Abstract
Ras gene (HRAS, NRAS, and KRAS) has been observed to be mutated and hyper-activated in a significant proportion of cancers. However, mutant Ras remains a challenging therapeutic target. Similarly, inhibition of targets upstream and downstream of Ras has shown limited clinical utility. There have been attempts to develop and deliver mutant K-Ras silencing RNAs either through their encapsulation in liposomes or nanoparticles. However, these approaches show very limited success due to the lack of stability of such carrier molecules alongside associated toxicity. There is a pressing need for the identification of better therapeutic targets for Ras or its associated pathways as well as improvements in the design of superior RNAi delivery systems to suppress mutant K-Ras. More than a decade ago, it was shown that aggregates of palmitoylated Ras isoforms (H-Ras and N-Ras) passage through the cytosol on rapidly moving nanosized particles (“rasosomes”). Fast forward a decade, considerable new knowledge has emerged in the area of small vesicles, microparticles, and exosomes. Exosomes are tiny vesicles and play a significant role in regulating cancer-related signaling pathways. Exosomes have also been studied as delivery vehicles to transport drugs, proteins, and microRNAs of choice for therapeutic purposes. K-Ras pathway proteins have been implicated in exosome biogenesis and extravasation processes. This review provides an update on the current knowledge related to K-Ras signaling and exosomes and also discusses how these tiny vesicles can be harnessed to successfully deliver the K-Ras silencing moieties.
Keywords: Kras, Ras, Mutant Kras, Exosomes, Vesicles, Therapeutic drug delivery, RNAi
1. Introduction
Since the discovery of Ras genes, there have been great initiatives put forth into understanding its functionality as an oncogene [1]. First observed in retroviral studies, the identification of the Ras superfamily was a critical discovery allowing researchers to better understand cell signaling mechanisms and advancing the knowledge of how cancers are affected by Ras mutations. There have been considerable efforts put into developing pharmaceutical strategies aimed at targeting Ras mutation-driven cancers [2]. The Ras gene family is conserved throughout all mammalian cells and contributes to a host of functions. One of the best understood functions is Ras involvement in tyrosine receptor kinase signaling and its downstream activation of the ERK/MAPK pathway (https://www.cancer.gov/research/key-initiatives/ras/ras-central/blog/2015/ras-pathway-v2). Given its role in regulating several different downstream pathways, it is not surprising to note that components of the Ras pathway play a significant role in exosome formation. More than a decade ago, studies have shown that aggregates of palmitoylated Ras isoforms (H-Ras and N-Ras) passage through the cytosol on rapidly moving nanosized particles (“rasosomes”) [3]. Emerging studies have also suggested that Ras may exert its signals by secreting through exosomes [4]. There is a compelling body of evidence indicating that cancer cells can exploit exosomes into sending their proteins, DNA and RNA to normal cells as a way to increase their tumorigenicity and proliferation, and to promote a pro-cancer phenotype within the human body [5] (and reviewed in [6]). Recently, there has been an increased effort put forth into exploring the relationship between Ras signaling and its role in exosome formation. Likewise, engineering naturally occurring exosomes to deliver Ras targeted therapeutics to specific tumor locations within the body is also being explored. Such an understanding of exosomes and the process of intracellular transport is key to engineering drug delivery vehicles that have the capability of targeting cancers with Ras mutations.
2. Ras and Cancer
Depending on the stimuli, either extracellular or intracellular, Ras proteins balance cellular fate between survival and programmed death [7]. A perturbation in the downstream pathway could potentially have deleterious consequences shifting the fate toward uncontrolled cell division and ultimately cancer development. In normally functioning cells, the pro-survival path involves receptor tyrosine kinase activation through autophosphorylation resulting in the association of the plasma membrane protein scaffold Ras/SOS/GREB2 (reviewed in another article of this thematic issue [8]). Further, stimulations with mitogens or growth factors result in the Ras GTPase substituting its bound GDP with a GTP signaling downstream effects to the serine/threonine kinases RAF, MEK, and ERK. ERK is re-localized to the nucleus and phosphor-ylates the transcription factor ELK1 to transcribe pro-survival genes like NF-kβ, PKB/AKT, as well as promoting the cell cycle to proceed out of G 0 resting phase. Intuitively this pro-survival process inactivates apoptotic proteins such as Bad and initiator caspase-9. During cellular shock or DNA damage, Ras signals the cell to favor apoptosis by activating pro-apoptotic factors like Rassf1 and Nore1 which in turn activates effector caspase-3 [7]. In cancerous cells, mutations affecting the pro-survival scheme allows for continuous Ras-GTP association resulting in increased activity of the cell cycle, increased cellular proliferation and a loss of cellular control. Mutations, mostly missense and point mutations, affecting Ras proteins are commonly found in many cancers and usually occur early in cancer progression. Mutations in KRas are commonly found in pancreatic, non-small cell lung and colon cancers contributing to their aggressive phenotypes. Extensive sequencing studies have shown Ras involvement in cancer development and it was found that upto 85% of pancreatic ductal adenocarcinoma cases, the most common type of pancreatic cancer, harbor point mutations of KRas along with TP53 and SMAD2 resulting in a shift toward deregulated glycolytic metabolism and increased autophagy [10,11]. The alternatively spliced K-Ras isoform, KRas4B, is the major mutational form which in turn causes increased cell proliferation and survival by the excessive cellular signaling. The Ras gene family is an ideal therapeutic target due to its involvement in ~30% of all human cancers. There have been hurdles in designing effective treatments for cancers harboring Ras mutations due to its functionally redundant pathways as well as the differences that each Ras isoform contributes to the tumor microenvironment. Due to the glaringly large mutation rate of K-Ras in pancreatic, lung and colon cancers and evidence of different functions within a cell compared to other Ras isoforms, K-Ras will be the primary focus of this review.
