Abstract
Microtubule-associated proteins (MAPs) are a functionally highly diverse class of proteins that help to adjust the shape and function of the microtubule cytoskeleton in space and time. For this purpose, MAPs structurally support microtubules, modulate their dynamic instability, or regulate the activity of associated molecular motors. The microtubule-binding domains of MAPs are structurally divergent, but often depend on electrostatic interactions with the negatively charged surface of the microtubule. This suggests that the surface exposure of positive charges rather than a certain structural fold is sufficient for a protein to associate with microtubules. Consistently, positively charged artificial objects have been shown to associate with microtubules and to diffuse along their lattice. Natural MAPs, however, show a more sophisticated functionality beyond lattice-diffusion. Here, we asked whether basic electrostatic interactions are sufficient to also support advanced MAP functionality. To test this hypothesis, we studied simple positively charged peptide sequences for the occurrence of typical MAP-like behavior. We found that a multivalent peptide construct featuring four lysine-alanine heptarepeats (starPEG-(KA7)4)—but not its monovalent KA7-subunits—show advanced, biologically relevant MAP-like behavior: starPEG-(KA7)4 binds microtubules in the low nanomolar range, diffuses along their lattice with the ability to switch between intersecting microtubules, and tracks depolymerizing microtubule ends. Further, starPEG-(KA7)4 promotes microtubule nucleation and growth, mediates depolymerization coupled pulling at plus ends, and bundles microtubules without significantly interfering with other proteins on the microtubule lattice (as exemplified by the motor kinesin-1). Our results show that positive charges and multivalency are sufficient to mimic advanced MAP-like behavior.
INTRODUCTION
The eukaryotic microtubule cytoskeleton is key to vital cellular processes such as intracellular transport (Verhey et al., 2011), cell migration (Dogterom and Koenderink, 2019), and mitosis (McIntosh, 2016). During these altogether highly dynamic processes, the shape and functionality of the microtubule cytoskeleton have to be continuously adjusted in space and time. In addition to molecular motors, these adjustment are aided by a multitude of microtubule-associated proteins (MAPs) that have the capacity to 1) modulate the dynamic instability of microtubules (Goodson and Jonasson, 2018) (i.e., the stochastic transition from growing to shrinking phases and vice versa; Brouhard and Rice, 2018), 2) regulate the activity of molecular motors (Dixit et al., 2008; Drechsler et al., 2014; Roberts et al., 2014), and 3) mediate the interaction of microtubules with each other (Scott et al., 1992; Bieling et al., 2010) and with particular cellular substructures (Goldberg, 2017; Monda and Cheeseman, 2018). Depending on their task, MAPs can associate with the microtubule lattice in general or target specific microtubule substructures such as the less dynamic minus end (Akhmanova and Hoogenraad, 2015) or the GTP-rich highly dynamic plus end (Akhmanova and Steinmetz, 2008).
The structures of the involved microtubule-binding domains (MTBD) can be highly diverse, suggesting that the capacity to bind microtubules has arisen independently on several occasions during cytoskeletal evolution:
– loops in α-helical stretches: during mitosis, the kinetochore-bound Ska-complex (consisting of Ska1, -2, and -3) couples chromosomes with the plus end of depolymerizing microtubules, facilitating the congression and subsequent anaphase separation of sister chromatids (Hanisch et al., 2006; Gaitanos et al., 2009; Welburn et al., 2009; Auckland et al., 2017). Microtubule binding occurs via a noncanonical winged-helix motif within Ska1 that consists of eight α-helical stretches followed by a C-terminal β-hairpin. Mutational analysis has shown that three basic patches within the interhelical loops (R155/R236/R245, K183/K184/K203/K206, and K217/K223/K226/K227) contribute to a multipartite microtubule-binding mode of Ska1 (Abad et al., 2014). A similar MTBD has been reported for PRC1, a microtubule cross-linker that organizes anti-parallel microtubule bundles in the spindle midzone/midbody during mitosis (Bieling et al., 2010; Lee et al., 2015). The MTBD of PRC1 consists of three α-helices that adopt a spectrin-like fold, exposing a conserved patch of basic amino acids (R377/K387/K390) within the loop between helices 1 and 2 that are essential for tubulin binding (Subramanian et al., 2010).
– α-helices: the centrosomal scaffolding protein CEP135 is essential to centriole biogenesis in human cells (Kleylein-Sohn et al., 2007). In Drosophila, CEP135 is involved in the initiation of the central microtubule pair assembly, an activity that requires a microtubule-binding and stabilizing activity associated with its N-terminus (Carvalho-Santos et al., 2012). Recent work described that the N-terminus of CEP135 dimerizes into a parallel coiled-coil exposing the basic amino acids K101/K104/K108 that are essential to microtubule binding (Kraatz et al., 2016).
– β-sheet/-hairpin: the actin cross-link factor 7 connects the actin and the microtubule cytoskeleton at tight junctions, keeping them dynamic during tissue morphogenesis (Kodama et al., 2003; Ma et al., 2017). Its MTBD consists of a central 5-strand β-sheet exposing a flexible β-hairpin that harbors the basic amino acids K53/R63/R66/R69/R75, which are essential to microtubule binding (Ma et al., 2017).
Thus, while structurally divergent, all these MTBDs strictly require the presence and surface exposure of basic amino acids. This suggests that the interaction between these MAPs and the microtubule lattice occurs primarily through electrostatic interactions with the exposed acidic C-termini (E-hooks) (Nogales et al., 1999; Okada and Hirokawa, 2000) of single tubulin subunits and does not crucially depend on a certain secondary/tertiary structure. In fact, microtubule binding can occur even in the absence of any complex protein folds, as demonstrated by the intrinsically disordered structural MAP tau (Kellogg et al., 2018) or by positively charged artificial nanoparticles that also bind and diffuse along the microtubule lattice (Minoura et al., 2010). Structural determinants, however, become important when MAPs target particular substructures of the microtubule such as the plus end (by the TOG-domain, e.g., XMAP215 or the CH-domain, e.g., EB1) or the minus end (by the CKK-domain, e.g., CAMSAP1). Microtubule binding by these specialized MAPs often occurs in between two neighboring microtubule protofilaments and often also depends on additional hydrophobic interactions (Slep and Vale, 2007; Maurer et al., 2012; Atherton et al., 2017).
In summary, the current literature suggests that any peptide/protein exhibiting a certain distribution of positive surface charges can bind to and diffuse along the microtubule lattice. Natural MAPs, however, exhibit a wide range of advanced functionality beyond lattice diffusion, which, among others, include modulation of microtubule dynamics (Goodson and Jonasson, 2018), nucleation/severing of microtubules (Roostalu et al., 2015; McNally and Roll-Mecak, 2018; Thawani et al., 2018), following the growing or shrinking microtubule plus ends (Akhmanova and Steinmetz, 2008; Maciejowski et al., 2017; Volkov et al., 2018), or coupling cellular structures to the dynamic microtubule ends (Monda and Cheeseman, 2018). The comparison of published MTBDs made above suggests that positive charges are sufficient to also support those advanced, biologically highly relevant MAP functions independently of the actual binding domain fold, but experimental evidence has been lacking so far.
To test this hypothesis, we designed simple, tuneable lysine-alanine peptide constructs and asked to what extent these positively charged constructs will mimic advanced MAP behavior. While we did not observe significant microtubule binding for short peptides (4–7 KA repeats), we show that a tetravalent construct, in which four (KA)7 peptides are linked via a branched polyethylene glycol (PEG) chain, shows significant binding in the low nanomolar range. Moreover, this construct mimics a wide range of MAP-like behavior as it 1) diffuses along the microtubule lattice and tracks depolymerizing microtubules ends, 2) nucleates microtubules and modulates their dynamics, 3) mediates force coupling to depolymerizing microtubule plus ends, and 4) cross-links and bundles microtubules without 5) affecting the biology of other microtubule-associated proteins (as exemplified by kinesin-1). We hence show a proof of principle that multivalent electrostatic interactions are indeed sufficient to support biologically relevant functionality of artificial MAPs (in vitro) and discuss possible applications for those constructs. On the basis of our observations, we further hypothesize that multivalency is a key feature of a subset of natural MAPs defining their functionality.
RESULTS
To test our hypothesis that the exposure of positive charges on a protein surface is sufficient to confer advanced MAP-like behavior, we have chosen a bottom-up approach asking whether simple positively charged peptides can bind to the microtubule lattice.
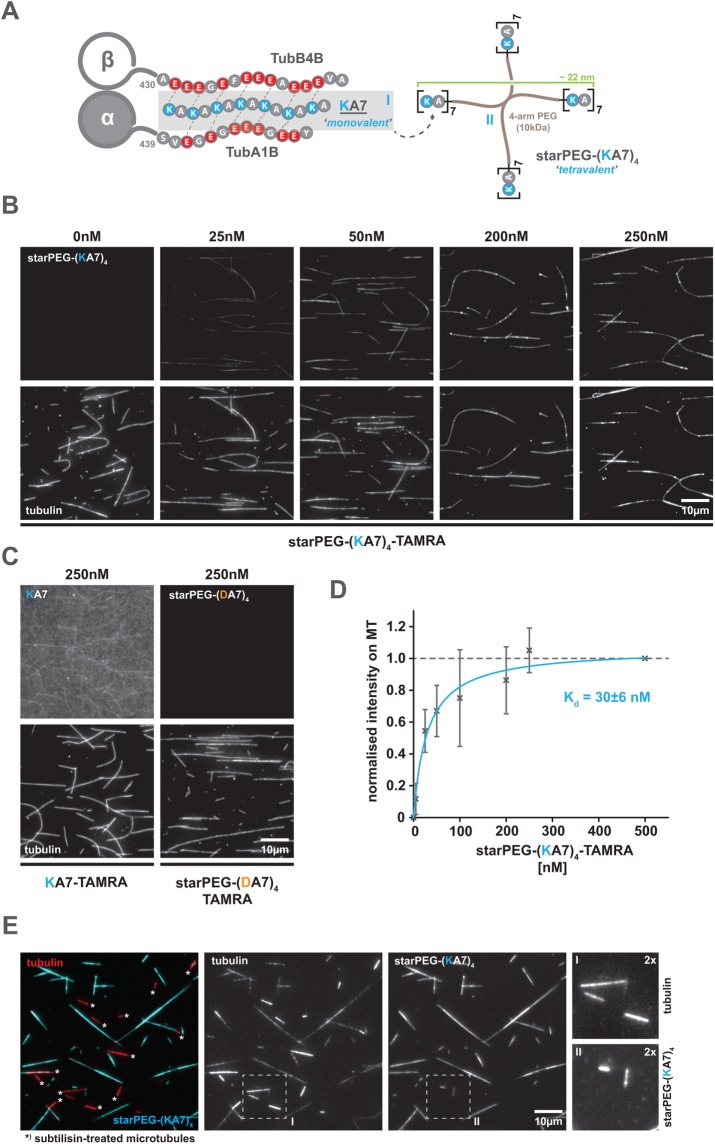
starPEG-(KA7) 4 binds microtubules in the low nanomolar range by electrostatic interactions with C-termini of tubulin
In a first approach, we synthesized short peptides of four to seven lysine-alanine repeats (KA4, KA5, KA6, KA7; see Figure 1A for a general scheme) and labeled them with 5-carboxytetramethylrhodamine (TAMRA) to visualize and quantify their microtubule-binding capacity on surface-bound, taxol-stabilized ATTO 647N-labeled microtubules using total internal reflection fluorescence (TIRF) microscopy. However, even at high nanomolar concentrations, the longest construct (i.e., 250 nM KA7) shows only weak microtubule-binding activity (Figure 1C, left). To increase the microtubule affinity of our peptide constructs, we then conjugated four KA7 units to a 10 kDa 4-arm PEG chain (starPEG). In the resulting multivalent construct (starPEG-(KA7)4-TAMRA; Figure 1A), 6.2 nm PEG arms spatially separate single KA7 units, allowing them to interact with the microtubule surface independently of each other (Figure 1A; Supplemental Figure S1). This tetravalent construct eventually binds taxol-stabilized microtubules with a Kd of 30 ± 6 nM (Figure 1, B and D, as well as Supplemental Figure S2 for higher-contrast versions of the 0 and 5 nM conditions) and saturation is reached at ∼200 nM starPEG-(KA7)4-TAMRA. A charge-inverted control construct (i.e., starPEG-(DA7)4-TAMRA), in contrast, does not show any microtubule binding even at high nanomolar concentrations (i.e., >250 nM) (Figure 1C and Supplemental Figure S2). We hence conclude that microtubule binding occurs neither via the PEG-backbone nor via the TAMRA dye, but likely depends on electrostatic interactions between the positively charged KA-repeats and the negatively charged microtubule surface.
