Abstract
Background
Increasing evidence has suggested the critical implication of microRNAs (miRNAs) in the initiation and progression of non-small cell lung cancer (NSCLC). Previous studies have shown the tumor-suppressive function of miR-1305 in cancer; however, the role of miR-1305 in NSCLC has not been fully understood.
Methods
The expression of miR-1305 in NSCLC was detected by RT-qPCR. The influence of miR-1305 on the growth of NSCLC cells was determined via Cell Counting Kit 8 (CCK-8), colony formation and FACS analysis. The targets of miR-1305 were predicted with the miRDB database. Luciferase reporter assay was performed to investigate the binding between miR-1305 and 3ʹ-UTR of MDM2. Western blot was applied to check the expression of MDM2 with miR-1305.
Results
Here, we found that miR-1305 was down-regulated in NSCLC tissues and cell lines. Decreased miR-1305 was significantly correlated with the metastasis and poor prognostics of NSCLC patients. Overexpression of miR-1305 inhibited the proliferation and migration and promoted the apoptosis of NSCLC cells. Bioinformatics and luciferase assay uncovered that the mouse/murine double minute 2 (MDM2) was a target of miR-1305. miR-1305 bound the 3ʹ-untranslated region (UTR) of MDM2 and decreased the expression of MDM2 in NSCLC cells. As MDM2 was a negative regulator of p53, decreased MDM2 by miR-1305 up-regulated the abundance of p53 in NSCLC cells. Restoration of MDM2 markedly attenuated the suppressive role of miR-1305 in the proliferation and migration of NSCLC cells.
Conclusion
The findings provided novel mechanism of miR-1305/MDM2 signaling in regulating the progression of NSCLC, suggesting miR-1305 as a promising target for the treatment of NSCLC.
Keywords: NSCLC, miR-1305, MDM2, p53
Introduction
Non-small cell lung cancer (NSCLC) is one of the most common types of cancers, which is a leading cause of mortalities around the world.1 Although advances in diagnostic technology, most of NSCLC patients are diagnosed at an advanced stage due to the lack of initial symptoms. Currently, surgery combined with radiotherapy and chemotherapy has been applied in the treatment of NSCLC; however, the 5-year survival rate of NSCLC patients still remains poor.2 Therefore, it is imperative to explore novel mechanisms that involved in the progression of NSCLC.
MicroRNAs (miRNAs) are a class of conserved small noncoding RNAs that suppress the expression of target mRNAs via binding the 3ʹ-untranslated region (UTR).3–6 MiRNAs participate in a broad range of biological processes, including cell proliferation and differentiation, embryonic development, and stress response. Interestingly, an increasing body of evidence showed the importance of miRNAs in regulating the progression of human cancers by acting as tumor suppressors or oncogenes.3,7–10 Recent studies found that dysfunction of miRNAs was associated with the progression and prognosis of NSCLC patients.11,12 For example, miR-221 promoted the proliferation and invasion of NSCLC cells via suppressing the expression of TIMP2, which suggested miR-221 was a promising target for the treatment of NSCLC.13 Overexpressed miR-203 in NSCLC was closely associated with the advanced progression of patients.14 Recent reports showed that miR-1305 played important roles in regulating the proliferation, differentiation and migration of stem cells.15–18 Due to the uncontrolled malignant behaviors of NSCLC, it would be necessary to evaluate the involvement of miR-1305 in NSCLC.
The mouse/murine double minute 2 (MDM2) was identified as a negative regulator of the tumor suppressor p53.19–22 MDM2 contains an N-terminal p53 interaction domain, central acidic domain, Zinc-finger domain and C-terminal RING domain.23 MDM2 interacted with p53 and triggered the ubiquitylation of p53 via the E3 ligase activity.23 Overexpression of MDM2 has been found in a variety of cancers.23,24 The important function of MDM2-p53 axis in the development of cancer led to the discovery of potential anti-cancer approaches that disrupted the interaction of p53 with MDM2 or targeted MDM2. Notably, MDM2 was also identified as the targets of miRNAs that modulated the growth of cancer cells.25,26 For example, recent study showed that miR-758-3p suppressed the proliferation, migration and invasion of hepatocellular carcinoma cells by targeting MDM2.27 Down-regulation of miR-194-5p conferred paclitaxel resistance in ovarian cancer cells by modulating the expression of MDM2.28 These findings suggested that targeting MDM2 by miRNAs might be a good strategy to interrupt the progression of cancers.
In this study, we found that miR-1305 was down-regulated in NSCLC tissues. Decreased expression of miR-1305 was significantly associated with the poor prognostics of NSCLC patients. Overexpressed miR-1305 inhibited the proliferation and migration of NSCLC cells. Further mechanism study uncovered that miR-1305 targeted MDM2 and decreased MDM2, which consequently led to the stabilization of p53. Our results provided insights into the importance of miR-1305/MDM2/p53 pathway in the development of NSCLC.
Materials And Methods
Patients And Tissues
Forty NSCLC patients were enrolled in this study at the People Hospital BaoJi city between January 2013 to November 2014. None of these patients received radiotherapy or chemotherapy before the surgery. Written informed consents from each participant were obtained prior to tissue collection. Paired NSCLC tissues and adjacent normal tissues were immediately snap-frozen and maintained at −80ºC. This study was performed according to the Helsinki Declaration and approved by the People Hospital BaoJi city.
Cell Culture
The NSCLC cell lines A549, H1299, H460, H157, H2106, H1650 and human normal bronchial epithelium cell line BEAS-2B were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in the DMEM medium containing 10% fetal bovine serum (FBS, Gibco, Thermo Fisher Scientific Inc., Waltham, MA, USA) and 1% penicillin-streptomycin solution (Gibco, Thermo Fisher Scientific Inc., Waltham, MA, USA). Cells were maintained in the incubator at 37ºC with 5% CO2.