3. Targeting K-Ras directly
Historically, targeting the constitutive GTPase activity of K-Ras in cancer cells was viewed as a promising and a viable option for cancer therapeutics. Theoretically, this should have been an ideal approach. Attempts to target the GTPase activity of Ras included the small GTPase inhibitor molecules FTIs (farnesyl transferase inhibitors) under the blanket assumption that all Ras proteins behave the same (reviewed in [12]). FTIs were designed to target the C-terminal lipid modifications (prenylations) that allow Ras to associate with the plasma membrane and disruption of these C-terminal lipid modifications should result in incorrect associations with the plasma membrane and inactivated downstream pathways. It was found that FTIs effectively targete H-Ras gernylgernylation associations on the membrane but did not prevent NRas or K-Ras membrane localization. K-Ras and H-Ras proteins have found ways to evade this FTI induced inhibition due to their alternate C-terminal modifications. Another GTPase inhibitor, GGTase, (geranylgeranyl transferase type I), has the ability to target the alternate prenylations which subsequently inhibit plasma membrane localization of K-Ras and N-Ras. K-Ras was also found to participate in an alternative pathway independent of other Ras subtypes causing hurdles in directly targeting Ras. To overcome the K-Ras evasion, small molecule inhibitors that process the C-terminal CAAX region on Ras after its dissociation from the plasma membrane, Rce1, and IcmT, were also explored [13]. In theory, these inhibitors should have promise due to the constitutive involvement in all of the Ras isoform processing. Unfortunately, one caveat with these small molecule inhibitors is there is little consideration about specificity to only mutant Ras proteins and limited evidence that the inhibitors can distinguish between mutant Ras and wild-type Ras in humans. To address this issue, Ras mutation-specific inhibitors are being developed and tested in a pre-clinical setting for various cancers [14].
4. Targeting mutant Kras through RNA interference
The idea of using nanoparticles as a more efficient and precise drug delivery system to the tumor sites have been extensively studied [15]. Nanoparticles are microscopic entities less than 100 nm in size and allow for adequate storage and transfer of biological warhead, particularly the small molecule drugs, to the cancerous site [16]. Nano-liposomes, are hollow transport vesicles made of plasma membrane phospholipids and cholesterol that exhibit various surface features including an electrostatically charged outer core, a variation of surface lipids being either hydrophobic or hydrophilic and varying membrane fluidity [17]. As with other siRNAs and small molecule drugs, nano-technology has also been exploited for the delivery of mutant K-Ras specific siRNAs. For example, in vivo mice models treated with liposomes packaged with cisplatin and the siKras, miR-34a, have been used in lung tumor models. Such an approach has shown promise by allowing tumors to increase the uptake of cisplatin and enhance toxicity at the tumor site [18]. In another study, siG12D-LODER™ was injected into patient tumors and released a siRNA drug against the mutant Kras (G12D) isoform in an open-label Phase 1/2a study in the first-line setting of locally advanced pancreatic cancer patients that were not eligible for surgical resection (ClincalTrials.gov identifier ). Evaluable responses were observed (Median Overall survival 15.12 months with a marked decrease in CA19–9 levels in 70% of the patients). Despite these promising results, there were acute toxicities in 5 of the 15 patients in this trial demonstrating serious adverse events (SAEs) [19]. Therefore, more work needs to be done in order to validate the utility of this approach in larger patient population. It is not completely clear whether liposomes are the best drug delivery system due to their inability to maintain long-term circulation through the body. On the other hand, the synthetic nanoparticles, although stable, have significant toxicities especially showing immunomodulatory effects resulting in excessive cytokine secretion that contributes to side effects of disease including autoimmune and neoplastic syndromes [28]. Alternatively, endogenously generated exosomes have no immunogenic activity, are highly stable and possess a longer circulatory life that is seemingly superior to synthetically engineered nanoparticle carriers. In the following sections, we will discuss the role of K-Ras in exosome biology and how these tiny vesicles are being harnessed for the development of effective therapeutics against this master oncogene.
5. Exosomes
Understanding the role of naturally occurring cellular exosomes is critical for gaining a better understanding of how their manipulation can evolve into cancer therapeutics. Naturally, exosomes develop during the progression of early to late endosomes that form into multivesicular bodies (MVBs) within the cell [20]. Proteins and exosomal contents are sorted through a specific protein family, endosomal sorting complexes required for transport (ESCRT), which cascade along the late endosome to sort the cargo into internalized and sealed off vesicles [21]. These multivesicular bodies can then disperse from the cell and move freely in the extracellular space, fuse with the plasma membrane and can either interact with other organ tissues or can be found in bodily fluids including but not limited to the breast milk [22], plasma [23] and urine [24]. The contents inside: proteins, miRNA, mRNA or DNA are released in a paracrine fashion and interact with the recipient cell or their surrounding microenvironment. Exosomes travel and subsequently associate with a recipient cell through a variety of mechanisms including differences in pH, osmotic stress or binding of cell ligands on the exosomal surface receptors allowing the exosomes to bind, via ligand-receptor binding or SNAREs, and disperse their contents in the new cellular location (reviewed in [25]).
Studies devoted to understanding exosomal function have gained momentum in recent years. This is due to the consistent observation that exosomes can modulate inter and intracellular signaling within most of the organs of the human body. Within the brain, exosomes have been shown to control the vascular network and maintain cellular homeostasis [26]. Recent research has suggested that viruses have the ability to propagate in an infected host cell through exosomal transport [27]. This led to the hypothesis that certain viruses, like the Epstein Barr virus, can manipulate normal host exosomes to carry pathogenic factors to alter the cellular microenvironment driving cancers and other diseases [28,29]. Studies have shown that proteins or nucleic acids secreted from exosomes can polarize normal cells into cancerous phenotype [30]. Not surprisingly, the interactions between exosomes and Ras signaling have also been studied. Prior to describing these interactions and subsequent future directions, in the below sections we review the role of Ras in exosome biogenesis.
