Abstract
Background:
Neurocognitive dysfunction remains prevalent among people living with HIV (PLWH), even after viral suppression on combination antiretroviral therapy (cART). We investigated associations between neuropsychological performance (NP) and patterns of circulating exosomal microRNA (exo-miRNA) expression in PLWH on cART.
Setting:
A cross-sectional examination of plasma exo-miRNA among PLWH on cART with systemic viral suppression and volunteers without HIV infection.
Methods:
31 PLWH who started cART during early infection (n=19) or chronic infection (n=12) participated in phlebotomy and an 11-test neuropsychological battery after >1 year on treatment. NP higher- or lower-performing participants were categorized based on normalized neuropsychological scores. Total RNA was extracted from purified exosomes of 31 PLWH and 5 volunteers without HIV and subject to small RNA sequencing. Differential expression of exo-miRNAs was examined and biological functions were predicted.
Results:
Eleven exo-miRNAs were up-regulated in NP lower-performing (n=18) relative to higher-performing PLWH (n=13). A high proportion of the differentiating exo-miRNA target the axon guidance KEGG pathway and neurotrophin tyrosine receptor kinase signaling Gene Ontology pathway. Differential expression analysis of exo-miRNAs between NP lower- (n=7) and higher-performing (n=12) PLWH within the early infection group alone confirmed largely consistent findings.
Conclusions:
Plasma exo-miRNA content differed between NP higher- and lower-performing PLWH. Several differentially expressed exo-miRNAs were predicted to be involved in inflammation and neurodegeneration pathways. Exo-miRNA in plasma may indicate cross-talk between the circulation and central nervous system and thus may be clinically relevant for neurocognitive dysfunction in PLWH.
Keywords: Exosomes, HIV-associated neurocognitive dysfunction, microRNA, axon guidance, inflammation
Introduction
Widespread use of combined anti-retroviral therapy (cART) has greatly reduced central nervous system (CNS)-related morbidity associated with HIV-1 infection; nonetheless, HIV-associated neurocognitive disorder (HAND) remains common, affecting an estimated 20–50% of people living with HIV (PLWH) and 19% of PLWH with well-suppressed HIV on cART [1, 2]. HAND represents a constellation of cognitive, motor, and behavioral symptoms with a wide spectrum of severity [3]. Detection of cerebrospinal fluid (CSF) HIV RNA and elevated markers of inflammation in the blood and CSF during suppressive cART suggests that persistent reservoirs of HIV in the CNS as well as inflammatory cross-talk between the two compartments may be implicated in clinically-relevant neuronal injury [4-6].
Exosomes have emerged as potential mediators of long-term immune activation and CNS perturbation during HIV infection [7]. Exosomes are cell-secreted lipid bilayer membrane microvesicles ranging from 30–150 nm in size that are released via exocytosis into a variety of body fluids, including the blood and CSF [8, 9]. Exosomes contain donor-cell derived lipids, proteins, messenger RNAs, and microRNAs that affect the functioning of target cells in the immediate microenvironment or at distant sites. Exosomal microRNAs (exo-miRNAs) regulate gene expression by post-transcriptionally controlling the translation and stability of their mRNA targets in recipient cells, thereby impacting cell proliferation, apoptosis and differentiation [10, 11].
Pro-inflammatory microRNAs, encapsulated in exosomes, may traffic between the CNS and the peripheral circulation, with systemically derived exosomes contributing to neurological sequelae in people living with chronic HIV infection and/or CNS-derived exosomes in the plasma reflecting the state of ongoing CNS processes. CNS inflammation promotes the ability of exosomes to cross the blood-brain barrier, facilitating the transport of pathologic or pro-inflammatory molecules between the blood and CSF [12]. Prior studies have implicated exosomal signaling in CNS pathologies. For example, pro-inflammatory exo-miRNAs have been shown to modulate microglial-mediated immune responses in neurodegenerative diseases including Parkinson and Alzheimer’s diseases [13, 14] and can reflect disease status in multiple sclerosis [15]. In studies of HIV, exosomes released from HIV-1 infected cells have been shown to induce quiescent CD4+ lymphocytes to produce HIV-1 as well as promote the release of pro-inflammatory cytokines from target monocyte-derived macrophages [16, 17].
Given that microRNAs are enriched in exosomes, in this study, we explore the association between exo-miRNAs and neurocognitive dysfunction during chronic HIV infection in people who have obtained viral suppression on cART. We hypothesized that there is a link between exosomal signaling patterns and neuropsychological testing (NP) performance as a clinical index of the effects of systemic inflammation on the brain as a target organ. This investigation of patterns of plasma exo-miRNAs in the pathogenesis of neurocognitive dysfunction during treated suppressed infection provides new potential targets for intervention for affected individuals.
Materials and Methods
Study participants.
Study participants included: participants from the Primary Infection CNS Events Study (PISCES) (University of California, San Francisco) enrolled during primary HIV infection (PHI), defined as within 12 months of HIV acquisition (n=19), participants from the HIV Associated Reservoirs and Comorbidities (HARC) study (Yale University) who initiated cART during chronic HIV infection (n=12), and HIV-uninfected participants without clinical diagnoses of neurologic disease who were recruited from the community (5 men, median age = 49 years, IQR = 48 – 53 years). For participants with HIV (n=31), plasma, bloodwork, lumbar puncture, and NP testing were performed after at least 1 year of cART initiation and systemic viral suppression (plasma viral load ≤ 40 copies/ml) were documented.
Neuropsychological testing.
