Abstract
Th17 cells are essential for the pathogenesis of inflammatory and autoimmune diseases. In the presence of TGF-β, the differentiation of Th17 cells can be induced by inflammatory cytokines, especially IL-6, which is mainly produced by antigen presenting cells (APCs); or IL-21, which is derived from T cells. IL-21 is required for IL-6-induced Th17 cell differentiation. However, the key regulators and underlying mechanisms for IL-21-induced Th17 differentiation is still elusive. Here we show that SMAD4 is a key regulator in IL-21-induced Th17 differentiation. SMAD4 deficient naïve T cells can differentiate into Th17 cells in the absence of TGF-β signaling, and these Th17 cells are pathogenic during EAE. SMAD4 represses Rorc mRNA transcription to constrain IL-21-induced Th17 differentiation in the absence of TGF-β signaling. While in the presence of TGF-β, SMAD4 losses its suppressive ability due to the degradation of SKI. Mutation of Y429A or A432E on SMAD4 disrupts the interaction of SKI from SMAD4 and eliminates SMAD4 mediated suppression of Th17 differentiation. SMAD4 is indispensable for SKI binding to Rorc promoter region to regulate Th17 differentiation. Moreover, activin can induce Th17 differentiation in combination with IL-21, and the process is also subjected to the control of SKI and SMAD4. This study therefore elucidates critical mechanism for IL-21-induced Th17 differentiation to indicate SKI and SMAD4 as potential therapeutic targets for treating autoimmune diseases.
Keywords: TGF-β, SKI, SMAD4, RORγt, Th17, T cell differentiation
Introduction
Autoimmune diseases, including rheumatoid arthritis, type I diabetes, multiple sclerosis, systemic lupus, Psoriasis, and inflammatory bowel disease, affect 5%–8% of the world population. T helper 17 (Th17) cells, defined by the production of cytokine IL-17 [1, 2], are essential for the pathogenesis of autoimmune disease[3, 4]. Naïve T cells can differentiate into Th17 cells in response to IL-6 and TGF-β through the induction of Th17 lineage-specific transcription factor RORγt [5–7]. IL-6 is dominantly produced by antigen presenting cells (APCs), including macrophages and dendritic cells (DCs) [8, 9].
Recently, we have identified and characterized SKI/SMAD4 as a key mechanism in constraining IL-6 induced Th17 differentiation through the repression of Rorc gene expression, and provided insights into the mechanism of IL-6-induced Th17 differentiation [10]. However, the IL-6 induced Th17 differentiation can be profoundly impaired by the blockade of IL-21, or by the deficiency of IL-21 or IL-21 receptor [11–14]. These studies suggest that IL-6 induce RORγt induction for Th17 differentiation largely depends on IL-21. IL-21 not only substantially enhances the IL-6-induced Th17 program in an autocrine manner, but also serves as a surrogate of IL-6 to induce Th17 differentiation [11–14]. Therefore, the elucidation of underlying mechanism for IL-21-induced Th17 differentiation would be crucial for understanding the regulation of Th17 differentiation.
IL-21 is a pleiotropic cytokine that regulates a wide range of cell types [15]. Distinct from IL-6, the IL-21 is mainly produced by T cells, including CD4+ T, CD8+ T and NKT cells [16]. The functional receptor of IL-21 is composed of IL-21R and common receptor gamma chain (γc, also known as CD132), which is shared by IL-2 family cytokines [17]. The receptor signal cascade leads to activation of multiple STATs, including STAT3, STAT1, and STAT5. With different cellular sources and receptor signals, IL-21 represents an alternative path to the initiation and augmentation of inflammatory and autoimmune diseases.
Although IL-21 alone can activate STAT3 signaling pathways [12–14], TGF-β is also required to synergize with IL-21 to induce the transcription of RORγt. A variety of TGF-β regulation mechanisms are involved in the efficient differentiation of Th17 cells [18]. However, it is unknown whether there is other ligand that can in concert with IL-21 to promote RORγt expression and Th17 differentiation. TGF-β superfamily member activin was reported degrade SKI and promote IL-6-induced Th17 differentiation [10] Therefore, it would be of interest to investigate if and how activin may contribute to IL-21-induced Th17 differentiation.
In this study, we attempted to decipher the key mechanism of IL-21-induced Th17 differentiation. We found that IL-21 can induce TGF-β-independent Th17 differentiation in SMAD4 deficient T cells, and these cells are pathogenic in mouse experimental autoimmune encephalomyelitis (EAE) disease model. We further investigated the role of SKI in cooperative with SMAD4 to regulate the IL-21-induced Th17 differentiation by modulating Rorc gene expression. Our study thus elucidated the underlying mechanism of IL-21-induced Th17 differentiation that is controlled by SKI and SMAD4. These results enhance the understanding of IL-21-induced Th17 differentiation and reveal SKI and SMAD4 complex as a universal molecular switch for TGF-β family promoted Th17 differentiation.