6. Exosome composition
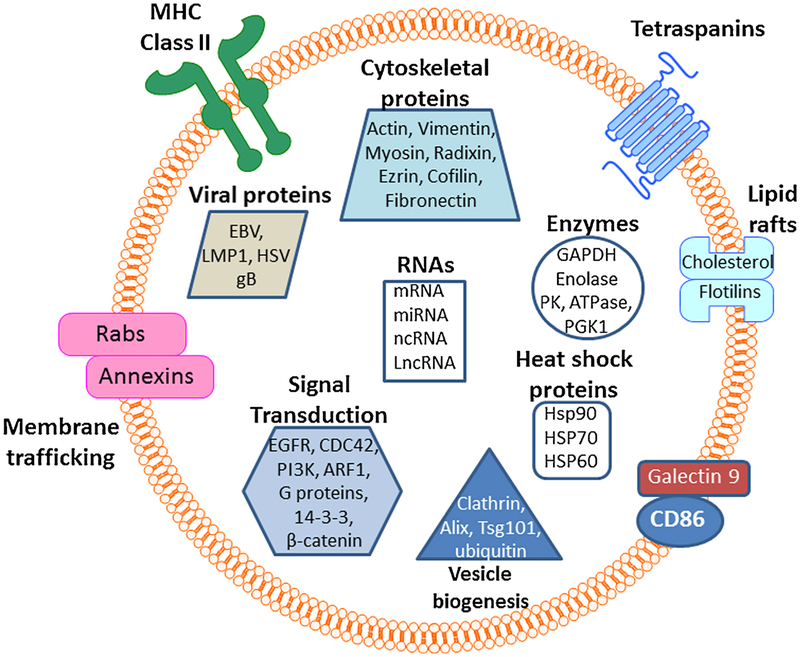
The exocarta database is an excellent resource to obtain the entire list of proteins, microRNAs, mRNAs and other exosome structural motifs that have been identified by various researchers in the field (http://exocarta.org/#). This is a continuously updated database that is populated with new entries frequently. At present, there are 286 research studies listed in this database that include 41,860 protein entries, 9769 proteins, 4946 mRNAs entries, 3408 mRNAs, 2838 miRNAs and 1116 lipid entries (searched on March 7th, 2018). Of notes, there is a significant fraction of the proteins, mRNAs, miRNAs, and lipids that are under the influence of the RAS network of proteins (Fig. 1). Below is the discussion of some of the Ras-related proteins that form the exosomes.
Fig. 1.
Exosome composition and content. Exosomes are bilyared vesicular structures that carry virtually all types of signal transduction proteins, structural proteins, lipids, enzymes, MHCs, RNAs, non-coding RNAs. RABs, EGFR, PI3K and CDC42 are all part of the Ras network and support exosome formation as well as are part of exosome structure.
7. Role of Ras family proteins in exosome biology
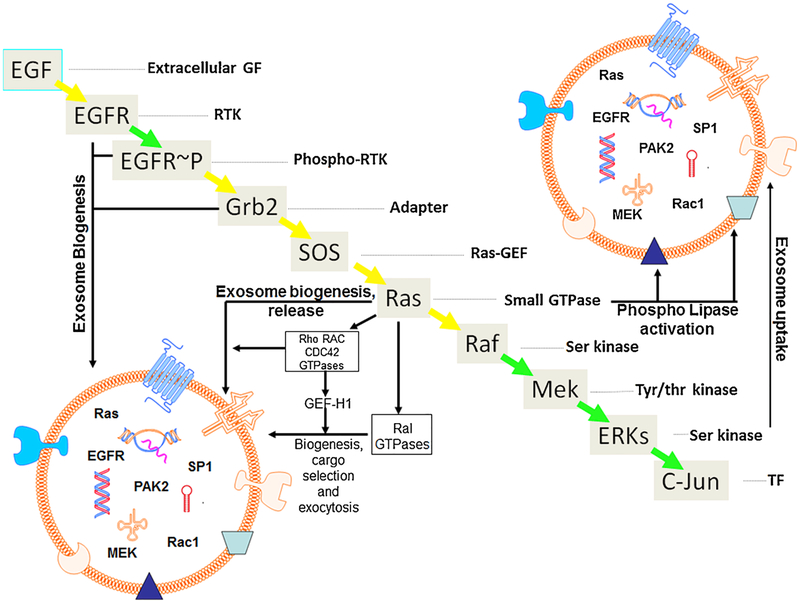
Emerging evidence suggests that Ras family proteins have a central role in the biology of exosomes (Fig. 1). Several downstream components of the Ras network have been shown to play a critical role in exosome biogenesis, maintenance, secretion and even cargo loading. In the following sections, some of the Ras family proteins and their role in exosome biology will be discussed (Fig. 2).
Fig. 2.
Role of Ras pathway proteins in exo-some biogenesis, cargo selection and release. Summary diagram showing different components of the Ras pathway and their roles in exosome biology. Exosomes are small vesicular structures that have a lipid bilayer. The bilayer is embedded with several macro and micromolecules including MHC class II, tetraspanins, immunoglobulins, receptors. These bilayered vesicles carry several types of cargoes such as mRNA, DNA, microRNAs etc. Ras, MAPK, CDC42, GTPases and their effectors such as p21 activated kinases have also been detected in exosomes. EGFR and EGFR-p are recognized for their role in exosome biogenesis and are also detected in intact exosomes. Ras and its downstream pathway proteins such as Rho, Rac and CDC42 GTPases, and their effectors are also known to promote exosome biogenesis, mediate cargo selection and induce exo-some release. Small GTPases have also been documented in stabilizing the phospholipases present in the exosome bilayer (shaded triangle). ERKs have been reported to play a role in facilitating exosome uptake.