For each PLWH, a 1.5-hour NP battery was performed. Participants were assessed across 5 common domains of neurocognitive functioning: motor (Timed Gait, Grooved Pegboard), executive function (Trail-Making A-B, Controlled Oral Word Association), processing speed (Digit Symbol, Stroop Tests), memory (Figure Delay), and learning (Hopkins Verbal Learning Test, Rey Auditory Verbal Learning) [18, 19]. Total-z scores were derived by averaging individual test z-scores across domains and externally normalizing individual raw test scores for age, gender, ethnicity and years of education to the general population [20-27]. Executive function-z scores were derived similarly, using only executive function domain tests. Study participants were divided into two groups, higher- and lower- performing, based on normalized total-z or executive function-z score of >0 and <0, respectively. Other studies have employed similar methods of studying participants by NP performance [28, 29].
Exosome isolation from plasma.
The isolation of exosomes was performed using a solution of 20% polyethylene glycol (PEG, Mn 6,000, Sigma-Aldrich, St. Louis, MO) as previously reported [30-32]. Briefly, the mixture of plasma and the solution (500 μl solution/500 μl plasma) was incubated at 4°C for 2 hours, followed by centrifugation at 13,000 rpm for 2 min to obtain the exosome pellet with two washes of 1x Dulbecco’s phosphate buffered saline (DPBS, Sigma-Aldrich).
Confirmation of exosomes.
Transmission electron microscopy (TEM) was performed to validate exosome morphology. DPBS-suspended exosomes were deposited on formvar carbon-coated EM grids. After negative staining with 2% uranyl acetate (pH 4), the grids were examined and imaged with a FEI TECNAI F20 FEG microscope running at 200 kV of accelerating voltage, the digital images were recorded with a FEI Eagle CCD camera (4k x 4k). Images of 100 representative vesicles were measured with ImageJ (https://imagej.nih.gov). Nanoparticle Tracking Analysis (NTA) was performed to measure exosomal size and concentration using a Nanosight LM10 instrument equipped with a 405 nm laser (NanoSight, Malvern Instruments) at 21°C. The Brownian movement of particles was tracked by NTA software (version 3.1, NanoSight). Quantitative ELISA assay was performed to measure exosome marker CD63 [33] using an ExoELISA-Ultra CD63 kit (System Biosciences Inc., cat. EXEL-ULTRA-CD63–1) following the manufacturer’s protocol [34]. The total protein concentration of each exosome suspension was measured using NanoDrop_1000 spectrophotometer with a wavelength 280 nm. DPBS exosome suspensions (normalized to 100 μg of protein) were plated and run in duplicate. The absorbance of exosomal CD63 was determined using a Biotek spectrophotometric 96-well microplate reader with a wavelength of 450 nm.
RNA preparation and library construction.
Total RNA was extracted from purified exosomes using SeraMir Exosome RNA purification kit (System Biosciences inc, cat. RA806A-1), and the quality and concentration of RNA were determined by Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA). Small RNA libraries were prepared using a NEBNext® multiplex small RNA library prep set for Illumina kit (New England BioLabs INC., Ipswich, MA) following the manufacturer’s instructions.
Next-generation small RNA-sequencing and analysis.
Single-end deep sequencing was performed on all cDNA libraries with a 75-nucleotide (nt) read length using HiSeq-2500 Genome Analyzer (Illumina, San Diego, CA). Adaptors and low quality regions were trimmed from raw sequences using btrim with options “−3 -P -l 15” [35]. The trimmed sequences were mapped to human genome (hg38) with BWA [36]. For microRNA annotation, miRBase v21 was used [37]. Differential gene expression analysis was performed using R package “DESeq2” [38]. MicroRNAs with base mean expression (BME) < 10 were excluded, and differential expression was defined as absolute log2(fold-change) > 1.0.
Functional analysis of predicted mRNA targets of differentially expressed exo-miRNAs.
To examine functional annotations of the mRNAs that the differentially expressed exo-miRNAs target, KEGG pathway and Gene Ontology (GO) analysis were performed using the DiANA tool of mirPath v.3 [39].
Measurement of BDNF in plasma.
The concentration of brain-derived neurotrophic factor (BDNF) was measured as a read-out of exo-miRNA activity on axonal modeling pathways. The concentration of free BDNF in plasma was determined using a commercial ELISA assay (R&D Systems, Inc., Minneapolis, MN,) following the manufacturer’s protocol. Correlation coefficients were generated between BDNF and both miR-30a-5p and miR-206 levels, two reported inhibitors of BDNF translation [40, 41].
Statistical analysis.
Fisher’s exact and Wilcoxon Rank Sum tests were employed to compare clinical characteristics. To test for the ability of exo-miRNA expression to distinguish participants’ NP status, principal component analysis (PCA) and receiver operating characteristic (ROC) analysis were performed. ROC analysis was performed in SAS v9.4. All other statistical analyses were performed in R v3.5.0.
Results
Participant characteristics
Characteristics for all study participants are summarized in Table 1. Of the 19 participants enrolled during PHI, the median time between estimated date of infection and treatment initiation was 0.6 years (IQR 0.2 – 1.8). Patients enrolled during PHI had median age 43 years (IQR 35.5–48), while patients enrolled during chronic infection had median age 58.5 years (IQR 51.75–62.5, p=0.0001). Based on total-z scores on NP testing, 13 participants with HIV were sorted into the NP higher-performing group (NP-higher, median total-z=0.3) and 18 into the NP lower-performing group (NP-lower, median total-z= −0.7). Based on executive function-z scores, 14 participants with HIV were sorted into executive function higher-performing (median z-score=0.44, 64% enrolled during PHI) and 16 into executive function lower-performing (median z-score= −0.49, 56% enrolled during PHI). Seven patients enrolled during PHI had incomplete learning and memory NP data, 1 patient had incomplete executive function data, and 2 patients enrolled during chronic infection had incomplete motor NP data; missing domains were not included in the total-z score calculations for these patients. Participants enrolled during PHI were more prevalent in the NP-higher group, while most of the participants enrolled during chronic infection were in the NP-lower group (p = 6.5 × 10−6). NP-lower participants had an overall longer duration of cART treatment (p=0.035), fewer years of education (p=6.0×10−4) and lower CSF protein level (p = 0.009) relative to NP-higher.