RNA Extraction And Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
The total RNA was extracted from NSCLC tissues and cell lines using the Trizol reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. Reverse transcription was performed with the PrimeScript RT Reagent (Takara, Japan). The expression of miR-1305 was quantified by quantitative PCR using the SYBR Green Master Mix (Tiangen Biotechnology, Beijing, China) on the 7500 Real-time PCR System (Applied Biosystems, Carlsbad, CA, USA). U6 RNA was detected as the internal control for normalization. The expression of miR-1305 was calculated using the 2−ΔΔCT method from three replicates. The primers used for the qPCR were designed as follows: miR-1305 forward: 5ʹ-ACAGGCCGGGACAAGTGCAATA and reverse, 5ʹ-GCTGTCAACGATACGCTACGTAACG; U6 forward, 5ʹ-AACGCTTCACGAATTTGCGT and reverse, 5ʹ-CTCGCTTCGGCAGCACA.
Cell Counting Kit-8 (CCK-8) Assay
The cell proliferation was determined using the Cell Counting kit-8 (CCK-8) with the guideline of manufacturer. Briefly, 1000 cells were plated into the 96-well plates and cultured overnight to allow the cells attached. And then cells were transfected with miR-1305 mimics or control miRNA for 0, 24, 48, 72 and 96 hrs as indicated. 10 μL of CCK-8 solution was added into the medium and incubated for 4 hrs at 37ºC. The absorbance of each well at 450 nm was measured with the microplate reader (Bio-Rad, USA).
Cell Apoptosis Analysis
Cells were transfected with miR-1305 mimics or control miRNA for 48 hrs. After that, cells were harvested by trypsinization and rinsed with pre-cold PBS. To evaluate the cell apoptosis, cells were resuspended in the binding buffer and stained with Annexin V-FITC and PI reagent (BD Biosciences, USA) in the darkness at room temperature (RT). The cell apoptosis was quantified with the flow cytometry (BD Biosciences, USA).
Western Blot
Cells were harvested after the transfection of miR-1305 and control miRNA for 48 hrs. Cells were lysed with the RIPA buffer (Beyotime, Shanghai, China) and the protein concentration was quantified using the BCA protein assay kit (Beyotime, Shanghai, China). Equal amount of protein was separated by SDS-PAGE and transferred onto the PVDF membrane (GE Healthcare, USA). After blocking with 5% non-fat milk, the membrane was incubated with the primary antibody overnight at 4ºC followed by the secondary HRP-conjugated antibody for 1 hr at RT. These antibodies used in this study including anit-MDM2 (ab38618; Abcam, USA), anti-p53 (ab32389; Abcam, USA) and anti-GADPH (ab9485; Abcam, USA) were commercially purchased. The protein bands were analyzed using the ImageQuant LAS 4000 min Imaging System (GE Healthcare, USA).
Luciferase Reporter Assay
The wild-type or mutant 3ʹ-UTR of MDM2 was separately inserted into the pGL3 luciferase reporter plasmid (Promega, Madison, WI, USA). NSCLC cells were co-transfected with miR-1305 mimics or control miRNA, pGL3-MDM2-WT or pGL3-MDM2-Mut using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, Inc., Waltham, MA, USA). After transfection for 48 hrs, the luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
Colony Formation
NSCLC cells transfected with control miRNA or miR-1305 were seeded into 6-well plate with approximately 1000 cells per well. After cultured for 2 weeks, colonies were washed with pre-cooled PBS and fixed with methanol at RT for 15 mins. The colonies were stained with 0.1% crystal violet at RT for 10 mins and the quantified with the light microscope.
BrdU Incorporation
The incorporation of BrdU of NSCLC cells expressing control miRNA or miR-1305 was determined using the BrdU Cell Proliferation Assay Kit (Millipore, Temecula, CA) according to the manufacturer’s instruction. Briefly, A549 and H460 cells were seeded into the 96-well plate with the density of 1000 cells per well. After cultured for 12 hrs, BrdU were added into the medium and incubated for 48 hrs. Cells were firstly fixed at RT for 30 mins followed by incubating with pre-diluted anti-BrdU monoclonal antibody for 1 hr at RT. After washing twice with washing buffer, cells were incubated with peroxidase-conjugated secondary antibody for 30 mins at RT. The cells were then incubated with TMB Peroxidase Substrate at RT for 30 mins in the darkness. The absorbance of each well was measured at 450 nm with the microplate reader.
Statistical Analysis
All the data were expressed as mean ± SD. The statistical analysis was performed using SPSS 19.0 (SPSS, Chicago, IL, USA). Differences were assessed by Student’s t test or one-way analysis of variance (ANOVA). P values of 0.05 or less were considered as statistical significance.
Results
MiR-1305 Was Down-Regulated In NSCLC
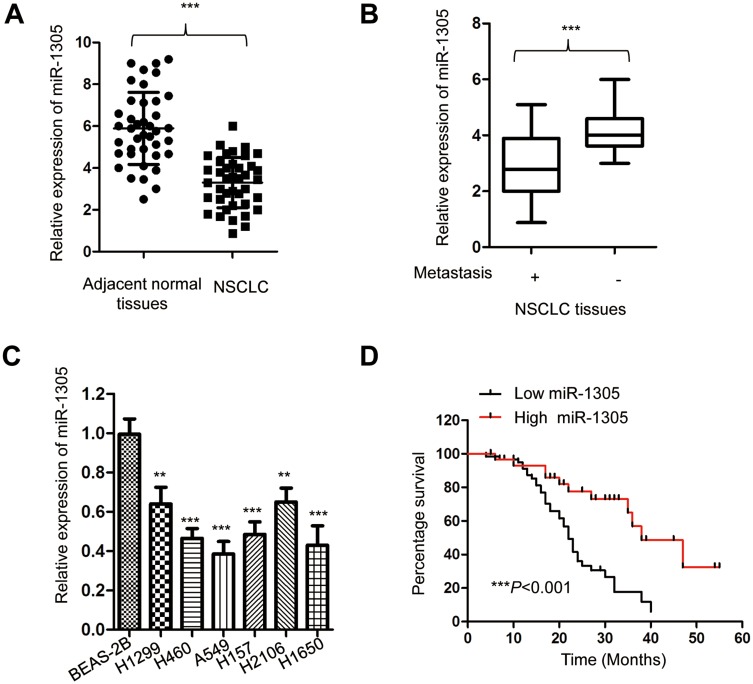
To evaluate the involvement of miR-1305 in the progression of NSCLC, the expression of miR-1305 in paired NSCLC tissues and adjacent normal tissues was detected using RT-qPCR. The data showed that miR-1305 expression in NSCLC tissues was significantly lower compared with that in adjacent non-tumor tissues (Figure 1A). The decreased expression of miR-1305 was also observed using the dbDEMC 2.0 database (http://www.picb.ac.cn/dbDEMC/search.php). Additionally, the expression of miR-1305 was also markedly down-regulated in NSCLC patients with metastasis compared with those without metastasis (Figure 1B). To further validate the aberrant expression of miR-1305 in NSCLC, the abundance of miR-1305 in NSCLC cell lines including A549, H1299, H460, H157, H2106 and H1650 as well as the normal cell BEAS-2B was detected. As presented in Figure 1C, the expression of miR-1305 was significantly decreased in NSCLC cell lines in comparison with that in the control cells (Figure 1C). To further explore the potential clinical significance of miR-1305 in NSCLC, another 90 NSCLC patients were divided into high-miR-1305 or low-miR-1305 group according to the mean value of miR-1305. The correlation between the expression of miR-1305 and 5-year overall survival of these patients was analyzed with the Log rank test. The data indicated that patients with lower level of miR-1305 had significantly shorter overall survival (OS) than those with higher miR-1305 expression (Figure 1D). These results suggested that down-regulated miR-1305 might be involved in the progression of NSCLC.
