Abstract
Purpose
To describe the coexistence of additional non-ocular genetic diseases in patients diagnosed with inherited retinal degenerations (IRDs).
Methods
The study was based on a retrospective chart review of patients diagnosed with IRD and additional rare systemic diseases. The chart review included the ophthalmic and genetic aspects of each patient. The ophthalmic examination included best-corrected visual acuity, biomicroscopic examination, cycloplegic refraction, retinal imaging (fundus photos, optical coherence tomography, and fundus autofluorescence), and electroretinography. Genetic testing included homozygosity mapping, whole exome sequencing, and Sanger sequencing.
Results
Fifteen index cases diagnosed with IRDs and one or more rare systemic diseases were identified. Six of the families were consanguineous. Of six patients with complete molecular diagnosis, four (66%) had pathogenic variants in two autosomal recessive (AR) disease genes, and of the total pathogenic variants identified, AR mutations were the most common (16/22, 72%). One patient was diagnosed with mutations in three different genes, underlying three distinct genetic conditions. Nine patients could have had an incorrect clinical diagnosis based on the clinical evaluation only (e.g., retinitis pigmentosa and hearing loss could have been diagnosed as Usher syndrome).
Conclusions
The common working paradigm for the ophthalmologist is combining the different symptoms observed in a patient into one unifying diagnosis. However, IRD is a strikingly heterogeneous condition, and may coincide with other genetic (and non-genetic) rare conditions. Establishing a correct diagnosis is important for the patients and their family members, as it enables prediction of disease prognosis, aids in tailoring the correct follow-up and treatment, and allows patients to pursue prenatal counseling and reproductive planning.
Introduction
Inherited retinal degenerations (IRDs) are a group of rare diseases, damaging photoreceptors and RPE cells, thus decreasing vision and visual functions. IRDs are one of the most genetically heterogeneous groups of disorders in humans. Non-syndromic IRD can be inherited as autosomal recessive (AR), autosomal dominant (AD), or X-linked (XL) [1]. To date, more than 200 genes have been implicated in the etiology of IRD (RetNet).
The most common form of IRD is retinitis pigmentosa (RP), with a worldwide prevalence of approximately 1/4,000. In RP, the disease process initially affects the rod photoreceptors to a more severe degree than the cones, and thus, clinical symptoms include night blindness, followed by visual field loss, often resulting in severe visual impairment [2]. In cone-rod dystrophy (CRD; the estimated prevalence is approximately 1/40,000), cone involvement initially exceeds that of the rods, and thus, early symptoms include reduced visual acuity, photophobia, and defective color vision [3]. Macular dystrophies (MDs) are a heterogeneous group of disorders characterized by progressive central vision loss, due to degeneration of photoreceptors in the macular region only. Stargardt disease (STGD), the most common form of hereditary MD, is characterized by macular atrophy and peripheral yellow–white flecks at the level of the RPE, and is caused by recessive mutations of the ABCA4 (NM_000350.2; OMIM: 601691) gene [4]. Another group of MDs is bestrophinopathies, caused by mutations in the BEST1 (NM_004183.3; OMIM: 607854) gene. Mutations in BEST1 can be inherited as AD (Best vitelliform macular dystrophy, BVMD), or as AR (autosomal recessive bestrophinopathy, ARB) [5].
Although in most cases of IRD the disease is limited to the eye (non-syndromic), more than 100 forms of syndromic IRD have been described [6,7] (OMIM). The most common is Usher syndrome (USH) characterized by the combination of RP and hearing loss (HL) [8]. In some cases of syndromic IRD, the retinal dystrophy may be the presenting symptom, and other systemic findings evolve during childhood, puberty, or later on in life. In other cases, the first identifiable symptom of the syndrome is non-ocular, and the retinal dystrophy is revealed only later in life.
We present 15 cases of patients who had a retinal dystrophy with one or more extraocular phenotypes. Following the clinician’s primary evaluation, some of these patients were clinically suspected to have syndromic IRD (such as USH), while in other cases, two or more unrelated diagnoses (an IRD and another disease) were suspected from the beginning. As part of the workup, the patients underwent molecular genetic evaluation, which proved the coexistence of two or more different diseases attributed to genetic mutations in two or more genes.
Methods
Patients and clinical evaluation
Twenty-five patients from 15 families were included in this study. Patients were ascertained by the inherited retinal degeneration clinic at Schneider Children’s Medical Center of Israel (SCMCI) and by the Laboratory of Visual Genetics at the Technion. The tenets of the Declaration of Helsinki were followed, the study adhered to the ARVO statement on human subjects, the study was approved by institutional review boards in participating Medical Centers, and written informed consent was obtained from all participants. Ophthalmic examination included best-corrected visual acuity (BCVA), biomicroscopic examination, cycloplegic refraction, retinal imaging (fundus photos, optical coherence tomography (OCT), and fundus autofluorescence), and electroretinography (ERG) testing in most patients. In addition, patients and their families were evaluated by clinical geneticists before and after the molecular diagnosis was made, and were advised about the relevant genetic testing needed, as well as the patient’s prognosis, further needed medical evaluation, and future family planning, accordingly.
Genetic analyses
Venous blood samples were obtained using K3EDTA vacuette tubes (Greiner Bio-One, Kremsmunster, Austria) and kept at 4 ºC for up to 48 h. Genomic DNA was extracted from venous blood samples using a high salt solution according to a standard protocol [9]. Homozygosity mapping in family 1 was performed with the Infinium Human Linkage-12 Genotyping BeadChip Kit (Illumina, Inc., San Diego, CA), which is capable of genotyping 6,090 highly informative single nucleotide polymorphisms (SNPs) with an average genetic distance of 0.58 cM across the human genome. Homozygous regions were calculated using HomozygosityMapper [10]. Chromosomal microarray analysis (CMA) in family 2 was performed using the CytoScanTM 750K Chip Kit (PE Applied Biosystems, Foster City, CA).