As mentioned above it was more than a decade ago when Rotblat and colleagues discovered that aggregates of only palmitoylated Ras isoforms H-Ras and N-Ras could diffuse through the cytoplasm while being encapsulated in small cytosolic nanoparticles aptly termed rasosomes [3]. Later on, the same group showed that the farnesyl group of Ras helps the interaction between the prenyl-binding pockets in galectin-1, −3, and cGMP phosphodiesterase delta [31,32]. Such compartmentalization of Ras contributes to the spatial organization and dependent signaling by promoting its redistribution and adding further selectivity to the signals emanating from this master oncogene. Using Total internal reflection fluorescence (TIRF) microscopy together with spatial analysis algorithm, Kofer-Geles and colleagues demonstrated that rasosome movement in close proximity to plasma membrane is restricted to distinctive areas that were termed rasosomal ‘hotspots’ [33]. These hotspots were shown to be localized between actin filament cages. These findings confirmed that molecular signals arising from plasma membrane hotspots are relayed to rasosomes, thereby serving as robust Ras signaling nodes that spread signals across the cell. This association was also linked to the prolonged occurrence of Ras signals in the plasma membrane. Working on identifying the sub-cellular origin of rasosomes, Grunwald and colleagues used photoactivatable (PA)-GFPH-Ras mutants and showed that rasosomes arise from the Golgi apparatus [34]. These studies also deciphered that newly released rasosomes have a biphasic movement pattern: (a) initial being energy-dependent,(b) followed by a random diffusion.
Triple SILAC quantitative proteomic analyses comparing exosomes from immortalized cells and non-small cell lung cancer cell lines identified 721 different proteins including mutant Kras, EGFR, GRB2, and SRC specific to NSCLC exosomes. These oncoproteins were shown to promote cellular proliferation in the target cells [35]. Rab proteins have been extensively studied for their diverse role and especially for their functions related to membrane trafficking. Rabs, utilize the guanine nucleotide-dependent switch mechanism to mediate several of the four major steps in membrane traffic that include budding of vesicles, delivery of vesicles, tethering of the vesicles, and fusion of vesicles [36]. Using an RNA interference screening methodology, Ostrowski and colleagues showed that five different Rab GTPases work in tandem to promote exosome secretion in HeLa cells [37]. More specifically, they showed that Rabs (27a and 27b) function in endosome attachment to the plasma membrane and Rab27a silencing regulated the size of multivesicular endosomes. On the other hand, Rab27b silencing had a more prominent effect on the redistribution of multivesicular endosomes towards the perinuclear region. Due in part to their role in trafficking, around 2010 Hsu and colleagues investigated the role of GTPases in exosome generation [38]. Using a neural system, they showed that inhibition of Rab35 (which is activated by its GAP the TBC1 domain family member 10 A) GTPase function leads to the accumulation of endosomal vesicles that blocks exosome secretion. Here Rab35 demonstrated localization to the surface of oligodendroglia in a GTP-dependent manner, thereby impacting density of vesicles suggesting its critical role in docking/tethering.
In a very recent study the impact of constitutive activation S1P signaling-induced filamentous actin (F-actin) formation on Rho family GTPases, including Cdc42/Rac1, and its consequence on exosomal maturation was evaluated using fluorescence energy transfer (FRET) methodology and also by capturing structural changes [39]. These studies revealed that S1P protein works by enhancing F-actin formation that was abrogated by small molecule inhibitors of Gβγ subunits (M119) and could be fully salvaged by the synchronized expression of constitutively active Cdc42 and Rac1. These results further confirmed that triggering of the S1P receptor and the consequent activation of the downstream G protein signaling to Gβγ subunits/Rho family GTPases-promoted sorting into exosomes.
Ras-related proteins (Ral) GTPases have been studied for their role as regulators of multivesicular body formation and exosome release. Using a nematode model and mammary tumor cells, Hyenne and colleagues showed that the Ral GTPases have a significant role in exosome biogenesis [40]. RAL-1 was shown to localize at the surface of secretory multivesicular bodies and was proven to help multivesicular formation and fusion to the plasma membrane. In the mammalian system, both RalA and RalB were found to be essential for the release of exosomes in cellular culture. The microtubule-associated RhoA-activating factor GEF-H1 has been shown to play a role in endo- and exocytic trafficking of the vesicles [41]. Such investigations led to the identification that GEF-H1 promotes RhoA upon stimulation of RalA GTPase, which in turn regulates the assembly of the exocytosis machinery. These studies, therefore provided conclusive proof for the RhoA mediated activation arising from the initial RalA signal during the trafficking of vesicles.
8. Mutant K-ras detection in exosomes
The first studies linking Ras to exosomes came in 2003 during the investigations on the role of interferon (IFN)-inducible proteins on normal cell growth and functions [42]. This report highlighted that IFN-inducible proteins are abundantly expressed in exosome-like structures. The same study also described that Ras-mediated alterations of mouse mast cells were associated with the down-regulation of IFN-inducible proteins thereby linking it to malignant transformation. In later years, Ji and colleagues evaluated the exosomes from NIH3T3 and Ras-transformed NIH-3T3 cells for differences in the protein expression [43]. In their report, certain unique differences between parent and Ras-transformed cells were observed. Milk fat globule EGF factor 8, Serpin B6, 14-3-3 isoforms, collagen alpha-1 (VI), the eukaryotic translation initiation factors elF-3 gamma and elF-5 A, and the guanine nucleotide-binding proteins such as v-Ha-Ras p21 protein were the prominent proteins that were found to be over-expressed in exosomes upon Ras-induced oncogenic transformation. Similarly, studies investigating the lipid-related proteins in exosomes revealed universal expression of a majority of Ras GTPase family proteins [44]. These GTPases were linked to activation of the phospholipases in exosomes. In glioblastoma models, Luhtala and colleagues demonstrated that the exosome like particles from an H-RasV12 myr-Akt mouse model for glioblastoma multiforme carried active Ras protein signal transduction, and also harbor active Ras. These authors showed that GTP binding of K-Ras was indispensable for its loading within exosomes [45].