RESULTS
SMAD4 deficiency led to IL-21-induced Th17 differentiation in the absence of TGF-β signaling
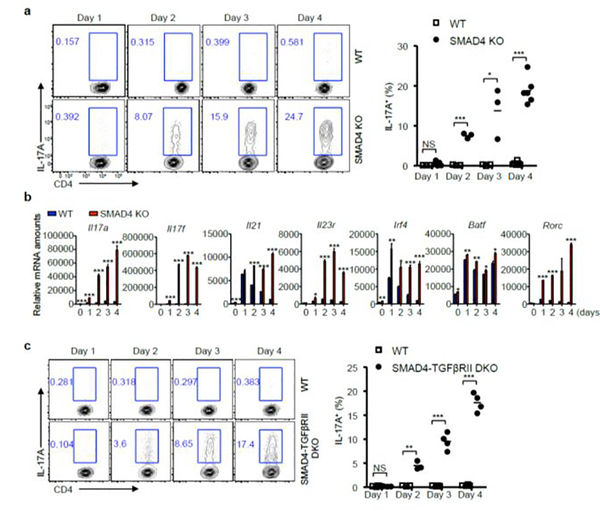
To investigate the possible involvement of SMAD4 in IL-21-induced Th17 differentiation, we cultured CD4+ T cells derived from wild-type (WT) and CD4-cre;Smad4fl/fl (SMAD4 KO) mice with IL-21. To prevent TGF-β signaling, we employed a widely used TGF-β receptor I pharmacological inhibitor SB525334. In the absence of TGF-β signaling, the WT cells did not differentiate into Th17 cells with IL-21 alone. However, the SMAD4 deficient T cells steadily differentiated into Th17 cells (Fig.1a). These Th17 cells are characterized by the upregulated expression of multiple Th17 related genes (Fig.1b). To rule out potential off-target effects of the inhibitor, and to further confirm that TGF-β signaling is indeed dispensable for IL-21-induced Th17 differentiation in SMAD4 deficient T cells, we generated CD4-cre;Smad4fl/fl;Tgfbr2fl/fl (SMAD4-TGFβRII DKO) mice to delete both SMAD4 and TGF-β receptor II. Consistent with the results of TGF-β receptor I inhibitor, the SMAD4 and TGF-β receptor II double deficient T cells also differentiated into Th17 cells with IL-21 alone (Fig.1c). These results suggest that SMAD4 plays a deciding role in IL-21-induced Th17 differentiation, and the deficiency of SMAD4 can offset the requirement of TGF-β for IL-21-induced Th17 generation.
Figure 1. IL-21-induced Th17 differentiation in SMAD4 deficient T cells.
(a) CD4+ T cells isolated from wild-type (WT) and Cd4-cre;Smad4fl/fl (SMAD4 KO) mice were cultured with IL-21 and TGF-β receptor I inhibitor. After 1, 2, 3, and 4 days, IL-17A+ cells were analyzed by flow cytometry. Representative results and statistical analysis are shown. (b) The relative mRNA amounts of Th17-related genes on day 4 were analyzed by qRT-PCR. (c) CD4+ T cells isolated from wild-type (WT) and CD4-cre;Smad4fl/fl;Tgfbr2fl/fl (SMAD4-TGFβRII DKO) mice were cultured with IL-21. After 1, 2, 3, and 4 days, IL-17A+ cells were analyzed by flow cytometry. Representative results and statistical analysis are shown.
IL-21-induced TGF-β-independent Th17 cells are encephalitogenic
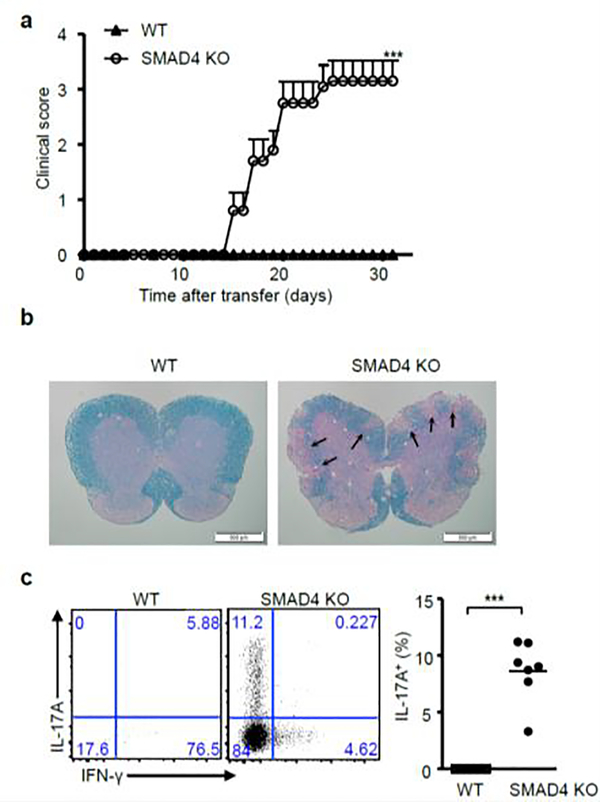
To examine the in vivo function of these IL-21-induced Th17 cells during experimental autoimmune encephalomyelitis (EAE), we crossed CD4-cre;Smad4fl/fl mice with 2D2 TCR transgenic mice to obtain SMAD4 deficient T cell with transgenic TCR that can recognize myelin oligodendrocyte glycoprotein (MOG) antigen. 2D2 TCR carrying CD4+ T cells from WT and SMAD4 deficient mice were cultured with IL-21 alone in the absence of TGF-β signaling and transferred into recipient mice. The SMAD4-deficient but not WT 2D2 T cells elicited severe EAE (Fig.2a), with immune pathology in the spinal cords (Fig.2b). IL-21-induced TGF-β-independent Th17 cells were found to infiltrate into spinal cord (Fig.2c) to contribute to the pathology during EAE.
Figure 2. Adoptive transferred EAE model.
(a) CD4+ T cells isolated from wild-type 2D2 (WT) and Cd4-cre;Smad4fl/fl 2D2 (SMAD4 KO) mice were cultured with IL-21 and TGF-β receptor I inhibitor, and transferred into irradiated wild-type recipient mice on day0. Pertussis toxin was i.p. injected on day 0 and day 2. The EAE onset is day15. After 31 days, clinical scores (Mean ± SEM, n=10), (b) pathological and (c) flow cytometry and statistical analysis of EAE are shown. Lesions were indicated by the arrows in the figure.