FIGURE 1:
The starPEG-(KA7)4 binds to microtubules at low nanomolar concentrations via electrostatic interactions with the microtubule e-hooks. (A) Schematic overview of the synthetic peptides (KA7, [I] left and starPEG(KA7)4, [II] right) used in this study. Scheme on the left shows the hypothetical interactions of a positively charged KA7 peptide with the negatively charged C-terminal extensions (e-hooks) of a tubulin heterodimer (α-, β-tubulin) exemplified by α-tubulin 1B and β-tubulin 4B. Not drawn to scale. (B) Binding of tetravalent starPEG-(KA7)4-TAMRA molecules (top panel row) to taxol-stabilized Atto647N-labeled microtubules (bottom panel row) at indicated concentrations. Please refer to Supplemental Figure S2A for an extended concentration range. (C) Binding of the charge-inverted starPEG-(DA7)4-TAMRA version (negative control) or a monovalent KA7 peptide to Atto647N-labeled microtubules. Arrangement as in B. Please refer to Supplemental Figure S2B for high-contrast pictures of 0 nM, 5 nM starPEG-(KA7)4-TAMRA, and 250 nM starPEG-(DA7)4-TAMRA. (D) Saturation binding curve of starPEG-(KA7)4-TAMRA showing the concentration-dependent fluorescence intensity of microtubule associated TAMRA normalized to saturating conditions at 500 nM. Error bars depict the SD of five independent experiments. Blue line is a Michaelis-Menten model fit. (E) Binding of starPEG(KA7)4-TAMRA (cyan) to brightly Atto647N-labeled subtilisin-treated (bright red, marked with an asterisk) and dimly Atto647N-labeled nontreated (dim red) CMP-CPP/taxol double-stabilized microtubules. From left to right: overlay, tubulin, starPEG-(KA7)4-TAMRA, twofold magnifications of boxed areas in the single color channels.
Assuming an electrostatic-binding mode, we would predict starPEG-(KA7)4-TAMRA microtubule binding to be salt- and pH-sensitive, as increasing the ionic strength of the buffer is expected to electrostatically shield the charges of the peptide (and the microtubule), while raising the pH to the pKa-value of the lysine side chain (pKa = 10.3) will discharge the KA7-repeats—both reducing the microtubule affinity of starPEG-(KA7)4-TAMRA. We hence tested the microtubule-binding affinity of 50 nM starPEG-(KA7)4-TAMRA in an ionic strength range from I = 180 mM to I = 350 mM (Supplemental Figure S3, left) and in a pH range from 6.8 to 9.0 (Supplemental Figure S3, right). In both cases, the microtubule affinity of starPEG-(KA7)4-TAMRA drops exponentially with increasing ionic strength or increasing pH, thereby supporting our initial assumption.
The electrostatic interaction of starPEG-(KA7)4-TAMRA with the microtubule is likely mediated by the exposed acidic C-termini of α- and ß-tubulin (see Figure 1A). To test this, we removed the C-termini by a subtilisin protease treatment (Alper et al., 2014) of GMP-CPP/taxol double-stabilized microtubules (Supplemental Figure S4), which completely abolishes microtubule binding of starPEG-(KA7)4-TAMRA (Figure 1E). Thus, in summary, we conclude from our binding studies that starPEG-(KA7)4 binds microtubules in the low nanomolar range by electrostatic interactions with C-termini of tubulin.
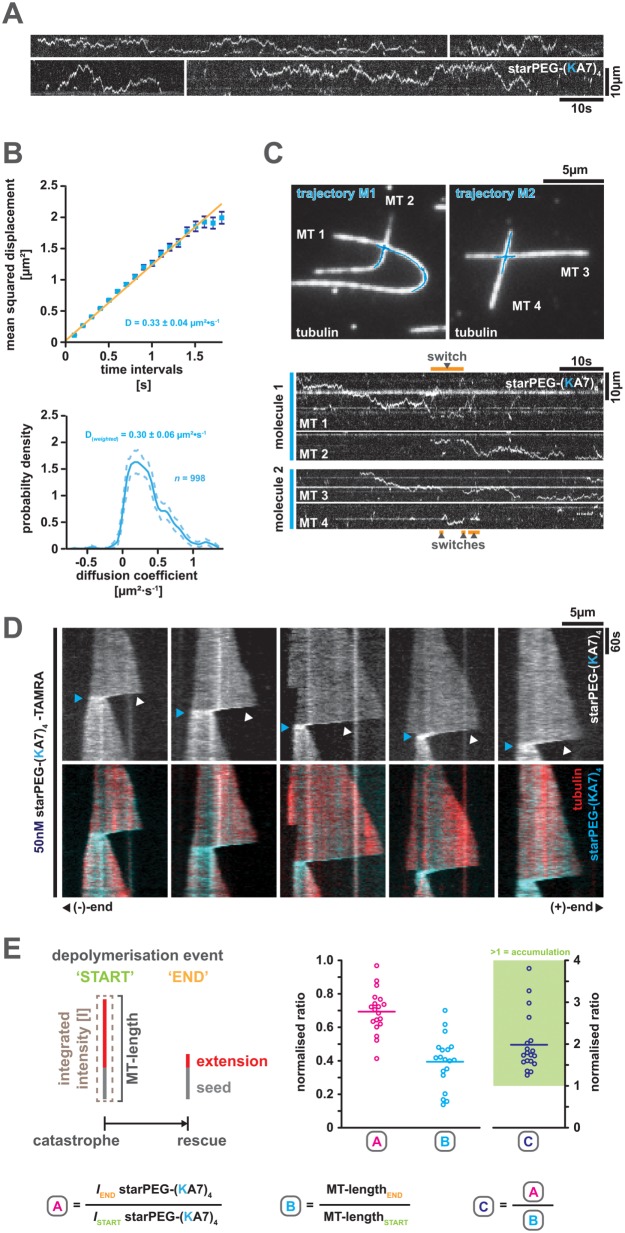
starPEG-(KA7) 4 diffuses along the microtubule lattice, switches between intersecting microtubules, and tracks depolymerizing microtubule plus ends
We next examined the behavior of single starPEG-(KA7)4-TAMRA molecules on taxol-stabilized ATTO 647N-labeled microtubules using TIRF microscopy. Molecules show extended episodes of diffusion along the microtubule lattice (Figure 2A) with a diffusion coefficient of ∼0.3 µm2s–1 (i.e., CVE: D = 0.30 µm2s–1 ± 0.06 and MSD: D = 0.33 ± 0.04, mean ± SEM, N = 998) (Figure 2B). Owing to their multivalency and the generous backbone dimensions (Figure 1A), single molecules are able to switch their microtubule track at microtubule–microtubule intersections, which occasionally involves a certain period of time, during which starPEG-(KA7)4-TAMRA molecules are associated with both intersecting microtubules at the same time (Figure 2C).
FIGURE 2:
starPEG-(KA7)4 diffuses along the microtubule lattice, switches tracks at microtubule intersections, and tracks the depolymerizing plus end of dynamic microtubules. (A) Representative kymographs showing the diffusion of 1 nM starPEG-(KA7)4-TAMRA along the lattice of taxol-stabilized microtubules at 25°C in BRB80. (B) Mean square displacement analysis (top) and covariance-based estimator of the diffusion coefficient D (bottom) of starPEG-(KA7)4-TAMRA at 25°C in BRB80. Error bars in MSD plot (top) are the SE. Data are fitted with a linear regression. The dashed line in the CVE plot (bottom) indicates the 95% confidence interval and the calculated mean D value is weighted for molecule trajectory length/data points. (C) Track-switching of single starPEG-(KA7)4-TAMRA at microtubule intersections. Top pictures show the geometry of intersecting taxol-stabilized Atto647N-labeled microtubules superimposed by the trajectories (blue) of single starPEG-(KA7)4-TAMRA molecules (i.e., M1 and M2) tracked by FIESTA. Bottom kymographs derived by line-scans along the respective substrate microtubules (MT1-4) show the diffusional movement of single starPEG-(KA7)4-TAMRA molecules (i.e., molecule 1 and molecule 2). Switch events are indicated by orange bars above and below the respective kymographs. (D) starPEG-(KA7)4-TAMRA (top row and cyan, bottom row) tracks the depolymerizing plus end of dynamic Alexa 647–labeled microtubules (red, bottom row) grown from GMP-CPP-stabilized seeds at 35°C. White arrowheads indicate end-tracking starPEG-(KA7)4-TAMRA molecules and blue arrowheads their maximal compression at the end of depolymerization events. (E) Tracking efficiency of starPEG-(KA7)4-TAMRA. Left: schematic definition of the parameters and time points used for the graphs on the right. Magenta data points show the fraction of total microtubule-associated starPEG-(KA7)4-TAMRA intensity that remains bound to the microtubules during individual tracking events (n = 19). Cyan data points show the relative microtubule length remaining at the end of individual tracking events. Blue data points are the normalized starPEG-(KA7)4-TAMRA intensity values divided by the normalized microtubule length values; values above 1 indicate accumulation of starPEG-(KA7)4-TAMRA in terms of intensity per length.
The starPEG-(KA7)4-TAMRA molecules also “track” the depolymerizing plus end of dynamic Alexa 674–labeled microtubules grown from dimly labeled biotinylated GMP-CPP-stabilized seeds as observed by TIRF microscopy (Figure 2D). Tracking, however, is not a result of a specific association of starPEG-(KA7)4-TAMRA with the plus end. Rather, the starPEG-(KA7)4-TAMRA molecules appear to move by biased diffusion in a wave preceding the depolymerizing microtubule end (Figure 2D; Supplemental Movie S1). Hence, we assume that starPEG-(KA7)4-TAMRA, in principle, is able to track a depolymerizing minus end as well. However, we could not observe any minus-end-tracking event, which might be explained by either the fast depolymerization rates or the short depolymerization distances at the minus end. In the first case, the peptide is lost from the end, as it is not able to keep up with the depolymerization rates by diffusion and in the latter case starPEG-(KA7)4-TAMRA does not accumulate sufficiently to be visible against the starPEG-(KA7)4-TAMRA background on the lattice (please note the delay after catastrophe until the signal of plus end tracking starPEG-(KA7)4-TAMRA becomes evident in Figure 2D).
Movie S1.