9. Mutant Kras and its consequence on exosomes mediated extracellular signaling
Cells containing wild-type K-Ras behave quite different from those with mutant K-Ras both phenotypically and molecularly. The differences are also observable within exosome-mediated signaling in cells harboring different K-Ras mutations. A comparison of exosomes secreted from mutant K-Ras vs wild-type Ras cell lines using proteomic analysis revealed several key differences in proteins content and in their abilities to mediate distinct signaling mechanisms. The mutant K-ras derived exosomes showed higher expression of oncogenic proteins including K-Ras, integrins, EGFR and Src among others [46]. Such high oncogenic protein content, particularly related to K-Ras pathway, showed a direct link to the transformation inducing capabilities in wild-type K-Ras cells. In another study, exosomes from Ras-transformed Madin-Darby Canine Kidney cells were reprogrammed with factors such as matrix metalloproteinase (MMP) MMP-1, MMP-14, MMP-19, a disintegrin and metalloproteinase domain-containing protein (ADAM) ADAM-10, ADAMTS1, integrin beta −1 (ITGB1), integrin alpha (ITGA3, and ITGA6), Y box binding protein 1 (YBX1), splicing factor 3B subunit 1 (SF3B1), splicing factor 3B subunit 3 (SF3B3), serine and arginine rich splicing factor 1 (SFRS1) and showed direct induction of epithelial-tomesenchymal transition (EMT) in recipient epithelial cells [47]. In pancreatic cancer cellular models, mutant K-Ras-expressing MiaPaCa-2 cells exhibited excessive pinocytosis which promoted transport of exosomes into the cells when compared with BxPC-3 pancreatic cancer cell line that carries wild-type K-Ras [48].
In studies investigating the tumor microenvironment, prostate cancer cell-derived exosomes that were abundant in H-Ras and K-Ras signaling further contained small RNAs such as microRNA-125b, microRNA-130b, and microRNA-155 as well Rab (G proteins) 1a, 1b, and 11a belonging to the Ras superfamily of GTPases. The presence of these miRNAs and Rab proteins were observed to induce aggressive tumors in secondary recipients [49]. Further, a recent study showed that the microbial metabolite Manumycin-A (MA), could inhibit exo-some formation and is released only by hormone refractory/castration-resistant prostate cancer (CRPC) and not normal prostate cells [50]. Such inhibition by MA was shown to be directly related to the targeting of Ras/Raf/ERK1/2 signaling and in part due to ERK-dependent inhibition of the oncogenic splicing factor hnRNP H1. Lee and colleagues showed that transformation of the human H-Ras oncogene in the rat epithelial cell resulted in the enhanced secretion of exosomes carrying chromatin-associated double-stranded DNA fragments that could promote proliferation in distinct recipient cells [51]. Similarly, Song and colleagues showed that the extended survival of tumor monocytes (that are precursors of tumor associated macrophages), occurs through cancer-derived exosomes. These exosomes were demonstrated to abundantly express activated Ras and extracellular signal-regulated kinases in the MAP kinase pathway, phosphorylated EGFR and human epidermal growth factor receptor 2 [52]. Lastly, using Ras-transformed MDCK cells it was demonstrated that ability of oncogenic cells to undergo EMT and impact distant endothelial cells via exosomes is through a mechanism involving Rho GTPase effectors Rac1/PAK2 that were postulated to be the basis of a metastatic niche [53].
10. Oncogenic K-ras in exosomes and cancer diagnostics
Several studies have highlighted the differences in exosomal content between cancer and normal cells thereby allowing for the potential advancement of current cancer detection methods based on the exosomal components. The role of oncogenic RAS in cancer and normal cellular exosome biogenesis and regulation has been clearly demonstrated. For example, in cervical cancer models, activating transcription factor 1 (ATF1) and RAS genes were detected in the systemically circulating blood exosomes of the corresponding murine models. Interestingly, this study revealed that ATF1 and RAS genes were found significantly elevated in tumors of primary and recurrent cervical cancer in the mouse model, and they were also detected in the blood exosomes making them excellent biomarkers for cancer monitoring [54]. In the quest for alternative pancreatic cancer diagnostics, mutations in K-Ras and p53 were detected in the exosomal DNA obtained from serum of pancreatic cancer patients as well as established cell lines [55]. In a follow-up study from the same group, the protein glypican-1 (GPC1) was found to be specifically enriched [GPC1(+)] on cancer-cell-derived exosomes in pancreatic cancer patients serum. This study was also able to correlate the levels of GPC1(+) in circulating exosomes with patient tumor load and overall survival who were eligible for surgical resection of tumors. Further the GPC1(+) positive circulating exosomes were also shown to possess specific K-Ras mutations [56]. Analysis of exosomal DNA demonstrated that a higher percentage of pancreatic patients with localized and early-stage disease exhibited detectable K-Ras mutations in exosome encapsulated DNA that was superior to that of the observed circulating DNA [57]. Nevertheless, this study also did detect mutations in K-Ras in normal/healthy individual derived exosomes.
11. MicroRNA sorting in exosomes and the impact on mutant Kras
Exosomes have been shown to harbor microRNAs. Some of these microRNAs can cross-talk with oncogenic Ras signaling. Cancer cells can maintain their oncogenic potential by exporting tumor suppressor mi-RNAs (particularly mutant K-Ras suppressing mi-RNAs) to the extracellular compartments through exosomes. For example, metastatic gastric cancer cell lines were shown to maintain their oncogenicity through secretion of Let-7 (a Ras regulating mi-RNA) through exosomes [58]. Investigations in paired colon cancer cell lines with differences in K-Ras mutational status revealed contrasting RNA profiles with over-expression of mir100 that was in contrast to the wild-type K-Ras cell line that showed enhanced mir10b indicating a K-Ras dependent sorting mechanism of these non-coding RNAs [59]. Studies have also revealed the direct role of Ras-MEK network in regulating the RNA-induced silencing complex (RISC) component Argonaute 2 (Ago2) thereby influencing microRNA secretion in exosomes [60]. The majority of exosomal DNA was shown to be in double stranded conformation and represent the entire genome in a study of leukimia, colorectal cancer and melanoma cell lines [61].