SMAD4 repressed RORγt expression during IL-21-induced Th17 differentiation
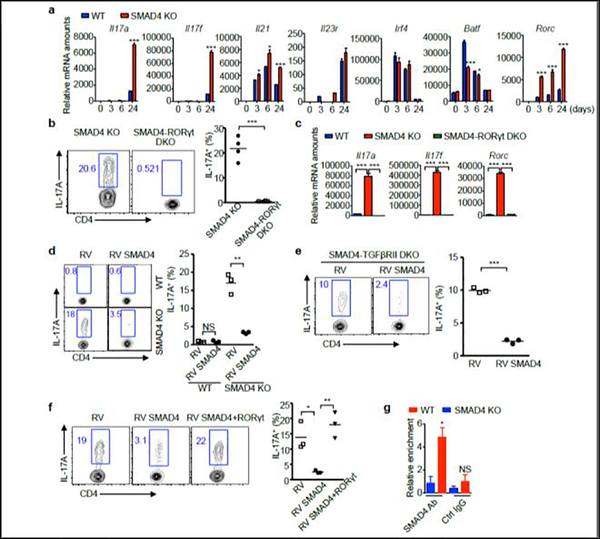
We next investigated how IL-21 induced TGF-β-independent Th17 program in SMAD4 deficient T cells. To reveal the primary effect of SMAD4 deficiency, we assessed Th17 related gene transcription within a short time range of cell culture. As early as three hours of IL-21 induction, the Rorc mRNA level was dramatically elevated, while other genes were either not detectable or showed no significant difference (Fig.3a). To validate whether the aberrant Rorc expression is responsible for the IL-21-induced TGF-β-independent Th17 cell differentiation, we generated CD4-cre;Smad4fl/fl;Rorc−/− (SMAD4-RORγt DKO) mice. Deletion of RORγt effectively abolished the IL-21-induced Th17 differentiation in SMAD4 deficient T cells, evidenced by flow cytometry and RT-qPCR (Fig.3b, 3c).
Figure 3. RORγt mediates IL-21-induced Th17 differentiation.
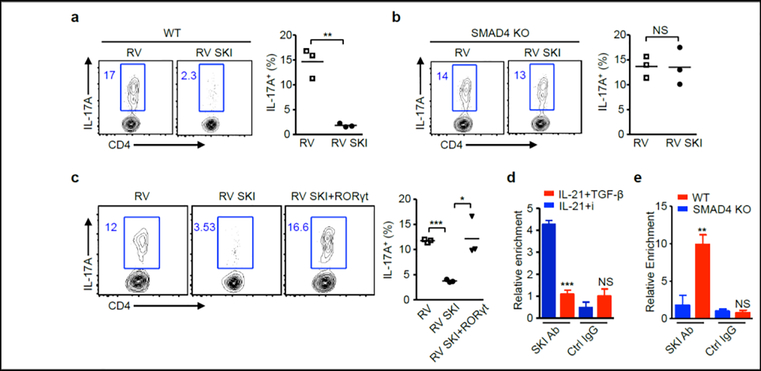
(a) Naïve CD4+ T cells isolated from wild-type (WT) and Cd4-cre;Smad4fl/fl (SMAD4 KO) mice were cultured with IL-21 and TGF-β receptor I inhibitor. After 3, 6, and 24 hours, the relative mRNA amounts of Th17-related genes were analyzed by qRT-PCR. (b) CD4+ T cells isolated from Cd4-cre;Smad4fl/fl (SMAD4 KO) and CD4-cre;Smad4fl/fl;Rorc−/− (SMAD4-RORγt DKO) mice were cultured with IL-21 and TGF-β receptor I inhibitor. After 4 days, IL-17A+ cells were analyzed by flow cytometry, (c) relative mRNA amounts of Rorc, Il17a and Il17f genes were analyzed by qRT-PCR. Representative results and statistical analysis are shown. (d) CD4+ T cells isolated from wild-type (WT) and Cd4-cre;Smad4fl/fl (SMAD4 KO) mice were cultured with IL-21 and TGF-β receptor I inhibitor, and then transduced with control retrovirus (RV) or SMAD4-expressing retrovirus (RV SMAD4). IL-17A+ cells were analyzed by flow cytometry. Representative results and statistical analysis are shown. (e) CD4+ T cells isolated from CD4-cre;Smad4fl/fl;Tgfbr2fl/fl (SMAD4-TGFβRII DKO) mice were cultured with IL-21, and then transduced with control retrovirus (RV) or SMAD4-expressing retrovirus (RV SMAD4). IL-17A+ cells were analyzed by flow cytometry. Representative results and statistical analysis are shown. (f) CD4+ T cells isolated from Cd4-cre;Smad4fl/fl (SMAD4 KO) mice were cultured with IL-21 and TGF-β receptor I inhibitor, and then transduced with control retrovirus (RV), SMAD4-expressing retrovirus (RV SMAD4), or SMAD4 and RORγt co-expressing retrovirus (RV SMAD4+RORγt). IL-17A+ cells were analyzed by flow cytometry. Representative results and statistical analysis are shown. (g) CD4+ T cells isolated from wild-type (WT) and Cd4-cre;Smad4fl/fl (SMAD4 KO) mice were cultured with IL-21 and TGF-β receptor I inhibitor, cells were harvested after 3 days. ChIP assay was performed with control IgG antibody or SMAD4 antibody. The relative enrichment of SMAD4 binding to the Rorc locus was determined.
Since SMAD4 deficiency causes IL-21-induced, TGF-β-independent Th17 differentiation, we examined whether overexpression of SMAD4 can reverse this phenomenon. Indeed, retrovirus-mediated SMAD4 expression substantially abolished IL-21-induced Th17 differentiation under the context of either TGF-β receptor I inhibition or TGF-β receptor II deletion (Fig. 3d, 3e). In addition, RORγt overrode SMAD4-mediated suppression (Fig.3f), suggesting SMAD4 targets Rorc gene transcription. Finally, the binding of SMAD4 at Rorc locus in IL-21 culture condition further explained the direct regulation of Rorc expression by SMAD4 (Fig.3g).