Behaviour of starPEG-(KA7)4-TAMRA (50 nM) on dynamic microtubules. Please note the accumulation of starPEG-(KA7)4-TAMRA on the microtubule lattice during depolymerisation indicated by an intensity increase of the TAMRA-signal.
starPEG-(KA7)4-TAMRA tracks the depolymerizing plus end with high efficiency given that in average only ∼30% of microtubule-bound starPEG-(KA7)4-TAMRA intensity is lost during one depolymerization event. Since the microtubules shorten on average by ∼60% during those events, end-tracking concentrates starPEG-(KA7)4-TAMRA molecules on the remaining microtubule lattice about twofold (Figure 2E). Once a rescue event occurs, starPEG-(KA7)4-TAMRA is unable to track the growing microtubule plus end but expands by diffusion onto the newly polymerized microtubule lattice (Supplemental Figure S5).
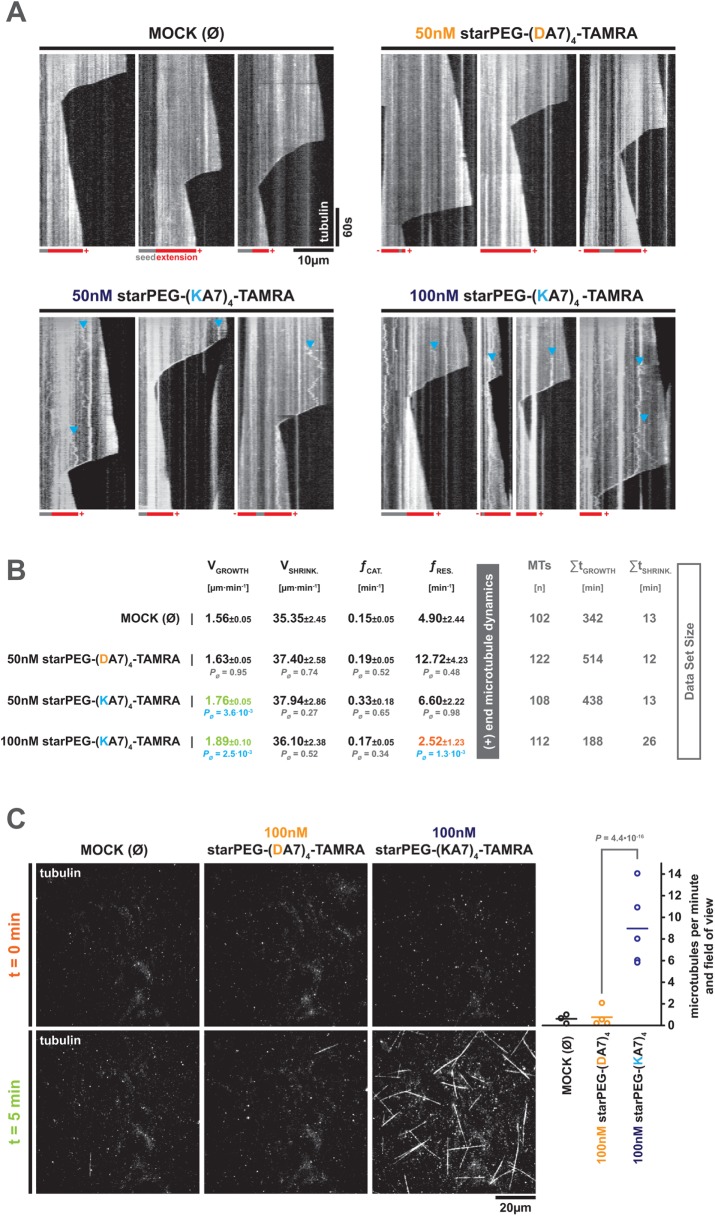
starPEG-(KA7) 4 promotes the growth and nucleation of microtubules
Since starPEG-(KA7)4-TAMRA tracks and accumulates at the depolymerizing microtubule plus end (i.e., the major site for addition and loss of tubulin dimers), we asked whether starPEG-(KA7)4—like other MAPs (Goodson and Jonasson, 2018)—is able to modulate microtubule dynamics. We therefore observed the dynamics of Alexa 647–labeled microtubules in the presence of 50 and 100 nM starPEG-(KA7)4-TAMRA by TIRF microscopy and compared them to the microtubule dynamics in the presence of 50 nM starPEG-(DA7)4-TAMRA or the absence of any peptide (buffer control, MOCK) (Figure 3A). While 50 nM starPEG-(DA7)4-TAMRA do not change microtubule dynamics compared with MOCK control, the addition of 50 or 100 nM starPEG-(KA7)4-TAMRA increases the growth velocity significantly. In addition, 100 nM but not 50 nM starPEG-(KA7)4-TAMRA significantly reduces the rescue frequency (Figure 3B).
FIGURE 3:
starPEG-(KA7)4 promotes microtubule growth and nucleation. (A) Kymographs showing the dynamicity of Alexa 647–labeled microtubules at 35°C in the presence of starPEG-(KA7)4-TAMRA or starPEG-(DA7)4-TAMRA at the indicated concentrations in BRB80. MOCK corresponds to an empty buffer control. Magenta/gray pictograms below the kymographs indicate the polarity of microtubules as judged by the dynamicity of the respective ends. Blue arrowheads indicate the diffusion of small tubulin aggregates along the lattice of dynamic microtubules that eventually convert into end-tracking events during episodes of depolymerization. (B) Parameters, describing the dynamics of microtubules at 35°C in the absence or presence of the indicated peptides at given concentrations in BRB80. Mean values of three independent experiments are given with their respective SE of the mean. The probability values were derived from a Mann–Whitney test. Total data set size is given on the right. (C) Spontaneous microtubule nucleation/growth of Alexa 647–labeled microtubules at 35°C in the absence or presence of the indicated peptides at the given concentrations in BRB80. Top row shows representative snapshots of a complete field of view (FOV, 81.4 × 81.4 µm) in the Alexa 647 channel at time 0 for the given conditions. Bottom row shows complete FOV images after 5 min incubation at 35°C. Please note that 0/5-min image pairs do not show the same FOV. For development of microtubule growth in the same FOV please refer to Supplemental Movie S2. Right graph shows the number of microtubules that nucleate de novo per FOV and minute at the given conditions. Data points indicate the mean value of individual experiments (10 FOVs each). Probability values were derived from a Mann–Whitney test.
On the basis of our observation that starPEG-(KA7)4-TAMRA efficiently promotes microtubule growth and due to its structural analogy (i.e., four microtubule-binding entities) to potent natural microtubule polymerases and nucleation factors such as XMAP215 (Brouhard et al., 2008; Widlund et al., 2011; Roostalu et al., 2015; Thawani et al., 2018), we hypothesized that starPEG-(KA7)4 might also exhibit some microtubule nucleation activity. To test this, we omitted the stabilized GMP-CPP seeds from our dynamic microtubule assay setup and observed the formation of microtubules in the absence of any peptide (MOCK) or in the presence of 100 nM starPEG-(DA7)4-TAMRA or starPEG-(KA7)4-TAMRA by TIRF microscopy. Microtubules were allowed to form for 5 min from subcritical concentrations of Alexa 647–labeled tubulin and were bound to the glass surface by an immobilized rigor kinesin-1 mutant (Nakata and Hirokawa, 1995; Rogers et al., 2001; see also Materials and Methods). The average number of microtubules per field of view (FOV) at t = 5 min was determined and corrected for the average number of microtubules FOV at the beginning of the experiment (t = 0 min). While hardly any new microtubules nucleate during the 5-min period in the absence of any peptide (0.6 ± 0.4 MT per minute and field of view ± SD) or in the presence of 100 nM starPEG-(DA7)4-TAMRA (0.8 ± 1.0), robust nucleation is evident in the presence of 100 nM starPEG-(KA7)4-TAMRA (9 ± 2.5) (Figure 3C; Supplemental Movie S2). Hence, starPEG-(KA7)4-TAMRA indeed acts as a nucleation factor.
Movie S2.
Microtubule nucleation in the absence/presence of the indicated peptides at the given concentrations. Behaviour of starPEG-(KA7)4-TAMRA (50 nM) on dynamic microtubules. Please note that a rigor kinesin-1 mutant was used to immediately surface immobilise nucleated microtubules.
starPEG-(KA7) 4 mediates force coupling with depolymerizing microtubule plus ends
In the presence of starPEG-(KA7)4-TAMRA, microtubules in our dynamic microtubule setup eventually picked up surface-associated tubulin aggregates that started to diffuse along the microtubule lattice and tracked depolymerizing plus ends (Figure 3A, blue arrowheads). As this behavior was not evident in the presence of starPEG-(DA7)4-TAMRA or in the absence of any peptide (Figure 3A), it is likely to originate from the microtubule binding and plus end tracking activity of starPEG-(KA7)4. We hence speculated that starPEG-(KA7)4 is able to couple microtubules (or other structures) to the (pulling) forces generated by depolymerizing microtubules, like it has, for example, been described for the plus end tracking Ndc80 and Ska-complexes at the kinetochore (Welburn et al., 2009; Volkov et al., 2018). Thus, we analyzed our dynamic microtubule data for depolymerizing events, during which the depolymerizing plus end traverses a second intersecting microtubule and asked whether the depolymerizing end manages to cross-link and pull the intersecting microtubule. Indeed, we observed frequent pulling events in the presence of 50 or 100 nM starPEG-(KA7)4-TAMRA (Figure 4, A and B; Supplemental Figure S6A; Supplemental Movies S3.1–S3.3 and S5.1–S5.3), but none in the presence of the negatively charged starPEG-(DA7)4-TAMRA or in the absence of any peptide (Figure 4B). Hence, pulling of intersecting microtubules crucially depends on the presence of starPEG-(KA7)4.
