Infect Dis 2, 427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright MH, Paape D, Storck EM, Serwa RA, Smith DF, and Tate EW, 2015. Global analysis of protein N-myristoylation and exploration of N-myristoyltransferase as a drug target in the neglected human pathogen Leishmania donovani. Chem Biol 22, 342–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Zheng Y, Li H, Luo X, He Z, Cao S, Shi Y, Zhao Q, Xue Y, Zuo Z, and Ren J, 2016. GPS-Lipid: a robust tool for the prediction of multiple lipid modification sites. Sci Rep 6, 28249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MC, Kostiuk MA, Martin DD, Perinpanayagam MA, Hak PG, Siddam A, Majjigapu JR, Rajaiah G, Keller BO, Prescher JA, Wu P, Bertozzi CR, Falck JR, and Berthiaume LG, 2010. Rapid and selective detection of fatty acylated proteins using omega-alkynyl-fatty acids and click chemistry. J Lipid Res 51, 1566–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.