Whole exome sequencing (WES) in families 2, 4, 5, 10, and 13 was performed using SureSelectXT Human All Exon V5 (Agilent Technologies, Santa Clara, CA) and HiSeq2500 or HiSeq4000 (Illumina, San Diego, CA). For family 12, a Roche NimbleGen Exome Enrichment Kit (Roche NimbleGen, Inc., Pleasanton, CA) was used for target enrichment.
Testing for specific mutations and verification of mutations identified with WES were performed by PCR amplification with specifically designed primers. PCR was performed on 50 ng of genomic DNA in a 25 µl reaction volume in the presence of 5X Readymix (LAROVA GmbH, Teltow, Germany) and 10 pmol of each forward and reverse primers. This was followed by direct sequencing with the BigDye Terminator Cycle Sequencing Kit on an ABI 3130xl Genetic Analyzer (PE Applied Biosystems). The primer sequences used for PCR amplification and Sanger sequencing are listed in Appendix 1. Information regarding the genes in which mutations were found is provided in Appendix 2.
Results
Family 1
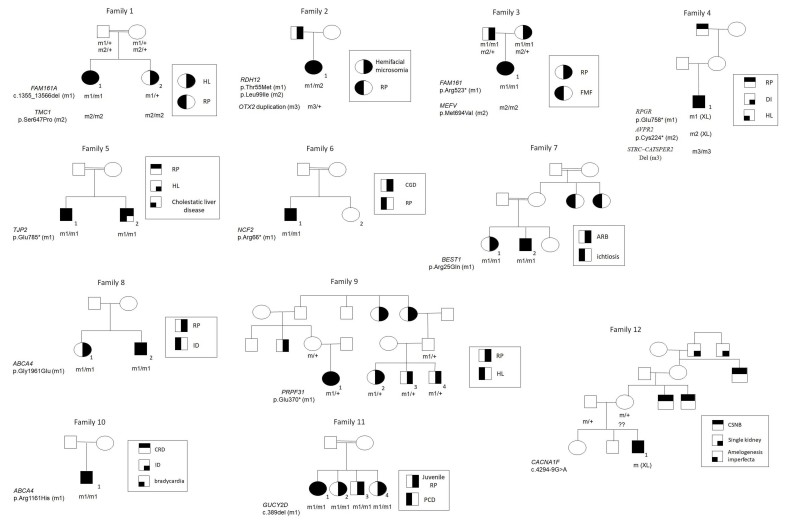
Family 1 is a consanguineous family of Moroccan Jewish origin. Patient 1–1 has RP and congenital profound neurosensory HL. This phenotypic combination led to an initial diagnosis of type 1 Usher syndrome (USH1). However, her sister (patient 1–2) had HL, without RP (Figure 1). Homozygosity mapping performed on individual 1–1 revealed many large homozygous regions. None of these regions included a known USH causative gene. However, two regions harbored known causative genes for AR nonsyndromic RP and for AR nonsyndromic HL: A 24 Mb interval located on chromosome 2 (61–85 Mb) included the FAM161A gene, and a 58 Mb interval located on chromosome 9 (31–89Mb) included the TMC1 gene. Sequencing of all coding exons of these two genes in patient 1–1 revealed homozygous mutations in both: FAM161A (NM_001201543; OMIM: 613596) harbored the c.1355_1356del mutation, which is one of the most common RP causative mutations in the Israeli Jewish population [11]; TMC1 (NM_138691; OMIM: 606706) harbored the p.Ser647Pro founder mutation, which is prevalent among Moroccan Jews [12] (Table 1). Testing of additional family members revealed that both parents were heterozygotes for both mutations, while patient 1–2 was homozygous for the TMC1 mutation, but heterozygote for the FAM161A mutation (Figure 1).
Figure 1.
Family pedigrees. Shown are 12 families segregating IRD and additional systemic diseases. Filled symbols represent affected individuals, whereas clear symbols represent unaffected individuals. A double line represents consanguinity. Patients recruited for this study are marked by numbers. In mutation names, * represents a termination codon. +, wild-type (wt) allele; m, mutant allele; RP, retinitis pigmentosa; HL, hearing loss; FMF, familial Mediterranean fever; DI, diabetes insipidus; CGD, chronic granulomatous disease; ARB, autosomal recessive bestrophinopathy; ID, intellectual disability; CRD, cone-rod dystrophy; PCD, primary ciliary dyskinesia; CSNB, congenital stationary night blindness.
Table 1. Patients with IRD and additional diagnoses included in this study.
| Family | Patient # (Gender) | Consanguinity | Suspected diagnosis (inheritance mode) | Actual diagnosis (inheritance mode) | First molecular finding | Second molecular finding | Third molecular finding | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 |
1
(F) |
Yes |
USH (AR) |
RP (AR) + HL (AR) |
FAM161A (NM_001201543; OMIM: 613596):
c.1355_1356del; p.Thr452Serfs*3 (hom)
(RP) |
TMC1 (NM_138691; OMIM: 606706):
c.1939T>C; p.Ser647Pro (hom)
(HL) |
NA |
Current study |
| 2 |
1
(F) |
No |
Syndromic RP (?) |
RP (AR) + hemifacial microsomia (AD) |
RDH12 (NM_152443; OMIM: 608830):
c.164C>T; p.Thr55Met (het)
c.295C>A; p.Leu99Ile (het)
(RP) |
OTX2 (NM_021728.3; OMIM: 600037): duplication
Arr[hg19]14q22.3 (57269186_57925544)dup (het)
(hemifacial microsomia) |
NA |
Current study |
| 3 |
1
(F) |
No |
FMF (AR) and RP (?) |
RP (AR)+ FMF (AR) |
FAM161A (NM_001201543; OMIM: 613596):
c.1567C>T; p.Arg523* (hom)
(RP) |
MEFV (NM_000243.2; OMIM: 608107):
c.2080A>G; p.Met694Val (hom)
(FMF) |
NA |
Current study |
| 4 |
1
(M) |
No |
Syndromic RP (?) |
RP (XL) + DI (XL) + HL (AR) |
RPGR (NM_001034853.1; OMIM: 312610):
c.2272G>T; p.Glu758* (hemi)
(RP) |
AVPR2 (NM_000054.5; OMIM: 300538):
c.672C>A; p.Cys224* (hemi)
(DI) |
STRC (NM_153700.2; OMIM: 606440) – CATSPER2 (NM_172095.3; OMIM: 607249) del (hom)
(HL) |
Current study |
| 5 |
1
(M) |
Yes |
Syndromic RP (?) |
RP (?) + HL (AD?) + cholestatic liver disease (AR) |
TJP2 (NM_004817; OMIM: 607709):
c.2353C>T; p.Glu785* (hom)
(cholestatic liver disease and HL) |
?
