Abstract
Cell size is important for cell physiology because it sets the geometric scale of organelles and biosynthesis. A number of methods exist to measure different aspects of cell size, but each has significant drawbacks. Here, we present an alternative method to measure the size of single human cells using a nuclear localized fluorescent protein expressed from a constitutive promoter. We validate this method by comparing it to several established cell size measurement strategies, including flow cytometry optical scatter, total protein dyes, and quantitative phase microscopy. We directly compare our fluorescent protein measurement with the commonly used measurement of nuclear volume and show that our measurements are more robust and less dependent on image segmentation. We apply our method to examine how cell size impacts the cell division cycle and reaffirm that there is a negative correlation between size at cell birth and G1 duration. Importantly, combining our size reporter with fluorescent labeling of a different protein in a different color channel allows measurement of concentration dynamics using simple wide-field fluorescence imaging. Thus, we expect our method will be of use to researchers interested in how dynamically changing protein concentrations control cell fates.
INTRODUCTION
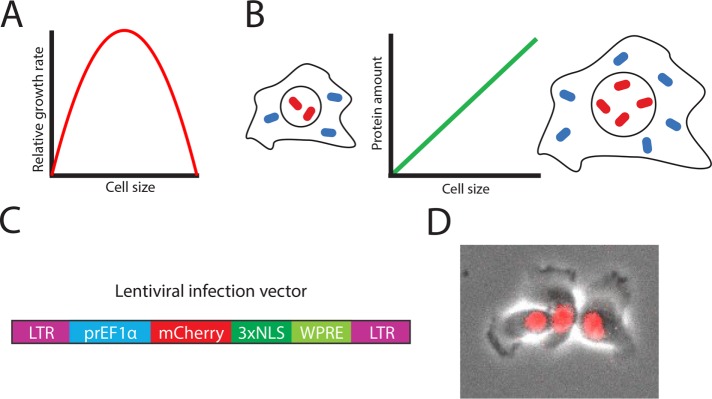
Cell size has an important effect on cellular physiology through its influence on biosynthesis, mitochondrial efficiency, and hormone secretion (Figure 1A; Smith, 1971; Pende et al., 2000; Miettinen and Björklund, 2016). Variation in cell size is one of the most noticeable differences between cells of different type and function (Ginzberg et al., 2015). However, this size variation is rarely emphasized by researchers, possibly because it has been difficult to measure. While a growing number of methods provide size information, either as the experiment’s primary objective or as an additional parameter during experiments designed for other purposes, there is a need for more convenient methods for measuring size during live-cell microscopy (Table 1).
FIGURE 1:
Measuring cell size using a constitutively expressed fluorescent protein. (A) Single-cell growth rate has an optimum as a function of cell size (Miettinen and Björklund, 2016). (B) Principle of size reporter: the amount of constitutively expressed proteins increases in proportion to cell size. (C) Schematic of the lentiviral infection vector. LTR denotes long terminal repeats, prEF1α denotes 1kb of the EF1α promoter, NLS denotes nuclear localization sequence, and WPRE denotes a woodchuck posttranscriptional regulatory element that boosts expression (Zufferey et al., 1999). (D) Representative composite phase and fluorescence image of HMEC cells expressing mCherry-NLS from an EF1α promoter.
TABLE 1:
Comparison of methods to measure cell size.
Further complicating accurate measurement of cell size is the ambiguity as to what exactly “size” means. In general, researchers mean one of three things: volume, dry mass, or protein content. Different techniques exist to measure each of these parameters, but all three mostly correlate and are thought to reflect size. That is, cells of a given type in a particular condition have constant ratios of mass to volume and of protein content to mass. However, some cells, including mitotic cells, chondrocytes, and cell cycle arrested budding yeast, dilute their dry mass so it is important to understand which parameter a particular technique is measuring (Cooper et al., 2013; Son et al., 2015; Zlotek-Zlotkiewicz et al., 2015; Neurohr et al., 2019).
The gold standard for cell volume measurements is considered to be the Coulter counter, which flows cells through a measurement chamber where they displace an isotonic solution to cause a volume-proportional change in electrical impedance (Gregg and Steidley, 1965). Coulter counters measure cell number and size in high throughput, but do not provide any additional information (such as amount of a protein of interest) and cannot make multiple measurements on the same cell over time (Tzur et al., 2009). A related single time point high-throughput technique is flow cytometry, in which cells pass one by one through a series of lasers. Flow cytometry can be used to collect multidimensional, high-throughput data when used in combination with immunofluorescence techniques labeling individual protein species. Flow cytometers also measure two optical properties—forward scatter (FSC) and side scatter (SSC)—that reflect cell size, shape, and granularity. Although commonly used as a proxy for size, evidence for a quantitative, linear relationship between FSC and cell size is sparse, and the relationship depends on cell type and flow cytometer machine and settings (Holme et al., 1988; Tzur et al., 2011; Shapiro, 2018). Calibration of size estimates from cytometry scatter is often done by comparison to fluorescent dyes that stain total cellular protein. These dyes, CFSE and SE-Alexa647, use reactive succinimidyl ester groups to react with free amino groups on proteins, so that the amount of staining corresponds to total cellular protein (Kafri et al., 2013; Shapiro, 2018). Importantly, these dyes do not provide quantitative information on total protein when used in live-cell imaging.
Live-cell microscopy provides a unique and rich source of single-cell data on cellular processes through time. Live-cell studies have been particularly impactful for studying the cell cycle, where molecular signaling and the passage of time both contribute to progression through cell cycle phases (Doncic et al., 2011; Doncic and Skotheim, 2013; Spencer et al., 2013; Atay et al., 2016; Cappell et al., 2016; Barr et al., 2017; Schwarz et al., 2018). Because cell size changes over time in growing cells and has been shown to be an important driver of cell cycle transitions, it is particularly important to accurately measure cell size in cell cycle experiments (DiTalia et al., 2007; Schmoller et al., 2015). Unfortunately, such studies, particularly in the irregularly shaped animal cells, have been limited by the challenge of accurately measuring cell size during live-cell microscopy.
The simplest strategy to determine cell size from imaging data is to directly measure cellular area, and then calculate cell volume by assuming that cells are spherical, cylindrical, ellipsoid, or some other regular geometry. While this strategy is effective in many bacteria, yeast, and round mitotic, trypsinized, or suspension animal cells, it is inadequate for adherent interphase animal cells due to their irregular shape. A straightforward extension of the geometric strategy is to instead measure nuclear area, calculate nuclear volume assuming a spherical nucleus, and then rely on the known proportionality relationship between nuclear volume and cellular volume, that is, the fixed karyoplasmic ratio, to estimate cell size (Jorgensen et al., 2007; Neumann and Nurse, 2007; Edens et al., 2013; Levy and Heald, 2016). Two disadvantages of this approach are that the correlation between nuclear and total cellular size is not perfect—for example, human cells’ karyoplasmic ratios are lower in differentiated cells, and higher in high-grade tumors (Kumar et al., 2015)—and that it is sensitive to small errors in nuclear area measurements. Nevertheless, it has been used successfully to demonstrate that cell cycle phase modulates cellular growth rate and to identify a role for p38 in maintenance of cell size homeostasis (Ginzberg et al., 2018; Liu et al., 2018).