FIGURE 4:
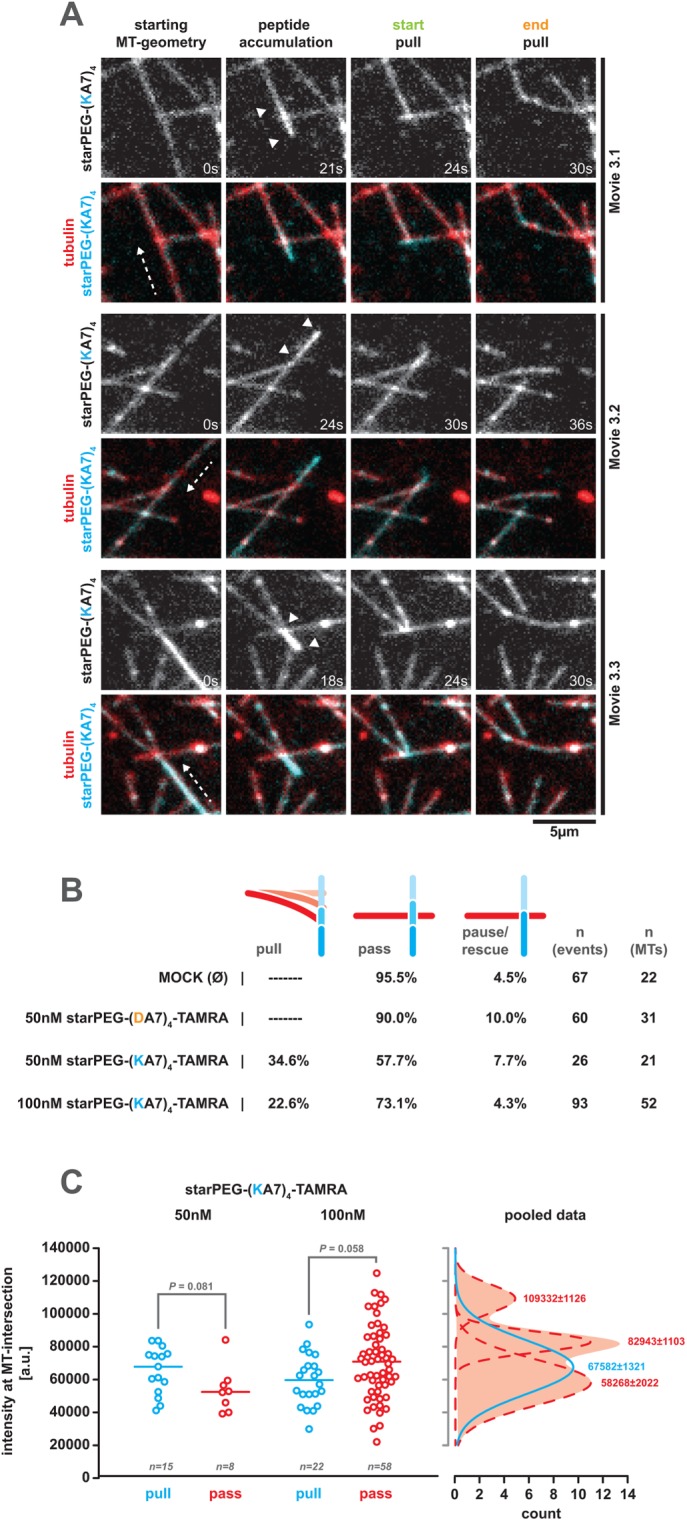
The starPEG-(KA7)4 mediates force coupling to depolymerizing microtubule plus ends. (A) Stills of Supplemental Movies S3.1–S3.3 showing steps of starPEG-(KA7)4-TAMRA-mediated pulling of intersecting microtubules by depolymerizing microtubule plus ends. The respective top rows show the starPEG-(KA7)4-TAMRA (50 nM) signal only; the bottom rows show the starPEG-(KA7)4-TAMRA (cyan) together with Alexa 647–labeled tubulin (red). White arrowheads indicate molecule accumulation during end tracking. (B) The occurrence in the percentage of possible outcomes of a depolymerizing plus end crossing a second intersecting microtubule (pause, pull, pause/rescue) in the dependency of the supplemented (amount of) molecules. Please note that during depolymerization, a single microtubule can interact consecutively with several intersecting microtubules (see also Supplemental Movie S5 and Supplemental Figure S6A for consecutive pulling events driven by the same depolymerizing microtubule). (C) Left graph: integrated starPEG-(KA7)4-TAMRA intensity (5-px diameter circle) at the intersection obtained from the frame when a passing (“pass,” red) or pulling (“pull,” blue) depolymerizing plus end traverses the intersecting microtubule at the indicated starPEG-(KA7)4-TAMRA concentrations. Probability values were derived from a Mann–Whitney test. Right graph: Gaussian fits to the pooled (i.e., 50 and 100 nM) intensity distribution of intersection bound starPEG-(KA7)4-TAMRA. Passing events are indicated in red, pulling events in blue. Peak values of the fits ± SE are indicated.
Movie S3.
Collage of 3 movies (3.1-3.3) showing the depolymerisation coupled pulling of intersecting dynamic microtubules cross-linked by 50 nM starPEG-(KA7)4-TAMRA. Left column shows the starPEG-(KA7)4-TAMRA signal only, the column on the right starPEG-(KA7)4-TAMRA (cyan) superimposed on the tubulin signal (red). See also Figure 4 and Supplementary Movie 1.
Movie S4.
Collage of 3 movies (4.1-4.3) showing bundling of dynamic microtubules by 50 nM starPEG-(KA7)4-TAMRA. Left column shows the starPEG-(KA7)4-TAMRA signal only, the column on the right starPEG-(KA7)4-TAMRA (cyan) superimposed on the tubulin signal (red). See also Figure 5 and Supplementary Movie 2. Blue arrowheads indicate microtubules before bundling, white arrowheads the final microtubule overlap.
Movie S5.
Collage of 3 movies (1.1-1.3) showing the depolymerisation coupled pulling of intersecting dynamic microtubules cross-linked by 50 nM starPEG-(KA7)4-TAMRA at a three-fold better time resolution as compared to Movie 3. Only the starPEG-(KA7)4-TAMRA signal is shown. Blue arrowheads and writing indicate single pulling events. Please note that the same starPEG-(KA7)4-TAMRA carrying depolymerising microtubule plus end can perform multiple sequential pulling events on different intersecting microtubules. See also Movie 3 and Supplementary Figure 6A.
In line with this, pulling events (Figure 4A) are initiated by the accumulation of starPEG-(KA7)4-TAMRA between the plus end and the microtubule intersection (Figure 4A, white arrowheads). Plus ends that once have accumulated sufficient starPEG-(KA7)4 are able to carry out multiple consecutive pulling events during the same shrinkage event (Supplemental Figure S6A; Supplemental Movies S5.1 and S5.2). Interestingly, the likelihood of pulling events to occur peaks at 50 nM starPEG-(KA7)4 (Figure 4B), suggesting that there is an optimal (local) concentration of starPEG-(KA7)4 for a pulling event to happen. To test this, we measured the starPEG-(KA7)4-TAMRA intensity at depolymerizing microtubule ends in the frames they traverse the intersecting microtubule (i.e., the first frame of a pull or pass event) and grouped them according to the outcome of the encounter (i.e., pull or pass). At 50 nM starPEG-(KA7)4, pulling microtubule plus ends exhibit a higher starPEG-(KA7)4-TAMRA accumulation than those just passing the intersecting microtubule (Figure 4C). Raising the starPEG-(KA7)4 concentration to 100 nM inverts this ratio and passing plus ends feature a higher amount of starPEG-(KA7)4-TAMRA (Figure 4C). Fitting Gaussians to the intensity distribution of the pooled data from both conditions reveals three discrete intensity subpopulations of passing plus ends (low, medium, and high intensity), while the starPEG-(KA7)4-TAMRA intensity at pulling ends is best described by a single Gaussian intensity profile. The maximum of the pulling end intensity distribution locates right in between the maxima of the low and medium intensity subpopulations of the passing end intensity distribution, thereby defining the optimal amount of end-bound starPEG-(KA7)4 that allows pulling events to occur (Figure 4C). In our interpretation, starPEG-(KA7)4-TAMRA amounts below the optimum are not capable of providing sufficient cross-linking and end-tracking activity to support pulling, while amounts above the optimum saturate all binding sites on both intersecting microtubules, thereby preventing any microtubule cross-linking needed for pulling to occur.
Given that end-tracking starPEG-(KA7)4 couples a second intersecting microtubule to the forces generated by a depolymerizing plus end, we next estimated the forces starPEG-(KA7)4 is able to transduce during a pulling event. For this we approximated the microtubule bending forces generated during pulling events that satisfy the small angle (≤30°) solution of the beam equation for a clamped rod whose free end is deflected by a transverse force applied to it (Howard, 2001). Then the bending force is given by
Using Fiji, we measured the length L of the deflected microtubule (i.e., the distance between the hinge point of the pulled microtubule and the microtubule intersection) and the deflection of the free tip y(L) (see scheme in Supplemental Figure S6B). Assuming the bending stiffness of a GTP-capped microtubule (i.e., EI = 26 Nm; Mickey and Howard, 1995), we calculated the respective bending forces for 11 events and corrected for the direction of the force applied by the depolymerizing microtubule according to: Fcorrected = F/cos (α), with α being the angle between the depolymerizing microtubule and a thought line that runs perpendicular to the bent microtubule, originating from its hinge point (see scheme in Supplemental Figure S6B). Calculated forces ranged between 1.6 and 8.4 pN with a mean of 4.2 ± 2.7 pN (mean ± SD, N = 11) (Supplemental Figure S6B) and thus fit into the range of forces that are coupled by natural MAPs (Akiyoshi et al., 2010; Miller et al., 2016; Helgeson et al., 2018; Volkov et al., 2018). As pulled microtubules occasionally were strongly deflected (e.g., beyond a small angle of 30°) and often stayed attached to the pulling microtubule after maximal deflection, our force approximation is conservative and likely represents the lower bound of the maximal forces that can be transduced by starPEG-(KA7)4.
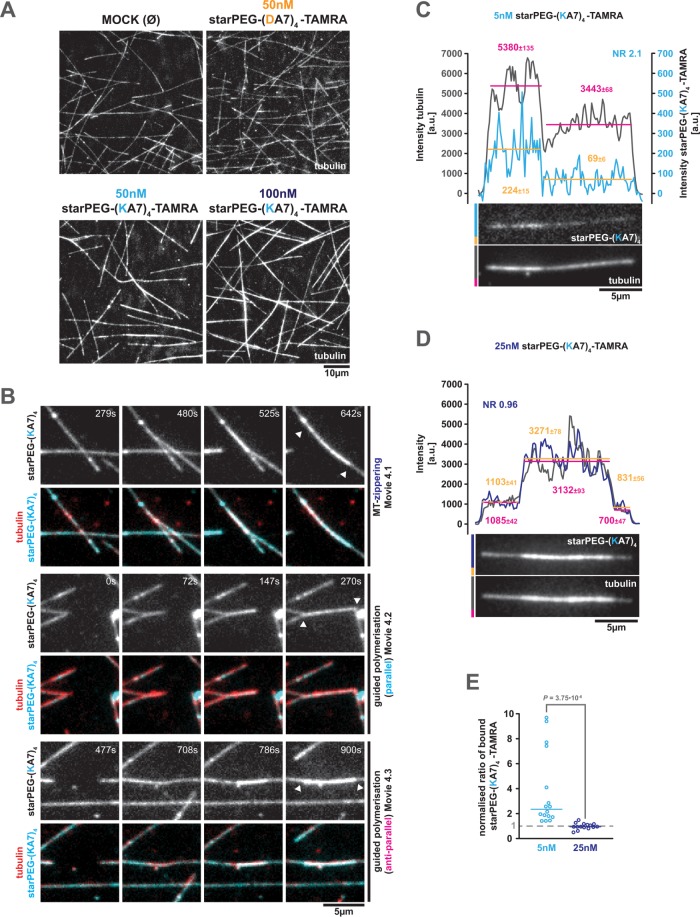
starPEG-(KA7) 4 accumulates in microtubule overlaps and drives the formation of microtubule bundles
The force coupling between intersecting microtubules by depolymerizing starPEG-(KA7)4-TAMRA-decorated microtubule plus ends already suggests that starPEG-(KA7)4 is a potent microtubule cross-linker—a feature we would expect due to its multivalent microtubule-binding interfaces (see Figure 1A). Hence, we asked whether starPEG-(KA7)4 also supports extensive microtubule cross-linking and bundling. In fact, we observed pronounced microtubule bundling during the time course of our dynamic microtubule assays in the presence of 50 or 100 nM starPEG-(KA7)4-TAMRA, but not in the presence of 50 nM starPEG-(DA7)4-TAMRA (Figure 5A). In general, starPEG-(KA7)4-TAMRA-driven microtubule bundling occurs by the lateral zippering of existing microtubules (Figure 5B, Supplemental Movies S6 and S4.1, and Supplemental Figure S7) or by guided (co-)polymerization of microtubules along each other, irrespective of their relative orientation (i.e., parallel or anti-parallel; Figure 5B; Supplemental Movies S4.2 and S4.3).