Figure 1.
miR-1305 was down-regulated in NSCLC. (A) Expression of miR-1305 in NSCLC tissues and paired adjacent normal tissues was detected by RT-qPCR. (B) The level of miR-1305 in NSCLC tissues with or without metastasis was compared. (C) Expression of miR-1305 in NSCLC cell lines and normal cells was examined by RT-qPCR. (D) Lower expression of miR-1305 was significantly correlated with the worse prognosis of NSCLC patients. **P<0.01; ***P<0.001.
Overexpression Of miR-1305 Inhibited The Proliferation And Promoted Apoptosis Of NSCLC Cells
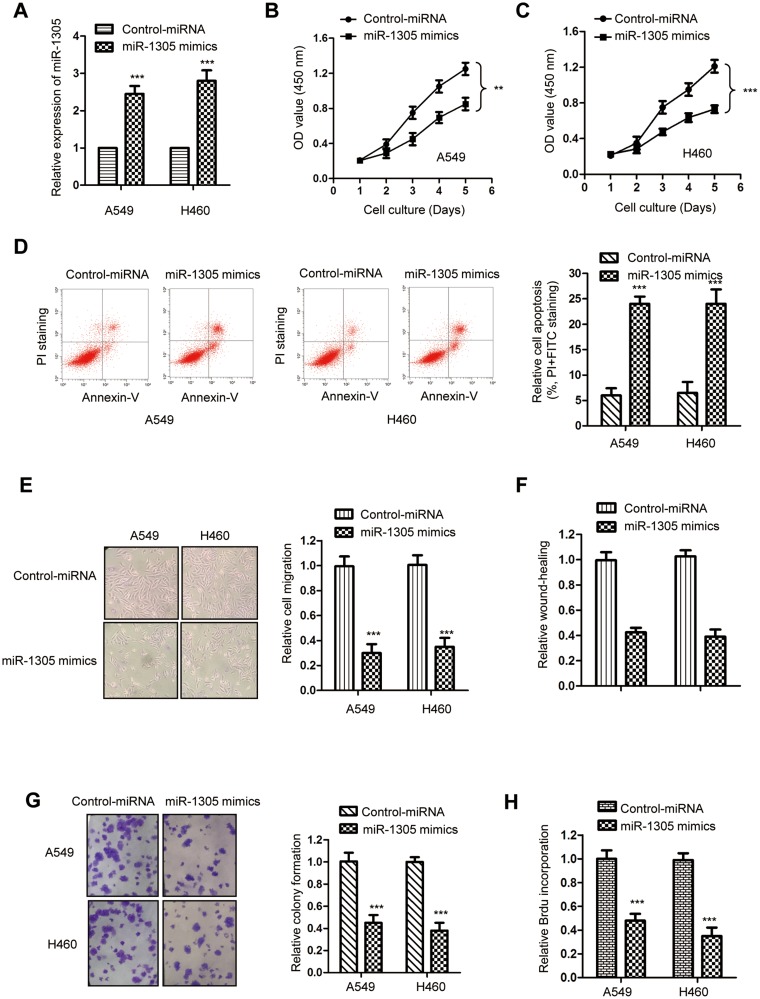
Because both A549 and H460 cells showed relative lower level of miR-1305 among the cells shown in Figure1C, these two cell lines were transfected with miR-1305 mimics or control miRNA to evaluate the influence of miR-1305 on the growth of NSCLC cells. The transfection efficiency of miR-1305 mimics was validated by RT-qPCR assay (Figure 2A), which showed the significantly increased level of miR-1305 with the transfection of miR-1305 mimics. The proliferation of NSCLC cells was determined by the CCK-8 assay. Overexpression of miR-1305 markedly inhibited the proliferation of both A549 and H460 cells (Figure 2B and C). The suppressive function of miR-1305 in regulating the growth of NSCLC cells was further evaluated by detecting the cell apoptosis. The data showed that overexpressed miR-1305 significantly increased the percentage of cells with both PI and annexin V-FITC staining, suggesting up-regulated apoptosis of both A549 and H460 cells (Figure 2D). To further study the inhibitory effect of miR-1305 in NSCLC, cell migration assay was performed by NSCLC cells transfected with miR-1305 mimics or control. The result showed that highly expressed miR-1305 significantly inhibited the migration of A549 and H460 cells compared with the mock group (Figure 2E). The wound-healing of NSCLC cells with overexpressed miR-1305 was obviously inhibited (Figure 2F). Additionally, the colony formation assay suggested the decreased formation of colonies with the overexpression of miR-1305 in NSCLC cells (Figure 2G). Consistently, highly expressed miR-1305 significantly reduced the BrdU incorporation of NSCLC cells (Figure 2H). Collectively, these findings suggested the tumor-suppressive role of miR-1305 in NSCLC.