Cell volumes can be reconstructed in three dimensions using confocal microscopy when the cell membrane is fluorescently labeled. Nuclear volumes can also be reconstructed using a fluorescent nuclear membrane dye. However, these methods are limited by phototoxicity in live-cell experiments, and rely on challenging 3D image segmentation (Bajcsy et al., 2015). Another microscopy-based approach that measures cell dry mass is quantitative phase microscopy (QPM). This technique detects the phase shift as light waves pass through a specimen whose refractive index differs from that of the medium. The specimen’s optical path length can then be calculated by comparing the phase shift through the specimen to a blank reference image. Because the specimen’s optical path length is proportional to its dry mass, QPM provides a per-pixel optical mass. However, QPM can be difficult to use because it requires a specialized camera that only works with some objective lenses and its accuracy depends on acquiring a good reference image and precisely defining cell boundaries (Barer, 1952; Mir et al., 2011; Zangle and Teitell, 2014).
Three novel techniques have recently expanded the repertoire for measuring cell size. Microchannel resonators contain a microfluidic channel suspended within a vibrating cantilever. When single suspended cells flow through the channel, they displace the medium and the overall mass of the cantilever increases by the cell’s buoyant mass. This buoyant mass increase is then measured by the change in vibration frequency of the cantilever. By embedding microchannel resonators within a more complex microfluidic setup, it is possible to measure individual cells’ buoyant masses at multiple timepoints (Godin et al., 2010; Son et al., 2012, 2015; Cermak et al., 2016). Furthermore, other techniques, such as fluorescence microscopy or single-cell RNA sequencing, can be added (Stevens et al., 2016; Cetin et al., 2017; Kimmerling et al., 2018). The microchannel resonator approach is powerful and precise but requires specialized microfluidic instrumentation, is low throughput, and is not easily applied to adherent cells (Park et al., 2008, 2010). Another new technique is fluorescence exclusion microscopy in which cells are grown in a medium containing fluorescent dextran. As long as the cells do not take up the fluorescent compound, the “background” fluorescence of a region can be measured. A cell displaces fluorescent media so that the amount of reduced fluorescence corresponds to cell volume. This approach does require cells to grow in a specialized low-ceiling device (Bottier et al., 2011; Zlotek-Zlotkiewicz et al., 2015; Cadart et al., 2018). Both microchannel resonators and fluorescence exclusion were used to show that cell density is nearly constant throughout the cell cycle, but that cells swell ∼20% during mitosis (Grover et al., 2011; Bryan et al., 2013; Son et al., 2015; Zlotek-Zlotkiewicz et al., 2015). A final option is to sidestep the difficulty of measuring irregular shapes by forcing animal cells to adopt more convenient morphologies. For example, cells can be grown in narrow channels requiring them to adopt a rod-shaped morphology, where length is directly proportional to volume (Varsano et al., 2017). However, it is unknown to what extent such shape manipulation affects cell physiology.
Here, we present an alternative strategy, inspired by work in yeast (DiTalia et al., 2007), that can be added to the toolkit of researchers seeking to measure animal cell size. Our approach relies on the fact that most proteins in a cell are produced proportionally to the total protein content of the cell (Figure 1B; Newman et al., 2006; Zhurinsky et al., 2010; Neurohr et al., 2019). In this article, we show that a constitutively expressed nuclear fluorescent protein can easily be used to measure animal cell size. We use this technique to understand size control at the G1/S transition, where we observe that small birth size correlates with long G1 duration and that large size during G1 predicts rapid S phase entry.
RESULTS
Construction of a prEF1α-mCherry-NLS size reporter cell line
Good candidate promoters for a fluorescent total protein reporter should be highly, ubiquitously, and constitutively expressed. Promoters for genes involved in protein translation frequently meet these criteria. We selected the promoter of the translation elongation factor EF1α because it has also been commonly used in lentiviral infection systems (Chang et al., 2011; Cheng et al., 2011; Schwarz et al., 2018). We used this promoter to drive expression of either of two nuclear localized fluorescent proteins: mCherry-NLS and E2-Crimson-NLS. We selected these fluorescent proteins due to their fast maturation, long lifespan, and compatibility with other fluorescent reporters of interest such as mAG-Geminin and an Rb-Clover fusion protein (Shaner et al., 2004; Strack et al., 2009). Localizing the fluorescent reporter to the nucleus facilitates image segmentation and measurement of total protein via wide-field fluorescence imaging.
To construct a cell line with a total protein reporter, we inserted a prEF1α-mCherry-NLS expression cassette into immortalized human mammary epithelial cells (HMECs) by lentiviral infection and confirmed bright nuclear expression of the fluorescent protein (Figure 1, C and D). Because we expected that expression variability due to gene copy number and location within the genome could be a major source of noise when comparing expression across cells, we sorted single cells by fluorescence-activated cell sorting (FACS) and expanded clones.
Next, we set about assessing how well mCherry expression reflected cell size within a clone. Because our approach works only in cells, it cannot be validated by measuring noncell objects of known sizes, such as beads. Therefore, because there is no single gold-standard method for size measurement (Table 1), we proceed to compare mCherry-NLS expression by several established methods.
Constitutively expressed mCherry-NLS correlates with scatter, nuclear volume, and total protein
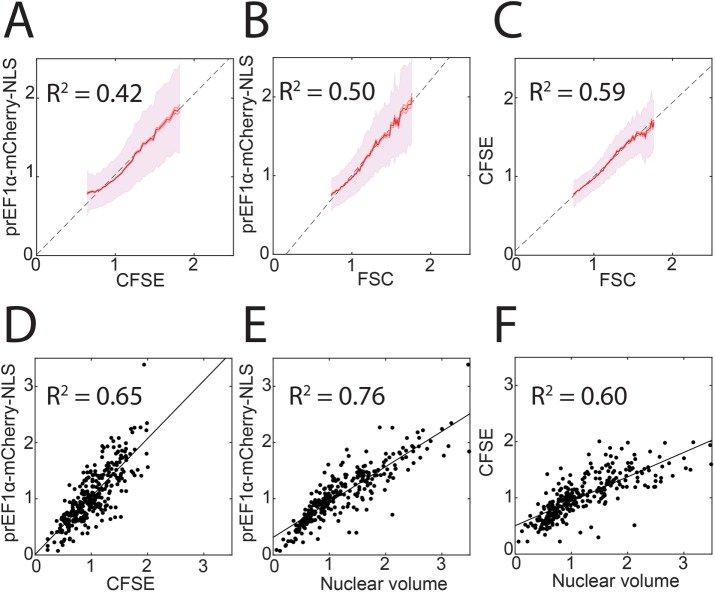
We incubated EF1α-mCherry-NLS HMECs with the protein dye CFSE and used flow cytometry to measure individual cells’ FSC, CFSE amount, and mCherry amount. We plotted each pair of measurements and performed a linear regression (Figure 2, A–C). The intercepts for all three lines were close to the origin, indicating that all three measurements are approximately proportional. We found similar coefficients of determination (R2 between 0.4 and 0.6) between all three pairs of measurements, suggesting that no one measurement is substantially noisier than the others. We compared these cells with HMECs expressing mCherry-NLS from the ACTB (beta-actin) promoter and determined that EF1α-mCherry-NLS was approximately eightfold brighter (in terms of median mCherry intensity) and more proportional to size (Supplemental Figure S1, A and B). To test whether our strategy also works in another cell type, we introduced EF1α-mCherry-NLS into K562 cells. We found that also in these cells, mCherry-NLS was proportional to FSC (Supplemental Figure S1C). Moreover, comparing similar plots across 10 clones of EF1α-mCherry-NLS K562 cells, we observed a positive relationship between median mCherry intensity in each clone and the coefficient of determination (Supplemental Figure S1D). These data support utilizing a highly expressed fluorescent signal to minimize autofluorescence and transcriptional noise and to maximize cell size measurement accuracy.