FIGURE 5:
starPEG-(KA7)4 bundles microtubules. (A) Representative images showing the microtubule organization of dynamic Alexa 647–labeled microtubules after 15 min incubation at 35°C in the absence or presence of the indicated peptides at given concentrations in BRB80. (B) Stills of Supplemental Movies S4.1–S4.3 showing different modes of starPEG-(KA7)4-TAMRA-mediated microtubule overlap formation; MT-zippering and (anti-)parallel guided polymerization. The respective top rows show the starPEG-(KA7)4-TAMRA (50 nM) signal only; the bottom rows show the starPEG-(KA7)4-TAMRA (cyan) together with Alexa 647–labeled tubulin (red). White arrowheads indicate the resulting microtubule overlaps. (C, D) Fluorescence images of starPEG-(KA7)4-TAMRA (top) on microtubule overlaps (bottom). Intensity profiles derived by line scans along the microtubule are shown in the graphs above. The starPEG-(KA7)4-TAMRA intensity profile is shown in light (5 nM) and dark blue (25 nM, average intensities in orange); the microtubule intensity profile is shown in gray (average intensities in magenta). Vertical lines depict the average intensity across the indicated range. Numbers are average ± SE. NR, normalized ratio (see also E) (E) Normalized intensity ratio of microtubule-bound starPEG-(KA7)4-TAMRA (at 5 and 25 nM), that is, the average starPEG-(KA7)4-TAMRA intensity on microtubule overlaps divided by the corresponding average intensity on single microtubules normalized by the same value ratio measured in the tubulin channel. Horizontal bars indicate median. Values above 1 mean accumulation within the overlap and values below 1 exclusion from the overlaps; n = 16 and 15, respectively. P value is derived from a Mann–Whitney test.
Movie S6.
Collage of 3 movies (2.1-2.3) showing bundling of dynamic microtubules by 50 nM starPEG-(KA7)4-TAMRA at a three-fold better time resolution as compared to Movie 4. Only the starPEG-(KA7)4-TAMRA signal is shown. Blue arrowheads indicate single zippering events, white arrowheads the final microtubule overlap. See also Movie 4 and Supplementary Figure 7.
Additionally, low-density starPEG-(KA7)4-TAMRA preferentially binds to or accumulates in overlaps of taxol-stabilized microtubules (Figure 5, C and E), indicating that the peptide has a higher affinity to microtubule overlaps as compared with single microtubules. As expected, this effect is lost near a half-saturating starPEG-(KA7)4-TAMRA concentration, at which single microtubules are evenly occupied and peptides on overlapping microtubules compete with peptides from the respective other microtubule for binding sites (Figure 5, D and E).
starPEG-(KA7) 4 does not interfere with kinesin-1 stepping on microtubules
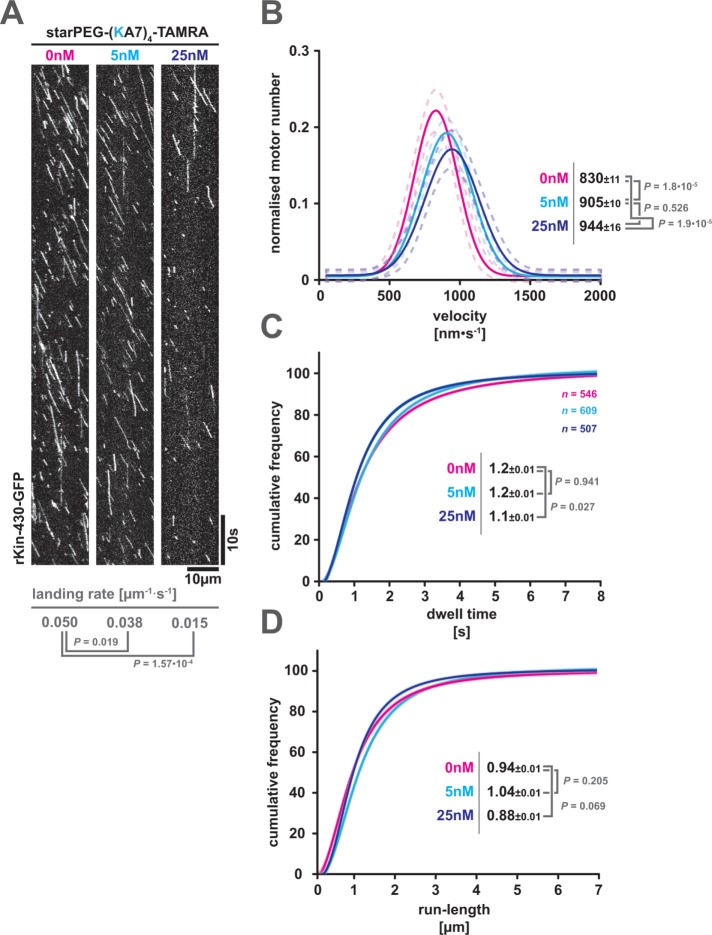
Having established the MAP-like properties of starPEG-(KA7)4, we sought to test its impact on natural MAPs, here represented by the conventional kinesin, kinesin-1. Kinesin-1 is a molecular motor that carries out the main load of plus end–directed microtubule-dependent cellular transport (Verhey et al., 2011). Kinesin motility is sensitive to physical roadblocks as the motor’s mean velocity, run length, and dwell time negatively correlate with the density of roadblocks on the microtubule. When encountering a static road block, single kinesin-1 motors enter a waiting state that lasts for ∼0.4 s and is resolved either by detachment of the motor or by continuation of walking (Schneider et al., 2015). When experiencing molecular crowding on the microtubule lattice by motile obstacles such as diffusive MAPs or other motors, kinesin-1 motors show a reduced mean landing rate, dwell time, velocity, and run length (Telley et al., 2009; Conway and Ross, 2014). Further, kinesin-1 motors are not able to penetrate into cohesive high lattice densities of MAPs (e.g., tau islands; Siahaan et al., 2019). Thus, this motor is a sensitive reporter for any obstacles (such as starPEG-(KA7)4) on the microtubule lattice.
We hence added increasing amounts of starPEG-(KA7)4-TAMRA (i.e., 5 and 25 nM) to a conventional stepping assay reconstituting the motility of constitutively active rat kinesin-1 (i.e., rKin430-GFP, lacking the autoinhibitory kinesin-1 tail) on taxol-stabilized microtubules. The starPEG-(KA7)4-TAMRA significantly decreases the landing rate of kinesin-1 in a concentration-dependent manner (Figure 6A), presumably by physically blocking landing sites or shielding the negative surface charge of the substrate microtubules. However, once a kinesin motor has successfully landed, its motility is not significantly affected by diffusing starPEG-(KA7)4-TAMRA molecules on the lattice. While, for unknown reasons, the velocity of kinesin-1 motors is slightly increased in the presence of starPEG-(KA7)4-TAMRA, this effect does not scale with the peptide concentration (Figure 6B) and is therefore unlikely to be caused by direct motor peptide interactions (like a road block mechanism). Furthermore, starPEG-(KA7)4-TAMRA does not significantly affect the motor’s average dwell time (Figure 6C) nor the average motor run length (Figure 6D) at any given concentration. We hence conclude that lattice-bound diffusive starPEG-(KA7)4 does not physically interfere with kinesin-1 motility.
FIGURE 6:
starPEG-(KA7)4 interferes with microtubule binding of kinesin-1 but not with its motility. (A) Kymographs showing the motility of GFP-labeled truncated rat kinesin-1 (rKin-430-GFP) on taxol-stabilized microtubule in the presence of starPEG-(KA7)4-TAMRA at 25°C and the indicated concentrations. Kinesin landing rates are given below the kymographs. Probability values are derived from a Mann–Whitney test. (B) Frequency distribution of kinesin-1 velocities in the presence of starPEG-(KA7)4-TAMRA at 0 nM (magenta), 5 nM (cyan), and 25 nM (blue). The dashed lines indicate the respective 95% confidence intervals. Numbers given are maxima of Gaussian fits ± SE. Probability values are derived from a Mann–Whitney test. (C, D) Cumulative frequency plots of kinesin-1 dwell time (C) and run length (D) in the presence of starPEG-(KA7)4-TAMRA at the indicated concentrations. Color coding of the data as in B. Numbers given are the respective half-maxima ± SE. Probability values are derived from a Mann–Whitney test.
DISCUSSION
starPEG-(KA7) 4 steps up to the performance of natural MAPs
With starPEG-(KA7)4, we here describe an artificial peptide that features many aspects of MAP functionality. We hence sought to compare its microtubule performance to those of natural MAPs. For a fair comparison, we searched the literature for a close structural and functional analogue of starPEG-(KA7)4, that is, a multivalent MAP with low structural complexity that relies mainly on electrostatic interactions to interact with the microtubule. Interestingly, we found it in the well-studied tau protein, a MAP involved in the pathogenesis of several neuropathies such as Alzheimer or Pick’s disease (Goedert et al., 2017). While KA7 peptides adopt a random coil structure (Wieduwild et al., 2013), tau is intrinsically disordered and contains four microtubule-binding repeats that mediate the longitudinal binding of the elongated tau peptide along the ridge of a protofilament (Kellogg et al., 2018).
The dissociation constant of starPEG-(KA7)4 (30 ± 6 nM) is comparable to the lowest of the reported tau dissociation constants ranging from 16 ± 5 nM (Makrides et al., 2004) to 1.1 µM (Gustke et al., 1994; see also Elie et al., 2015) for full-length tau. Both peptide and protein have a similar diffusion coefficient, that is, 0.3 ± 0.03 µm2s–1 for starPEG-(KA7)4 at 25°C and 160 mM ionic strength and 0.336 ± 0.062 µm2s–1 for tau at 35°C and 124 mM ionic strength (Hinrichs et al., 2012). Like starPEG-(KA7)4, tau can also switch microtubules at intersections (Hinrichs et al., 2012). Furthermore, tau also shows nucleation activity (Gustke et al., 1994) and bundles microtubules in vivo (Chung et al., 2016) and in vitro (the latter, however, only at high concentrations in the lower millimolar range; Scott et al., 1992). Finally, the concentration-dependent increase in microtubule growth by starPEG-(KA7)4 and tau is nearly identical (Ramirez-Rios et al., 2016).
In summary, the artificial starPEG-(KA7)4 steps up to the performance of a natural MAP, showing biologically relevant functionality such as microtubule cross-linking, microtubule bundling, microtubule nucleation, microtubule growth promotion, tracking of depolymerizing microtubule ends, and force coupling to depolymerizing ends. We therefore consider starPEG-(KA7)4—to the best of our knowledge—as a first prototype of an artificial MAP.
Multivalency as a critical parameter for MAP functionality
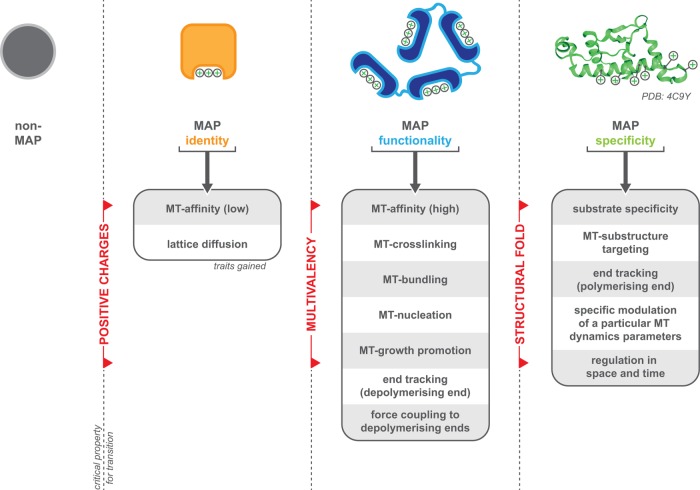
The simplicity of starPEG-(KA7)4 allows us to speculate the origin of MAP functions that are relevant for biological processes and to identify key features that define a MAP (Figure 7). Our work and that of others show that electrostatic interactions with the acidic tubulin C-termini are sufficient to mediate microtubule binding and allow lattice diffusion. Both properties set the ”identity” of a MAP, as they allow proteins (Subramanian et al., 2010; Abad et al., 2014; Kraatz et al., 2016; Ma et al., 2017), (single KA7-) peptides (this study), or even artificial beads (Minoura et al., 2010) to associate with a microtubule, although binding and diffusion, per se, are not sufficient to carry out any relevant biological functions (apart from being a roadblock or targeting factor for other proteins).