Figure 2.
Overexpressed miR-1305 inhibited the growth of NSCLC cells. (A) The transfection of miR-1305 was confirmed by RT-qPCR. (B, C) The proliferation of A549 and H460 cells transfected with miR-1305 mimics or control miRNA was determined with the CCK-8 assay. (D) The apoptosis of A549 and H460 cells transfected with miR-1305 was significantly increased. (E) Overexpression of miR-1305 decreased the migration of NSCLC cells. (F) Overexpression of miR-1305 inhibited the wound-healing of NSCLC cells. (G) The colony formation of NSCLC cells was decreased with the up-regulation of miR-1305. (H) Highly expressed miR-1305 significantly reduced the BrdU incorporation of both A549 and H460 cells. **P<0.01; ***P<0.001.
Down-Regulation Of miR-1305 Promoted The Proliferation Of NSCLC Cells
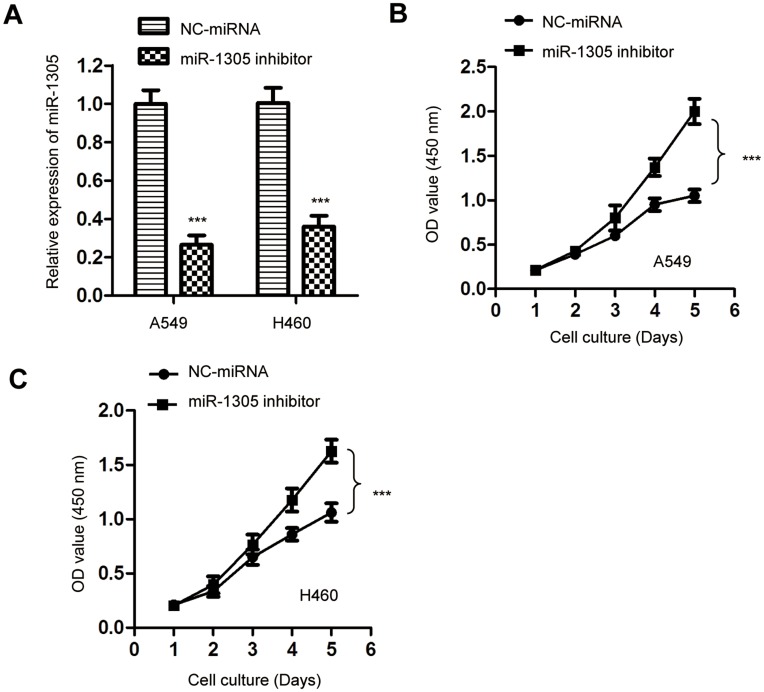
To further confirm the suppressive role of miR-1305 in regulating the proliferation of NSCLC cells, the expression of miR-1305 was down-regulated by transfecting miR-1305 inhibitor into both A549 and H460 cells. The knockdown efficiency of miR-1305 was validated by RT-qPCR (Figure 3A). The effect of miR-1305 down-regulation on the proliferation of NSCLC cells was determined by the CCK-8 assay. The result showed that depletion of miR-1305 significantly promoted the proliferation of both A549 and H460 cells (Figure 3B and C). These results further confirmed the suppressive role of miR-1305 in modulating the malignancy of NSCLC.
Figure 3.
Depletion of miR-1305 promoted the proliferation of NSCLC cells. (A) A549 and H460 cells were transfected with NC-miRNA or miR-1305 inhibitor and the knockdown of miR-1305 was validated by RT-qPCR. (B, C) Down-regulation of miR-1305 significantly promoted the proliferation of both A549 and H460 cells. ***P<0.001.
MDM2 Was A Target Of miR-1305 In NSCLC Cells
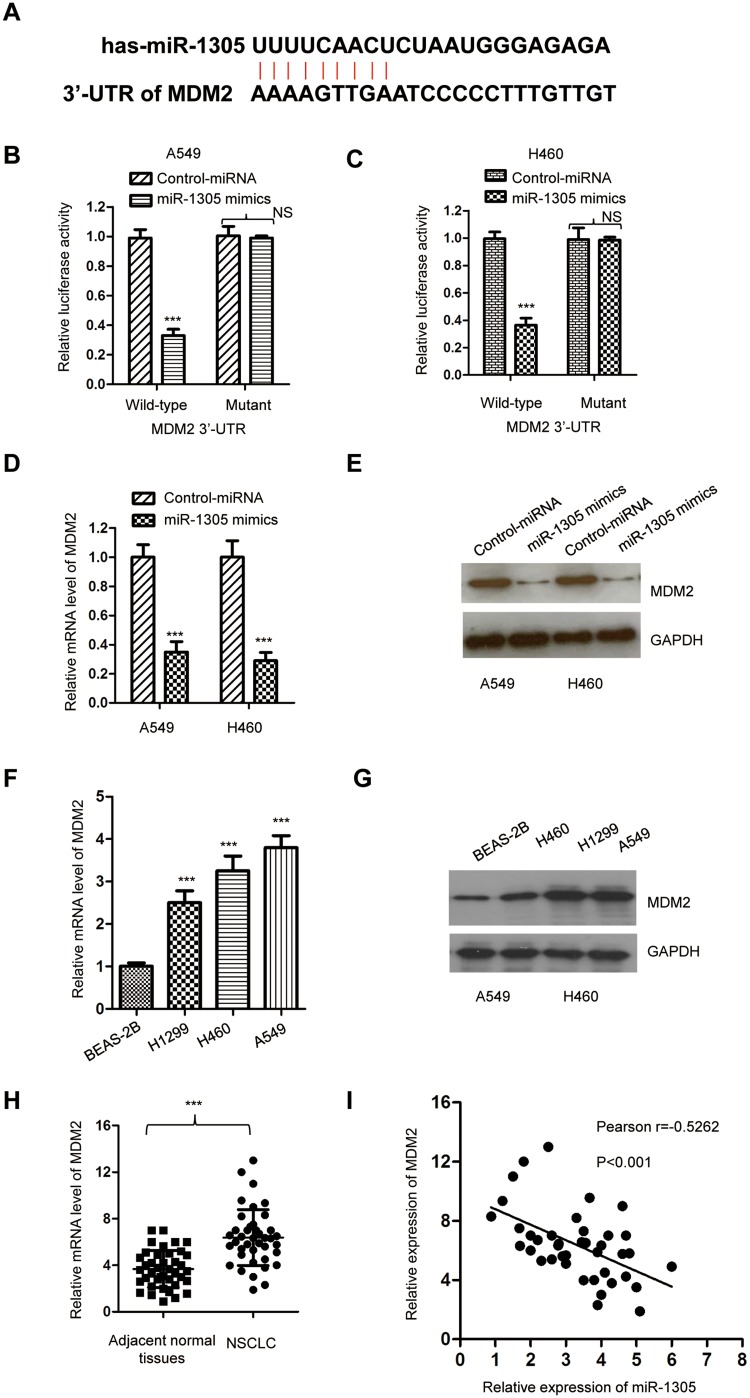
To clarify the functional mechanism of miR-1305 in NSCLC, the potential targets of miR-1305 were searched with the miRDB database (http://mirdb.org/). We found that the 3ʹ-UTR of MDM2 containing the complementary binding sites of miR-1305 as presented in Figure 4A. To confirm this prediction, luciferase reporters containing wild-type or mutant 3ʹ-UTR of MDM2 were constructed and transfected into the NSCLC cells in the presence of miR-1305 mimics or control miRNA. The data showed that miR-1305 mimics significantly repressed the luciferase activity of WT MDM2 3ʹ-UTR; however, only very slight effect on the mutant reporter was observed (Figure 4B and C). To determine whether the binding of miR-1305 with the 3ʹ-UTR affected the mRNA stability of MDM2, both A549 and H460 cells were transfected with miR-1305 mimics or control miRNA, and the mRNA level of MDM2 was detected by RT-qPCR. The result indicated that miR-1305 repressed the mRNA level of MDM2 in NSCLC cells (Figure 4D). Consistently, the protein expression of MDM2 was also checked with the overexpression of miR-1305, which suggested decreased level of MDM2 compared with cells