Abstract
Background:
Timely and sensitive diagnosis of gastric cancer is crucial for efficient treatment and survival of the patients. microRNAs have been considered as diagnostic biomarkers in different type of cancers including gastric cancer. In the present study, the expression profile of four microRNAs, miR-103, miR-107, miR-194 and miR-210 were evaluated in patients with intestinal-type of gastric cancer (IGC) in order to assess their diagnosis utility as noninvasive biomarkers.
Methods:
A total number of 100 plasma samples from patients with gastric cancer and healthy controls were obtained and total RNA was extracted using a commercial monophasic solution of phenol and guanidium thiocyanate. Reverse transcription (RT) reactions were performed by specific stem-loop RT primers and M-MuLV RT-enzyme. The expression patterns of microRNAs were assessed using reverse transcription quantitative real-time PCR (RT-qPCR) method and the expression of SNORD47 RNA was used as the reference for normalization.
Results:
The results indicate that the plasma levels of miR-107, miR-194, and miR-210 were significantly lower in patients. Receiver operating characteristic (ROC) curve analysis showed that the patients could be distinguished from healthy individuals at the cutoff levels of 0.504, 0.266, and 0.394 of miR-107, miR-194, and miR-210, respectively. On the other hand, the expression levels of these miRNAs were not significantly different in different clinicopathological stages of the disease.
Conclusion:
These findings suggest that the plasma levels of miR-107, miR-194 and miR-210 were downregulated in patients with ICG and propose these molecules as potential non-invasive biomarkers for detection of IGC.
Keywords: Biomarker, diagnosis, gastric cancer, MicroRNA, RT-qPCR
Introduction
Although gastric cancer is considered as the third most frequent cancer-related death worldwide, it remains the major cause of cancer mortality, with nearly 10,000 deaths per year in Iran (Fitzmaurice et al., 2015). One of most the important features of this type of cancer is the absence of clinical symptoms in the early stages of disease. More than 80% of the diagnosis of this type of cancer is made at the late stages of the disease (Siegel et al., 2011). Timely and correct diagnosis of gastric cancer in the early stages is crucial for efficacy of the treatment which results in higher survival of patients. Although traditional endoscopy and microscopic examinations have important roles in diagnosis of gastric cancer, these methods are invasive and inappropriate for regular screening (Liu et al., 2011). On the other hand, conventional biomarkers such as carcino-embryonic antigen (CEA) and carbohydrate antigens 19-9 and 125 (CA 19-9 and CA 125) do not have adequate sensitivity and specificity in the early diagnosi (Yin et al., 2012). MicroRNAs (miRNAs) have the capability to be considered as diagnostic biomarkers for cancer (Wang et al., 2016a). These non-coding small RNAs regulate gene expression at the post-transcriptional level and play important roles in many aspects of biology, including tissue development and degeneration, apoptosis, response to cellular stresses, and etiology and progression of various diseases (Filipowicz et al., 2008; Mondanizadeh et al., 2015). Furthermore, the expression of miRNAs is frequently deregulated in cancer and they may act as either oncogenes or tumor suppressors (Rotkrua et al., 2013). Several reports have shown that miRNAs are stable in plasma and serum and that circulating molecules display unique profiles for each tumor. The stability of miRNAs in circulation might be due to their protection by exosome and microvesicle, Argonaute2 (Ago2), and high-density lipoprotein (Filipowicz et al., 2008; Tsujiura et al., 2010; Rotkrua et al., 2013). It was also shown that miRNAs can be used as molecular biomarkers for the diagnosis and progression evaluation of several cancers (Cortez et al., 2011; Song and Meltzer, 2012). A few studies have evaluated the expression profiles of different miRNAs in gastric cancer and tried to use them to distinguish patients from healthy individuals (Zhang et al., 2008; Xie et al., 2016). In a study conducted in mice model of diffuse-type gastric cancer (DGC), Rotkrua et al. assessed four miRNAs, miR-103, miR-107, miR-194 and miR-210, using microarray data, and reported that they are significantly overexpressed in the animal model(Rotkrua et al., 2013). It was proposed that these miRNAs can be used as novel non-invasive biomarkers for the early detection of DGC.