FIGURE 7:
Scheme summarizing the critical properties (red) that confer a protein MAP identity (orange), MAP functionality (blue), and MAP specificity (green). Boxes summarize the traits associated with each aspect. Please also refer to the Discussion.
Linking multiple positively charged microtubule-binding entities together, however, constitutes—based on our starPEG-(KA7)4 results—a quantum leap in terms of functionality. Suddenly, the peptides show microtubule affinity in the nanomolar range, cross-link and bundle microtubules, support microtubule growth, and are able to link objects (i.e., in this case another microtubule) to depolymerizing microtubule ends. This suggest that multivalency—as defined by the capacity to bind the microtubule by multiple, spatially separated and independent microtubule-binding entities—is another key feature, one that sets the “functionality” of a MAP. We note, however, that the dependency on multivalent microtubule-binding entities might be restricted to MAPs that primarily rely on electrostatic interactions for microtubule binding (e.g., such as Ska1; Widlund et al., 2011; Abad et al., 2014) and appears not to be strictly essential to MAPs that feature additional, hydrophobic interactions with the microtubule lattice (e.g., XMAP215; Slep and Vale, 2007).
The concept of multivalency can be found intrinsically in several natural MAPs (e.g., tau and XMAP215; Widlund et al., 2011; Kellogg et al., 2018), but is also often achieved by the formation of higher-order complexes (e.g., Ska and Ndc80 complexes; Jeyaprakash et al., 2012; Volkov et al., 2018). Besides the aforementioned tau protein, the microtubule polymerase and nucleation factor XMAP215/ch-TOG is another prototype for intrinsic multivalency. It features five N-terminal tumor overexpressed gene (TOG) domains, each of which is capable to bind free α-/β-tubulin, and an additional positively charged microtubule lattice domain in between TOG domains 4 and 5 (Widlund et al., 2011). Full polymerase activity of XMAP215 requires the presence of TOG 1 and 2 as well as an additional microtubule-binding entity (i.e., the lattice binding domain [ Widlund et al., 2011] or the laterally binding TOG 5 domain [ Byrnes and Slep, 2017])—and thus a trivalent protein. Full microtubule nucleation activity of XMAP215 in contrast requires the full set of TOG domains and thus a pentavalent protein (Thawani et al., 2018). A common theme of both, microtubule polymerization and nucleation, is the idea that XMAP215 uses its C-terminal end beyond TOG 4 to bind to the growing microtubule end or a conucleation factor (i.e., the gamma tubulin ring complex), while recruiting free tubulin from solution using TOG 1-4 (Al-Bassam and Chang, 2011; Thawani et al., 2018) alleviating its incorporation into the microtubule lattice or the nascent microtubule seed. A similar mechanism could also account for the nucleation activity and growth-promoting activity of starPEG-(KA7)4. Being tetravalent, starPEG-(KA7)4 could bring together four tubulin dimers alleviating the nucleation of a microtubule seed, while end-bound peptide could promote incorporation of free tubulin subunits from solution into the growing lattice. This however, requires some affinity of starPEG-(KA7)4 to free tubulin dimers—an aspect we unfortunately could not address in this study due to technical issues (i.e., detection) associated with the small size of the peptide.
A good example for multivalency through complex formation is the kinetochore resident Ndc80 complex, which couples chromosomes to the dynamic plus ends of k-fiber microtubules. Single Ndc80 complexes associate with the microtubule lattice already at low picomolar concentrations, but fail to track depolymerizing plus ends, unless they form a higher-order complex with the kinetochore components CENP-T and MIS12 in a 3:1:1 (Ndc80:CENP-T:MIS12) ratio (Volkov et al., 2018), associate with the—as well microtubule-associated—Ska complex (Helgeson et al., 2018; Schmidt et al., 2012), or are artificially oligomerized (Powers et al., 2009).
Although starPEG-(KA7)4 phenocopies most of the functionality that can be found in natural MAPs, it completely lacks those traits that define the specificity of a MAP. Due to its structural simplicity, starPEG-(KA7)4 will bind to any other negatively charged biomolecule surfaces (e.g., phospholipid bilayers or the DNA phosphoribose backbone) and therefore lacks any substrate specificity. In fact, the KA7-PEG system has been previously used to noncovalently cross-link other negatively charged polymers, including highly sulphated polysaccharides such as heparin or conductive polymer PEDOT:PSS, resulting in stable hydrogel networks (Wieduwild et al., 2013; Xu et al., 2018). Further, starPEG-(KA7)4 does not recognize the tubulin nucleotide state nor any particular substructure of the microtubule (e.g., plus ends, minus ends) and therefore also fails to track the growing microtubule end. Further, it is not capable of modulating any specific microtubule dynamics parameter other than microtubule growth, and finally, it is not possible to regulate the activity of starPEG-(KA7)4 in space or time by, for example, posttranslational modifications. Hence, all these biological-relevant specificity traits must be encoded by a third critical property, which is likely the structural fold that defines the local environment of a microtubule-binding entity and their spatial alignment with each other or the substrate.
The potential of artificial MAPs
To test the importance of particular structural folds, future work should therefore try to transform functional artificial MAPs—like starPEG-(KA7)4—into selective artificial MAPs by the addition of defined but limited local environments and structural elements. This requires the development of an easily modifiable modular artificial MAP system. For this, starPEG-(KA7)4 will be a good starting point, as previous studies with artificial KA-repeat-bearing peptides already showed that KA-repeat-mediated electrostatic interactions are easily tuned by the number or spacing of repeats (Wieduwild et al., 2013) and not disturbed by the addition of further sequences to the KA motif (Tondera et al., 2017). Such a modular system eventually may be used to establish a library of engineered “one feature—one function” reference artificial MAPs. This library would enable us to make functional predictions of uncharacterized natural MAPs and, more importantly, to design and produce modular artificial MAPs tailored to fulfill a specific task. Modular artificial MAPs, designed for the task and cheap to produce in larger amounts and maybe even switchable, will be valuable tools to probe and modulate microtubule biology in vivo and in vitro.
However, even in its current state, starPEG-(KA7)4 can serve as a versatile, easily modifiable, and cheap to produce tool for nanotechnical applications that aim to modulate the microtubule lattice, modify microtubule interacting proteins, or functionally engineer microtubules for nanotechnological applications (see Supplemental Figure S8). On the basis of the functionality of starPEG-(KA7)4, we think it is suitable for 1) the dynamic and reversible fluorescence-labeling of microtubules in vitro, 2) the reversible physical and electrostatic screening of microtubules in vitro, 3) the targeting of modifying enzymes to the microtubule lattice in vitro, 4) processivity modification of molecular motors, 5) functionalization of microtubules as plasmonic waveguides, or 6) measuring depolymerization-coupled pulling forces in vivo.
MATERIALS AND METHODS
Peptide synthesis and labeling
General peptide synthesis.
For peptide synthesis, all required chemicals were purchased from IRIS Biotech GmbH (Marktredwitz, Germany) unless otherwise specified. All peptides were prepared using standard Fmoc chemistry on a solid-phase with 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate activation on an automated solid-phase peptide synthesizer (ResPep SL, Intavis, Cologne, Germany). For good peptide quality, each amino acid was coupled twice with fivefold excess, and all nonreacted amino groups were capped with acetic anhydride, the same way 5(6)-TAMRA was coupled to the N-terminus of the peptides on the resin. The peptide was cleaved off the resin with trifluoroacetic acid/triisopropyl silane/water/dithiothreitol (DTT) (90[vol/vol]:5[vol/vol]:2.5[vol/vol]:2.5[m/vol]) for 1.5 h. The product was precipitated and washed with ice-cold diethyl ether. Peptide purification was performed via reverse-phase high-pressure liquid chromatography (HPLC) on a preparative HPLC (Waters, Milford, MA) equipped with a preparative C18 column (AXIA 100A, bead size 10 µm, 250 × 30 mm, Phenomenex, Torrance, CA). Purity was confirmed by electrospray ionization mass spectrometry (ACQUITY TQ Detector; Waters, Milford, MA).
Synthesis of peptide-PEG conjugate.
Peptide-starPEG conjugate synthesis was conducted via Michael addition reactions between maleimide-terminated starPEG (10 kDa) and cysteine-terminated KA7 peptides. Both components were solved in phosphate-buffered saline, pH 7.4, and mixed in a molar ratio of 1:5 (star-PEG:peptide). The reaction mixture was quickly sealed and stirred (MR Hei-Standard, Heidolph, Schwabach, Germany) at 750 rpm at room temperature overnight. Uncoupled peptides and salt were removed from the reaction mixture by dialyzing the peptide-star PEG product for 2 d against water using tubing with an 8-kDa cut-off. In general, the final peptide products were lyophilized and kept at –20°C for long-term storage.
For TIRF experiments, the respective peptides were rehydrated in BRB80, pH 6.8 (i.e., 80 mM PIPES, pH 6.8, 1 mM MgCl2, 1 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid [EGTA]) and stored as single-use aliquots at –20°C.
General TIRF-microscopy
TIRF microscopy experiments in this study were performed on an Axio Observer Z1 microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) equipped with 488/532/642 nm laser lines (Omicron-Laserage, Rodgau-Dudenhofen, Germany) and an iXon X3 EMCCD (Andor Technology, Belfast, UK). Image data were acquired using an alpha Plan-Apochromat 63×/1.46 Oil objective (Zeiss) with a 1.6-fold auxiliary magnification—achieving a final pixel size of 159 ± 3 nm. For temperature control, the objective was equipped with a water-supplied heating ring connected to a thermostatic circulator (F-25-MC; Julabo, Seelbach, Germany).
Assays on stabilized microtubules
Microtubule preparation.
Taxol-stabilized microtubules were grown at 37°C for 1 h from 30 µM Atto647N-labeled pig brain tubulin (labeled:unlabeled = 1:20) in BRB80, pH 6.8, supplemented with 4 mM MgCl2, 1 mM GTP (Jena Bioscience GmbH, Jena, Germany), and 4.8% (vol/vol) dimethyl sulfoxide (DMSO). Microtubules were sedimented at room temperature for 30 min at 17,000 × g and resuspended in warm (i.e., 37ºC) BRB80 supplemented with 10 µM taxol (Sigma-Aldrich, Hamburg, Germany). Microtubules were stored in the dark at room temperature and kept for a maximal 48 h. GMP-CPP-stabilized microtubule were grown 37°C for 2 h from 2 µM Atto647N-labeled pig brain tubulin (labeled:unlabeled = 1:4 [bright] or 1:20 [dim]) in BRB80, 1 mM GMP-CPP (Jena Bioscience). Microtubules were sedimented at room temperature for 30 min at 17,000 × g and resuspended in warm BRB80. For subtilisin treatment, GMP-CPP microtubules were polymerized and sedimented as above and resuspended in BRB80 supplemented with 10 µm taxol and 200 µg/ml subtilisin A (Sigma-Aldrich). Microtubules were incubated for 1 h at 30°C before protease digest was stopped by the addition of 10 mM phenylmethanesulfonyl fluoride (Sigma-Aldrich). Microtubules were sedimented as above and resuspended in warm BRB80. The samples were split and analyzed on a 4–12/% precast SDS–PAGE gel (ThermoFisher, Langenselbold, Germany) or directly used in the TIRF assay.