Table 1.
Participant Characteristics, All Participants and PHI Participants Alone.
| NP Higher- Performing Among All Participants |
NP Lower- Performing Among All Participants |
p-value | NP Higher- Performing Among PHI Participants |
NP Lower- Performing Among PHI Participants |
p-value | |
|---|---|---|---|---|---|---|
| N, participants | 13 | 18 | 12 | 7 | ||
| Clinical Characteristics | ||||||
| Proportion from primary infection study, UCSF (%) | 12/13 (92.3%) |
7/18 (38.9%) |
6.5×10−16a | 12/12 (100%) |
7/7 (100%) |
|
| Proportion from chronic infection study, Yale (%) | 1/13 (7.7%) |
11/18 (61.1%) |
||||
| Total-z score | 0.3 (0.2 - 0.8) |
−0.7 (−1.7 - −0.5) |
9.7×10−9b | 0.4 (0.04 - 0.7) |
−0.5 (−0.7 - −0.2) |
4.0×10−5 b |
| Time on cART, years | 1.2 (1.1 - 1.9) |
6.3 (1.1 - 21.2) |
0.035 b | 1.2 (1.1 - 1.8) |
1.1 (1.1 - 1.6) |
0.711 b |
| Duration pre-ART infection, years | Unavailable | Unavailable | 0.5 (0.3 - 1.9) |
0.7 (0.2 - 1.8) |
0.773 b | |
| Age, years | 45 (42 - 48) |
51 (44 - 61) |
0.118 b | 45 (41 - 48) | 36 (35 - 46) | 0.400 b |
| Male, number (%) | 13/13 (100%) |
17/18 (94%) |
12/12 (100%) |
7/7 (100%) |
||
| Education, years | 16 (15 - 18) |
13 (12 - 15) |
6.0×10−4b | 16 (15 - 18) |
16 (14 - 16) |
0.273 b |
| Bloodwork | ||||||
| CD4 count, cells/μL | 641 (371 - 780) |
595 (432 - 858) |
0.498 b | 592 (368.3 - 729.5) |
676 (572 - 898) |
0.167 b |
| Blood neopterin, nmol/L | 6.12 (4.9 - 8.7) |
6.43 (5.2 - 7.7) |
0.983 b | 5.7 (4.9 - 8.8) |
6.6 (4.6 - 7.7) |
0.586 b |
| Log10 plasma viral load, copies/μL | All participants had log10 plasma viral load at the lower limit of detection of each institution | |||||
| Lumbar Puncture | ||||||
| CSF WBC, cells/μL | 2 (1 - 3) |
1 (0 - 2) |
0.09 b | 2 (1 - 3) |
1 (0 - 2) |
0.258 b |
| CSF protein, mg/dL | 39.5 (35.5 - 48.5) |
29.5 (24.3 - 36.8) |
0.009 b | 39.5 (35.5 - 48.5) |
33.0 (29.5 - 39.5) |
0.219 b |
= Fisher’s exact test;
= Wilcoxon Rank Sum test, medians (IQR) displayed
Abbreviations: NP = neuropsychological, cART = combined anti-retroviral therapy, CSF = cerebrospinal fluid, WBC = white blood cells
Characterization of circulating exosomes in plasma
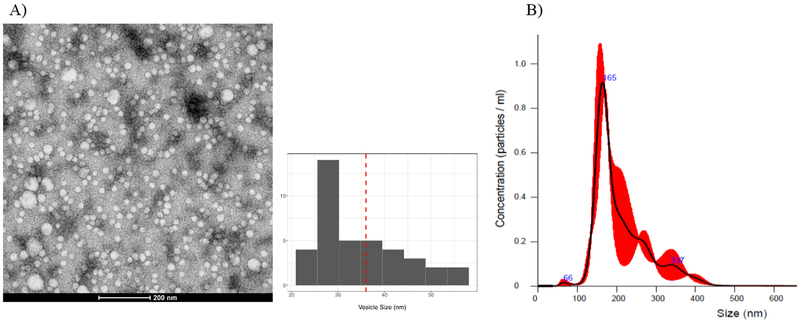
TEM images showed that the morphology of PEG-purified exosomes was vesicular with diameters ranging from 20–70 nm (Figure 1A). NTA, which is based on the principle of Brownian motion of particles in liquid, demonstrated that the majority of purified vesicles ranged 110–210 nm in diameter and were approximately 1010-1011 particles per mL of plasma (Figure 1B). The smaller sizes of exosomes measured under TEM vs. NTA may be due to sample preparation and dehydration for TEM imaging as well as lower sensitivity of NTA to detect vesicles in the 20–60nm range [42, 43]. ELISA assay confirmed that all purified exosomes were CD63-positive and revealed no significant difference in exosome abundance, quantified by CD63 ELISA signal, between the NP-higher (median=1.56×1010) versus NP-lower groups (median=1.73×1010, p=0.569) or between cohorts enrolled during PHI (median=1.55×1010) versus chronic infection (median=1.78×1010, p=0.371).
Figure 1. Characterization of plasma exosomes.
A) Representative Transmission Electron Microscopy (TEM) image of purified exosomes from plasma using PEG-based solution; scale bar: 200 nm. The histogram represents the distribution of vesicle sizes in the TEM image, with an average size of 36.7nm (dashed red line). (B). Representative NanoSight Tracking Analysis (NTA) graph showing the size (x-axis) and concentration (y-axis) of diluted exosomes (1:1000) in 1x PBS solution. Red shading indicates +/− 1 standard error of the mean.