FIGURE 2:
Comparison of constitutively expressed fluorescent protein with other cell size metrics. (A–C) Binned means and standard deviations for prEF1α-mCherry-NLS, CFSE, and FSC as measured by flow cytometry. CFSE is a total protein dye and FSC is the optical forward scatter (see Materials and Methods). Each metric was normalized to its own median, and bins are shown for the middle 95% of the data. Least-squares regression lines and coefficients of determination are also shown. N > 30,000 cells. (D–F) Single-cell measurements made using wide-field fluorescence microscopy, least-squares linear regression, and coefficients of determination for the indicated metrics. N = 303 cells.
Next, we returned to EF1α-mCherry-NLS HMECs and examined them by fluorescence microscopy rather than flow cytometry. In this experiment, we measured nuclear volume (calculated as [nuclear area]3/2) and performed a pairwise comparison with mCherry-NLS and with CFSE total protein dye fluorescence signals (Figure 2, D–F). Once again, we observed that all three intercepts were close to the origin and that the coefficients of determination were similar, suggesting that none of the three measurements is substantially inferior to the other two.
Constitutively expressed mCherry-NLS size estimates are less sensitive than nuclear volume to variation in image segmentation
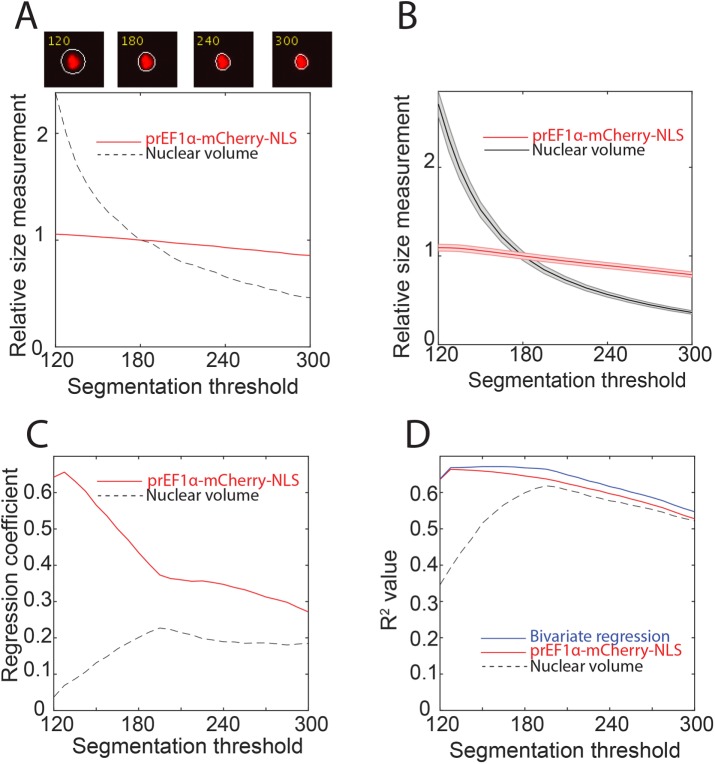
Because nuclear volume and mCherry-NLS total intensity are the two most experimentally straightforward approaches to measuring live cell size, we sought to compare them head-to-head. One pitfall of fluorescence microscopy is that it is necessary to distinguish foreground objects (cells or nuclei) from background by segmenting the image, often by applying a brightness threshold. We examined how varying our nuclear segmentation threshold (mCherry pixel intensity) affected our measurement of nuclear volume and mCherry-NLS total intensity. One representative nucleus segmented at different thresholds is shown in Figure 3A. Nuclear volume for this cell was highly sensitive to threshold choice, especially at less stringent thresholds, while mCherry-NLS total intensity was robust to threshold choice. To confirm this observation, we applied different thresholds to all the cells in our images and saw a similar pattern (Figure 3B). The reason for this phenomenon can be seen in the representative nucleus images. We can be confident that the region segmented at the most stringent threshold is truly part of the nucleus, while the peripheral halo that expands as the threshold decreases is less certain to be truly part of the nucleus. Adding or subtracting a single pixel from the uncertain periphery of the segmented area has an equivalently large effect on nuclear area or volume as adding or subtracting a pixel from the high-confidence central region. However, because the vast majority of the mCherry fluorescence is contained in the high-confidence central region (because the vertical diameter of the nucleus is largest there), changes in the segmentation threshold have only a small effect on the total mCherry fluorescence measured. Thus, mCherry fluorescence is less sensitive than nuclear volume to changes in segmentation.
FIGURE 3:
Sensitivity of nuclear volume and EF1α-mCherry-NLS to the segmentation threshold. (A) Top panels, a single nucleus segmented at four different intensity thresholds. Bottom panel, nuclear volume and total mCherry-NLS measurements for that cell at each threshold. Measurements were normalized to their value when the segmentation threshold is 180 (see Materials and Methods). (B) Mean and associated SE of nuclear volume and mCherry-NLS measurements for a cell population at different intensity thresholds. Measurements were normalized to the mean value when the segmentation threshold is 180. N = 379 cells. (C, D) Analysis of the correlation of nuclear volume and mCherry-NLS with CFSE total protein dye. Each measurement was normalized to its mean, and bivariate regression was performed using nuclear volume and mCherry-NLS as predictor variables, and using CFSE as the response variable. N = 303 cells. (C) Plot of the regression coefficient (i.e., slope) for each predictor variable as a function of segmentation threshold in a bivariate regression indicating that mCherry-NLS contributes more to predicting CFSE. (D) Plot of the coefficient of determination as a function of segmentation threshold for the bivariate regression alongside the coefficient of determination for each single variable regression.
To confirm that this result was not an artifact of using the same mCherry fluorescent reporter to calculate the nuclear volume and to define the region of interest in which the mCherry total nuclear intensity was measured, we performed a similar experiment using the DNA stain 4’,6-diamidino-2-phenylindole (DAPI) to segment the nucleus. Just as with mCherry, it is necessary but difficult to define the appropriate segmentation threshold in the DAPI channel. We tested a variety of DAPI segmentation thresholds and found that, consistent with our prior findings, the estimated nuclear volume was sensitive to the segmentation threshold, while mCherry fluorescence within the region of interest was not (Supplemental Figure S2).
To further compare nuclear volume and mCherry fluorescence, we stained cells with the total protein dye CFSE and again segmented the nuclei at various thresholds. At each threshold, we performed a bivariate regression with CFSE as the response variable and nuclear volume and mCherry as the predictor variables. Both nuclear volume and mCherry were normalized to their mean values. The regression coefficient—that is, the slope with respect to the predictor variable—for the mCherry protein reporter was higher than for the nuclear volume, indicating that CFSE intensity in the bivariate regression was predominantly predicted by our protein reporter. This effect is enhanced at less stringent segmentation thresholds (Figure 3C). Moreover, adding nuclear volume to the regression only marginally improves the coefficient of determination with CFSE compared with mCherry alone (Figure 3D). Taken together, these data support preferential use of our EF1α-mCherry-NLS construct over nuclear volume to report total cellular protein content.