expressing control miRNA (Figure 4E). These results suggested that miR-1305 repressed the expression of MDM2 by targeting the 3ʹ-UTR of MDM2. Consist of the down-regulation of miR-1305 in NSCLC cells, both the mRNA and protein levels of MDM2 were higher in NSCLC cell lines than the control normal cells (Figure 4F and G). Meanwhile, the expression of MDM2 in NSCLC tissues and paired adjacent normal tissues was also detected. The result showed that MDM2 was significantly up-regulated in NSCLC tissues compared with the adjacent normal tissues (Figure 4H). The correlation between the expression of miR-1305 and MDM2 was further analyzed using Spearman test, which indicated that the level of miR-1305 was significantly negatively correlated with that of MDM2 in NSCLC tissues (Figure 4I). These findings demonstrated that MDM2 was target of miR-1305 in NSCLC.
Figure 4.
MDM2 was targeted by miR-1305. (A) Predicted binding sites of miR-1305 with the 3ʹ-UTR of MDM2 mRNA. (B, C) Luciferase activity of WT or mutant MDM2 3ʹ-UTR reporter was determined in A549 and H460 cells transfected with miR-1305 mimics or control miRNA. Renilla luciferase activity was also detected for normalization. (D, E) Overexpression of miR-1305 decreased both the mRNA and protein levels of MDM2 in A549 and H460 cells. (F, G) The level of MDM2 in NSCLC cell lines was increased compared with the normal cell. (H) The mRNA levels of MDM2 in paired NSCLC and adjacent normal tissues were detected by RT-qPCR. (I) The expression of MDM2 in NSCLC tissues was negatively correlated with that of miR-1305. ***P<0.001.
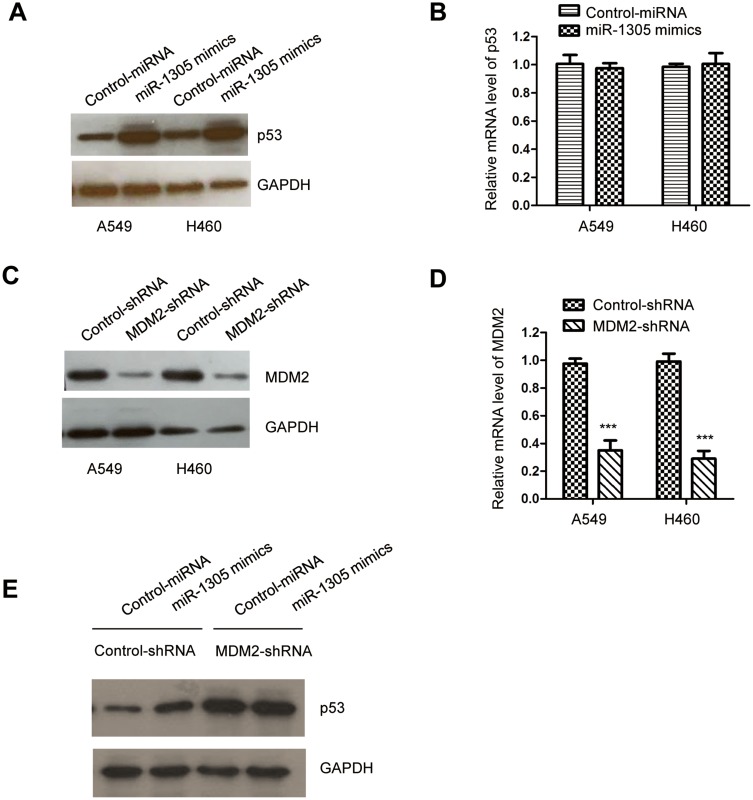
Decreased MDM2 Induced By miR-1305 Stabilized p53 In NSCLC Cells
MDM2 is reported as an E3 ubiquitin ligase of p53, which mediates the polyubiquitylation of p53 and induces 28S proteasome-dependent degradation of p53. Because miR-1305 targeted MDM2 and negatively regulated MDM2 in NSCLC, we then evaluated the level of p53 with the overexpression of miR-1305 in A549 and H460 cells. The results showed that transfection of miR-1305 increased the level of p53 in NSCLC cells (Figure 5A). Moreover, the mRNA level of p53 was also detected by RT-qPCR; however, no obvious change was observed for the mRNA level of p53 with the overexpression of miR-1305 (Figure 5B). To further determine whether the up-regulation of p53 was obtained via MDM2, both A549 and H460 cells were transfected with MDM2-shRNA. The down-regulation of MDM2 was validated by RT-qPCR and Western blot, respectively. The results suggested that the expression of MDM2 was significantly decreased in NSCLC cells by inducing MDM2-shRNA (Figure 5C and D). p53 expression was detected in A549 and H460 cells harboring MDM2-shRNA or control-shRNA. The data indicated that overexpression of miR-1305 up-regulated the expression of p53 in cells with control-shRNA, while depletion of MDM2 abolished the effect of miR-1305 on p53 (Figure 5E). These findings suggested that miR-1305 targeted MDM2 and stabilized p53 in NSCLC cells.