TGF-β signaling disrupted SMAD4-mediated suppression on IL-21-induced Th17 differentiation through SKI
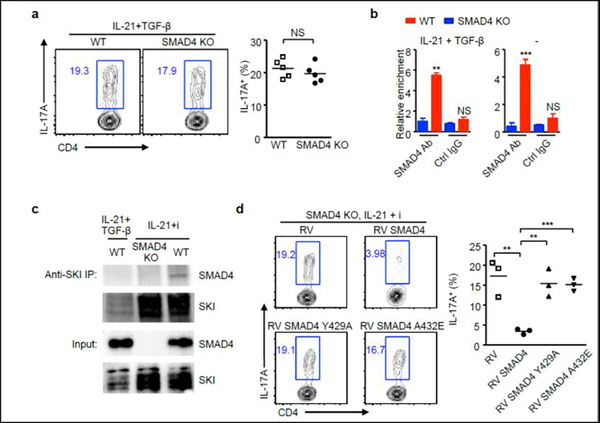
Next, we investigated whether SMAD4 could suppress IL-21-induced Th17 differentiation in the presence of TGF-β signaling. Interestingly, The presence of TGF-β signaling completely restored SMAD4-suppressed Th17 cells polarized with IL-21 (Fig.3d and Fig.4a). We then hypothesized that TGF-β signaling might be able to translocate SMAD4 off the proximal promoter region of Rorc. However, that was not the case. The ChIP results showed that SMAD4 bound to Rorc promoter in the presence of TGF-β signaling (Fig.4b). Moreover, the chromatin binding of SMAD4 at Rorc locus appeared to be cytokine independent, as the SMAD4 binding remains when IL-21 and TGF-β was absent (Fig.4b).
Figure 4. TGF-β disrupts SKI-SMAD4 interaction to facilitate IL-21-induced Th17 differentiation.
(a) CD4+ T cells isolated from wild-type (WT) and Cd4-cre;Smad4fl/fl (SMAD4 KO) mice were cultured with IL-21 and TGF-β. After 4 days, IL-17A+ cells were analyzed by flow cytometry. Representative results and statistical analysis are shown. (b) CD4+ T cells isolated from wild-type (WT) and Cd4-cre;Smad4fl/fl (SMAD4 KO) mice were cultured in the presence of absence of IL-21 plus TGF-β, cells were harvested after 3 days. ChIP assay was performed with control IgG antibody or SMAD4 antibody. The relative enrichment of SMAD4 binding to the Rorc locus was determined. (c) CD4+ T cells isolated from wild-type (WT) and Cd4-cre;Smad4fl/fl (SMAD4 KO) mice were cultured under indicated conditions, cells were harvested after 3 days.
Immunoprecipitation was performed with SKI antibody. Protein expression of SMAD4 and SKI was detected by immunoblotting. (d) CD4+ T cells isolated from Cd4-cre;Smad4fl/fl (SMAD4 KO) mice were cultured with IL-21 and TGF-β receptor I inhibitor (i), and then transduced with control retrovirus (RV), or retroviruses expressing wild-type form (RV SMAD4) or mutant form SMAD4 (RV SMAD4 Y429A or RV SMAD4 A432E). IL-17A+ cells were analyzed by flow cytometry. Representative results and statistical analysis are shown.
TGF-β signaling might disrupt the functional protein interaction of SMAD4 during IL-21-induced Th17 differentiation. We previously reported that protein SKI can interact with SMAD4 to regulate IL-6-induced Th17 differentiation, and degraded by TGF-β signaling [10]. We found SKI interacted with SMAD4 under IL-21 culturing condition and TGF-β substantially diminished SKI protein and its interaction with Smad4 (Fig.4c).
To assess whether the SKI interaction is critical in SMAD4-mediated suppression of IL-21-induced Th17 differentiation, we repeated SMAD4 suppression experiment with two previously characterized SMAD4 mutants that are unable to interact with SKI [10]. Under IL-21-induced Th17 polarizing condition in SMAD4 deficient T cells, unlikely WT, the SMAD4 mutants were ineffective to suppress Th17 differentiation (Fig.4d). Collectively, these results suggest that TGF-β signaling triggers the disruption of SKI to enable IL-21-induced Th17 differentiation.
SMAD4 was required for SKI mediated suppression of IL-21-induced Th17 differentiation
SKI antagonizes the TGF-β function in IL-6-induced Th17 differentiation [10]. However, it is unknown if exogenous SKI can suppress IL-21-induced Th17 differentiation in the presence of TGF-β. Interestingly, ectopic SKI expression substantially repressed IL-21-induced Th17 differentiation in WT (Fig.5a), but not in SMAD4 deficient T cells (Fig.5b). These results suggest SKI represses IL-21-induced Th17 differentiation through SMAD4-dependent mechanism. In addition, overexpression of RORγt overcame the SKI suppression on IL-21-induced Th17 differentiation, suggesting SKI controls RORγt expression (Fig.5c). ChIP results showed that SKI bound to Rorc locus (Fig.5d). As SKI does not contain predicted DNA binding domain, its DNA binding is likely mediated through transcription factors that it binds, e.g. SMAD4 (Fig.4c). Indeed, we found that although SKI bound to the Rorc locus in WT cells, it failed to do so in SMAD4 deficient T cells. These results demonstrate that SKI suppresses IL-21-induced Th17 differentiation by directly binding to Rorc through SMAD4.
Figure 5. SKI requires SMAD4 to modulate Rorc gene transcription.
(a) CD4+ T cells isolated from wild-type (WT) or (b) Cd4-cre;Smad4fl/fl (SMAD4 KO) mice were cultured with IL-21 and TGF-β, and then transduced with control retrovirus (RV) or SKI-expressing retrovirus (RV SKI). IL-17A+ cells were analyzed by flow cytometry. Representative results and statistical analysis are shown. (c) CD4+ T cells isolated from wild-type (WT) mice were cultured with IL-21 and TGF-β, and then transduced with control retrovirus (RV), SKI-expressing retrovirus (RV SKI), or SKI and RORγt co-expressing retrovirus (RV SKI+RORγt). IL-17A+ cells were analyzed by flow cytometry. Representative results and statistical analysis are shown. (d) CD4+ T cells isolated from wild-type (WT) mice were cultured under indicated conditions, cells were harvested after 3 days. ChIP assay was performed with control IgG antibody or SKI antibody. The relative enrichment of SKI binding to the Rorc locus was determined. (e) CD4+ T cells isolated from wild-type (WT) and Cd4-cre;Smad4fl/fl (SMAD4 KO) mice were cultured with IL-21 and TGF-β receptor I inhibitor, cells were harvested after 3 days. ChIP assay was performed with control IgG antibody or SKI antibody. The relative enrichment of SKI binding to the Rorc locus was determined.