(RP) |
NA |
Current study |
| 6 |
1
(M) |
Yes |
CGD and RP (XL) (AKA
contiguous gene deletion
syndrome, McLeod phenotype) |
RP (?) + CGD (AR) |
NCF2 (NM_001127651.2; OMIM: 608515):
c.196C>T; p.Arg66* (hom)
(CGD) |
?
(RP) |
NA |
Current study |
| 7 |
2
(M) |
Yes |
ARB (AR) + ichtiosis (AR) |
ARB (AR) + ichtiosis (AR) |
BEST1 (NM_004183.3; OMIM: 607854):
c.74G>A; p.Arg25Gln (hom)
(ARB) |
?
(ichtiosis) |
NA |
Current study |
| 8 |
2
(M) |
No |
RP (AR) + ID (?) |
RP (AR) + ID (?) |
ABCA4 (NM_000350.2; OMIM: 601691):
c.5882G>A; p.Gly1961Glu (hom)
(RP) |
?
(ID) |
NA |
Current study |
| 9 |
1
(F) |
No |
USH (AR) |
RP (AD) + HL (?) |
PRPF31 (NM_015629; OMIM: 606419):
c.1108G>T; p.Glu370* (het)
(RP) |
?
(HL) |
NA |
Current study |
| 10 |
1
(M) |
No |
Danon Disease (XL) |
CRD (AR) + ID (?) + bradycardia (?) |
ABCA4 (NM_000350.2; OMIM: 601691):
c.G3482G>A; p.Arg1161His (hom)
(CRD) |
?
(ID) |
?
(bradycardia) |
Current study |
| 11 |
1
(F) |
Yes |
Ciliopathy (AR) |
Juvenile RP (AR) + PCD (?) |
GUCY2D (NM_000180; OMIM: 600179):
c.389del; p.Pro130Leufs*36 (hom)
(juvenile RP) |
?
(PCD) |
NA |
Current study |
| 12 |
1
(M) |
No |
CSNB (XL) + single kidney (?) + amelogenesis imperfect (?) |
CSNB (XL) + single kidney (?) + amelogenesis imperfect (?) |
CACNA1F (NM_005183; OMIM: 300110):
c.4294-9G>A (IVS36-9G>A)
(CSNB) |
?
(single kidney) |
?
(amelogenesis imperfect) |
Current study |
| 13 |
1
(F) |
Yes |
USH (AR) |
USH (AR)+ RP (AR) |
MYO7A (NM_000620; OMIM: 276903):
c.2308delC; p.Asn769fsX4 (hom)
(USH) |
PDE6B (NM_000283; OMIM: 180072):
c.1417delC; p.Leu473fsX16 (hom)
(RP) |
NA |
[24] |
| 14 and 15 | 1 and 1 | No | ESCS (AR) + Early-onset OPMD (AD) | ESCS (AR) + Early-onset OPMD (AD) | NRL (NM_006177.3; OMIM: 162080): c.91C>T; p.Arg31* (hom) (ESCS) | PABPN1 (NM_004643; OMIM: 602279): (GCN)13 (hom) (OPMD) | NA | [25] |
AD, autosomal dominant; AR, autosomal recessive; ARB, autosomal recessive bestrophinopathy; CGD, chronic granulomatous disease; CRD, cone-rod dystrophy; CSNB, congenital stationary night blindness; DI, diabetes insipidus; ESCS, enhanced S-cone syndrome; FMF, Familial Mediterranean Fever; het, heterozygote; hom, homozygote; ID, intellectual disability; NA, not applicable; PCD, primary ciliary dyskinesia; OPMD, oculopharyngeal muscular dystrophy, RP, Retinitis Pigmentosa; USH, Usher Syndrome; XL, X-linked; *, termination codon
Family 2
Patient 2–1 is a Jewish female, born to non-consanguineous parents of North African/Turkish origin. She was referred for ophthalmic evaluation at the age of 16 years, due to suboptimal visual acuity, even with optical correction. The first thing noticed were her unusual facial features, which included bilateral microtia with a hearing aid in the left ear, and profound micrognathia. The ophthalmic exam was positive for bilateral lipodermoid lesions on the temporal aspect of the bulbar conjunctiva. BCVA was 20/40 in oculus dexter (OD) and 20/50 in oculus sinister (OS). The fundus exam revealed optic disc pallor, macular atrophy, multiple bone spicules in retinal midperiphery, and vessel attenuation, leading to a diagnosis of RP. The combination of RP and facial features led us to consider a syndromic form of RP. However, CMA revealed a heterozygous 656 kb duplication on chromosome 14q22.3, which includes exons 1–3 of the OTX2 gene (NM_021728.3; OMIM: 600037; Appendix 3). OTX2 duplications are a known cause of AD hemifacial microsomia [13]. This mutation was likely inherited from the patient’s father, who had similar facial features, according to descriptions by the family; unfortunately, he was unavailable for clinical or genetic evaluation. In addition, WES revealed that she was compound heterozygous for two mutations in the RDH12 gene (NM_152443; OMIM: 608830), a known causative gene for arRP (Table 1). Both mutations in RDH12 were previously identified in Jewish patients of North African and Oriental descent with RP [14] (Figure 1).