Constitutively expressed mCherry-NLS measurements are less noisy than nuclear volume measurements in live cells tracked over time
Having established that constitutively expressed mCherry-NLS reflects size in populations of cells at a single moment in time, we turned our attention to live-cell microscopy to examine size dynamics in single cells. We imaged EF1α-mCherry-NLS HMECs for 48–72 h, segmented the nuclei, and tracked them over time. We examined the growth of single cells by looking at single-cell traces of nuclear volume or mCherry intensity over time (Supplemental Movie 1 and Figure 4, A and B). We then compared the variability of nuclear volume versus mCherry total intensity within single-cell traces. Because nuclear volume and total protein content are only expected to change slowly over time, a variation in the signal over shorter time scales is presumably due to measurement and/or segmentation error. To estimate the magnitude of these sources of error, we approximated each trace with overlapping piecewise linear fits and summed the squared residuals from each timepoint (Figure 4, C and D). We found significantly more timepoint-to-timepoint variability in nuclear volume than in mCherry total intensity (Figure 4E). As in Figure 3, this result is because small changes in image segmentation have a large effect on nuclear volume but not on total mCherry measurements. We also used our live imaging data to investigate whether cell cycle phase affects mCherry-NLS expression. We categorized cells by Geminin expression as G1 or S/G2 and plotted mCherry-NLS versus nuclear volume for all individual measurements. We did not observe any detectable change in this relationship as a function of cell cycle phase, though as expected cells in S/G2 were larger (Figure 4F).
FIGURE 4:
Nuclear volume and mCherry-NLS size measurements for live single cells tracked through time. (A, B) Single-cell traces over time. Measurements were normalized to the mean of all the data. (C, D) Representative single-cell traces along with a piecewise fit to quantify timepoint-to-timepoint variability (see Materials and Methods). Each trace was normalized to its mean. (E) Plot of an error metric: the mean and SE of the sum of squared residuals from the piecewise linear fits to each cell as in C and D. (F) All individual single-cell, single-timepoint measurements of nuclear volume and mCherry-NLS were plotted and categorized as G1 or S/G2 according to the cell’s position in the cell cycle as determined by Geminin-mAG (see Materials and Methods).
Movie S1.
Time lapse imaging and tracking of prEF1α-mCherry-NLS HMECs. White outlines show automated nuclear segmentation, and the example cell shown in Figure 4C-D is outlined in cyan.
Constitutively expressed mCherry-NLS correlates with dry mass measurements
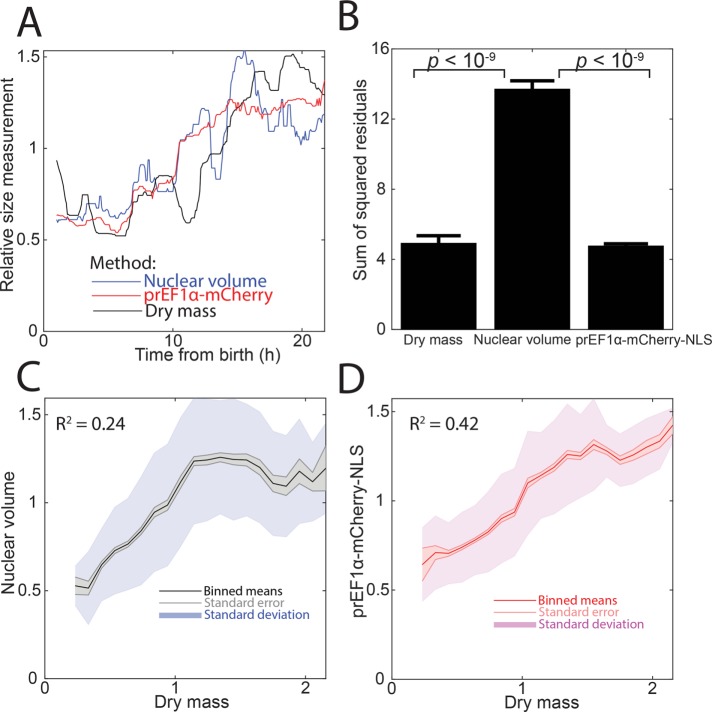
Next, we sought to compare our mCherry-NLS reporter with QPM that measures cell dry mass. In live cells tracked through time, we measured dry mass, mCherry total intensity, and nuclear volume. We found that mCherry-NLS and dry mass were less noisy than nuclear volume (Figure 5, A and B). Note that because accurate automated segmentation (as in Figure 4) was not possible in these quantitative phase images, all three measurements for this experiment are from manual segmentation. When we compared measurements across multiple cells, dry mass correlated better with mCherry than with nuclear volume. However, unlike with CFSE, these curves do not intercept close to the origin, highlighting the challenge of precisely taring quantitative phase data (Figure 5, C and D). On the basis of these experiments and those described in the preceding section, we concluded that mCherry intensity continues to reflect cell size during live-cell imaging.
FIGURE 5:
Comparison of nuclear volume, mCherry-NLS, and dry mass measurements. Dry mass was measured by quantitative phase microscopy (see Materials and Methods). (A) Representative trace of a single cell tracked through time. Each measurement type was normalized to its median. (B) Plot of an error metric: the mean and SE of the sum of squared residuals from a piecewise linear fit, as in Figure 4, C and D. N = 8 cells. (C, D) Correlation of nuclear volume or mCherry-NLS with cell dry mass. Binned means, SE of the mean, and SD of the data are shown. N > 2000 measurements.
Constitutively expressed E2-Crimson-NLS enables measurement of G1 size control
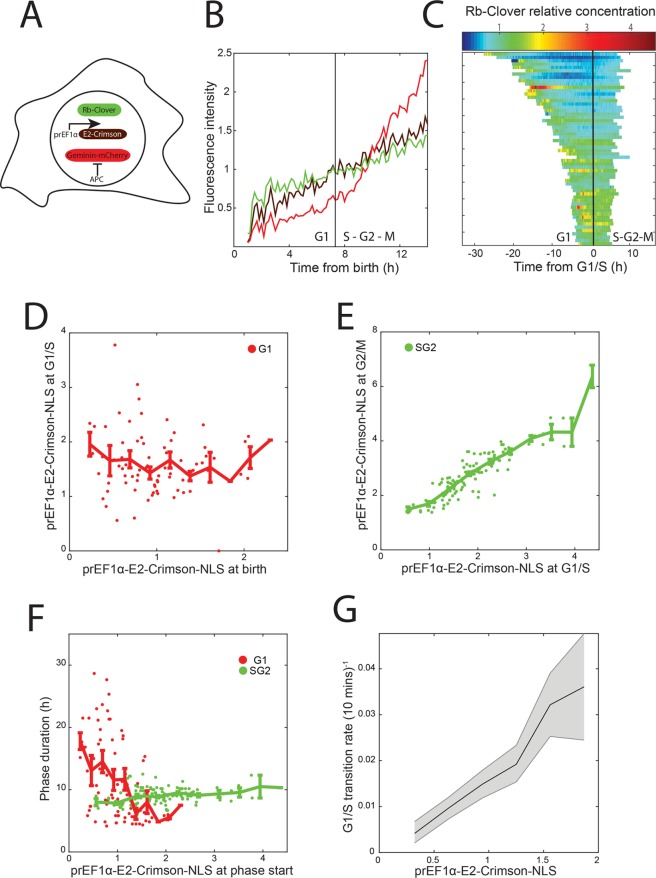
Finally, we applied our protein content measurements to describe cell growth and the concentration dynamics of key cell cycle regulators through the cell cycle, which we anticipate will be a frequent application of our method. To show how our size reporter can be used to measure concentration dynamics of a key cell cycle regulator, we integrated our size reporter into cells expressing the G1/S inhibitor protein Rb (retinoblastoma protein) tagged with Clover green fluorescent protein from its endogenous locus. RB was tagged using CRISPR/Cas9 as described in Zatulovskiy et al. (2018). We infected RB-Clover cells with lentivirus particles containing a Geminin-mCherry reporter construct to define the G1 to S phase transition by the accumulation of red fluorescence (Sakaue-Sawano et al., 2008). Owing to using a red fluorophore as a cell cycle phase reporter, we switched to far-red prEF1α-E2-Crimson-NLS to measure size (Figure 6A). This color-swapped reporter also correlates well with other size metrics (Supplemental Figure S3). We were able to track cells and make fluorescence measurements in three colors without phototoxicity (Figure 6B and Supplemental Figure S4). Rb protein concentration can then be measured as the ratio of Clover to E2-Crimson-NLS total fluorescence and plotted as a function of cell cycle phase (Figure 6C). We observed that Rb concentration decreases during G1 and increases again starting in S phase, consistent with Zatulovskiy et al. (2018).