Activin promotes IL-21-induced Th17 differentiation
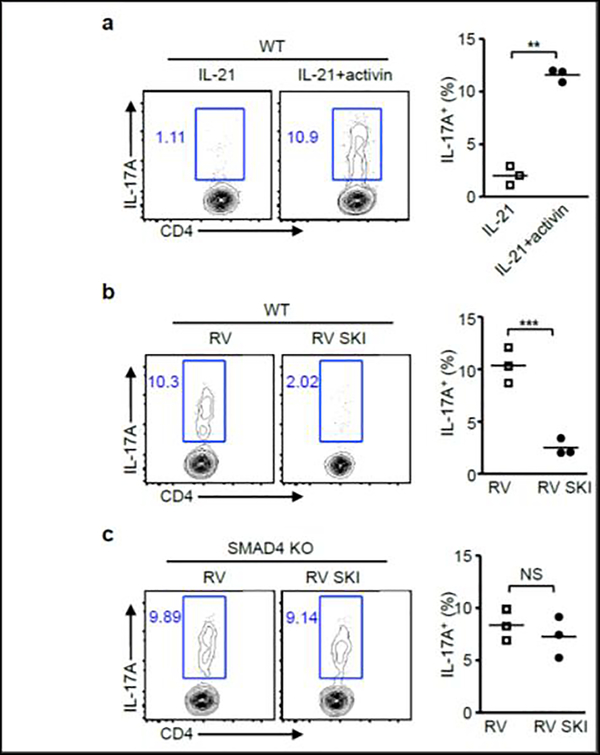
One of the most important roles for TGF-β signaling in IL-6-induced Th17 differentiation is to degrade SKI [10, 18]. As a TGF-β superfamily member, activin shares similar signaling with TGF-β, and is also capable of degrade SKI[10]. We then addressed whether activin could promote IL-21-induced Th17 differentiation. We found that WT T cells cultured with IL-21 plus activin differentiated into Th17 cells (Fig.6a). In addition, these Th17 cells can be effectively suppressed by SKI expression (Fig.6b). To explore the possible involvement of SMAD4 during this process, we performed same experiment with SMAD4 deficient T cells. SKI overexpression was unable to repress Th17 differentiation in SMAD4 deficient T cells, suggesting SMAD4 is required for SKI to regulate IL-21 and activin induced Th17 differentiation.
Figure 6. Activin cooperates with IL-21 to promote Th17 differentiation through SKI.
(a) CD4+ T cells isolated from wild-type (WT) mice were cultured with IL-21 and activin. After 4 days, IL-17A+ cells were analyzed by flow cytometry. Representative results and statistical analysis are shown. (b) CD4+ T cells isolated from wild-type (WT) or (c) Cd4-cre;Smad4fl/fl (SMAD4 KO) mice were cultured with IL-21 and activin, and then transduced with control retrovirus (RV) or SKI-expressing retrovirus (RV SKI). IL-17A+ cells were analyzed by flow cytometry. Representative results and statistical analysis are shown.
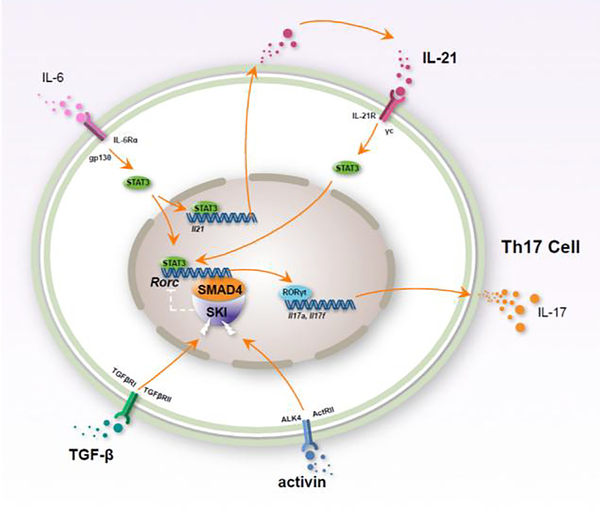
In summary, we uncovered an important mechanism for IL-21-induced Th17 differentiation and established a SKI/SMAD4 centered Th17 differentiation model (Fig.7).
Figure 7. Schematic diagram of IL-21-induced Th17 differentiation.
IL-21 and IL-6-induced autocrine IL-21 activates STAT3 to potentiate Rorc expression in T cell. Additional TGF-β or activin signal disrupts SKI and SMAD4 interaction, and reverses SKI/SMAD4 imposed repression on Rorc transcription. The expression of Th17 lineage transcription factor RORγt results in the production of Th17 cell signature cytokine IL-17.