Family 3
The index patient (patient 3–1) is a female, born to nonrelated Jewish parents of North African origin (Figure 1). Both parents were diagnosed with advanced RP. Their daughter was diagnosed with familial Mediterranean fever (FMF) and treated with colchicine. FMF, an AR disease caused by mutations in the MEFV gene (NM_000243.2; OMIM: 608107), is common in the Israeli population and predominantly affects North African Jews [15], with a carrier frequency of approximately 1:5 [16]. Patient 3–1 was found to be homozygous for the common MEFV founder mutation p.Met694Val. At the age of 10 years, during an episode of headaches and vomiting, she was referred to the eye clinic for a fundus examination to rule out papilledema and elevated intracranial pressure. Upon exam, BCVA was 20/20 in each eye, and the retinal midperiphery was noted to have mild hypopigmented lesions and few bone spicules. ERG showed no rod response, with severely decreased cone response, thus confirming a diagnosis of RP. Molecular testing confirmed she was homozygous for the p.Arg523* mutation in FAM161A, which is a prevalent RP causative mutation among Jews [11].
Family 4
A 2.5-year-old male, born to non-consanguineous Ashkenazi Jewish parents, came to the clinic due to progressive myopia and nyctalopia. His medical history was positive for diabetes insipidus (DI) and HL, thus raising the possibility of syndromic IRD. On his exam, BCVA was 20/30 in both eyes, and the fundus exam seemed normal. Family history revealed that his maternal grandfather had been diagnosed with RP (Figure 1). WES revealed that the patient harbors three mutations, each causing one of his diseases (DI, HL, and RP; Table 1). Two mutations are XL: a novel nonsense mutation, p.Glu758*, in RPGR (NM_001034853.1; OMIM: 312610; Appendix 3), and a novel nonsense mutation, p.Cys224*, in AVPR2 (NM_000054.5; OMIM: 300538; causing RP and DI, respectively). The third mutation is a large homozygous deletion on chromosome 15q15, involving the STRC (NM_153700.2; OMIM: 606440) and CATSPER2 (NM_172095.3; OMIM: 607249) genes. STRC mutations are a known cause of AR HL [17]. Deletions in STRC-CATSPER2 have been previously described in several patients with AR deafness-infertility syndrome (DIS) [18-20]. Due to the patient’s young age, his fertility was not assessed.
Family 5
This is a consanguineous Muslim Arab family (parents are first cousins). The index patient (5–1) had liver transplantation during childhood due to cholestatic liver disease. In addition, he had progressive HL and RP, which presented as nyctalopia since childhood, with progressive visual fields loss and visual deterioration. On exam (at the age of 25 years), BCVA was counting finger (CF) in each eye. Full-field ERG (ff-ERG) was non-recordable for cone and rod responses. His brother (patient 5–2) is also affected by cholestatic liver disease and RP. Data regarding his hearing status were not available (Figure 1). WES of patient 5–1 identified a novel homozygous nonsense mutation, p.Glu785*, in the TJP2 gene (NM_004817; OMIM: 607709), which is known to cause AR cholestatic liver disease [21] and AD progressive HL [22] (Appendix 3). A mutation causing RP was not found.
Family 6
This is a consanguineous Israeli Muslim Arab family (parents are double cousins). Patient 6–1 was clinically diagnosed at the age of 2 years with chronic granulomatous disease (CGD), and underwent bone marrow transplantation. At the age of 7 years, he was referred for ophthalmic evaluation due to visual disturbances. On exam, BCVA was 20/80 OD and 20/50 OS. The fundus exam showed perivascular chorioretinal punched out lesions, and perivascular pigmentary changes. ERG was consistent with rod-cone disease, i.e., RP (Figure 1). The combination of CGD and RP raised the possibility of the XL contiguous gene deletion syndrome known as the McLeod phenotype [23]. However, genetic analysis revealed that in patient 6–1 CGD is inherited as an AR trait, caused by a homozygous nonsense mutation in the NCF2 gene (NM_001127651.2; OMIM: 608515; Table 1). This mutation, p.Arg66*, has been previously reported in patients with CGD [24,25]. The genetic cause of RP remains unknown.
Family 7
This is a consanguineous Muslim Arab family. The parents are healthy. Their older daughter (patient 7–1) was diagnosed at the age of 17 years with a bestrophinopathy, and the parents were advised to bring in her siblings for ophthalmic exams. One of the siblings (patient 7–2; age 14 years) was asymptomatic at presentation, with BCVA of 20/20 in each eye. However, on the fundus exam he was noted to have mild foveal RPE changes. OCT showed a small subfoveal accumulation. In addition, he had ichthyosis presenting with severe perioral and hand skin atrophy. Both siblings were found to have a previously reported homozygous missense mutation in the BEST1 gene (NM_004183.3; OMIM: 607854), p.Arg25Gln [26], thus confirming a diagnosis of ARB (Table 1). A causative mutation for ichthyosis was not found; however, intrafamilial marriage for several generations in this family, and the occurrence of additional affected individuals in previous generations, supports an AR inheritance mode (Figure 1).
Family 8
Family 8 is a non-consanguineous Jewish family of Yemenite origin. One of the daughters (patient 8–1) has severe RP, due to homozygosity for a known and common mutation (p.Gly1961Glu) in ABCA4 (NM_000350.2; OMIM: 601691). Her intelligence is normal. Her brother (patient 8–2) is intellectually disabled. He was referred to the eye clinic for vision evaluation at the age of 11 years, because his teachers suspected he had impaired vision. On exam, BCVA was 20/80 OD, 20/160 OS, with exotropia in the left eye. The fundus exam showed pallor of the optic disc, arterial narrowing, abnormal macular appearance with RPE changes, and no bone spicules. FF-ERG demonstrated a significant decrease in rod and cone responses, thus confirming a diagnosis of RP. Genetic analysis confirmed homozygosity for the p.Gly1961Glu mutation in ABCA4. The cause of the intellectual disability (ID) remains unknown.