Assay setup.
Anti–β-tublin antibodies (mouse, SAP.4G5, Sigma-Aldrich) at 15 µg/ml were allowed for 10 min to bind to the silylated glass surface of a flow cell. The surface then was blocked for 1 h with 1% Pluronic F-127 (Sigma-Aldrich). Surplus Pluronic F-127 was washed out using BRB80 and Atto647N-labeled microtubules were allowed to bind to the anti–β-tublin antibodies until the desired density was reached (<1 min). Nonbound microtubules were removed by BRB80 supplemented with 10 µM Taxol and the flow cell was perfused with the assay mix, which had been precleared at room temperature for 2 min at 17,000 × g. The assay mix contained starPEG-(KA7)4-TAMRA, starPEG-(DA7)4-TAMRA, and/or truncated, GFP-labeled rat kinesin-1 (rKin1–430-GFP) at the indicated concentrations in 80 mM PIPES, pH 6.8, 1 mM MgCl2, 1 mM EGTA, 1 mM ATP (Jena Bioscience, only in kinesin motility assay), 10 µM taxol, 50 µg⋅ml–1 casein, 10 mM DTT, 20 mM glucose, 0.2 mg⋅ml–1 glucose oxidase (Serva Electrophoresis GmbH, Heidelberg, Germany), and 20 µg⋅ml–1 catalase (Sigma-Aldrich). Evaluation of the ionic strength and pH dependency of starPEG-(KA7)4-TAMRA microtubule binding was carried out in 25 mM PIPES, pH 6.8 (BRB25), ionic strength adjusted with 150–300 mM NaCl, or in pH-adjusted 50 mM Tris that has been adjusted to the ionic strength of BRB80 using 145–180 mM NaCl. The flow channels were sealed with VALAP (Vaseline/lanoline/paraffine, 1:1:1 wt/wt) and imaged at 25°C for a maximal 30 min. To minimize experimental variations, a full set of the respective experiments was conducted on the same day using the same stock solutions in flow channels on the same glass coverslip.
Imaging the diffusion of starPEG-(KA7) 4.
To describe the diffusion of starPEG-(KA7)4-TAMRA along the microtubule lattice, 5-min movies were recorded in a continuous stream with 100-ms exposure using the 532-nm laser line at 40 mW. The position of the substrate microtubules was determined by taking a snapshot before each movie using the 642-nm laser line at 5.6 mW and 40-ms exposure time. To minimize superimposition of multiple molecule traces for subsequent automated tracking, starPEG-(KA7)4-TAMRA was imaged at 1 nM concentration. For microtubule binding by starPEG-(KA7)4, flow chambers were assembled as described above and incubated for 15 min on the microscope at 25°C. Then, snapshots of the tubulin (642 nm) and the starPEG-(KA7)4 channel (532 nm) were taken at random positions within the flow chamber using the above-mentioned specifications. To assess the motility of rKin1–430-GFP (Rogers et al., 2001) in the presence of starPEG-(KA7)4-TAMRA, 5-min movies were recorded in a continuous stream with 100-ms exposure using the 488-nm laser line at 5 mW. The position of the substrate microtubules was determined by taking a snapshot before each movie using the 642-nm laser line at 5.6 mW and 40- to 100-ms exposure time. The amount of starPEG-(KA7)4-TAMRA on each microtubule was recorded by taking a snapshot before each movie using the 532–nm laser line at 40 mW and 100-ms exposure time.
Analyzing the diffusion of starPEG-(KA7) 4.
Tracking of single starPEG-(KA7)4-TAMRA molecules and calculation of the diffusion coefficient D were carried out by FIESTA v1.6 (Ruhnow et al., 2011) using the following settings (alterations from standard settings only): threshold—relative intensity, height, 1; connecting—maximum velocity, 5000 nm⋅s–1; weights, 100% position; tracks—minimum length, 5 frames; maximum break, three frames; maximum angle, 180°. Resulting tracks were manually validated based on the starPEG-(KA7)4-TAMRA signal in the corresponding kymographs. Microtubule affinity of starPEG-(KA7)4—the integrated total intensity of microtubule-bound starPEG-(KA7)4-TAMRA and the length of individual microtubules were measured in ImageJ. Overlapping or bundled microtubules were not considered for analysis. For each condition, the mean intensity per micrometer was calculated and corrected for the chamber-specific background mean values off the microtubules as well as for the microtubule-specific background in the 532-nm channel (empty buffer control). To compare different sets of experiments, individual data sets were normalized to saturating conditions at 500 nM. Data fitting was done in Origin 9 using the Michaelis-Menten fit function. Data fitting for Supplemental Figure S3 was done in OriginPro 2015 using an exponential decay fit. Kinesin-1 motility—all rKin430 parameters (i.e., landing rate, velocity, dwell time, and run length) were determined manually from individual kymographs using ImageJ. Data fitting was done in Origin 9 using Gaussian and log-normal fits.
Dynamic microtubule assays
Seed preparation.
Short microtubule seeds were grown at 37°C for 60 min from 18.5 µM biotinylated, Alexa 647–labeled pig brain tubulin (biotinylated:labeled:unlabeled = 1:2:47) in BRB80, pH 6.8, supplemented with 1 mM GMP-CPP (Jena Biosciences) and 1 mM DTT. The seed mix was stored in the dark at room temperature and kept for a maximum of 48 h.
Assay setup.
Anti-biotin antibodies (goat, B3640, Sigma-Aldrich) at 70 µg/ml were allowed for 15 min to bind to the silylated glass surface of a flow cell. The surface then was blocked for 1 h with 1% Pluronic F-127. Surplus Pluronic F-127 was washed out using BRB80 and biotinylated, dimly Alexa 647–labeled, GMP-CPP-stabilized tubulin seeds were allowed to bind to the anti-biotin antibodies until the desired density was reached (<1 min). Nonbound seeds were removed by BRB80 and the flow cell was perfused with the assay mix, which had been precleared for 2 min at room temperature in a tabletop centrifuge at 17,000 × g. The assay mix contained 14 µM Alexa 647–labeled pig brain tubulin (labeled:unlabeled = 1:14) and starPEG-(KA7)4-TAMRA or starPEG-(DA7)4-TAMRA at the indicated concentrations in 80 mM PIPES, pH 6.8, 4 mM MgCl2, 1 mM EGTA, 1 mM GTP, 0.27% (wt/vol) methyl-cellulose, 0.8 mg⋅ml–1 casein, 5 mM DTT, 67 mM glucose, 0.5 mg⋅ml–1 glucose oxidase, and 0.27 mg⋅ml–1 catalase. The flow channels were sealed with VALAP and imaged at 35°C for maximal 30 min. All experiments were performed using the same, prealiquoted labeled tubulin preparation. For each condition/concentration, three independent experiments have been carried out. To minimize experimental variations, a full set of experiments (i.e., MOCK, 50 mM starPEG-(DA7)4-TAMRA, and 50/100 mM starPEG-(KA7)4-TAMRA) was conducted on the same day using the same stock solutions in four flow channels on the same glass coverslip.
Imaging.
Single-color time-lapse movies were recorded with one frame per second using the 647-nm laser line at 100-ms exposure to visualize the Alexa 647–labeled tubulin. Dual-color time-lapse movies were recorded with one frame every 3 s using the 647-nm laser line at 100-ms exposure to visualize the Alexa 647–labeled tubulin and the 564-nm laser line at 100-ms exposure to visualize starPEG-(KA7)4-TAMRA.
Analyzing microtubule dynamics.
Movies were analyzed and processed with ImageJ. Kymographs were produced by the MultipleKymograph plug-in (www.embl.de/eamnet/html/body_kymograph.html). Parameters of microtubule dynamicity were extracted manually from kymographs using ImageJ. The following definitions were used: catastrophe frequency, number of catastrophe events divided by the total time of growth; rescue frequency, number of rescue events divided by the total time of shrinkage. The probability values given in Figure 3B were derived from a Mann–Whitney U test. The SE of mean was calculated from the mean of the respective values per microtubule. To quantify the starPEG-(KA7)4 intensity during pulling events the integrated intensity of a five-pixel diameter circle was measured in the frame the depolymerizing plus end traverses the intersecting microtubules and locally corrected for the background. Data fitting shown in Figure 4C was done in OriginPro 2015 using the multiple peak Gaussian fit tool. The probability values given in Figure 3C were derived from a Mann–Whitney U test.
Nucleation assay.
For the nucleation assay, we omitted the microtubule seeds from our dynamic microtubule assay setup and treated the glass surface of our flow cells in a different manner. First, a His-tagged and truncated rigor variant of the rat kinesin-1 (i.e., rKin1-430, T93N-His6) (Nakata and Hirokawa, 1995; Rogers et al., 2001) was allowed to bind the silylated glass surface for 15 min at ∼600 nM concentration. The surface was then blocked for 1 h with 1% Pluronic F-127 and for 30 min with 0.8 mg⋅ml–1 casein. Flow chambers were rinsed with BRB80, perfused with assay mix (as above), and sealed with VALAP. Flow channels were imaged at 35°C using the 647-nm laser line at 75- to 100-ms exposure to visualize the Alexa 647–labeled tubulin. First, snapshots from 10 random positions were taken, also allowing for temperature equilibration. Then, a 5-min time-lapse movie was recorded with one frame per second. Finally, snapshots were again taken from 10 random positions within the flow cell. For quantitative analysis, microtubules FOV were manually counted using ImageJ. Since we aimed to quantify the de novo nucleation of microtubules and we cannot formally exclude that starPEG-(KA7)4 facilitates templated nucleation on the lattice of already existing microtubules, microtubule bundles that formed in the presence of starPEG-(KA7)4 were counted as one microtubule only. Thus, the actual number of microtubules nucleated de novo in the presence of starPEG-(KA7)4 is higher than the apparent values given in Figure 3C.
Supplementary Material
Acknowledgments
We are grateful to Ulrike Hofmann and Corina Bräuer for technical assistance as well as to Friedrich W. Schwarz for initial observations of microtubule cross-linking by starPEG-(KA7) 4. This work was supported by the Technische Universität Dresden (Y.Z. and S.D.), by the German Research Foundation through SFB1027, and by the Cluster of Excellence Center for Advancing Electronics Dresden (S.D.) as well as by the China Scholar Council (Y.X.).
Abbreviations used:
- CVE
covariance-based estimator
- DTT
dithiothreitol
- FOV
field of view
- HPLC
high-pressure liquid chromatography
- MAP
microtubule-associated protein
- MSD
mean square displacement
- MTBD
microtubule-binding domain
- PEG
polyethylene glycol
- starPEG
four arm PEG chain
- TAMRA
carboxytetramethylrhodamine
- TIRF
total internal reflection fluorescence
- TOG
tumor overexpressed gene.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-05-0247) on October 10, 2019.