12. Targeting mutant K-Ras through exosomes
Aside from their role in cancer diagnostics and predictions for sensitivity or resistance to treatments, exosomes can be also be utilized in cancer therapeutics. There are several reviews that highlight the therapeutic potential of exosomes as carriers of anti-cancer warheads. Here we will highlight a specific case where proof of concept study was performed to show delivery of direct K-Ras targeted therapies to influence K-Ras pathway proteins using exosomes in cancer cells. In the first study of its kind, Raghu Kalluri’s group demonstrated that exosomes derived from normal fibroblast-like mesenchymal cells could be engineered to carry a short hairpin RNA specific to the commonly observed oncogenic KrasG12D mutation [62]. When compared to lysosome based RNAi, the exosome loaded KrasG12D siRNA (iExosome) demonstrated much-enhanced efficacy in cell lines and animal tumor models of pancreatic cancer. Molecularly the iExosome treatment showed marked suppression of K-Ras downstream effector pathways ERK only in Panc1 cell line with KrasG12D mutations and not MiaPaCa-2 or BxPC-3 (with KrasG12C or wild-type Kras, respectively). iExosomes were also effective in reducing tumor growth of the Panc1 orthotopic xeno-graft and prolonged the survival of Ptf1acre/+; LSL-KrasG12D/+; Tgfbr2Lox/Lox (KTC) mice model. The retention of exosomes in circulation was shown to be through their protection by CD47 allowing prolonged delivery of the KrasG12D short hairpin RNA. These initial proof-of-concept studies indicate that exosomes can be harnessed to tame mutant K-Ras.
13. Conclusions
Ras is mutated in ˜30–40% of all cancers and more frequently found to be altered in some of the difficult to treat cancers such as pancreatic ductal adenocarcinoma. Ras genes encode for proteins that play a central role in the biology of several critical growth regulatory pathways. As demonstrated in this review, Ras and its related proteins have an important role in exosomal secretion, maintenance, and cargo selection. Emerging studies indicate that these tiny particles can be utilized to target mutant K-Ras by delivering selective therapeutics. However, further-in-depth studies are needed to fully understand the critical role of mutant K-Ras pathway proteins in exosome biology that would help design better strategies that may revolutionize cancer therapeutic approaches as a whole. These studies will also help expanding beyond mutant K-Ras to treat other oncogene-driven cancers as well. This will help to effectively tame Ras pathway for better treatment outcome in some of the most difficult to treat cancers that are dependent on mutations in Kras.
Acknowledgements
Work in the lab of ASFAR S. AZMI is supported by NIH/NCI R37 CA215427–02. The authors acknowledge the support from SKY Foundation Inc. for pancreatic cancer research.
References
- [1].Bar-Sagi D, Ras proteins: biological effects and biochemical targets (review), Anticancer Res. 9 (1989) 1427–1437. [PubMed] [Google Scholar]
- [2].Papke B, Der CJ, Drugging RAS: know the enemy, Science 355 (2017) 1158–1163. [DOI] [PubMed] [Google Scholar]
- [3].Rotblat B, Yizhar O, Haklai R, Ashery U, Kloog Y, Ras and its signals diffuse through the cell on randomly moving nanoparticles, Cancer Res. 66 (2006) 1974–1981. [DOI] [PubMed] [Google Scholar]
- [4].Zhang S, Zhang Y, Qu J, Che X, Fan Y, Hou K, Guo T, Deng G, Song N, Li C,Wan X, Qu X, Liu Y, Exosomes promote cetuximab resistance via the PTEN/Akt pathway in colon cancer cells, Braz. J. Med. Biol. Res 51 (2017) e6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fu Q, Zhang Q, Lou Y, Yang J, Nie G, Chen Q, Chen Y, Zhang J, Wang J,Wei T, Qin H, Dang X, Bai X, Liang T, Primary tumor-derived exosomes facilitate metastasis by regulating adhesion of circulating tumor cells via SMAD3 in liver cancer, Oncogene 37 (47) (2018) 6105–6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W, Exosomes in cancer: small particle, big player, J. Hematol. Oncol 8 (2015) 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cox AD, Der CJ, The dark side of Ras: regulation of apoptosis, Oncogene 22 (2003) 8999–9006. [DOI] [PubMed] [Google Scholar]
- [8].Khan AQ, Travers JB, Kemp MG, Roles of UVA radiation and DNA damage responses in melanoma pathogenesis, Environ. Mol. Mutagen (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cox AD, Der CJ, Ras history: the saga continues, Small GTPases 1 (2010) 2–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bryant KL, Mancias JD, Kimmelman AC, Der CJ, KRAS: feeding pancreatic cancer proliferation, Trends Biochem. Sci 39 (2014) 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Marin-Ramos NI, Ortega-Gutierrez S, Lopez-Rodriguez ML, Blocking Ras inhibition as an antitumor strategy, Semin. Cancer Biol (2018). [DOI] [PubMed] [Google Scholar]
- [13].Yang WS, Yeo SG, Yang S, Kim KH, Yoo BC, Cho JY, Isoprenyl carboxyl methyltransferase inhibitors: a brief review including recent patents, Amino Acids 49 (2017) 1469–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Scott AJ, Lieu CH, Messersmith WA, Therapeutic approaches to RAS mutation, Cancer J. 22 (2016) 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Min Y, Caster JM, Eblan MJ, Wang AZ, Clinical translation of nanomedicine, Chem. Rev 115 (2015) 11147–11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R, Nanocarriers as an emerging platform for cancer therapy, Nat. Nanotechnol 2 (2007) 751–760. [DOI] [PubMed] [Google Scholar]