Figure 5.
miR-1305 overexpression up-regulated p53 in NSCLC. (A) The protein level of p53 in A549 and H460 cells with overexpressed miR-1305 was detected by Western blot. (B) The mRNA abundance of p53 with highly expressed miR-1305 was investigated by RT-qPCR. (C, D) Knockdown efficiency of MDM2 with the transfection of MDM2-shRNA was confirmed by RT-qPCR and Western blot, respectively. (E) miR-1305 was transfected into A549 and H460 cells that expressing depleted MDM2, and the protein level of p53 was detected. ***P<0.001.
MiR-1305 Regulated The Proliferation Of NSCLC Cells Through Targeting MDM2
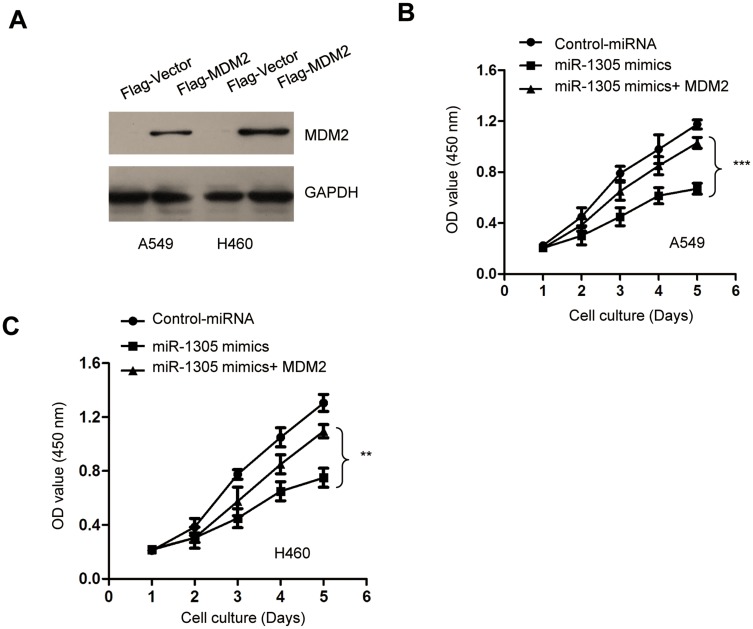
To investigate the tumor-suppressive role of miR-1305 in NSCLC by down-regulating MDM2, both A549 and H460 cells were transfected with pcDNA-Flag-MDM2 and miR-1305 mimics. The transfection efficiency of MDM2 was confirmed by Western blot analysis (Figure 6A). CCK-8 assay demonstrated that overexpression of miR-1305 decreased the proliferation of NSCLC cells, and transfection of MDM2 significantly reversed miR-1305-induced suppression of both A549 and H460 cell proliferation (Figure 6B and C). These results demonstrated that miR-1305 targeted MDM2 and regulated the malignant behaviors of NSCLC cells.
Figure 6.
Restoration of MDM2 abrogated the inhibitory effect of miR-1305 on the proliferation of NSCLC cells. (A) Restoration of MDM2 was confirmed by Western blot with anti-Flag antibody. (B, C) The proliferation of A549 and H460 cells transfected with miR-1305 and MDM2 was determined by CCK-8 assay. **P<0.01; ***P<0.001.
Discussion
Increasing evidence has indicated that miRNAs function as important regulators in a variety of biological processes, including cell proliferation, differentiation and migration.29,30 The critical roles of miRNAs in the development of NSCLC were demonstrated in recent decades, suggesting miRNAs as promising targets to disrupt the progression of NSCLC.31–33 In this study, miR-1305 was down-regulated in NSCLC tissues and cell lines. Decreased expression of miR-1305 was significantly correlated with the advanced progression and worse prognosis of NSCLC patients. These findings suggested miR-1305 might be a promising diagnostic biomarker and target for the treatment of NSCLC.
Previous studies have uncovered the essential functions of miR-1305 in regulating the progression of cancer.15 MiR-1305 inhibited the Akt signaling, suppressing the self-renewal and tumorigenesis of liver cancer stem cells.15 Recent findings also demonstrated that restoration of miR-1305 relieved the inhibitory effect of nicotine on the proliferation, migration and osteogenic differentiation of periodontal ligament-derived stem cells.16 In the current study, miR-1305 expression was decreased in NSCLC. Overexpression of miR-1305 significantly reduced the proliferation, migration and up-regulated the apoptosis of NSCLC cells. These results suggested the tumor-suppressive function of miR-1305 in regulating the malignancy of NSCLC. Based on the in vitro inhibitory evidence of miR-1305 in NSCLC, it is imperative to investigate the potential of miR-1305 as a target of NSCLC treatment in vivo via the gene delivery technologies.