FIGURE 6:
Analysis of the relationship between cell size and cell cycle progression. (A) Schematic of the three fluorescent reporters expressed in single cells. Rb-Clover fluorescent fusion protein was expressed from one of the two endogenous RB alleles. A Geminin-mCherry reporter was used to identify the G1/S transition (see Materials and Methods). (B) Representative trace of the three fluorescent reporters from cell birth to mitosis. Each measurement type was normalized to its mean. (C) Plot of Rb-Clover relative protein concentration (calculated as Rb-Clover amount divided by E2-Crimson-NLS amount). Cells were aligned at the time of their G1/S transitions. (D) Plot of the amount of E2-Crimson-NLS at birth vs. the amount at G1/S. Data were normalized to the mean amount of E2-Crimson-NLS at birth and are shown with binned means ± SE of the mean. (E) Plot of the amount of E2-Crimson-NLS at G1/S vs. the amount at G2/M. Data were normalized to the mean amount of E2-Crimson-NLS at birth and are shown with binned means ± SE of the mean. (F) Plot of the duration of G1 or S/G2 phases vs. E2-Crimson-NLS amount at birth or at G1/S, respectively. N = 77 and 122 cells for G1 and S/G2 durations, respectively. (G) Calculation of the rate at which cells stochastically pass the G1/S transition as a function of E2-Crimson-NLS amount (see Materials and Methods). The fraction of measurements in each E2-Crimson-NLS bin that correspond to a G1/S transition at that frame is shown, along with 5th and 95th percentile confidence intervals from bootstrap resampling of each bin 1000 times. Bins are shown for the middle 90% of size measurements and were normalized to the overall mean. N = 290 transition events from >18,000 measurements.
It has previously been observed in several mammalian cell lines that cells adjust the amount of time they spend in G1 phase to compensate for differences in birth size, while the amount of time spent in S/G2 phases is unaffected (Cadart et al., 2018). To investigate size control at G1/S in HMEC cells, we measured E2-Crimson total intensity ∼2.5 h after birth (to allow time for nuclear reimport of E2-Crimson), at the G1/S transition as determined using Geminin-mCherry, and ∼1.5 h before cytokinesis. We observed that smaller-born cells grow more during G1 than larger-born cells so that all cells are a similar size at the G1/S transition (Figure 6D). Conversely, during S/G2, cells add a similar amount of mass regardless of their size at G1/S so that cell size at mitotic entry was highly correlated with cell size at G1/S (Figure 6E). We further observed that G1 length is negatively correlated with size at birth, while S/G2 length is remarkably constant (Figure 6F). As predicted by this model, we observed that larger cells enter S phase at a higher rate than smaller cells (Figure 6G). These results highlight the importance of G1 for maintaining cell size homeostasis, consistent with other reports (Killander and Zetterberg, 1965; Varsano et al., 2017; Ginzberg et al., 2018; Liu et al., 2018; Zatulovskiy et al., 2018). Intriguingly, our data also suggest that the regulatory network that governs the G1/S transition can be modulated by cell size, though they do not directly point to a mechanism.
DISCUSSION
Here, we show that a constitutively expressed fluorescent protein provides a convenient metric of cell size. We validated this method by showing that integrated fluorescence intensity correlates well with more established size measurements including forward scatter from flow cytometry, CFSE total protein dye, nuclear volume, and QPM measurements of dry mass. Importantly, we found that fluorescent reporter total intensity, nuclear volume, CFSE, and forward scatter were not only linearly related, but proportionally related so that the intercepts are close to the origin. While a linear relationship would mean that any metric could be used to rank cells in order of size, a proportional relationship implies that any can be used also to identify fold differences in sizes between cells. We further showed that fluorescence intensity is less sensitive to segmentation differences and less noisy than nuclear volume. Finally, we measured cell size over time to recapitulate several prior observations: that Rb protein concentration falls during G1, that time spent in G1 is negatively correlated with birth size, that time spent in S/G2 is uncorrelated with cell size, and that larger cells have a higher rate of progression through the G1/S transition.
Our method is limited by several factors. First, it requires generating a clonal cell population expressing the fluorescent reporter of choice. Second, it occupies a fluorescence channel and therefore may not be compatible with experiments in which more than two or three other color channels are being used. Third, it relies on the scaling of individual proteins with cell size. While this principle has been consistently demonstrated in the normal cell size range, it may not work for abnormally large cells (Neurohr et al., 2019). Finally, our method provides only a relative, not an absolute, measurement of cell size. However, by combining our method with an orthogonal approach to measure the average absolute size of the population, it would be possible to calculate individual absolute cell sizes. Despite these limitations, we expect that our straightforward strategy for measuring cell size will enable other investigators to investigate how cell size and growth affect other areas of cell biology. Importantly, the combination of our size reporter with another fluorescently labeled protein allows the measurement of that protein’s relative concentration using a common wide-field microscope. This could be especially impactful for studies determining how dynamic protein concentrations control key cellular events.
MATERIALS AND METHODS
Cell culture conditions and cell lines
Cells were cultured at 37°C with humidified 5% CO2. Nontransformed hTERT1-HMECs were obtained from the laboratory of Stephen Elledge at Harvard Medical School (Solimini et al., 2012) and cultured in Mammary Epithelial Growth Medium (Lonza CC-3150). To reduce background fluorescence in imaging experiments we used the same medium without phenol red (Lonza CC-3153 basal medium supplemented with growth factors and other components from the CC-4136 kit). Medium was refreshed every 24 h during imaging experiments to mitigate phototoxicity and nutrient depletion. Endogenously tagged RB-Clover cells were generated as described in Zatulovskiy et al. (2018). HEK 293T cells (for producing lentivirus) were cultured in DMEM containing l-glutamine, 4.5 g/l glucose, and sodium pyruvate (Corning), supplemented with 10% fetal bovine serum (FBS; Corning) and 1% penicillin/streptomycin. K562 cells were a gift from Ravi Majeti’s lab at Stanford University and were generally cultured in RPMI 1640 medium (HyClone) supplemented with 10% FBS and 1% penicillin/streptomycin. For 3 wk after single-cell sorting, K562 clones were expanded in IMDM medium (Life Technologies) supplemented with 20% FBS and 1% penicillin/streptomycin.
Fluorescent reporter cell lines
Fluorescent reporters (mCherry-3xNLS, E2-Crimson-NLS, mAG-hGeminin, and mCherry-hGeminin) were cloned into the CSII-EF-MCS lentiviral vector backbone as in Schwarz et al. (2018). The CSII vector, the lentiviral packaging vector dr8.74, and the lentiviral envelope vector VSVg were transfected into HEK 293T cells with TurboFect (Life Technologies). Transfected HEK 293T cells were incubated for 2 d before the virus-laden medium was transferred to HMECs or K562 cells. At least 3 d after infection, single cells positive for the fluorescent protein of interest were sorted by FACS and clonally expanded.