Family 9
Patient 9–1 was born to healthy non-related Jewish parents of Iraqi and Tunisian origin. She had HL and used hearing aids since 9 months of age. She was referred for ophthalmic evaluation at the age of 2 years due to her mother’s concern of nyctalopia. On exam, BCVA was fix and follow in each eye. The fundus exam revealed an abnormal macular reflex and bone spicules in the midperiphery. ERG showed no rod response and significantly attenuated cone response, thus leading to a diagnosis of RP. The combination of RP and HL led to an initial diagnosis of USH. However, further questioning revealed the existence of non-syndromic RP in several maternal relatives (Figure 1). The mode of segregation in the extended family appeared to be AD with reduced penetrance, and a heterozygous mutation in the PRPF31 (NM_015629; OMIM: 606419) gene (p.Glu370*) was previously detected in some of the affected individuals. Genetic testing of patient 9–1 confirmed the presence of the same heterozygous mutation (Appendix 3). The cause of the HL currently remains unknown.
Family 10
Family 10 is a non-consanguineous Jewish family of mixed origin. Patient 10–1 was seen at the age of 34 years. Medical history was positive for pacemaker insertion at the age of 25 years due to bradycardia. In addition, he had mild ID. He was referred to the ophthalmic clinic due to reduced vision. BCVA was CF OD, and 20/200 OS. FF-ERG demonstrated significantly reduced cone response and slightly reduced rod response, suggesting a cone-rod dystrophy (CRD; Figure 1). The combination of cardiac disease, cognitive impairment, and retinopathy led to a possible diagnosis of XL Danon disease [27]. The causative gene of Danon disease, LAMP2 (NM_002294.2; OMIM: 309060), was sequenced, but no mutation was found. WES analysis revealed that the patient was homozygous for a known missense mutation in the ABCA4 gene (p.Arg1161His), which confirmed a diagnosis of arCRD. The causes of cardiac disease and cognitive impairment remain unknown.
Family 11
This is a consanguineous Muslim Arab family. Patient 11–1 was diagnosed with early-onset RP (her ERG was non-recordable at the age of 18 years). In addition, she was diagnosed with primary ciliary dyskinesia (PCD). The combination of these two phenotypes initially raised a suspicion of a ciliopathy, affecting the lung and the retina. However, further questioning of the family revealed that three additional siblings were affected by early-onset RP, but without PCD (Figure 1). In addition, in RP-affected individuals from the same village, we previously identified a known mutation (c.389del) in GUCY2D (NM_000180; OMIM: 600179) [28]. Genetic testing confirmed homozygosity for this mutation in all four siblings. The cause of PCD in patient 11–1 remains unknown.
Family 12
This is a non-consanguineous Jewish family of Ashkenazi origin. Patient 12–1 was followed up in the ophthalmology clinic due to suboptimal BCVA even with refractive correction (20/40 OD and 20/30 OS). His mother reported he had difficulty in night vision. In addition, he had a single kidney and amelogenesis imperfecta. His family history was positive for nyctalopia in three maternal male relatives, and a single kidney in his mother’s grandfather and his brother (suggestive of an XL inheritance pattern for both traits; Figure 1). WES detected a novel single nucleotide variant located at position −9 of CACNA1F (NM_005183; OMIM: 300110) intron 36. This variant (c.4294–9G>A) is rare (not present in the Genome Aggregation Database, gnomAD), and was predicted to generate a novel acceptor splice site, 7 bp upstream of the original splice site of intron 36 (Human Splicing Finder). The use of this novel acceptor site will generate a 7-bp insertion, leading to a frameshift (Appendix 3). Mutations in CACNA1F are associated with XL congenital stationary night blindness (OMIM: 300071). The causes of the kidney and teeth phenotypes are currently unknown (Table 1).
Families 13, 14, and 15
These families were previously described in detail elsewhere. Patient 13–1 is homozygous for pathogenic mutations in two IRD-related genes (MYO7A (NM_000620; OMIM: 276903) and PDE6B (NM_000283; OMIM: 180072), causing syndromic and non-syndromic RP [29]. Patients 14–1 and 15–1 have a combination of non-syndromic IRD, caused by homozygosity for a recessive mutation in NRL (NM_006177.3; OMIM: 162080), and early-onset oculopharyngeal muscular dystrophy, caused by homozygosity for a dominant mutation in PABPN1 (NM_004643; OMIM: 602279) [30].
Discussion
The incidence of a second genetic disease in a patient who has a genetically proven disease is approximately 5% [31,32]. Although multiple molecular diagnoses in one patient have been described previously [31-33], most reports described patients with non-ocular diseases. In this paper, we demonstrated that this phenomenon exists among patients with IRDs as well. In some cases (patients 1–1, 9–1, 6–1, and 10–1), the various phenotypes presented by the patients blended into one known syndrome (USH1, the McLeod syndrome, and Danon disease, respectively). In all these cases, the actual diagnosis could have been missed due to a satisfactory clinical diagnosis of a known syndrome, and only the genetic testing enlightened the presence of two or more distinct phenotypes. In other cases, there seemed to be no link between the different phenotypes, as was proven with molecular testing.
The common working paradigm for the ophthalmologist is combining the different symptoms observed in a patient into one unifying diagnosis. Thus, a patient presenting to an ophthalmologist with a retinal dystrophy and some other rare systemic finding would most probably be suspected to have syndromic IRD. However, IRD is a strikingly heterogeneous condition, associated with mutations in more than 200 genes. Although the contribution of each one of these genes to the overall prevalence of IRD is relatively small, the total number of pathogenic mutations in all IRD causative genes is relatively elevated in the general population, and it is estimated that approximately one in four to five individuals may be a carrier of an IRD causative mutation [34]. Consequently, IRD has a relatively high prevalence of 1:2,000–1,5:000 in various populations [35-37], and may coincide with other genetic (and non-genetic) rare conditions.
The co-occurrence of IRD with another condition is most likely to appear when the other condition is relatively frequent. For example, HL (either congenital or acquired) is the most common neurosensory disorder in humans. Congenital HL occurs in one out of 1,000 infants, with approximately 60% of the cases due to genetic causes [38]. ID is estimated to affect 1% to 8% of individuals worldwide, with at least 40% due to genetic defects [39]. Therefore, co-occurrence of IRD with HL or with ID (as seen in families 1, 4, 5, 8, 9, and 10) is not uncommon.