Differential exo-miRNA expression between neuropsychological higher- and lower- performance groups
Expression levels of exo-miRNAs identified by NGS were compared between NP-higher and NP-lower groups (Supplementary Table 1). PCA performed on the exo-miRNA expression data demonstrated that NP-higher and NP-lower separated into distinct groups, particularly along the 2nd component (Figure 2A). After correcting for multiple comparisons using False Discovery Rate (FDR<0.1), we found 11 exo-miRNAs were significantly differentially expressed between the groups (miR-206, miR-193b-5p, miR-193a-5p, miR-30a-5p, miR-216b-3p, miR-499a-5p, miR-499b-3p, miR-708–3p, miR-1183, miR-375 and miR-483–5p). All 11 differentially expressed exo-miRNAs were upregulated in NP-lower relative to NP-higher (Figure 2B). Exo-miRNA expression was also compared between PLWH with higher- versus lower-executive function performance. Three exo-miRNA (miR-216b-3p, miR-148a-3p, and miR-504–5p) were significantly upregulated in participants with lower executive function performance relative to higher performance by p<0.05.
Figure 2. Distinct exo-miRNA expression patterns between neuropsychological lower- and higher-performing groups.
A) Principle component analysis (PCA) plot based on small RNA-seq data, where the red and blue points represent participants with higher and lower neuropsychological (NP) performance, respectively. (B) Volcano plot of differential micoRNA expression between lower versus higher NP performance groups. The black points indicate exo-miRNA with differential expression FDR <0.1, and the grey points represent no significant difference between the groups. X-axis represents log2(fold-change) of exo-miRNA (lower vs. higher NP performance), and y-axis represents the –log10(p-value) of differences in exo-miRNA expression (lower vs. higher NP performance). (C) Predictive performance (ROC analysis) of the differentially expressed exo-miRNAs in distinguishing lower from higher NP performance.
To evaluate the ability of differentially expressed circulating exo-miRNAs to distinguish NP performance groups, we conducted receiver operating characteristic (ROC) analyses with the 11 identified exo-miRNAs. The 11 differentially expressed exo-miRNAs between NP-higher and NP-lower showed high area under the curve (AUC) with a value of 0.93 (95% CI: 0.83 −1.00) in distinguishing the performance groups (Figure 2C).
Biological role of differentially expressed exo-miRNAs
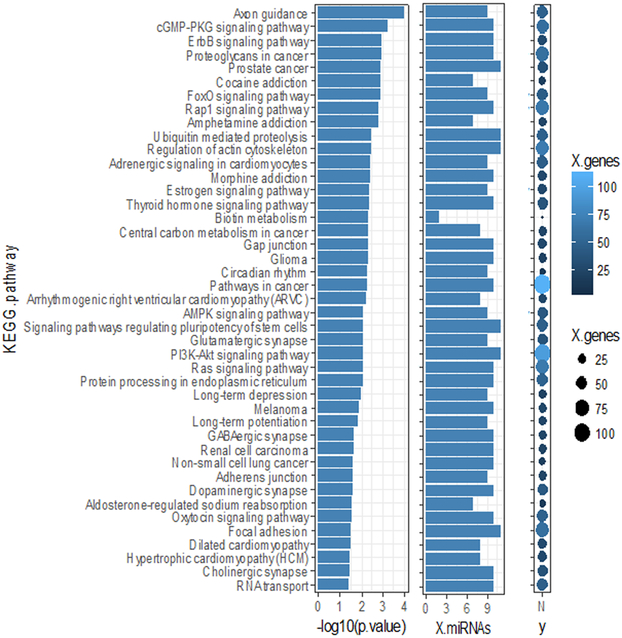
To better understand the biological relevance of the differentially expressed microRNAs between NP performance groups, we performed canonical KEGG pathway and GO enrichment analyses using the 11 differentially expressed exo-miRNAs. Axon guidance ranked highest of the enriched KEGG pathways (Figure 3), with 9 of the 11 identified exo-miRNAs targeting genes in this pathway.
Figure 3. KEGG pathway analysis for differentially expressed exo-miRNAs between neuropsychological lower- and higher-performing groups.
Predicted KEGG pathways enriched with the 11 significantly differentially expressed exo-miRNAs (lower- vs. higher-neuropsychological (NP) performing, FDR<0.1). −1og10(p.value) indicates the statistical significance that the pathway is enriched with gene targets of the microRNAs, X.mRNA is the number of exo-miRNAs targeting genes in the enriched pathway, and X.genes is the number of genes in the pathway targeted by the differentially expressed microRNAs, represented by the color and size of the dot
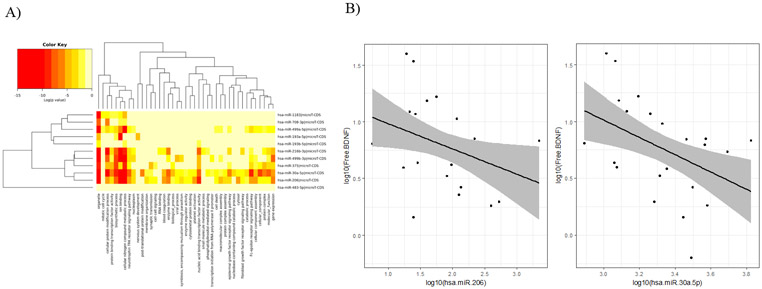
GO analysis demonstrated that genes involved in organelle function, ion binding, cellular nitrogen compound metabolic process, cellular protein modification, biosynthesis, and neurotrophin tyrosine receptor kinase (TRK) signaling were predicted targets of the 11 differentially expressed exo-miRNAs (Figure 4A). Two identified exo-miRNAs, miR-30a-5p and miR-206, are known inhibitors of BDNF translation. Measurement of BDNF in the 31 plasma samples revealed a significant negative correlation between both exosomal miR-206 and miR-30a-5p expression and BDNF levels (Figure 4B). Pearson correlation coefficients were −0.50 (95% CI: [−0.72, −0.16]; p value=0.004) and −0.47 (95% CI: [−0.91, −0.13]; p value=0.008), respectively. There was no difference in BDNF levels between the NP-higher (7.64, range 1.43–15.4) and NP-lower (5.57, range 0.64–39.0) groups (p=0.514). There was no significant correlation between BDNF levels and CD4+/CD8+ cell count ratios (p=0.16) or CD8+ cell counts (p=0.60) across all participants.