Cell imaging
Cells were seeded the day before imaging on 35-mm glass-bottom dishes (MatTek) at low density. The cells were kept overnight in standard culture conditions before being transferred to a Zeiss Axio Observer Z1 microscope equipped with an incubation chamber to maintain the cells at 37°C with humidified 5% CO2. For live-cell experiments, fluorescence and brightfield or phase images were collected every 10 min for 48–72 h using a Zyla 5.5 sCMOS camera, a 10× Plan-APOCHROMAT 0.45 NA or 10× Plan-NEOFLUAR 0.30 NA objective, a Colibri.2 light source, and an automated stage, all controlled by μManager software (Edelstein et al., 2014). These objectives have thick enough focal planes to collect emitted light from the entire nucleus without z-stacks. We did not observe significant photobleaching during our experiments (Supplemental Figure S5). For total protein dye experiments, cells were incubated with CFSE (Thermo Fisher; C34554) at 1:2500 dilution for 15 min at 37°C. For DAPI experiments, cells were fixed with 4% paraformaldehyde for 10 min at room temperature, permeabilized with 0.2% Triton X-100 for 15 min at 4°C, and stained with 300 nM DAPI for 5 min at room temperature before imaging.
Flow cytometry and cell sorting
HMECs were grown on dishes to ∼70% confluence, harvested by trypsinization and centrifugation, and then resuspended in phosphate-buffered saline (PBS). To generate clonal cell lines, cells were sorted using a BD FACSAria machine. For total protein dye experiments, cells were incubated with CFSE (Thermo Fisher; C34554) at 1:4000 dilution for 15 min at 37°C. K562 cells were harvested by centrifugation and resuspended in PBS. For all analyses, flow cytometry was performed using a BD LSRII.UV instrument, and then single cells were gated using FlowJo. Forward scatter area (FSC-A), CFSE area, and mCherry or E2-Crimson area were each normalized to the population median. For clarity, bins are shown for the middle 95% of the data due to wide variance and nonlinearities in measurements for cells of extreme sizes. Plots were made with the MATLAB function “shadedErrorBar.”
Fluorescence image analysis
Images were quantified using custom MATLAB scripts (Mathworks). In MATLAB, cell nuclei were segmented using the mCherry, E2-Crimson, or DAPI channels by Gaussian filtering, thresholding with manually chosen parameters, and opening and closing of the segmented regions using the MATLAB functions imopen and imclose. Nuclear volume was calculated as the segmented nuclear area (in pixels) to the 3/2 power. We also performed our analyses using nuclear area rather than nuclear volume with qualitatively similar results (unpublished data). For CFSE experiments, complete cells were similarly segmented using the CFSE channel, and additional thresholds were applied to omit cell debris from subsequent analysis. For all imaging experiments, total pixel intensity in the segmented region was calculated after applying a location-dependent intensity adjustment to account for vignetting due to our large camera field of view, which caused the center of an image to be illuminated more brightly than the periphery. To make the adjustment we used an image of dissolved fluorescein in solution to measure effective illumination as a function of position in the field of view. Then, each pixel in each image of cells was divided by the difference between the effective illumination intensity and the darkfield intensity at that pixel location (Supplemental Figure S6), as in Bottier et al. (2011). Finally, each segmented object’s local background was subtracted to account for fluorescence of the cell medium.
Segmentation threshold analysis
A range of possible thresholds that yielded plausible nuclear segmentations was manually chosen, and nuclei were automatically segmented at each possible threshold. An mCherry threshold of 180 was judged to be the closest to manual segmentation and was therefore used as the reference point. Nuclear volume and mCherry total intensity were calculated at each possible threshold and normalized to their respective values for the cell of interest at the reference threshold (for Figure 3A) or to the mean value for all cells at the reference threshold (for Figure 3B). For regression analysis, each metric was normalized to its own mean. At each possible threshold, uni- and bivariate regression was performed using either nuclear volume, mCherry total intensity, or both as predictor variables and CFSE total intensity as the response variable. The regression coefficients in Figure 3C represent the change in normalized CFSE total intensity per unit change in normalized nuclear volume or mCherry intensity. The coefficients of determination in Figure 3D represent the goodness of fit for the bivariate regression compared with each univariate regression. The DAPI segmentation in Supplemental Figure S2 was performed the same way, except a threshold of 900 was used as the reference point. The DAPI-segmented region was used to calculate both the nuclear volume and the mCherry total intensity.
Live-cell imaging analysis
Nuclei were manually tracked over time with the aid of a custom MATLAB graphical user interface. During tracking, in cases where automated segmentation failed to separate neighboring nuclei, the fused object was split by applying a watershed algorithm while decreasing the size of the morphological structuring element until the desired number of distinct nuclei were segmented. During subsequent analysis, additional noise, for instance due to segmentation errors or nearby cell debris, was corrected as follows. At timepoints where the cell was not segmented at all, the average of the measurements at the timepoints before and after the missing timepoint was used. Then, the measurement at each timepoint was compared with a moving median with window size equal to nine frames. If the difference between the actual measurement and the moving median was more than half the magnitude of the moving median, the moving median was used instead. Finally, measurements from the first hour after birth were omitted on account of low-confidence segmentation while the cells reimported fluorescent protein into their postmitotic nuclei, and measurements from the last 2 h before cytokinesis were omitted on account of nuclear envelope breakdown. For analysis of timepoint-to-timepoint measurement noise, each trace was normalized to its own mean and then decomposed into an overlapping series of 2-h segments. A line was fitted to the data points within each 2-h segment. Then, the residuals were calculated for each segment, squared, and summed across all segments. The resulting sum of squared residuals was calculated for each nuclear volume or mCherry intensity trace, and this noise metric was compared for the two measurement types using a two-sided t test.
QPM
QPM was performed with assistance from Phasics (Saint-Aubin, France). Images were acquired using a wavefront SID4Bio camera using Quadriwave Lateral Shearing Interferometry, then processed using the manufacturer’s SID4Bio software according to the procedures in Popescu et al. (2008) and Aknoun et al. (2015). Briefly, a quadratic fit was applied to an empty reference frame and this fit was subtracted from the image being analyzed. These images were imported into ImageJ and manually segmented and tracked. Then, each object’s local background was subtracted. For these experiments, a 20× Plan-Apochromat 0.8 NA objective lens was used and images were collected every 5 min. Because manual segmentation resulted in significant timepoint-to-timepoint variability, Figure 5A shows moving medians with a window size of 17 frames (∼1.5 h). As in Figure 4, C–E, for Figure 5B we performed a piecewise linear fit to the raw data, but used overlapping 1-h rather than 2-h segments to address noise in the data as described above. The sums of squared residuals for each measurement type were compared by analysis of variance and a multicomparison t test. The correlations shown in Figure 5, C and D, were calculated by treating each single-cell, single-timepoint measurement as an independent data point.