DISCUSSION
Th17 cells play crucial roles in immune response [19, 20]. IL-6 has been recognized as a key cytokine inducing Th17 generation [6, 7, 21]. IL-21 is highly expressed in Th17 cells and is recognized as a self-promoting Th17 cytokine [11, 12, 14]. Interestingly, deletion of RORγt does not affect the IL-21 mRNA level during Th17 differentiation[22], which is consistent with the observation that IL-21 is induced during the initiation phase of Th17 cells. It is therefore plausible that, IL-21 induction might be independent of RORγt. IL-21 deficiency impairs IL-6-induced Th17 differentiation in vitro and in vivo, suggesting IL-21 is an essential inducer for Th17 generation rather than merely a Th17 signature cytokine [12, 13]. Moreover, IL-21[23], but not IL-6[24, 25], can induce the naïve human CD4+ T cells differentiate into Th17 cells. Uncovering the mechanism underlying IL-21-induced Th17 differentiation is therefore imperative for understanding the generation of Th17 cells. In this study, we identified SKI and SMAD4 complex as a key molecular switch controlling the IL-21-induced Th17 differentiation through the repression of RORγt expression. Given the fact that both IL-6 and IL-21 can induce the activation of STAT3 signaling[13, 26–29], it is currently not clear whether STAT3 is directly participated in the regulation mechanism of SKI and SMAD4.
IL-21 is a versatile cytokine under physiological conditions, it affects cell differentiation, proliferation, maturation, and apoptosis [15]. IL-21 signaling is important in many Th17 related inflammatory diseases model including type I diabetes[30]. Both IL-21−/− mice and IL-21R−/− 2D2 mice have less EAE [12, 31]. However, there are also studies showed that IL-21 signaling is not essential in some other cases of autoimmune diseases [32–34]. As a pleiotropic cytokine, IL-21 can induce IL-10 in Th17 cells [35], and support Th2 response [34]. Collectively, the function of IL-21 is not limited to inducing Th17 [16].
Different cellular sources of IL-6 and ways of its presentation result in varied IFN-γ expression in Th17 cells [36]. IL-17+IFN-γ+ T cells are pathogenic during inflammation and autoimmunity [37]. These Th17 cells co-express Th1 and Th17 lineage-specific transcription factors T-bet and RORγt [38]. The generation of pathogenic Th17 cells is context-dependent [39, 40]. IL-21-induced and IL-6-induced Th17 cells may be different in their pathogenicity. Therefore, comprehensive characterization for IL-21-induced Th17 cell function would be of interest.
Previous studies have demonstrated an important role of TGF-β signaling in Th17 differentiation [18]. Our recent works as well as this study recognize SMAD4 as a critical regulator that can regulate T cell functions independent of TGF-β signaling[10, 41]. These studies highlight SMAD4 in T cell mediated autoimmunity, anti-tumor response, anti-virus function[42] in a non-canonical TGF-β signaling manner. Under physiological conditions, other TGF-β superfamily cytokines may also regulate SKI and SMAD4 especially in local environment devoid of TGF-β. In this study, we identified activin exerted a similar function as TGF-β in promoting IL-21-induced Th17 differentiation through SKI and SMAD4. This observation agrees with a synergistic role for activin and TGF-β in promoting Treg cells. Therefore, TGF-β superfamily members may have overlapping, additive and/or synergistic function in immune regulation. What and how TGF-β superfamily members contribute to T cell regulation is therefore a question warrants further investigation to lay the foundation for drug development and effective treatment of inflammatory diseases [43, 44].
METHODS
Mice
All Smad4fl/fl, Tgfbr2fl/fl, Cd4-cre, and Rorc−/− mice were on C57BL/6 background. All mice were housed and bred in SPF (specific pathogen-free) animal facility at the University of North Carolina at Chapel Hill. All mouse related experiments were approved by the Institution Animal Care and Use Committee of the University of North Carolina.
Flow cytometry
Cells were re-stimulated for 4 hours with 50ng/ml PMA, 1μM ionomycin and 2μg/ml brefeldin A. Fluorescence-conjugated antibodies (Biolegend), anti-CD4 (GK1.5), anti-IFN-γ (XMG1.2), anti-Thy1.1 (OX-7), and anti-IL-17A (TC11–18H10.1) were used for flow cytometry analysis. All cells were analyzed on FACSCanto (BD Biosciences).
T cell culture, differentiation
Naïve CD4+ T cells were sorted from the spleens and peripheral lymph nodes of mice on day 0. Cells were then cultured with RPMI medium containing 10% FBS and 1% antibiotics in 24-well plates coated with 10 μg/ml anti-CD3 (145–2C11, BioXCell) and 10 μg/ml anti-CD28 (37.51, BioXCell). Cytokines for cells culture with the following concentration were used in the indicated conditions: 50 ng/ml recombinant IL-21 (Biolegend), 1 ng/ml TGFβ1 (R&D systems), 20 μg/ml anti-IFN-γ, 20 μg/ml anti-IL-4, and 100 ng/ml recombinant human activin (Biolegend). 10 μM TGF-β receptor I inhibitor SB525334 (Selleckchem) was used in indicated conditions. Cells were transduced with retrovirus on day 1 and analyzed by flow cytometry on day 4.
EAE
T-cell adoptive transfer EAE follows previously reported protocol [10], CD4+ T cells cultured with IL-21 and TGF-β receptor I inhibitor were transferred into irradiated wild-type recipient mice on day 0. Pertussis toxin (200ng) was intraperitoneally injected on day 0 and day 2. The EAE clinical scores were recorded based on the following criteria: 1, limp tail; 2, poor righting ability and/or partial hind-limb paralysis; 3, total hind-limb paralysis; 4, hind-limb paralysis + 75% of body paralysis; 5, moribund; 6, dead. Mice were closely monitored and euthanized. Spinal cords were stained with luxol fast blue for demyelination analysis. Spinal cord infiltrated T cells were isolated using standard Percoll gradients method and analyzed by flow cytometry following previously reported protocol [45].
Immunoprecipitation
Cells were lysed with protein extract buffer (20 mM HEPES, pH 7.9, with 1.5 mM MgCl2, 0.42 M NaCl, 0.2 mM EDTA, 25% (v/v) glycerol, and 1% Triton X-100) in the presence of protease inhibitor, diluted with dilution buffer (20 mM HEPES, pH 7.9, with 1.5 mM MgCl2, 0.2 mM EDTA, 10 mM KCl, and 25% (v/v) glycerol), and then processed with Bioruptor. Protein supernatant was collected and incubated with SKI antibody conjugated with magnetic beads at 4°C overnight. The immunocomplex was washed with washing buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% NP40 and 150 mM NaCl), and eluted with Laemmli sample buffer. Proteins were analyzed by immunoblotting with the following antibodies: anti-SKI (H-329, Santa Cruz), and anti-SMAD4 (D3M6U, CST).