Figure 4. TRK signaling is a target pathway of differentially expressed exo-miRNAs between neuropsychological lower- and higher-performing groups.
(A) Heatmap of Gene Ontology (GO) enrichment analysis, where the color key of log(p value) indicates the statistical significance that the GO category (column) is enriched with a given exo-miRNA (row). (B) Plasma BDNF levels negatively correlate with exosomal miR-206 and exosomal miR-30a-5p expression. The black line is the line of best fit between the two markers, and the grey ribbon represents the 95% confidence interval of the line of best fit. Log10 expression of the exo-miRNA is on the x-axis, and log10 levels of plasma BDNF are on the y-axis.
Differential exo-miRNA expression between NP higher- and lower-performing groups in the PHI group alone
Our analysis of differentially expressed exo-miRNAs between the NP-higher and NP-lower groups encompassed two distinct populations: participants from the San Francisco, USA, area, treated during early HIV infection, and HARC participants from the New Haven, USA, area, initially treated during chronic infection. In order to ensure that our findings were not driven by demographic differences between these two populations, differential exo-miRNA expression analyses were performed between NP-higher and -lower performing participants within the PHI study alone, where participants were distributed more evenly between NP-higher and -lower than the chronic infection group.
Using the same threshold of total-z score greater than or less than 0, 12 participants were classified as NP-higher (median total-z=0.4) and 7 participants were classified as NP-lower (median total-z= −0.5) within the PHI study alone. There was no significant difference in clinical characteristics between the NP groups (Table 1).
Differential expression analyses revealed 15 exo-miRNAs that were significantly differentially expressed (p<0.05) between NP-higher and -lower within this subset of participants. 5 exo-miRNAs (miR-454–3p, miR-548k, let-7a-5p, let-7e-5p, and let-7f-5p) were upregulated in NP-higher, while 10 exo-miRNAs (miR-30d-3p, miR-125b-5p, miR-193a-5p, miR-4742–3p, miR-4755–3p, miR-141–3p, miR-125b-2–3p, miR-205–5p, miR-1183, miR-708–3p) were upregulated NP-lower (Supplementary Figure 1, Supplementary Table 2). 3/10 exo-miRNAs that were upregulated in NP-lower (miR-193a-5p, miR-1183, and miR-708–3p) within the PHI-only group were also upregulated in NP-lower in the combined PHI and chronic infection group analysis. These 3 exo-miRNAs, differentially expressed in both analyses, demonstrated AUC of 0.86 (95% CI: 0.73–0.99) in distinguishing NP performance groups within the PHI cohort.
KEGG pathway analysis revealed that the axon guidance pathway was again significantly enriched for the 15 differentially expressed exo-miRNAs between the NP-higher and -lower groups in this PHI group, targeting 54 genes in the pathway (Supplementary Table 3). The 3 exo-miRNAs differentially expressed in both analyses were specifically implicated in the phosphatidylinositol signaling (p=0.01) and glycan degradation (p=0.0016) KEGG pathways and were predicted to target genes involved in nervous system development (p=1.5E-06), neurotrophin TRK signaling (1.6E-04) and ion binding (p=6.69E-20) by GO analysis.
Differential exo-miRNA expression between individuals with and without HIV infection
In an exploratory analysis, we compared exo-miRNA expression between a small group of HIV-negative individuals (n=5) and participants with HIV (n=31) to investigate whether the 11 exo-miRNA associated with NP status would differentiate by infection status. 25 exo-miRNAs were found to be differentially expressed (p<0.05) between the participants with and without HIV infection (Supplementary Table 4). miR-375, which was found to be up-regulated in NP-lower HIV-infected participants was also found to be 5.7-fold higher in HIV-infected participants relative to HIV-uninfected participants. Otherwise, the differential exo-miRNA expression between infection status did not overlap with those that were found when stratifying groups by NP testing.
Discussion
In this study, we demonstrated that PLWH with higher versus lower neuropsychological performance have different circulating exosomal microRNA content. Our analysis of exo-miRNA transcriptomic data revealed that the expression of 11 exo-miRNAs in plasma is associated with lower neuropsychological performance in PLWH, and certain processes, including axon guidance, ERBB signaling and TRK receptor signaling, are predicted targets of these 11 microRNAs. Three of these exo-miRNAs, along with the axon guidance KEGG pathway, were further validated in the exo-miRNA expression analysis within the group of PHI participants alone, suggesting that these exo-miRNA may be implicated in cognitive function during chronic HIV infection that is not specific to the timing of ART initiation.
Several of the differentially expressed exo-miRNAs have been implicated in inflammatory pathways, suggesting that exo-miRNAs may play a role in regulating inflammation during chronic HIV infection. For example, miR-483–5p and miR-30a-5p were up-regulated in NP-lower individuals. miR-483–5p has been shown to attenuate the host anti-viral immune response following hepatitis C infection through down-regulation of NF-kB, while miR-30a-5p has been shown to enhance anti-viral inflammation via up-regulation of IFN-1 [44, 45]. The interplay between anti- and pro-inflammatory exo-miRNAs may contribute to damaging, prolonged inflammation in PLWH as well as downstream clinical sequelae such as HAND, although this was not reflected by a difference in blood neopterin concentration by NP group.