To explore the underlying mechanism responsible for miR-1305-mediated antitumor function in NSCLC, the targets of miR-1305 in NSCLC were studied. Interestingly, MDM2, a well-documented negative regulator of p53, was identified as a target of miR-1305. MDM2 mediated the polyubiquitylation of p53 and induced 28S proteasome-dependent degradation of p53.34 Overexpressed MDM2 was found in a variety of human cancers and associated with the poor prognostics of cancer patients.24,35 An increasing body of evidence has reported that MDM2 was regulated by numerous miRNAs in cancers.36–40 For example, miR-194-5p targeted MDM2 and modulated the paclitaxel resistance in ovarian cancer cells.28 MiR-518 inhibited the growth of cancer cells by inducing apoptosis via targeting MDM2.41 In the current study, miR-1305 bound the 3ʹ-UTR of MDM2 and inhibited the expression of MDM2 in NSCLC cells. Consistently, decreased MDM2 by miR-1305 up-regulated the expression of p53. Restoration of MDM2 significantly reversed the inhibitory effect of miR-1305 on the proliferation of NSCLC cells. These results suggested the novel mechanism of miR-1305/MDM2/p53 signaling in suppressing the growth of NSCLC cells.
In conclusion, our results uncovered the critical function of miR-1305 in NSCLC. MiR-1305 was down-regulated in NSCLC and associated with the poor prognostics of NSCLC patients. Overexpression of miR-1305 inhibited the proliferation of NSCLC cells via targeting MDM2 and up-regulated p53. These findings demonstrated that miR-1305 may be a potential and promising target in designing the treatment strategy for NSCLC.
Disclosure
The authors report no conflicts of interests in this work.
References
- 1.Bradbury P, Sivajohanathan D, Chan A, et al. Postoperative adjuvant systemic therapy in completely resected non-small-cell lung cancer: a systematic review. Clin Lung Cancer. 2017;18(3):259–273 e258. doi: 10.1016/j.cllc.2016.07.002 [DOI] [PubMed] [Google Scholar]
- 2.Vergnenegre A, Chouaid C. Review of economic analyses of treatment for non-small-cell lung cancer (NSCLC). Expert Rev Pharmacoecon Outcomes Res. 2018;18(5):519–528. doi: 10.1080/14737167.2018.1485099 [DOI] [PubMed] [Google Scholar]
- 3.Tiwari A, Mukherjee B, Dixit M. MicroRNA key to angiogenesis regulation: miRNA biology and therapy. Curr Cancer Drug Targets. 2018;18(3):266–277. doi: 10.2174/1568009617666170630142725 [DOI] [PubMed] [Google Scholar]
- 4.Lai EC. Two decades of miRNA biology: lessons and challenges. Rna. 2015;21(4):675–677. doi: 10.1261/rna.051193.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang TC, Pinto SM, Pandey A. Proteomics for understanding miRNA biology. Proteomics. 2013;13(3–4):558–567. doi: 10.1002/pmic.201200339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchan JR, Parker R. Molecular biology. The two faces of miRNA. Science. 2007;318(5858):1877–1878. doi: 10.1126/science.1152623 [DOI] [PubMed] [Google Scholar]
- 7.Zahid KR, Su M, Khan ARR, et al. Systems biology based meth-miRNA-mRNA regulatory network identifies metabolic imbalance and hyperactive cell cycle signaling involved in hepatocellular carcinoma onset and progression. Cancer Cell Int. 2019;19:89 eCollection 2019. doi: 10.1186/s12935-019-0804-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Z, Shi K, Yang S, et al. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17(1):147. doi: 10.1186/s12943-018-0897-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunz M, Pittroff A, Dandekar T. Systems biology analysis to understand regulatory miRNA networks in lung cancer. Methods Mol Biol. 2018;1819:235–247. doi: 10.1007/978-1-4939-8618-7_11 [DOI] [PubMed] [Google Scholar]
- 10.Najafi A, Tavallaei M, Hosseini SM. A systems biology approach for miRNA-mRNA expression patterns analysis in non-small cell lung cancer. Cancer Biomark. 2016;16(1):31–45. doi: 10.3233/CBM-150538 [DOI] [PubMed] [Google Scholar]
- 11.Zhou Q, Huang SX, Zhang F, et al. MicroRNAs: a novel potential biomarker for diagnosis and therapy in patients with non-small cell lung cancer. Cell Prolif. 2017;50(6). doi: 10.1111/cpr.12394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Florczuk M, Szpechcinski A, Chorostowska-Wynimko J. miRNAs as biomarkers and therapeutic targets in non-small cell lung cancer: current perspectives. Target Oncol. 2017;12(2):179–200. doi: 10.1007/s11523-017-0478-5 [DOI] [PubMed] [Google Scholar]
- 13.Yin Z, Xu M, Li P. miRNA-221 acts as an oncogenic role by directly targeting TIMP2 in non-small-cell lung carcinoma. Gene. 2017;620:46–53. doi: 10.1016/j.gene.2017.04.007 [DOI] [PubMed] [Google Scholar]
- 14.Chen T, Xu C, Chen J, et al. MicroRNA-203 inhibits cellular proliferation and invasion by targeting Bmi1 in non-small cell lung cancer. Oncol Lett. 2015;9(6):2639–2646. doi: 10.3892/ol.2015.3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei X, You X, Zhang J, Zhou C. MicroRNA-1305 inhibits the stemness of LCSCs and tumorigenesis by repressing the UBE2T-dependent Akt-signaling pathway. Mol Ther Nucleic Acids. 2019;16:721–732. doi: 10.1016/j.omtn.2019.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Liu HL. Restoration of miR-1305 relieves the inhibitory effect of nicotine on periodontal ligament-derived stem cell proliferation, migration, and osteogenic differentiation. J Oral Pathol Med. 2017;46(4):313–320. doi: 10.1111/jop.12492 [DOI] [PubMed] [Google Scholar]