ChIP
Cells were cross-linked with 1% formaldehyde and processed by following the protocol of Upstate Biotechnology. Anti-IgG (sc-2027, Santa Cruz), anti-SMAD4 (EP618Y, Abcam), and anti-SKI (H-329, Santa Cruz) were used for immunoprecipitation. Quantitative RT-PCR was performed to detect the binding at Rorc promoter region. The primers sequences were: GGGGAGAGCTTTGTGCAGAT and AGTAGGGTAGCCCAGGACAG.
Statistics
Statistical analysis was performed by two-tailed unpaired Student’s t-test. P values were indicated with *, *P < 0.05, **P < 0.01, and ***P < 0.001. NS indicates not significant. Mean ± SD was used unless indicated otherwise. Results from biologically independent experiments were used for statistics.
Highlights.
SMAD4 deficiency led to IL-21-induced Th17 differentiation in the absence of TGF-β signaling
IL-21-induced TGF-β-independent Th17 cells are encephalitogenic
SMAD4 repressed RORγt expression during IL-21-induced Th17 differentiation
TGF-β signaling disrupted SMAD4-mediated suppression on IL-21-induced Th17 differentiation through SKI
SMAD4 was required for SKI mediated suppression of IL-21-induced Th17 differentiation
Activin promotes IL-21-induced Th17 differentiation
ACKNOWLEDGEMENT
We thank E. Robertson and E. Bikoff for Smad4fl/fl mice, H. Moses for Tgfbr2fl/fl mice, D. Littman for Rorc−/− mice. This study was supported by NIH/NIAID (AI097392; AI123193), National Multiple Sclerosis Society (RG-1802-30483) and Yang Family Biomedical Scholars Award (Y.Y.W).
Footnotes
DECLARATION OF INTEREST
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C, A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17, Nature immunology, 6 (2005) 1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT, Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages, Nature immunology, 6 (2005) 1123–1132. [DOI] [PubMed] [Google Scholar]
- [3].Singh RP, Hasan S, Sharma S, Nagra S, Yamaguchi DT, Wong D, Bh H, Hossain A, Th17 cells in inflammation and autoimmunity, Autoimmunity reviews, DOI 10.1016/j.autrev.2014.08.019(2014). [DOI] [PubMed] [Google Scholar]
- [4].Miossec P, Korn T, Kuchroo VK, Interleukin-17 and type 17 helper T cells, The New England journal of medicine, 361 (2009) 888–898. [DOI] [PubMed] [Google Scholar]
- [5].Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR, The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells, Cell, 126 (2006) 1121–1133. [DOI] [PubMed] [Google Scholar]
- [6].Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK, Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells, Nature, 441 (2006) 235–238. [DOI] [PubMed] [Google Scholar]
- [7].Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B, TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells, Immunity, 24 (2006) 179–189. [DOI] [PubMed] [Google Scholar]
- [8].Van Snick J, Interleukin-6: an overview, Annual review of immunology, 8 (1990) 253–278. [DOI] [PubMed] [Google Scholar]
- [9].Tanaka T, Narazaki M, Kishimoto T, IL-6 in inflammation, immunity, and disease, Cold Spring Harbor perspectives in biology, 6 (2014) a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang S, Takaku M, Zou L, Gu AD, Chou WC, Zhang G, Wu B, Kong Q, Thomas SY, Serody JS, Chen X, Xu X, Wade PA, Cook DN, Ting JPY, Wan YY, Reversing SKI-SMAD4-mediated suppression is essential for TH17 cell differentiation, Nature, 551 (2017) 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Korn T, Bettelli E, Gao W, Awasthi A, Jager A, Strom TB, Oukka M, Kuchroo VK, IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells, Nature, 448 (2007) 484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, Dong C, Essential autocrine regulation by IL-21 in the generation of inflammatory T cells, Nature, 448 (2007) 480–483. [DOI] [PubMed] [Google Scholar]
- [13].Wei L, Laurence A, Elias KM, O’Shea JJ, IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner, The Journal of biological chemistry, 282 (2007) 34605–34610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR, IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways, Nature immunology, 8 (2007) 967–974. [DOI] [PubMed] [Google Scholar]
- [15].Leonard WJ, Wan CK, IL-21 Signaling in Immunity, F1000Research, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tian Y, Zajac AJ, IL-21 and T Cell Differentiation: Consider the Context, Trends in immunology, 37 (2016) 557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mehta DS, Wurster AL, Grusby MJ, Biology of IL-21 and the IL-21 receptor, Immunological reviews, 202 (2004) 84–95. [DOI] [PubMed] [Google Scholar]
- [18].Zhang S, The role of transforming growth factor beta in T helper 17 differentiation, Immunology, 155 (2018) 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tesmer LA, Lundy SK, Sarkar S, Fox DA, Th17 cells in human disease, Immunological reviews, 223 (2008) 87–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Stockinger B, Omenetti S, The dichotomous nature of T helper 17 cells, Nature reviews. Immunology, 17 (2017) 535–544. [DOI] [PubMed] [Google Scholar]
- [21].Korn T, Bettelli E, Oukka M, Kuchroo VK, IL-17 and Th17 Cells, Annual review of immunology, 27 (2009) 485–517. [DOI] [PubMed] [Google Scholar]
- [22].Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, Watowich SS, Tian Q, Jetten AM, Dong C, T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma, Immunity, 28 (2008) 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yang L, Anderson DE, Baecher-Allan C, Hastings WD, Bettelli E, Oukka M, Kuchroo VK, Hafler DA, IL-21 and TGF-beta are required for differentiation of human T(H)17 cells, Nature, 454 (2008) 350–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, Basham B, Smith K, Chen T, Morel F, Lecron JC, Kastelein RA, Cua DJ, McClanahan TK, Bowman EP, de Waal Malefyt R, Development, cytokine profile and function of human interleukin 17-producing helper T cells, Nature immunology, 8 (2007) 950–957. [DOI] [PubMed] [Google Scholar]