One of the main factors that increase the risk for intrafamilial clustering of multiple hereditary diseases is consanguinity, which increases the risk of homozygosity for rare recessive alleles. In the present study, six of 15 families were consanguineous. Most striking is patient 13–1, who has a combination of RP and HL, and is homozygous for pathogenic mutations causing USH1 and nonsyndromic RP. Therefore, RP in this patient is caused simultaneously by two distinct genetic defects [29]. In addition, specific recessive mutations are frequent in certain populations due to founder effects. For example, mutations in FAM161A are the second most frequent cause of RP in Israel [40], and are mainly frequent among North African Jews. FMF is common in the Israeli population, affecting predominantly North African Jews, with a carrier frequency of 1:5 in this population [15,16]. Therefore, the chances of having FMF and (FAM161A-related) RP in a Jewish individual of North African descent (such as patient 3–1) are considerably fair, even in the absence of consanguinity. Nevertheless, multiple rare genetic diagnoses occur in non-consanguineous families as well. The most striking example is patient 4–1, with three distinct phenotypes due to three distinct genetic alterations.
In a recent study analyzing 101 patients with multiple genetic diagnoses, the most commonly observed pattern was pathogenic variants in two AD disease genes, and variants in AD disease genes totaled 54.1% of the total observed variants [31]. Results in the present cohort are different. Of six patients with complete molecular diagnosis, four (66%) had pathogenic variants in two AR disease genes, and of the total pathogenic variants identified, AR mutations were the most common (16/22, 72%). This is probably due to the high consanguinity rates in the Israeli population, which make AR inheritance the most common cause of IRD and other genetic conditions in this population [37].
New diagnostic technologies which emerged in recent years, and specifically next-generation sequencing (including WES), dramatically improved our capacity to molecularly diagnose patients with various genetic conditions, including patients affected by multiple conditions, such as the ones presented here. In all families presented in this study, syndromic IRD was ruled out by identification of at least one genetic defect underlying one of the observed phenotypes. However, an obvious limitation of this study is that not all patients had a full genetic workup. WES was performed on six patients; in three of them, the genetic cause of all phenotypic components was identified (patients 2–1, 4–1, and 13–1), while in the other three patients, only a partial genetic diagnosis was established (patients 5–1, 10–1, and 12–1). Nine additional patients were tested for specific founder mutations, based on the patients’ clinical symptoms and ethnic background. This testing led to complete genetic diagnosis in four patients (1–1,3–1, 14–1, and 15–1), and to partial diagnosis in five patients (6–1, 7–2, 8–2, 9–1, and 11–1). Performing WES on these five patients will most probably enable the identification of additional genetic defects.
In summary, establishing a correct diagnosis is important for patients and their family members. It enables understanding of the natural history course, and the prediction of disease prognosis, and it aids in tailoring correct follow-up and treatment. It also allows patients to pursue prenatal counseling and reproductive planning. Although the pattern recognition skill often leads ophthalmologists to the correct diagnosis, clinicians should be aware that a differential diagnosis does exist. The existence of features that do not fit a single diagnosis, atypical features of a suspected genetic syndrome, or parental consanguinity should raise the index of suspicion for the possibility of multiple cooccurring diagnoses.
Acknowledgments
We are grateful to the patients for their participation in this study. We thank Dror Sharon for his helpful contribution. The study was supported by research grants from the Israeli Ministry of Health (3–12583) and from the Foundation Fighting Blindness (BR-GE-0518–0734-TECH) to TB and NGC.
Appendix 1.PCR primers.
To access the data, click or select the words “Appendix 1.”
Appendix 2.Function of genes included in the study.
To access the data, click or select the words “Appendix 2.”
Appendix 3. Novel mutations reported in this study.
To access the data, click or select the words “Appendix 3.” (A) Chromosomal Microarray Analysis of chromosome 14q22.3 in patient 2–1, showing a heterozygous 656Kb duplication, which includes exons 1–3 of the OTX2 gene. (B) Integrative Genomics Viewer (IGV) visualization of part of RPGR exon 15 (ORF15) in patient 4–1 (reverse strand), showing a homozygous C>A transversion, leading to the c.2272G>T; p.Glu758* mutation. (C) Nucleotide sequence traces of TJP2 exon 16 in a non-carrier individual (wt) and in patient 5–1, who is homozygote for the c.2353C>T; p.Glu785* mutant allele (hom). The exon-intron boundary is marked. (D) IGV visualization and nucleotide sequence trace of part of PRPF31 exon 11 in patient 9–1, showing a heterozygote G>T transversion, leading to the c.1108G>T; p.Glu370* mutation. (E) IGV visualization (reverse strand) and nucleotide sequence traces of CACNA1F intron 36 in patient 12–1, who is hemizygote for the c.4294–9G>A (IVS37–9G>A) mutant allele. The intron-exon boundary is marked. The original and the newly-formed splice acceptor sites are underlined.