- 17.Jin S, Collin J, Zhu L, et al. A novel role for miR-1305 in regulation of pluripotency-differentiation balance, cell cycle, and apoptosis in human pluripotent stem cells. Stem Cells. 2016;34(9):2306–2317. doi: 10.1002/stem.2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng TK, Huang L, Cao D, et al. Cigarette smoking hinders human periodontal ligament-derived stem cell proliferation, migration and differentiation potentials. Sci Rep. 2015;5:7828. doi: 10.1038/srep07828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joerger AC, Fersht AR. The p53 pathway: origins, inactivation in cancer, and emerging therapeutic approaches. Annu Rev Biochem. 2016;85:375–404. doi: 10.1146/annurev-biochem-060815-014710 [DOI] [PubMed] [Google Scholar]
- 20.Tollini LA, Zhang Y. p53 regulation goes Live-Mdm2 and MdmX Co-Star: lessons learned from mouse modeling. Genes Cancer. 2012;3(3–4):219–225. doi: 10.1177/1947601912454732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baek S, Kutchukian PS, Verdine GL, et al. Structure of the stapled p53 peptide bound to Mdm2. J Am Chem Soc. 2012;134(1):103–106. doi: 10.1021/ja2090367 [DOI] [PubMed] [Google Scholar]
- 22.Sun Y. Targeting E3 ubiquitin ligases for cancer therapy. Cancer Bio Ther. 2003;2(6):623–629. [PubMed] [Google Scholar]
- 23.Lucia H, Borivoj V. HDM2 and HDMX proteins in human cancer. Klin Onkol. 2018;31(Supplementum2):63–70. doi: 10.14735/amko20182S63 [DOI] [PubMed] [Google Scholar]
- 24.Shaikh MF, Morano WF, Lee J, et al. Emerging role of MDM2 as target for anti-cancer therapy: a review. Ann Clin Lab Sci. 2016;46(6):627–634. [PubMed] [Google Scholar]
- 25.Tian XM, Luo YZ, He P, et al. Inhibition of invasion and migration of prostate cancer cells by miRNA-509-5p via targeting MDM2. Genet Mol Res. 2017;16(1). doi: 10.4238/gmr16019195 [DOI] [PubMed] [Google Scholar]
- 26.Kong Q, Shu N, Li J, Xu N. miR-641 functions as a tumor suppressor by targeting MDM2 in human lung cancer. Oncol Res. 2018;26(5):735–741. doi: 10.3727/096504017X15021536183490 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Jiang D, Cho W, Li Z, et al. MiR-758-3p suppresses proliferation, migration and invasion of hepatocellular carcinoma cells via targeting MDM2 and mTOR. Biomed Pharmacother. 2017;96:535–544. doi: 10.1016/j.biopha.2017.10.004 [DOI] [PubMed] [Google Scholar]
- 28.Nakamura K, Sawada K, Miyamoto M, et al. Downregulation of miR-194-5p induces paclitaxel resistance in ovarian cancer cells by altering MDM2 expression. Oncotarget. 2019;10(6):673–683. doi: 10.18632/oncotarget.26586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosseinahli N, Aghapour M, Duijf PHG, et al. Treating cancer with microRNA replacement therapy: a literature review. J Cell Physiol. 2018;233(8):5574–5588. doi: 10.1002/jcp.26514 [DOI] [PubMed] [Google Scholar]
- 30.Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35(1):3–11. doi: 10.1055/s-0034-1397344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arab A, Karimipoor M, Irani S, et al. Potential circulating miRNA signature for early detection of NSCLC. Cancer Genet. 2017;216–217:150–158. doi: 10.1016/j.cancergen.2017.07.006 [DOI] [PubMed] [Google Scholar]
- 32.Pastorkova Z, Skarda J, Andel J. The role of microRNA in metastatic processes of non-small cell lung carcinoma. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2016;160(3):343–357. doi: 10.5507/bp.2016.021 [DOI] [PubMed] [Google Scholar]
- 33.Shao Y, Liang B, Long F, Jiang S-J. Diagnostic microRNA biomarker discovery for non-small-cell lung cancer adenocarcinoma by integrative bioinformatics analysis. Biomed Res Int. 2017;2017:2563085. doi: 10.1155/2017/2563085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S, Zhao Y, Aguilar A, Bernard D, Yang C-Y. Targeting the MDM2-p53 protein-protein interaction for new cancer therapy: progress and challenges. Cold Spring Harb Perspect Med. 2017;7(5):a026245. doi: 10.1101/cshperspect.a026245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urso L, Calabrese F, Favaretto A, Conte P, Pasello G. Critical review about MDM2 in cancer: possible role in malignant mesothelioma and implications for treatment. Criti Rev Oncol Hematol. 2016;97:220–230. doi: 10.1016/j.critrevonc.2015.08.019 [DOI] [PubMed] [Google Scholar]
- 36.Wu D, Niu X, Tao J, et al. MicroRNA-379-5p plays a tumor-suppressive role in human bladder cancer growth and metastasis by directly targeting MDM2. Oncol Rep. 2017;37(6):3502–3508. doi: 10.3892/or.2017.5607 [DOI] [PubMed] [Google Scholar]
- 37.Zhang C, Liu J, Tan C, et al. microRNA-1827 represses MDM2 to positively regulate tumor suppressor p53 and suppress tumorigenesis. Oncotarget. 2016;7(8):8783–8796. doi: 10.18632/oncotarget.7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan Y, Peng Y, Ou Y, Jiang Y. MicroRNA-610 is downregulated in glioma cells, and inhibits proliferation and motility by directly targeting MDM2. Mol Med Rep. 2016;14(3):2657–2664. doi: 10.3892/mmr.2016.5559 [DOI] [PubMed] [Google Scholar]
- 39.Huang K, Tang Y, He L, Dai Y. MicroRNA-340 inhibits prostate cancer cell proliferation and metastasis by targeting the MDM2-p53 pathway. Oncol Rep. 2016;35(2):887–895. doi: 10.3892/or.2015.4458 [DOI] [PubMed] [Google Scholar]
- 40.Shen J, Niu W, Zhou M, et al. MicroRNA-410 suppresses migration and invasion by targeting MDM2 in gastric cancer. PLoS One. 2014;9(8):e104510. doi: 10.1371/journal.pone.0104510 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Feng C, Xian Q, Liu S. Micro RNA-518 inhibits gastric cancer cell growth by inducing apoptosis via targeting MDM2. Biomed Pharmacother. 2018;97:1595–1602. doi: 10.1016/j.biopha.2017.11.091 [DOI] [PubMed] [Google Scholar]