- [25].Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, Sallusto F, Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells, Nature immunology, 8 (2007) 942–949. [DOI] [PubMed] [Google Scholar]
- [26].Harris TJ, Grosso JF, Yen HR, Xin H, Kortylewski M, Albesiano E, Hipkiss EL, Getnet D, Goldberg MV, Maris CH, Housseau F, Yu H, Pardoll DM, Drake CG, Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity, Journal of immunology, 179 (2007) 4313–4317. [DOI] [PubMed] [Google Scholar]
- [27].Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, Shevach EM, O’Shea J J, Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation, Immunity, 26 (2007) 371–381. [DOI] [PubMed] [Google Scholar]
- [28].Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C, STAT3 regulates cytokine-mediated generation of inflammatory helper T cells, The Journal of biological chemistry, 282 (2007) 9358–9363. [DOI] [PubMed] [Google Scholar]
- [29].Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, Takahashi H, Sun HW, Kanno Y, Powrie F, O’Shea JJ, Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis, Immunity, 32 (2010) 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Spolski R, Kashyap M, Robinson C, Yu Z, Leonard WJ, IL-21 signaling is critical for the development of type I diabetes in the NOD mouse, Proceedings of the National Academy of Sciences of the United States of America, 105 (2008) 14028–14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lee Y, Mitsdoerffer M, Xiao S, Gu G, Sobel RA, Kuchroo VK, IL-21R signaling is critical for induction of spontaneous experimental autoimmune encephalomyelitis, The Journal of clinical investigation, 125 (2015) 4011–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Coquet JM, Chakravarti S, Smyth MJ, Godfrey DI, Cutting edge: IL-21 is not essential for Th17 differentiation or experimental autoimmune encephalomyelitis, Journal of immunology, 180 (2008) 7097–7101. [DOI] [PubMed] [Google Scholar]
- [33].Sonderegger I, Kisielow J, Meier R, King C, Kopf M, IL-21 and IL-21R are not required for development of Th17 cells and autoimmunity in vivo, European journal of immunology, 38 (2008) 1833–1838. [DOI] [PubMed] [Google Scholar]
- [34].Frohlich A, Marsland BJ, Sonderegger I, Kurrer M, Hodge MR, Harris NL, Kopf M, IL-21 receptor signaling is integral to the development of Th2 effector responses in vivo, Blood, 109 (2007) 2023–2031. [DOI] [PubMed] [Google Scholar]
- [35].Spolski R, Kim HP, Zhu W, Levy DE, Leonard WJ, IL-21 mediates suppressive effects via its induction of IL-10, Journal of immunology, 182 (2009) 2859–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Heink S, Yogev N, Garbers C, Herwerth M, Aly L, Gasperi C, Husterer V, Croxford AL, Moller-Hackbarth K, Bartsch HS, Sotlar K, Krebs S, Regen T, Blum H, Hemmer B, Misgeld T, Wunderlich TF, Hidalgo J, Oukka M, Rose-John S, Schmidt-Supprian M, Waisman A, Korn T, Trans-presentation of IL-6 by dendritic cells is required for the priming of pathogenic TH17 cells, Nature immunology, 18 (2017) 74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kamali AN, Noorbakhsh SM, Hamedifar H, Jadidi-Niaragh F, Yazdani R, Bautista JM, Azizi G, A role for Th1-like Th17 cells in the pathogenesis of inflammatory and autoimmune disorders, Molecular immunology, 105 (2019) 107–115. [DOI] [PubMed] [Google Scholar]
- [38].Yang Y, Weiner J, Liu Y, Smith AJ, Huss DJ, Winger R, Peng H, Cravens PD, Racke MK, Lovett-Racke AE, T-bet is essential for encephalitogenicity of both Th1 and Th17 cells, The Journal of experimental medicine, 206 (2009) 1549–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ahern PP, Schiering C, Buonocore S, McGeachy MJ, Cua DJ, Maloy KJ, Powrie F, Interleukin-23 drives intestinal inflammation through direct activity on T cells, Immunity, 33 (2010) 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wa YY, O’Connor W Jr., Rongvaux A, Van Rooijen N, Haberman AM, Iwakura Y, Kuchroo VK, Kolls JK, Bluestone JA, Herold KC, Flavell RA, Control of TH17 cells occurs in the small intestine, Nature, 475 (2011) 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gu AD, Zhang S, Wang Y, Xiong H, Curtis TA, Wan YY, A critical role for transcription factor Smad4 in T cell function that is independent of transforming growth factor beta receptor signaling, Immunity, 42 (2015) 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lewis GM, Wehrens EJ, Labarta-Bajo L, Streeck H, Zuniga EI, TGF-beta receptor maintains CD4 T helper cell identity during chronic viral infections, The Journal of clinical investigation, 126 (2016) 3799–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yang J, Sundrud MS, Skepner J, Yamagata T, Targeting Th17 cells in autoimmune diseases, Trends in pharmacological sciences, 35 (2014) 493–500. [DOI] [PubMed] [Google Scholar]
- [44].Patel DD, Kuchroo VK, Th17 Cell Pathway in Human Immunity: Lessons from Genetics and Therapeutic Interventions, Immunity, 43 (2015) 1040–1051. [DOI] [PubMed] [Google Scholar]
- [45].Pino PA, Cardona AE, Isolation of brain and spinal cord mononuclear cells using percoll gradients, Journal of visualized experiments : JoVE, DOI 10.3791/2348(2011). [DOI] [PMC free article] [PubMed] [Google Scholar]