References
- 1.Duncan JL, Pierce EA, Laster AM, Daiger SP, Birch DG, Ash JD, Iannaccone A, Flannery JG, Sahel JA, Zack DJ, Zarbin MA, Foundation Fighting Blindness Scientific Advisory Board Inherited retinal degenerations: current landscape and knowledge gaps. Transl Vis Sci Technol. 2018;7:6. doi: 10.1167/tvst.7.4.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verbakel SK, van Huet RAC, Boon CJF, den Hollander AI, Collin RWJ, Klaver CCW, Hoyng CB, Roepman R, Klevering BJ. Non-syndromic retinitis pigmentosa. Prog Retin Eye Res. 2018;66:157–86. doi: 10.1016/j.preteyeres.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Thiadens AAHJ, Phan TML, Zekveld-Vroon RC, Leroy BP, Van Den Born LI, Hoyng CB, Klaver CCW, Roosing S, Pott JWR, Van Schooneveld MJ, Van Moll-Ramirez N, Van Genderen MM, Boon CJF, Den Hollander AI, Bergen AAB, De Baere E, Cremers FPM, Lotery AJ. Clinical course, genetic etiology, and visual outcome in cone and cone-rod dystrophy. Ophthalmology. 2012;119:819–26. doi: 10.1016/j.ophtha.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 4.Tanna P, Strauss RW, Fujinami K. Stargardt disease: clinical features, molecular genetics, animal models and therapeutic options. Br J Ophthalmol. 2017;101:25–30. doi: 10.1136/bjophthalmol-2016-308823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson AA, Guziewicz KE, Lee CJ, Kalathur RC, Pulido JS, Marmorstein LY, Marmorstein AD. Bestrophin 1 and retinal disease. Prog Retin Eye Res. 2017;58:45–69. doi: 10.1016/j.preteyeres.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Werdich XQ, Place EM, Pierce EA. Systemic diseases associated with retinal dystrophies. Semin Ophthalmol. 2014;29:319–28. doi: 10.3109/08820538.2014.959202. [DOI] [PubMed] [Google Scholar]
- 7.Dias MF, Joo K, Kemp JA, Fialho SL, da Silva Cunha A, Woo SJ, Kwon YJ. Molecular genetics and emerging therapies for retinitis pigmentosa: Basic research and clinical perspectives. Prog Retin Eye Res. 2018;63:107–31. doi: 10.1016/j.preteyeres.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Friedman TB, Schultz JM, Ahmed ZM, Tsilou ET, Brewer CC. Usher syndrome: hearing loss with vision loss. Adv Otorhinolaryngol. 2011;70:56–65. doi: 10.1159/000322473. [DOI] [PubMed] [Google Scholar]
- 9.Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17:8390. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. HomozygosityMapper–an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–9. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bandah-Rozenfeld D, Mizrahi-Meissonnier L, Farhy C, Obolensky A, Chowers I, Pe’Er J, Merin S, Ben-Yosef T, Ashery-Padan R, Banin E, Sharon D. Homozygosity mapping reveals null mutations in FAM161A as a cause of autosomal-recessive retinitis pigmentosa. Am J Hum Genet. 2010;87:382–91. doi: 10.1016/j.ajhg.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brownstein Z, Friedman LM, Shahin H, Oron-Karni V, Kol N, Abu Rayyan A, Parzefall T, Lev D, Shalev S, Frydman M, Davidov B, Shohat M, Rahile M, Lieberman S, Levy-Lahad E, Lee MK, Shomron N, King MC, Walsh T, Kanaan M, Avraham KB. Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in middle eastern families. Genome Biol. Genome Biol. 2011;12:R89. doi: 10.1186/gb-2011-12-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zielinski D, Markus B, Sheikh M, Gymrek M, Chu C, Zaks M, Srinivasan B, Hoffman JD, Aizenbud D, Erlich Y. OTX2 duplication is implicated in hemifacial microsomia. PLoS One. 2014;9:e96788. doi: 10.1371/journal.pone.0096788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanany M, Allon G, Kimchi A, Blumenfeld A, Newman H, Pras E, Wormser OS, Birk O, Gradstein L, Banin E, Ben-Yosef T, Sharon D. Carrier frequency analysis of mutations causing autosomal-recessive-inherited retinal diseases in the Israeli population. Eur J Hum Genet. 2018;26:1159–66. doi: 10.1038/s41431-018-0152-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shohat M. Familial Mediterranean fever. University of Washington (Seattle): GeneReviews®; 1993. [Google Scholar]
- 16.Stoffman N, Magal N, Shohat T, Lotan R, Koman S, Oron A, Danon Y, Halpern GJ, Lifshitz Y, Shohat M. M694V homozygotes in the Israeli North African Jewish community. Eur J Hum Genet. 2000;8:307–10. doi: 10.1038/sj.ejhg.5200446. [DOI] [PubMed] [Google Scholar]
- 17.Verpy E, Masmoudi S, Zwaenepoel I, Leibovici M, Hutchin TP, Del Castillo I, Nouaille S, Blanchard S, Lainé S, Popot J-L, Moreno F, Mueller RF, Petit C. Mutations in a new gene encoding a protein of the hair bundle cause non-syndromic deafness at the DFNB16 locus. Nat Genet. 2001;29:345–9. doi: 10.1038/ng726. [DOI] [PubMed] [Google Scholar]
- 18.Knijnenburg J, Lesnik Oberstein SA, Frei K, Lucas T, Gijsbers AC, Ruivenkamp CA, Tanke HJ, Szuhai K. A homozygous deletion of a normal variation locus in a patient with hearing loss from non-consanguineous parents. J Med Genet. 2009;46:412–7. doi: 10.1136/jmg.2008.063685. [DOI] [PubMed] [Google Scholar]