Deep sequencing of exo-miRNAs allowed us to take an unbiased approach to detecting biologic processes that are differentially regulated in PWLH with different NP performance. Interestingly, a high proportion of exo-miRNAs in both the full group and subset group analyses were predicted to target genes in the axon guidance KEGG pathway. Integrated transcriptomic analyses of mRNAs and microRNAs in the frontal cortex have suggested that dysregulated axon guidance plays a key role in HIV-mediated neurodegeneration [46]. These results suggest that exo-miRNAs may traffic between the periphery and CNS, interfering with neuron repair pathways or reflecting ongoing degenerative processes associated with HAND. We were particularly interested in the executive function NP domain due to the known involvement of the frontal cortex in HIV-mediated neurodegeneration. We found that 3 exo-miRNA, including miR-148a-3p, which has been shown to promote apoptosis by targeting Bcl-2, were more highly expressed in PLWH with lower executive function performance [47]. miR-216b-3p was upregulated in both NP-lower and in the cohort with lower executive function, suggesting that this exo-miRNA may specifically contribute to deficits in executive function in PLWH. In addition, processing speed and motor function, two domains tested by our standardized neuropsychological exams, are commonly compromised in participants with HAND due to damage to myelinated white matter tracts [48, 49]. Defects in axon guidance would impede maintenance normal functioning of these tracts.
Exo-miRNAs up-regulated in NP-lower participants were also predicted to be involved in neurotrophin TRK signaling by GO analysis, and two of these microRNAs, miR-30a-5p and miR-206, have previously been shown to specifically inhibit translation of an important TRK ligand, BDNF [50-53]. We were able to corroborate this association by showing that there were significant negative correlations between plasma BDNF and both exosomal miR-30a-5p or miR-206. BDNF and its high-affinity receptor, tropomysin-related kinase B (TrkB), are important mediators of axon guidance and synaptic plasticity. Functional suppression of BDNF leads to the deficits of long-term synaptic potentiation, a major cellular mechanism underlying learning and memory [54]. Therefore, up-regulation of miR-30a-5p and miR-206 observed in NP-lower participants may cause neuropsychological deficits in PLWH by impeding BDNF signaling, and in turn, axon guidance.
In studies not focused on CNS deficits, it has been consistently shown that HIV infection alters the microRNA content of exosomes [45, 55, 56]. We similarly found significant differences in circulating exo-miRNA expression between individuals with and without HIV infection. These differences motivated our choice to focus on HIV-infected participants alone for our study of neuropsychological function. Elucidating whether the 11 exo-miRNA associated with neurocognitive status in PLWH are specific to HIV infection requires a larger, controlled study of uninfected volunteers with neuropsychological testing.
This study was an effort to investigate the relationship of exo-miRNA signaling and clinical sequelae of chronic HIV after viral suppression, and there are limitations to our analyses. Primarily, a higher proportion of participants who initiated cART during chronic infection sorted into the NP lower-performing group compared to those who started cART during early infection. Although NP tests have been standardized and validated to control for education level, age, and other demographic variables, performing differential exo-miRNA analysis in the more demographically homogenous PHI cohort alone was an important validation step. It is highly suggestive that three differentially expressed exo-miRNAs in both analyses are implicated in pathways of inflammation and neuronal function; however, repeating this analysis in a larger cohort of demographically homogenous individuals would strengthen this finding. Additionally, our study included only one female patient, and studying a more balanced cohort would increase the generalizability of these results. Second, without enriching for neuronal-derived exosomes (NDEs) in the plasma, we cannot know that the exosomes examined in this study would traffic to or from the CNS. Several studies have investigated NDEs specifically in the circulation as read-outs of CNS biology in neurocognitive disorders which could be employed in future work [28, 57, 58]. Nonetheless, this study provided a snapshot of biologic processes occurring in PLWH that may have widespread effects across various organs.
In summary, we found there was a distinct exosomal microRNA pattern that distinguished lower versus higher NP performance groups in PLWH. The differentially expressed exo-miRNAs were predicted to be involved in inflammation and neurodegeneration pathways. These findings suggest that circulating exo-miRNAs may reflect processes ongoing in the central nervous system in PLWH in the setting of durable viral suppression. Exo-miRNA content may serve as a useful diagnostic tool for individuals with chronic HIV infection on cART and provide further insight into potential targetable mechanisms of NP sequelae in this population.
Supplementary Material
Supplementary Figure 1. Validation of differentially expressed exo-miRNAs between neuropsychological lower- and higher-performing groups in the PHI study alone. The red points indicate exo-miRNAs with differential expression p < 0.05, and the grey points represent no significant difference between the neuropsychological (NP) groups. X-axis represents log2(fold-change) of exosomal microRNAs (lower vs. higher NP performance), and y-axis represents the –log10(p-value) of differences exo-miRNA expression (lower vs. higher NP performance).
Supplementary Table 1. All differentially expressed exo-miRNAs between neuropsychological higher- and lower-performing groups.
Supplementary Table 2. All differentially expressed exo-miRNAs between neuropsychological higher- and lower-performing groups within the PHI study alone.
Supplementary Table 3. KEGG Pathway enrichment analysis for differentially expressed exo-miRNAs in the PHI study alone.
Supplementary Table 4. All differentially expressed exo-miRNAs between participants with and without HIV infection.
Acknowledgments
Conflicts of Interest and Sources of Funding: KR and SS report that ViiV Healthcare, Inc. contributes study medications to a clinical trial co-directed by Drs. Spudich and Robertson outside of this work. The remaining authors have no conflicts of interest to disclose. This project was supported by grants R21MH110260, R01MH081772 and K23MH74466.