Considering the fact that the major type of gastric cancer in Iranian population is the intestinal-type (IGC), in the current study, the plasma levels of miR-103, miR-107, miR-194 and miR-210 were compared between IGC patients and healthy controls, using reverse transcription quantitative real-time PCR (RT-qPCR), to evaluate their diagnostic utility as noninvasive biomarkers for IGC detection.
Materials and Methods
Patients and samples
A total of 100 plasma samples, including 50 preoperative samples from patients with IGC and 50 samples from healthy individuals, were enrolled in this study. The IGC samples were obtained from Iran Tumor Bank (Cancer Institute, Imam Khomeini Hospital, Tehran Iran), which were related to 22 female and 28 male, with a median age of 56 years (range, 29 to 80 years). The patients who had no sign of metastasis at the time of specimen collections and had not been received cancer-related treatment were enrolled in this study. The specimens of the patients who received any type of cancer treatment or diagnosed with proven metastasis of tumor to other organs were excluded during sample collection. The pathological classifications of GC patients were according to the IGCC/TMN staging system (Wittekind et al., 2002). Other clinical characteristics of the patients are detailed in Table 1.
Table 1.
Baseline Characteristics of Patients and Control Groups
| Parameter | Patients Number (%) | Healthy controls Number (%) |
|---|---|---|
| Gender | ||
| Female | 22 (44) | 22 (44) |
| Male | 28 (56) | 28 (56) |
| Age | ||
| Mediam | 56 | 52 |
| Range | 29-80 | 26-65 |
| TNM stage | ||
| I | 8 | |
| II | 10 | |
| III | 20 | |
| IV | 12 | |
| Haematogenous metastasis (M) | ||
| No metastasis (M0) | 38 | |
| Metastasis (M1) | 12 | |
| Lymph node metastasis (N) | ||
| No metastasis (N0) | 18 | |
| Metastasis (N1-N3) | 32 | |
| Depth of invasion (T) | ||
| T1 | 2 | |
| T2 | 11 | |
| T3 | 37 | |
The samples from healthy individuals were obtained from Arak Reference Laboratory (Arak University of Medical Sciences, Arak, Iran) and they were matched with the samples in the patient group, based on the age and gender criteria (Table 1). All samples were stored at -80°C prior to analysis. The study was approved by the Ethics Committee of Arak University of Medical Sciences (Ethics number, 2143).
RNA Extraction
Total RNA was extracted using RNX-Plus reagent (SinaClon, Iran) with some modifications to the manufacturer`s protocol, to optimize the reagent for isolation of small RNAs. Briefly, 200 μl of plasma was mixed with 1ml RNX-Plus solution and incubated at room temperature for 5 min. After phase separation by adding 200 μl chloroform and incubation at 4°C for 5 min, the aqueous phase was separated and mixed with 2 volumes of absolute ethanol and incubated at -20°C overnight. After centrifugation at 4°C for 60 min, the RNAs were eluted in 20 μl RNase-free water and the extracted RNA was used directly for reverse transcription step.
Reverse transcription
Stem-loop primers were used for reverse transcription of mature miRNAs (Chen et al., 2005).The sequences of Stem-loop and gene-specific primers are summarized in Table 2. For cDNA synthesis, about 1μg of RNA was mixed with 1 pmol of specific stem-loop RT primers and incubated at 75°C for 5 min. A mix containing 100 units of M-MuLVRT-enzyme (Vivantis, Malaysia), 1x RT-buffer, and 400 μMdNTP were added to the tube. Finally, RT reaction mixtures were incubated at 25°Cfor 15 min, 37°C for 15 min, 42°C for 45 min, and 10 min at 75°C, in a thermal cycler instrument (Eppendorf, Germany). The resulting cDNAs were stored at -20°C prior to RT-qPCR analyses.
Table 2.