REFERENCES
- Abad MA, Medina B, Santamaria A, Zou J, Plasberg-Hill C, Madhumalar A, Jayachandran U, Redli PM, Rappsilber J, Nigg EA, et al (2014). Structural basis for microtubule recognition by the human kinetochore Ska complex. Nat Commun , 2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova A, Hoogenraad CC. (2015). Microtubule minus-end-targeting proteins. Curr Biol , R162–R171. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Steinmetz MO. (2008). Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol , 309. [DOI] [PubMed] [Google Scholar]
- Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S. (2010). Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature , 576–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Chang F. (2011). Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol , 604–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper JD, Decker F, Agana B, Howard J. (2014). The motility of axonemal dynein is regulated by the tubulin code. Biophys J , 2872–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton J, Jiang K, Stangier MM, Luo Y, Hua S, Houben K, van Hooff JJE, Joseph A-P, Scarabelli G, Grant BJ, et al (2017). A structural model for microtubule minus-end recognition and protection by CAMSAP proteins. Nat Struct Mol Biol , 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auckland P, Clarke NI, Royle SJ, McAinsh AD. (2017). Congressing kinetochores progressively load Ska complexes to prevent force-dependent detachment. J Cell Biol , 1623–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Telley IA, Surrey T. (2010). A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell , 420–432. [DOI] [PubMed] [Google Scholar]
- Brouhard GJ, Rice LM. (2018). Microtubule dynamics: an interplay of biochemistry and mechanics. Nat Rev Mol Cell Biol , 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. (2008). XMAP215 is a processive microtubule polymerase. Cell , 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes AE, Slep KC. (2017). TOG–tubulin binding specificity promotes microtubule dynamics and mitotic spindle formation. J Cell Biol , 1641–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Machado P, Alvarez-Martins I, Gouveia SM, Swadhin CJ, Duarte P, Amado T, Branco P, Freitas MC, Silva STN, et al (2012). BLD10/CEP135 Is a microtubule-associated protein that controls the formation of the flagellum central microtubule pair. Dev Cell , 412–424. [DOI] [PubMed] [Google Scholar]
- Chung PJ, Song C, Deek J, Miller HP, Li YL, Choi MC, Wilson L, Feinstein SC, Safinya CR. (2016). Tau mediates microtubule bundle architectures mimicking fascicles of microtubules found in the axon initial segment. Nat Commun , 12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway L, Ross JL. (2014). Kinesin motor transport is altered by macromolecular crowding and transiently associated microtubule-associated proteins. arXiv:1409.3455.
- Dixit R, Ross JL, Goldman YE, Holzbaur ELF. (2008). Differential regulation of dynein and kinesin motor proteins by tau. Science , 1086–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogterom M, Koenderink GH. (2019). Actin–microtubule crosstalk in cell biology. Nat Rev Mol Cell Biol , 38–54. [DOI] [PubMed] [Google Scholar]
- Drechsler H, McHugh T, Singleton MR, Carter NJ, McAinsh AD. (2014). The Kinesin-12 Kif15 is a processive track-switching tetramer. Elife , e01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elie A, Prezel E, Guerin C, Denarier E, Ramirez-Rios S, Serre L, Andrieux A, Fourest-Lieuvin A, Blanchoin L, Arnal I. (2015). Tau co-organizes dynamic microtubule and actin networks. Sci Rep , 9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanos TN, Santamaria A, Jeyaprakash AA, Wang B, Conti E, Nigg EA. (2009). Stable kinetochore–microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J , 1442–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Eisenberg DS, Crowther RA. (2017). Propagation of Tau aggregates and neurodegeneration. Annu Rev Neurosci , 189–210. [DOI] [PubMed] [Google Scholar]
- Goldberg MW. (2017). Nuclear pore complex tethers to the cytoskeleton. Semin Cell Dev Biol , 52–58. [DOI] [PubMed] [Google Scholar]
- Goodson HV, Jonasson EM. (2018). Microtubules and microtubule-associated proteins. Cold Spring Harb Perspect Biol , a022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustke N, Trinczek B, Biernat J, Mandelkow EM, Mandelkow E. (1994). Domains of tau-protein and interactions with microtubules. Biochemistry-US , 9511–9522. [DOI] [PubMed] [Google Scholar]
- Hanisch A, Silljé HH, Nigg EA. (2006). Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J , 5504–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgeson LA, Zelter A, Riffle M, MacCoss MJ, Asbury CL, Davis TN. (2018). Human Ska complex and Ndc80 complex interact to form a load-bearing assembly that strengthens kinetochore-microtubule attachments. Proc Natl Acad Sci USA , 2740–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs MH, Jalal A, Brenner B, Mandelkow E, Kumar S, Scholz T. (2012). Tau protein diffuses along the microtubule lattice. J Biol Chem , 38559–38568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J. (2001). Mechanics of Motor Proteins and the Cytoskeleton, Sunderland, MA: Sinauer Associates.
- Jeyaprakash AA, Santamaria A, Jayachandran U, Chan YW, Benda C, Nigg EA, Conti E. (2012). Structural and functional organization of the Ska complex, a key component of the kinetochore-microtubule interface. Mol Cell , 274–286. [DOI] [PubMed] [Google Scholar]
- Kellogg EH, Hejab NMA, Poepsel S, Downing KH, DiMaio F, Nogales E. (2018). Near-atomic model of microtubule-tau interactions. Science , 1242–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof Y-D, Nigg EA. (2007). Plk4-induced centriole biogenesis in human cells. Dev Cell , 190–202. [DOI] [PubMed] [Google Scholar]
- Kodama A, Karakesisoglou I, Wong E, Vaezi A, Fuchs E. (2003). ACF7: An essential integrator of microtubule dynamics. Cell , 343–354. [DOI] [PubMed] [Google Scholar]
- Kraatz S, Guichard P, Obbineni JM, Olieric N, Hatzopoulos GN, Hilbert M, Sen I, Missimer J, Gonczy P, Steinmetz MO. (2016). The human centriolar protein CEP135 contains a two-stranded coiled-coil domain critical for microtubule binding. Structure , 1358–1371. [DOI] [PubMed] [Google Scholar]
- Lee K-Y, Esmaeili B, Zealley B, Mishima M. (2015). Direct interaction between centralspindlin and PRC1 reinforces mechanical resilience of the central spindle. Nat Commun , 7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YL, Yue JP, Zhang Y, Shi CZ, Odenwald M, Liang WGG, Wei Q, Goel A, Gou XW, Zhang JM, et al (2017). ACF7 regulates inflammatory colitis and intestinal wound response by orchestrating tight junction dynamics. Nat Commun , 15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J, Drechsler H, Grundner-Culemann K, Ballister ER, Rodriguez-Rodriguez JA, Rodriguez-Bravo V, Jones MJK, Foley E, Lampson MA, Daub H, et al (2017). Mps1 regulates kinetochore-microtubule attachment stability via the ska complex to ensure error-free chromosome segregation. Dev Cell , 143–156.e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrides V, Massie MR, Feinstein SC, Lew J. (2004). Evidence for two distinct binding sites for tau on microtubules. Proc Natl Acad Sci USA , 6746–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. (2012). EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell , 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR. (2016). Mitosis. Cold Spring Harb Perspect Biol . [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally FJ, Roll-Mecak A. (2018). Microtubule-severing enzymes: from cellular functions to molecular mechanism. J Cell Biol , 4057–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickey B, Howard J. (1995). Rigidity of microtubules is increased by stabilizing agents. J Cell Biol , 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MP, Asbury CL, Biggins S. (2016). A TOG protein confers tension sensitivity to kinetochore-microtubule attachments. Cell , 1428–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoura I, Katayama E, Sekimoto K, Muto E. (2010). One-dimensional Brownian motion of charged nanoparticles along microtubules: a model system for weak binding interactions. Biophys J , 1589–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monda JK, Cheeseman IM. (2018). The kinetochore–microtubule interface at a glance. J Cell Sci , jcs214577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T, Hirokawa N. (1995). Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J Cell Biol , 1039–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH. (1999). High-resolution model of the microtubule. Cell , 79–88. [DOI] [PubMed] [Google Scholar]
- Okada Y, Hirokawa N. (2000). Mechanism of the single-headed processivity: Diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc Natl Acad Sci USA , 640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL. (2009). The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell , 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Rios S, Denarier E, Prezel E, Vinit A, Stoppin-Mellet V, Devred F, Barbier P, Peyrot V, Sayas C, Avila J, et al (2016). Tau antagonizes end-binding protein tracking at microtubule ends through a phosphorylationdependent mechanism. Mol Biol Cell , 2924–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Goodman BS, Reck-Peterson SL. (2014). Reconstitution of dynein transport to the microtubule plus end by kinesin. Elife , e02641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers KR, Weiss S, Crevel I, Brophy PJ, Geeves M, Cross R. (2001). KIF1D is a fast non‐processive kinesin that demonstrates novel K-loop-dependent mechanochemistry. EMBO J , 5101–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Cade NI, Surrey T. (2015). Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat Cell Biol , 1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhnow F, Zwicker D, Diez S. (2011). Tracking single particles and elongated filaments with nanometer precision. Biophys J , 2820–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, Monnier N, Markus M, Oberer M, Milligan RA, Bathe M, et al (2012). The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell , 968–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Korten T, Walter Wilhelm J, Diez S. (2015). Kinesin-1 motors can circumvent permanent roadblocks by side-shifting to neighboring protofilaments. Biophys J , 2249–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CW, Klika AB, Lo MMS, Norris TE, Caputo CB. (1992). Tau protein induces bundling of microtubules in vitro: Comparison of different tau isoforms and a tau protein fragment. J Neurosci Res , 19–29. [DOI] [PubMed] [Google Scholar]
- Siahaan V, Krattenmacher J, Hyman AA, Diez S, Hernández-Vega A, Lansky Z, Braun M. (2019). Kinetically distinct phases of tau on microtubules regulate kinesin motors and severing enzymes. Nat Cell Biol , 1086–1092. [DOI] [PubMed] [Google Scholar]
- Slep KC, Vale RD. (2007). Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, EB1. Mol Cell , 976–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Wilson-Kubalek EM, Arthur CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, Kapoor TM. (2010). Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell , 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley IA, Bieling P, Surrey T. (2009). Obstacles on the microtubule reduce the processivity of kinesin-1 in a minimal in vitro system and in cell extract. Biophys J , 3341–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thawani A, Kadzik RS, Petry S. (2018). XMAP215 is a microtubule nucleation factor that functions synergistically with the γ-tubulin ring complex. Nat Cell Biol , 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondera C, Wieduwild R, Röder E, Werner C, Zhang Y, Pietzsch J. (2017). In vivo examination of an injectable hydrogel system crosslinked by peptide–oligosaccharide interaction in immunocompetent nude mice. Adv Funct Mater , 1605189. [Google Scholar]
- Verhey KJ, Kaul N, Soppina V. (2011). Kinesin assembly and movement in cells. Annu Rev Biophys , 267–288. [DOI] [PubMed] [Google Scholar]
- Volkov VA, in’t Veldt PJH, Dogterom M, Musacchio A. (2018). Multivalency of NDC80 in the outer kinetochore is essential to track shortening microtubules and generate forces. Elife , e36764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JPI, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, Cheeseman IM. (2009). The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell , 374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund PO, Stear JH, Pozniakovsky A, Zanic M, Reber S, Brouhard GJ, Hyman AA, Howard J. (2011). XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc Natl Acad Sci USA , 2741–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieduwild R, Tsurkan M, Chwalek K, Murawala P, Nowak M, Freudenberg U, Neinhuis C, Werner C, Zhang Y. (2013). Minimal peptide motif for non-covalent peptide-heparin hydrogels. J Am Chem Soc , 2919–2922. [DOI] [PubMed] [Google Scholar]
- Xu Y, Yang X, Thomas AK, Patsis PA, Kurth T, Kräter M, Eckert K, Bornhäuser M, Zhang Y. (2018). Noncovalently Assembled Electroconductive Hydrogel. ACS Appl Mater Interfaces , 14418–14425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.