Cell cycle analysis
The Rb-Clover, E2-Crimson-NLS, and Geminin-mCherry traces shown in Figure 6B were normalized to their means for that trace so that they could be plotted on the same axes. The G1/S transition was automatically detected by an algorithm that looked for surpassing a threshold level of Geminin-mCherry, a kink in the Geminin-mCherry signal, and a local maximum of the second derivative of the signal. An equivalent procedure was used for Geminin-mAG in Figure 4D. We note that the E2-Crimson fluorescence was also excited by the 555-nm LED used for mCherry fluorescence. Therefore, in order to distinguish the accumulation of Geminin-mCherry beginning at G1/S from constitutive accumulation of the E2-Crimson size reporter, the Geminin-mCherry trace was divided by the E2-Crimson trace so that the mCherry signal is flat in G1, where there is expected to be no stable Geminin protein. The raw Geminin-mCherry values and the G1/S transition point identified by this method are shown in Figure 6B. Birth size was defined as the median size between 120 and 180 min after birth (to allow time for cells to reimport fluorescent protein into their postmitotic nuclei), while G1/S size was defined as the median size in the 50 min surrounding G1/S, and G2/M size was defined as the median size in the 100–150 min preceding cytokinesis (to avoid complications from nuclear envelope breakdown). Rb-Clover concentrations per unit E2-Crimson were calculated as Rb-Clover total intensity divided by the median value of the appropriate size metric within a five-frame window. For plotting the single-cell traces, Rb-Clover concentrations were normalized to the mean concentration for all measurements across all cells. For the G1/S rate experiment, automated segmentation and tracking with Aivia software (DRVision Technologies) were used to supplement manually tracked cells. Total E2-Crimson intensity was measured for all cells at all timepoints up through the G1/S transition, and each measurement was annotated with 0 or 1 for whether G1/S occurred there. Measurements were binned by E2-Crimson intensity and the fraction of measurements within that bin corresponding to a G1/S transition was calculated, along with 5th and 95th percentile confidence intervals calculated by bootstrapping.
Supplementary Material
Acknowledgments
We thank members of the Skotheim laboratory for discussion and comments on the article. We thank Flor Medina, Valentin Genuer, and Sherazade Aknoun from Phasics for the QPM measurements. We thank Madhav Mani from Northwestern University for helpful suggestions on image analysis. We thank Anna Rajaratnam for assistance with cell tracking. We thank the Elledge laboratory at Harvard Medical School for the HMEC cell line and the Majeti laboratory at Stanford for the K562 cell line. We thank Nicholas Buchler and Stefano Di Talia for helpful comments on the article. This work was supported by the National Institutes of Health (NIH) through R01 Grant no. GM-115479 and T32 Grant no. GM-007365 (D.F.B.). Cell sorting and flow cytometry analysis for this project were done on instruments in the Stanford Shared FACS Facility: flow cytometry data were collected on the LSRII.UV instrument, and sorting was performed on the Falstaff instrument, both of which were obtained using NIH S10 Shared Instrument Grants (Grant no. S10RR027431 and Grant no. S10RR027431, respectively).
Abbreviations used:
- CFSE
carboxyfluorescein succinimidyl ester
- DAPI
4’,6-diamidino-2-phenylindole
- EF1α
eukaryotic translation elongation factor 1 alpha
- FACS
fluorescence-activated cell sorting
- FSC
forward scatter
- HMECs
human mammary epithelial cells
- NLS
nuclear localization sequence
- QPM
quantitative phase microscopy
- SSC
side scatter
- WPRE
woodchuck posttranscriptional regulatory element
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-03-0171) on October 10, 2019.
REFERENCES
- Aknoun S, Savatier J, Bon P, Galland F, Abdeladim L, Wattellier B, Monneret S. (2015). Living cell dry mass measurement using quantitative phase imaging with quadriwave lateral shearing interferometry: an accuracy and sensitivity discussion. J Biomed Opt , 126009. [DOI] [PubMed] [Google Scholar]
- Atay O, Doncic A, Skotheim JM. (2016). Switch-like transitions insulate network motifs to modularize biological networks. Cell Syst , 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajcsy P, Simon MH, Florczyk SJ, Simon JG, Juba D, Brady MC. (2015). A method for the evaluation of thousands of automated 3D stem cell segmentations. J Microsc-Oxford , 363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barer R. (1952). Interference microscopy and mass determination. Nature , 169366b0. [DOI] [PubMed] [Google Scholar]
- Barr AR, Cooper S, Heldt FS, Butera F, Stoy H, Mansfeld J, Novák B, Bakal C. (2017). DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression. Nat Commun , 14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottier C, Gabella C, Vianay B, Buscemi L, Sbalzarini IF, Meister J-J, Verkhovsky AB. (2011). Dynamic measurement of the height and volume of migrating cells by a novel fluorescence microscopy technique. Lab Chip , 3855–3863. [DOI] [PubMed] [Google Scholar]
- Bryan AK, Hecht VC, Shen W, Payer K, Grover WH, Manalis SR. (2013). Measuring single cell mass, volume, and density with dual suspended microchannel resonators. Lab Chip , 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadart C, Monnier S, Grilli J, Sáez PJ, Vastava N, Attia R, Terriac E, Baum B, Cosentino-Lagomarsino M, Piel M. (2018). Size control in mammalian cells involves modulation of both growth rate and cell cycle duration. Nat Commun , 3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappell SD, Chung M, Jaimovich A, Spencer SL, Meyer T. (2016). Irreversible APCCdh1 inactivation underlies the point of no return for cell-cycle entry. Cell , 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak N, Olcum S, Delgado FF, Wasserman SC, Payer KR, Murakami M, Knudsen SM, Kimmerling RJ, Stevens MM, Kikuchi Y, et al (2016). High-throughput measurement of single-cell growth rates using serial microfluidic mass sensor arrays. Nat Biotechnol , 1052–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetin AE, Stevens MM, Calistri NL, Fulciniti M, Olcum S, Kimmerling RJ, Munshi NC, Manalis SR. (2017). Determining therapeutic susceptibility in multiple myeloma by single-cell mass accumulation. Nat Commun , 1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-W, Cheng W-C, Chen C-R, Shu W-Y, Tsai M-L, Huang C-L, Hsu IC. (2011). Identification of human housekeeping genes and tissue-selective genes by microarray meta-analysis. PLoS One , e22859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W-C, Chang C-W, Chen C-R, Tsai M-L, Shu W-Y, Li C-Y, Hsu IC. (2011). Identification of reference genes across physiological states for qRT-PCR through microarray meta-analysis. PLoS One , e17347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KL, Oh S, Sung Y, Dasari RR, Kirschner MW, Tabin CJ. (2013). Multiple phases of chondrocyte enlargement underlie differences in skeletal proportions. Nature , 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTalia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. (2007). The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature , 947. [DOI] [PubMed] [Google Scholar]
- Doncic A, Falleur-Fettig M, Skotheim JM. (2011). Distinct interactions select and maintain a specific cell fate. Mol Cell , 528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncic A, Skotheim JM. (2013). Feedforward regulation ensures stability and rapid reversibility of a cellular state. Mol Cell , 856–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. (2014). Advanced methods of microscope control using μManager software. J Biol Methods , 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens LJ, White KH, Jevtic P, Li X, Levy DL. (2013). Nuclear size regulation: from single cells to development and disease. Trends Cell Biol , 151–159. [DOI] [PubMed] [Google Scholar]