- [17].Bozzuto G, Molinari A, Liposomes as nanomedical devices, Int. J. Nanomed 10 (2015) 975–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Adams BD, Parsons C, Slack FJ, The tumor-suppressive and potential therapeutic functions of miR-34a in epithelial carcinomas, Expert Opin. Ther. Targets 20 (2016) 737–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Golan T, Khvalevsky EZ, Hubert A, Gabai RM, Hen N, Segal A, Domb A,Harari G, David EB, Raskin S, Goldes Y, Goldin E, Eliakim R, Lahav M,Kopleman Y, Dancour A, Shemi A, Galun E, RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients, Oncotarget 6 (2015) 24560–24570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Azmi AS, Bao B, Sarkar FH, Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review, Cancer Metastasis Rev. 32 (2013) 623–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schmidt O, Teis D, The ESCRT machinery, Curr. Biol 22 (2012) R116–R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hock A, Miyake H, Li B, Lee C, Ermini L, Koike Y, Chen Y, Maattanen P,Zani A, Pierro A, Breast milk-derived exosomes promote intestinal epithelial cell growth, J. Pediatr. Surg 52 (2017) 755–759. [DOI] [PubMed] [Google Scholar]
- [23].Baranyai T, Herczeg K, Onodi Z, Voszka I, Modos K, Marton N, Nagy G,Mager I, Wood MJ, El Andaloussi S, Palinkas Z, Kumar V, Nagy P, Kittel A, Buzas EI, Ferdinandy P, Giricz Z, Isolation of exosomes from blood plasma: qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods, PLoS One 10 (2015) e0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Street JM, Koritzinsky EH, Glispie DM, Star RA, Yuen PS, Urine exosomes: an emerging trove of biomarkers, Adv. Clin. Chem 78 (2017) 103–122. [DOI] [PubMed] [Google Scholar]
- [25].Prada I, Meldolesi J, Binding and fusion of extracellular vesicles to the plasma membrane of their cell targets, Int. J. Mol. Sci (2016) 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Janas AM, Sapon K, Janas T, Stowell MH, Janas T, Exosomes and other extracellular vesicles in neural cells and neurodegenerative diseases, Biochim. Biophys. Acta 1858 (2016) 1139–1151. [DOI] [PubMed] [Google Scholar]
- [27].Chahar HS, Bao X, Casola A, Exosomes and their role in the life cycle and pathogenesis of RNA viruses, Viruses 7 (2015) 3204–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Teow SY, Liew K, Khoo AS, Peh SC, Pathogenic role of exosomes in epstein-barr virus (EBV)-associated cancers, Int. J. Biol. Sci 13 (2017) 1276–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Teow SY, Nordin AC, Ali SA, Khoo AS, Exosomes in human immunodeficiency virus type I pathogenesis: threat or opportunity? Adv. Virol 2016 (2016) 9852494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Antonyak MA, Li B, Boroughs LK, Johnson JL, Druso JE, Bryant KL,Holowka DA, Cerione RA, Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells, Proc. Natl. Acad. Sci. U. S. A 108 (2011) 4852–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ashery U, Yizhar O, Rotblat B, Elad-Sfadia G, Barkan B, Haklai R, Kloog Y, Spatiotemporal organization of Ras signaling: rasosomes and the galectin switch, Cell. Mol. Neurobiol 26 (2006) 471–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ashery U, Yizhar O, Rotblat B, Kloog Y, Nonconventional trafficking of Ras associated with Ras signal organization, Traffic 7 (2006) 119–126. [DOI] [PubMed] [Google Scholar]
- [33].Kofer-Geles M, Gottfried I, Haklai R, Elad-Zefadia G, Kloog Y, Ashery U, Rasosomes spread Ras signals from plasma membrane’ hotspots’, Biochim. Biophys. Acta 1793 (2009) 1691–1702. [DOI] [PubMed] [Google Scholar]
- [34].Grunwald A, Gottfried I, Cox AD, Haklai R, Kloog Y, Ashery U, Rasosomes originate from the Golgi to dispense Ras signals, Cell Death Dis. 4 (2013) e496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Clark DJ, Fondrie WE, Yang A, Mao L, Triple SILAC quantitative proteomic analysis reveals differential abundance of cell signaling proteins between normal and lung cancer-derived exosomes, J. Proteomics 133 (2016) 161–169. [DOI] [PubMed] [Google Scholar]
- [36].Grosshans BL, Ortiz D, Novick P, Rabs and their effectors: achieving specificity in membrane traffic, Proc. Natl. Acad. Sci. U. S. A 103 (2006) 11821–11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF,Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M,Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C, Rab27a and Rab27b control different steps of the exosome secretion pathway, Nat. Cell Biol 12 (2010) 19–30. [DOI] [PubMed] [Google Scholar]
- [38].Hsu C, Morohashi Y, Yoshimura S, Manrique-Hoyos N, Jung S, Lauterbach MA,Bakhti M, Gronborg M, Mobius W, Rhee J, Barr FA, Simons M, Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C, J. Cell Biol 189 (2010) 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kajimoto T, Mohamed NNI, Badawy SMM, Matovelo SA, Hirase M,Nakamura S, Yoshida D, Okada T, Ijuin T, Nakamura SI, Involvement of Gbetagamma subunits of Gi protein coupled with S1P receptor on multivesicular endosomes in F-actin formation and cargo sorting into exosomes, J. Biol. Chem 293 (2018) 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hyenne V, Apaydin A, Rodriguez D, Spiegelhalter C, Hoff-Yoessle S, Diem M,Tak S, Lefebvre O, Schwab Y, Goetz JG, Labouesse M, RAL-1 controls multi-vesicular body biogenesis and exosome secretion, J. Cell Biol 211 (2015) 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pathak R, Dermardirossian C, GEF-H1: orchestrating the interplay between cytoskeleton and vesicle trafficking, Small GTPases 4 (2013) 174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Brem R, Oraszlan-Szovik K, Foser S, Bohrmann B, Certa U, Inhibition of proliferation by 1–8U in interferon-alpha-responsive and non-responsive cell lines, Cell. Mol. Life Sci 60 (2003) 1235–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ji H, Erfani N, Tauro BJ, Kapp EA, Zhu HJ, Moritz RL, Lim JW,Simpson RJ, Difference gel electrophoresis analysis of Ras-transformed fibroblast cell-derived exosomes, Electrophoresis 29 (2008) 2660–2671. [DOI] [PubMed] [Google Scholar]