- 19.Avidan N, Tamary H, Dgany O, Cattan D, Pariente A, Thulliez M, Borot N, Moati L, Barthelme A, Shalmon L, Krasnov T, Ben-Asher E, Olender T, Khen M, Yaniv I, Zaizov R, Shalev H, Delaunay J, Fellous M, Lancet D, Beckmann JS. CATSPER2, a human autosomal nonsyndromic male infertility gene. Eur J Hum Genet. 2003;11:497–502. doi: 10.1038/sj.ejhg.5200991. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Malekpour M, Al-Madani N, Kahrizi K, Zanganeh M, Lohr NJ, Mohseni M, Mojahedi F, Daneshi A, Najmabadi H, Smith RJ. Sensorineural deafness and male infertility: a contiguous gene deletion syndrome. J Med Genet. 2007;44:233–40. doi: 10.1136/jmg.2006.045765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrotta M, Strautnieks S, Papouli E, Rushton P, Clark BE, Parry DA, Logan CV, Newbury LJ, Kamath BM, Ling S, Grammatikopoulos T, Wagner BE, Magee JC, Sokol RJ, Mieli-Vergani G. University of Washington Center for Mendelian Genomics, Smith JD, Johnson CA, McClean P, Simpson MA, Knisely AS, Bull LN, Thompson RJ. Mutations in TJP2 cause progressive cholestatic liver disease. Nat Genet. 2014;46:326–8. doi: 10.1038/ng.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh T, Pierce SB, Lenz DR, Brownstein Z, Dagan-Rosenfeld O, Shahin H, Roeb W, McCarthy S, Nord AS, Gordon CR, Ben-Neriah Z, Sebat J, Kanaan M, Lee MK, Frydman M, King MC, Avraham KB. Genomic duplication and overexpression of TJP2/ZO-2 leads to altered expression of apoptosis genes in progressive nonsyndromic hearing loss DFNA51. Am J Hum Genet. 2010;87:101–9. doi: 10.1016/j.ajhg.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watkins CE, Litchfield J, Song E, Jaishankar GB, Misra N, Holla N, Duffourc M, Krishnaswamy G. Chronic granulomatous disease, the McLeod phenotype and the contiguous gene deletion syndrome-a review. Clin Mol Allergy. 2011;9:13. doi: 10.1186/1476-7961-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noack D, Rae J, Cross AR, Munoz J, Salmen S, Mendoza JA, Rossi N, Curnutte JT, Heyworth PG. Autosomal recessive chronic granulomatous disease caused by novel mutations in NCF-2, the gene encoding the p67-phox component of phagocyte NADPH oxidase. Hum Genet. 1999;105:460–7. doi: 10.1007/s004390051131. [DOI] [PubMed] [Google Scholar]
- 25.Teimourian S, de Boer M, Roos D. Molecular basis of autosomal recessive chronic granulomatous disease in iran. J Clin Immunol. 2010;30:587–92. doi: 10.1007/s10875-010-9421-6. [DOI] [PubMed] [Google Scholar]
- 26.Marquardt A, Stohr H, Passmore LA, Kramer F, Rivera A, Weber BH. Mutations in a novel gene, VMD2, encoding a protein of unknown properties cause juvenile-onset vitelliform macular dystrophy (Best’s disease). Hum Mol Genet. 1998;7:1517–25. doi: 10.1093/hmg/7.9.1517. [DOI] [PubMed] [Google Scholar]
- 27.Prall FR, Drack A, Taylor M, Ku L, Olson JL, Gregory D, Mestroni L, Mandava N. Ophthalmic manifestations of Danon disease. Ophthalmology. 2006;113:1010–3. doi: 10.1016/j.ophtha.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 28.Perrault I, Rozet JM, Calvas P, Gerber S, Camuzat A, Dollfus H, Chatelin S, Souied E, Ghazi I, Leowski C, Bonnemaison M, Le Paslier D, Frezal J, Dufier JL, Pittler S, Munnich A, Kaplan J. Retinal-specific guanylate cyclase gene mutations in Leber’s congenital amaurosis. Nat Genet. 1996;14:461–4. doi: 10.1038/ng1296-461. [DOI] [PubMed] [Google Scholar]
- 29.Goldenberg-Cohen N, Banin E, Zalzstein Y, Cohen B, Rotenstreich Y, Rizel L, Basel-Vanagaite L, Ben-Yosef T. Genetic heterogeneity and consanguinity lead to a “double hit”: Homozygous mutations of MYO7A and PDE6B in a patient with retinitis pigmentosa. Mol Vis. 2013;19:1565–71. [PMC free article] [PubMed] [Google Scholar]
- 30.Newman H, Blumen SC, Braverman I, Hanna R, Tiosano B, Perlman I, Ben-Yosef T. Homozygosity for a recessive loss-of-function mutation of the NRL gene is associated with a variant of enhanced S-cone syndrome. Invest Ophthalmol Vis Sci. 2016;57:5361–71. doi: 10.1167/iovs.16-19505. [DOI] [PubMed] [Google Scholar]
- 31.Posey JE, Harel T, Liu P, Rosenfeld JA, James RA, Coban Akdemir ZH, Walkiewicz M, Bi W, Xiao R, Ding Y, Xia F, Beaudet AL, Muzny DM, Gibbs RA, Boerwinkle E, Eng CM, Sutton VR, Shaw CA, Plon SE, Yang Y, Lupski JR. Resolution of disease phenotypes resulting from multilocus genomic variation. N Engl J Med. 2017;376:21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Veeraraghavan N, Hawes A, Chiang T. Molecular findings among patients referred for clinical whole- exome sequencing. JAMA. 2014;312:1870–9. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurolap A, Orenstein N, Kedar I, Weisz Hubshman M, Tiosano D, Mory A, Levi Z, Marom D, Cohen L, Ekhilevich N, Douglas J, Nowak CB, Tan WH, Baris HN. Is one diagnosis the whole story? Patients with double diagnoses. Am J Med Genet A. 2016;170:2338–48. doi: 10.1002/ajmg.a.37799. [DOI] [PubMed] [Google Scholar]
- 34.Nishiguchi KM, Rivolta C. Genes associated with retinitis pigmentosa and allied diseases are frequently mutated in the general population. PLoS One. 2012;7:e41902. doi: 10.1371/journal.pone.0041902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bunker CH, Berson EL, Bromley WC, Hayes RP, Roderick TH. Prevalence of retinitis pigmentosa in maine. Am J Ophthalmol. 1984;97:357–65. doi: 10.1016/0002-9394(84)90636-6. [DOI] [PubMed] [Google Scholar]
- 36.Haim M. Prevalence of retinitis pigmentosa and allied disorders in Denmark: III. Hereditary pattern. Acta Ophthalmol. 1992;70:615–24. doi: 10.1111/j.1755-3768.1992.tb02142.x. [DOI] [PubMed] [Google Scholar]
- 37.Sharon D, Banin E. Nonsyndromic retinitis pigmentosa is highly prevalent in the Jerusalem region with a high frequency of founder mutations. Mol Vis. 2015;21:783–92. [PMC free article] [PubMed] [Google Scholar]
- 38.Raviv D, Dror AA, Avraham KB. Hearing loss: a common disorder caused by many rare alleles. Ann N Y Acad Sci. 2010;1214:168–79. doi: 10.1111/j.1749-6632.2010.05868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ropers HH. Genetics of early onset cognitive impairment. Annu Rev Genomics Hum Genet. 2010;11:161–87. doi: 10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- 40.Shalev SA. Characteristics of genetic diseases in consanguineous populations in the genomic era: lessons from Arab communities in North Israel. Clin Genet. 2019;95:3–9. doi: 10.1111/cge.13231. [DOI] [PubMed] [Google Scholar]