References
- 1.Eggers C, et al. , HIV-1-associated neurocognitive disorder: epidemiology, pathogenesis, diagnosis, and treatment. Journal of Neurology, 2017. 264(8): p. 1715–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garvey L, Surendrakumar V, and Winston A, Low rates of neurocognitive impairment are observed in neuro-asymptomatic HIV-infected subjects on effective antiretroviral therapy. HIV Clin Trials, 2011. 12(6): p. 333–8. [DOI] [PubMed] [Google Scholar]
- 3.Clifford DB and Ances BM, HIV-associated neurocognitive disorder. The Lancet. Infectious diseases, 2013. 13(11): p. 976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulfhammer G, et al. , Persistent central nervous system immune activation following more than 10 years of effective HIV antiretroviral treatment. Aids, 2018. 32(15): p. 2171–2178. [DOI] [PubMed] [Google Scholar]
- 5.van Zoest RA, et al. , Structural Brain Abnormalities in Successfully Treated HIV Infection: Associations With Disease and Cerebrospinal Fluid Biomarkers. J Infect Dis, 2017. 217(1): p. 69–81. [DOI] [PubMed] [Google Scholar]
- 6.Eden A, et al. , Asymptomatic Cerebrospinal Fluid HIV-1 Viral Blips and Viral Escape During Antiretroviral Therapy: A Longitudinal Study. J Infect Dis, 2016. 214(12): p. 1822–1825. [DOI] [PubMed] [Google Scholar]
- 7.Ellwanger JH, Veit TD, and Chies JAB, Exosomes in HIV infection: A review and critical look. Infect Genet Evol, 2017. 53: p. 146–154. [DOI] [PubMed] [Google Scholar]
- 8.Thery C, Zitvogel L, and Amigorena S, Exosomes: composition, biogenesis and function. Nat Rev Immunol, 2002. 2(8): p. 569–79. [DOI] [PubMed] [Google Scholar]
- 9.Welton JL, et al. , Cerebrospinal fluid extracellular vesicle enrichment for protein biomarker discovery in neurological disease; multiple sclerosis. J Extracell Vesicles, 2017. 6(1): p. 1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valadi H, et al. , Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology, 2007. 9: p. 654. [DOI] [PubMed] [Google Scholar]
- 11.Lu C, et al. , miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood, 2011. 117(16): p. 4293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CC, et al. , Elucidation of Exosome Migration across the Blood-Brain Barrier Model In Vitro. Cellular and molecular bioengineering, 2016. 9(4): p. 509–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Idda ML, et al. , Noncoding RNAs in Alzheimer’s disease. Wiley Interdiscip Rev RNA, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pogue AI and Lukiw WJ, Up-regulated Pro-inflammatory MicroRNAs (miRNAs) in Alzheimer’s disease (AD) and Age-Related Macular Degeneration (AMD). Cell Mol Neurobiol, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebrahimkhani S, et al. , Exosomal microRNA signatures in multiple sclerosis reflect disease status. Scientific Reports, 2017. 7(1): p. 14293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arenaccio C, et al. , Exosomes from human immunodeficiency virus type 1 (HIV-1)-infected cells license quiescent CD4+ T lymphocytes to replicate HIV-1 through a Nef- and ADAM17-dependent mechanism. J Virol, 2014. 88(19): p. 11529–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadiu I, et al. , Biochemical and biologic characterization of exosomes and microvesicles as facilitators of HIV-1 infection in macrophages. J Immunol, 2012. 189(2): p. 744–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gold JA, et al. , Longitudinal Characterization of Depression and Mood States Beginning in Primary HIV Infection. AIDS and Behavior, 2014. 18(6): p. 1124–1132. [DOI] [PubMed] [Google Scholar]
- 19.Peluso MJ, et al. , Cerebrospinal Fluid and Neuroimaging Biomarker Abnormalities Suggest Early Neurological Injury in a Subset of Individuals During Primary HIV Infection. The Journal of Infectious Diseases, 2013. 207(11): p. 1703–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heaton R, Miller SW, Taylor MJ, and Grant I, Revised comprehensive norms for an expanded Halstead–Reitan Battery: demographically adjusted neuropsychological norms for African American and Caucasian adults. Psychological Assessment Resources, Inc, 2004. Odessa, FL, USA. [Google Scholar]
- 21.Brandt J, a.B RHB, The Hopkins verbal learning test: revised. Psychological Assessment Resources, 2001. [Google Scholar]
- 22.Wechsler D, WMS-III: administration and scoring manual. Psychological Corporation Harcourt Brace & Co, 1997. San Antonio, TX. [Google Scholar]
- 23.Stroop J, Studies of interference in serial verbal reactions J Exp Psychol, 1935. 18: p. 643–662. [Google Scholar]
- 24.Comalli P, Wapner S, and Werner H, Interference effects of Stroop color-word test in childhood, adulthood, and aging. J Genet Psychol 1962. 100: p. 47–53. [DOI] [PubMed] [Google Scholar]
- 25.Tombaugh T, Kozak J, and Rees L, Normative data stratified by age and education for two measures of verbal fluency: FAS and animal naming. Arch Cin Neuropsychol, 1999. 14: p. 167–177. [PubMed] [Google Scholar]
- 26.Gladsjo J, Schuman CC, Evans JD, Peavy GM, Miller SW, and and R Heaton, Norms for letter and category fluency: demographic corrections for age, education, and ethnicity. Assessment, 1999. 6(147–178). [DOI] [PubMed] [Google Scholar]
- 27.Ruff R, a.P SB, Gender- and age-specific changes in motor speed and eye-hand coordination in adults: normative values for the finger tapping and grooved pegboard tests. Percept Mot Skills, 1993. 76(1219–1230). [DOI] [PubMed] [Google Scholar]
- 28.Sun B, et al. , Blood neuron-derived exosomes as biomarkers of cognitive impairment in HIV. Aids, 2017. 31(14): p. F9–f17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wright EJ, et al. , Factors associated with neurocognitive test performance at baseline: a substudy of the INSIGHT Strategic Timing of AntiRetroviral Treatment (START) trial. HIV Med, 2015. 16 Suppl 1: p. 97–108. [DOI] [PubMed] [Google Scholar]
- 30.Zeringer E, et al. , Strategies for isolation of exosomes. Cold Spring Harb Protoc, 2015. 2015(4): p. 319–23. [DOI] [PubMed] [Google Scholar]
- 31.Alvarez ML, et al. , Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int, 2012. 82(9): p. 1024–32. [DOI] [PubMed] [Google Scholar]