- [44].Subra C, Grand D, Laulagnier K, Stella A, Lambeau G, Paillasse M, De Medina P.,Monsarrat B, Perret B, Silvente-Poirot S, Poirot M, Record M, Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins, J. Lipid Res 51 (2010) 2105–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Luhtala N, Aslanian A, Yates III JR, Hunter T, Secreted glioblastoma nanovesicles contain intracellular signaling proteins and active ras incorporated in a farnesylation-dependent manner, J. Biol. Chem 292 (2017) 611–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Demory Beckler M, Higginbotham JN, Franklin JL, Ham AJ, Halvey PJ,Imasuen IE, Whitwell C, Li M, Liebler DC, Coffey RJ, Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS, Mol. Cell Proteomics 12 (2013) 343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tauro BJ, Mathias RA, Greening DW, Gopal SK, Ji H, Kapp EA,Coleman BM, Hill AF, Kusebauch U, Hallows JL, Shteynberg D, Moritz RL, Zhu HJ, Simpson RJ, Oncogenic H-ras reprograms Madin-Darby canine kidney (MDCK) cell-derived exosomal proteins following epithelial-mesenchymal transition, Mol. Cell Proteomics 12 (2013) 2148–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nakase I, Kobayashi NB, Takatani-Nakase T, Yoshida T, Active macropinocytosis induction by stimulation of epidermal growth factor receptor and oncogenic Ras expression potentiates cellular uptake efficacy of exosomes, Sci. Rep 5 (2015) 10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Abd Elmageed ZY, Yang Y, Thomas R, Ranjan M, Mondal D, Moroz K, Fang Z, Rezk BM, Moparty K, Sikka SC, Sartor O, Abdel-Mageed AB, Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes, Stem Cells 32 (2014) 983–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Datta A, Kim H, Lal M, McGee L, Johnson A, Moustafa AA, Jones JC,Mondal D, Ferrer M, Abdel-Mageed AB, Manumycin A suppresses exosome bio-genesis and secretion via targeted inhibition of Ras/Raf/ERK1/2 signaling and hnRNP H1 in castration-resistant prostate cancer cells, Cancer Lett. 408 (2017) 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lee TH, Chennakrishnaiah S, Audemard E, Montermini L, Meehan B, Rak J, Oncogenic ras-driven cancer cell vesiculation leads to emission of double-stranded DNA capable of interacting with target cells, Biochem. Biophys. Res. Commun 451 (2014) 295–301. [DOI] [PubMed] [Google Scholar]
- [52].Song X, Ding Y, Liu G, Yang X, Zhao R, Zhang Y, Zhao X, Anderson GJ, Nie G, Cancer cell-derived exosomes induce mitogen-activated protein kinase-dependent monocyte survival by transport of functional receptor tyrosine kinases, J. Biol. Chem 291 (2016) 8453–8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gopal SK, Greening DW, Hanssen EG, Zhu HJ, Simpson RJ, Mathias RA, Oncogenic epithelial cell-derived exosomes containing Rac1 and PAK2 induce angiogenesis in recipient endothelial cells, Oncotarget 7 (2016) 19709–19722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Shi Y, Wang W, Yang B, Tian H, ATF1 and RAS in exosomes are potential clinical diagnostic markers for cervical cancer, Cell Biochem. Funct 35 (2017) 477–483. [DOI] [PubMed] [Google Scholar]
- [55].Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, Koch M, Zhang J, Weitz J,Chin L, Futreal A, Kalluri R, Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer, J. Biol. Chem 289 (2014) 3869–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J,LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R, Glypican-1 identifies cancer exosomes and detects early pancreatic cancer, Nature 523 (2015) 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Allenson K, Castillo J, San Lucas FA, Scelo G, Kim DU, Bernard V, Davis G,Kumar T, Katz M, Overman MJ, Foretova L, Fabianova E, Holcatova I,Janout V, Meric-Bernstam F, Gascoyne P, Wistuba I, Varadhachary G,Brennan P, Hanash S, Li D, Maitra A, Alvarez H, High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients, Ann. Oncol 28 (2017) 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y,Muramatsu K, Fukuda Y, Ogura S, Yamaguchi K, Mochizuki T, Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line, PLoS One 5 (2010) e13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, Weaver AM, Vickers K, Prasad N, Levy S, Zhang B, Coffey RJ, Patton JG, KRAS-dependent sorting of miRNA to exosomes, Elife 4 (2015) e07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].McKenzie AJ, Hoshino D, Hong NH, Cha DJ, Franklin JL, Coffey RJ, Patton JG, Weaver AM, KRAS-MEK signaling controls Ago2 sorting into exosomes, Cell Rep. 15 (2016) 978–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, Zheng Y,Hoshino A, Brazier H, Xiang J, Williams C, Rodriguez-Barrueco R, Silva JM,Zhang W, Hearn S, Elemento O, Paknejad N, Manova-Todorova K, Welte K,Bromberg J, Peinado H, Lyden D, Double-stranded DNA in exosomes: a novel biomarker in cancer detection, Cell Res. 24 (2014) 766–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ,Kalluri R, Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer, Nature 546 (2017) 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]