- Ginzberg M, Chang N, D’Souza H, Patel N, Kafri R, Kirschner MW. (2018). Cell size sensing in animal cells coordinates anabolic growth rates and cell cycle progression to maintain cell size uniformity. Elife , e26957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzberg MB, Kafri R, Kirschner M. (2015). On being the right (cell) size. Science , 1245075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin M, Delgado FF, Son S, Grover WH, Bryan AK, Tzur A, Jorgensen P, Payer K, Grossman AD, Kirschner MW, Manalis SR. (2010). Using buoyant mass to measure the growth of single cells. Nat Methods , 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg EC, Steidley KD. (1965). Electrical counting and sizing of mammalian cells in suspension. Biophys J , 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover WH, Bryan AK, Diez-Silva M, Suresh S, Higgins JM, Manalis SR. (2011). Measuring single-cell density. Proc Natl Acad Sci USA , 10992–10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holme S, Heaton A, Konchuba A, Hartman P. (1988). Light scatter and total protein signal distribution of platelets by flow cytometry as parameters of size. J Lab Clin Med , 223–231. [PubMed] [Google Scholar]
- Jorgensen P, Edgington NP, Schneider BL, Rupeš I, Tyers M, Futcher B. (2007). The size of the nucleus increases as yeast cells grow. Mol Biol Cell , 3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R, Levy J, Ginzberg MB, Oh S, Lahav G, Kirschner MW. (2013). Dynamics extracted from fixed cells reveal feedback linking cell growth to cell cycle. Nature , 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killander D, Zetterberg A. (1965). A quantitative cytochemical investigation of the relationship between cell mass and initiation of DNA synthesis in mouse fibroblasts in vitro. Exp Cell Res , 12–20. [DOI] [PubMed] [Google Scholar]
- Kimmerling RJ, Prakadan SM, Gupta AJ, Calistri NL, Stevens MM, Olcum S, Cermak N, Drake RS, Pelton K, De Smet F, et al (2018). Linking single-cell measurements of mass, growth rate, and gene expression. Genome Biol , 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Abbas AK, Aster JC. (2015). Neoplasia. In: Robbins and Cotran Pathologic Basis of Disease, Philadelphia: Elsevier Saunders, 270. [Google Scholar]
- Levy DL, Heald R. (2016). Biological scaling problems and solutions in amphibians. Cold Spring Harb Perspect Biol , a019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Ginzberg MB, Patel N, Hild M, Leung B, Li Z, Chen Y-C, Chang N, Wang Y, Tan C, et al (2018). Size uniformity of animal cells is actively maintained by a p38 MAPK-dependent regulation of G1-length. Elife , e26947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen TP, Björklund M. (2016). Cellular allometry of mitochondrial functionality establishes the optimal cell size. Dev Cell , 370–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M, Wang Z, Shen Z, Bednarz M, Bashir R, Golding I, Prasanth SG, Popescu G. (2011). Optical measurement of cycle-dependent cell growth. Proc Natl Acad Sci USA , 13124–13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann FR, Nurse P. (2007). Nuclear size control in fission yeast. J Cell Biol , 593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurohr GE, Terry RL, Lengefeld J, Bonney M, Brittingham GP, Moretto F, Miettinen TP, Pontano Vaites L, Soares LM, Paulo JA, et al (2019). Excessive cell growth causes cytoplasm dilution and contributes to senescence. Cell , 1083–1093.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. (2006). Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature , 840. [DOI] [PubMed] [Google Scholar]
- Park K, Jang J, Irimia D, Sturgis J, Lee J, Robinson PJ, Toner M, Bashir R. (2008). ‘Living cantilever arrays’ for characterization of mass of single live cells in fluids. Lab Chip , 1034–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, Millet LJ, Kim N, Li H, Jin X, Popescu G, Aluru N, Hsia JK, Bashir R. (2010). Measurement of adherent cell mass and growth. Proc Natl Acad Sci USA , 20691–20696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende M, Kozma SC, Jaquet M, Oorschot V, Burcelin R, Marchand-Brustel Y, Klumperman J, Thorens B, Thomas G. (2000). Hypoinsulinaemia, glucose intolerance and diminished β-cell size in S6K1-deficient mice. Nature , 994. [DOI] [PubMed] [Google Scholar]
- Popescu G, Park Y, Lue N, Best-Popescu C, Deflores L, Dasari RR, Feld MS, Badizadegan K. (2008). Optical imaging of cell mass and growth dynamics. Am J Physiol , C538–C544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al (2008). Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell , 487–498. [DOI] [PubMed] [Google Scholar]
- Schmoller KM, Turner J, Kõivomägi M, Skotheim JM. (2015). Dilution of the cell cycle inhibitor Whi5 controls budding-yeast cell size. Nature , 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz C, Johnson A, Kõivomägi M, Zatulovskiy E, Kravitz CJ, Doncic A, Skotheim JM. (2018). A precise Cdk activity threshold determines passage through the restriction point. Mol Cell , 253–264.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol , 1567–1572. [DOI] [PubMed] [Google Scholar]
- Shapiro HM. (2018). Practical Flow Cytometry, Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- Smith U. (1971). Effect of cell size on lipid synthesis by human adipose tissue in vitro. J Lipid Res , 65–70. [PubMed] [Google Scholar]
- Solimini NL, Xu Q, Mermel CH, Liang AC, Schlabach MR, Luo J, Burrows AE, Anselmo AN, Bredemeyer AL, Li MZ, et al (2012). Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science , 104–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S, Kang J, Oh S, Kirschner MW, Mitchison TJ, Manalis S. (2015). Resonant microchannel volume and mass measurements show that suspended cells swell during mitosis. J Cell Biol , 757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son S, Tzur A, Weng Y, Jorgensen P, Kim J, Kirschner MW, Manalis SR. (2012). Direct observation of mammalian cell growth and size regulation. Nat Methods , 910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SL, Cappell SD, Tsai F-C, Overton WK, Wang CL, Meyer T. (2013). The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell , 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens MM, Maire CL, Chou N, Murakami MA, Knoff DS, Kikuchi Y, Kimmerling RJ, Liu H, Haidar S, Calistri NL, et al (2016). Drug sensitivity of single cancer cells is predicted by changes in mass accumulation rate. Nat Biotechnol , 1161–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack RL, Hein B, Bhattacharyya D, Hell SW, Keenan RJ, Glick BS. (2009). A rapidly maturing far-red derivative of DsRed-Express2 for whole-cell labeling. Biochemistry , 8279–8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur A, Kafri R, LeBleu VS, Lahav G, Kirschner MW. (2009). Cell growth and size homeostasis in proliferating animal cells. Science , 167–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur A, Moore JK, Jorgensen P, Shapiro HM, Kirschner MW. (2011). Optimizing optical flow cytometry for cell volume-based sorting and analysis. PLoS One , e16053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varsano G, Wang Y, Wu M. (2017). Probing mammalian cell size homeostasis by channel-assisted cell reshaping. Cell Rep , 397–410. [DOI] [PubMed] [Google Scholar]
- Zangle TA, Teitell MA. (2014). Live-cell mass profiling: an emerging approach in quantitative biophysics. Nat Methods , nmeth.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatulovskiy E, Berenson DF, Topacio BR, Skotheim JM. (2018). Cell growth dilutes the cell cycle inhibitor Rb to trigger cell division. BioRxiv 470013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurinsky J, Leonhard K, Watt S, Marguerat S, Bähler J, Nurse P. (2010). A coordinated global control over cellular transcription. Curr Biol , 2010–2015. [DOI] [PubMed] [Google Scholar]
- Zlotek-Zlotkiewicz E, Monnier S, Cappello G, Berre M, Piel M. (2015). Optical volume and mass measurements show that mammalian cells swell during mitosis. J Cell Biol , 765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Donello J, Trono D, Hope T. (1999). Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol , 2886–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.