- 32.Li P, et al. , Progress in Exosome Isolation Techniques. Theranostics, 2017. 7(3): p. 789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kowal J, et al. , Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A, 2016. 113(8): p. E968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartjes TA, et al. , Extracellular Vesicle Quantification and Characterization: Common Methods and Emerging Approaches. Bioengineering (Basel, Switzerland), 2019. 6(1): p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong Y, Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics, 2011. 98(2): p. 152–3. [DOI] [PubMed] [Google Scholar]
- 36.Li H and Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 2009. 25(14): p. 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kozomara A and Griffiths-Jones S, miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research, 2014. 42(D1): p. D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Love MI, Huber W, and Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 2014. 15(12): p. 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vlachos IS, et al. , DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res, 2015. 43(W1): p. W460–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darcq E, et al. , MicroRNA-30a-5p in the prefrontal cortex controls the transition from moderate to excessive alcohol consumption. Mol Psychiatry, 2015. 20(10): p. 1219–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varendi K, Matlik K, and Andressoo JO, From microRNA target validation to therapy: lessons learned from studies on BDNF. Cell Mol Life Sci, 2015. 72(9): p. 1779–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dragovic RA, et al. , Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine : nanotechnology, biology, and medicine, 2011. 7(6): p. 780–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vestad B, et al. , Size and concentration analyses of extracellular vesicles by nanoparticle tracking analysis: a variation study. Journal of extracellular vesicles, 2017. 6(1): p. 1344087–1344087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shwetha S, et al. , Circulating miRNA profile in HCV infected serum: novel insight into pathogenesis. Scientific Reports, 2013. 3: p. 1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma Y, et al. , The Coronavirus Transmissible Gastroenteritis Virus Evades the Type I Interferon Response through IRE1α-Mediated Manipulation of the MicroRNA miR-30a-5p/SOCS1/3 Axis. Journal of Virology, 2018. 92(22): p. e00728–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou L, et al. , A parallel genome-wide mRNA and microRNA profiling of the frontal cortex of HIV patients with and without HIV-associated dementia shows the role of axon guidance and downstream pathways in HIV-mediated neurodegeneration. BMC Genomics, 2012. 13: p. 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H, et al. , MiR-148a promotes apoptosis by targeting Bcl-2 in colorectal cancer. Cell Death And Differentiation, 2011. 18: p. 1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ketzler S, et al. , Loss of neurons in the frontal cortex in AIDS brains. Acta Neuropathologica, 1990. 80(1): p. 92–94. [DOI] [PubMed] [Google Scholar]
- 49.Oh SW, et al. , Altered White Matter Integrity in Human Immunodeficiency Virus-Associated Neurocognitive Disorder: A Tract-Based Spatial Statistics Study. Korean journal of radiology, 2018. 19(3): p. 431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mellios N, et al. , A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum Mol Genet, 2008. 17(19): p. 3030–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Croce N, et al. , NPY modulates miR-30a-5p and BDNF in opposite direction in an in vitro model of Alzheimer disease: a possible role in neuroprotection? Mol Cell Biochem, 2013. 376(1–2): p. 189–95. [DOI] [PubMed] [Google Scholar]
- 52.Tian N, Cao Z, and Zhang Y, MiR-206 decreases brain-derived neurotrophic factor levels in a transgenic mouse model of Alzheimer’s disease. Neuroscience bulletin, 2014. 30(2): p. 191–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee ST, et al. , miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann Neurol, 2012. 72(2): p. 269–77. [DOI] [PubMed] [Google Scholar]
- 54.Shen K and Cowan CW, Guidance molecules in synapse formation and plasticity. Cold Spring Harb Perspect Biol, 2010. 2(4): p. a001842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aqil M, et al. , The HIV Nef protein modulates cellular and exosomal miRNA profiles in human monocytic cells. Journal of Extracellular Vesicles, 2014. 3(1): p. 23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Madison MN and Okeoma CM, Exosomes: Implications in HIV-1 Pathogenesis. Viruses, 2015. 7(7): p. 4093–4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abu-Rumeileh S, et al. , The CSF neurofilament light signature in rapidly progressive neurodegenerative dementias. Alzheimer’s Research & Therapy, 2018. 10(1): p. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mustapic M, et al. , Plasma Extracellular Vesicles Enriched for Neuronal Origin: A Potential Window into Brain Pathologic Processes. Frontiers in Neuroscience, 2017. 11(278). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Validation of differentially expressed exo-miRNAs between neuropsychological lower- and higher-performing groups in the PHI study alone. The red points indicate exo-miRNAs with differential expression p < 0.05, and the grey points represent no significant difference between the neuropsychological (NP) groups. X-axis represents log2(fold-change) of exosomal microRNAs (lower vs. higher NP performance), and y-axis represents the –log10(p-value) of differences exo-miRNA expression (lower vs. higher NP performance).
Supplementary Table 1. All differentially expressed exo-miRNAs between neuropsychological higher- and lower-performing groups.
Supplementary Table 2. All differentially expressed exo-miRNAs between neuropsychological higher- and lower-performing groups within the PHI study alone.
Supplementary Table 3. KEGG Pathway enrichment analysis for differentially expressed exo-miRNAs in the PHI study alone.
Supplementary Table 4. All differentially expressed exo-miRNAs between participants with and without HIV infection.