Primers Used for Reverse-Transcription and RT-qPCR Assay of the Target miRNAs
| Target | Primer sequences (5`-3`) |
|---|---|
| miR-103 | Sa: 5`-GTCGTATCGAGAGCAGGGTCCGAGGTATTCGCACTCGATACGACCAAGGCA-3` |
| Fb: 5`-GCTTCTTTACAGTGCTGCC-3` | |
| miR-107 | S: 5`-GTCGTATCGAGAGCAGGGTCCGAGGTATTCGCACTCGATACGACAGCAGCA-3` |
| F: 5`-GCCCTGTACAATGCTGC-3` | |
| miR-194 | S: 5`-GTCGTATCGAGA GCAGGGTCCGAGGTATT CGCACTCGATACGACTC CACAT-3` |
| F: 5`-TGTAACAGCAACTCCATGTG-3` | |
| miR-210 | S: 5`-GTCGTATCGAGAGCAGGGTCCGAGGTATT CGCACTCGATA CGACCAGTGTG-3` |
| F: 5`-CTGCCCACCGCACACTG-3` | |
| SNORD47 | S: 5`-GTCGTATGCAGAGCAGGGTCCGAGGTATTCGCACTGCATACGACAACCTCA-3` |
| F: 5`-ATCACTGTAAAACCGTTCCA-3` | |
| Common Reverse | 5`-AGAGCAGGGTCCGAGGT-3` |
a , Stem-loop gene-specific reverse-transcriptase primers; b , Specific forward primers for RT-qPCR amplification
Quantitative real-time PCR
Expression of SNORD47 (U47) RNA was used as endogenous reference gene for normalization of miRNAs expressions. The sequences of forward and the universal reverse primers are summarized in Table 2. RT-qPCR reactions were performed in a LightCycler®96 instrument(Roche, Germany) using the QuantiNova SYBR Green PCR Kit (Qiagen, Germany).The amplification profile consisted of a single cycle of enzyme activation at 95°C for 2 min followed by 40 cycles of denaturation at 95°C for 5 sec and annealing/extension at 60°C for 15 sec, with a single fluorescence acquisition at channel green. Melting curve analysis was carried out following amplification with a ramp rate of 0.2°C/second to 96°Cwith continuous fluorescence acquisition. The relative expression was calculated by the comparative Cq method using the relative expression software tool (REST) (Pfaffl et al., 2002).
Statistical methods
RT-qPCR data were analyzed by REST 2009 and the data in graphs were expressed as the mean ± SE. The differences between groups were compared by analysis of variance (ANOVA). Receiver operating characteristic (ROC) curve analysis was performed to evaluate the diagnostic value of the plasma miRNA levels for IGC. All statistical analyses were calculated using SPSS software (version 16; SSPS Inc., 184 Chicago). P-values of <0.05 were considered to indicate statistical significance.
Results
Evaluation of RT-qPCR for measuring miRNAs plasma levels
The results of RT-qPCR analyses showed that in comparison to healthy individuals, the plasma levels of miR-107 (0.147 fold, P= 0.002), miR-194 (0.117 fold, P= 0.001), and miR-210 (0.159 fold, P= 0.002) were significantly lower in IGC patients. However, the expression of miR-103 exhibited no significant difference between the healthy controls and IGC patients (0.889 fold, P= 0.77) (Figure 1).
Figure 1.
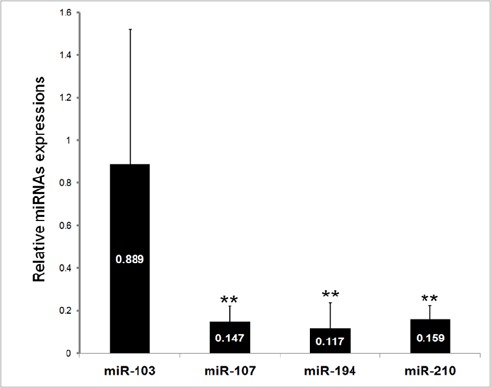
Expression Levels of the miR-103, miR-107, miR-195, and miR-210 in IGC Patients. Error bars indicate the standard error of the mean. **P < 0.01, significantly different from control group.
Expression of the miRNAs in different clinicopathological status
The plasma levels of miR-107, miR-194, and miR-210 were analyzed in patients with different TNM stages. As shown in Figure 2, the expression levels of the three miRNAs were not significantly different among patients with stages I-IV of IGC. Nevertheless, in comparison to control group, patients with stages I-IV for miR-107 (P< 0.01) and stages II-IV for miR-194 (P< 0.01) and miR-210 (P< 0.05) had reduced plasma levels. The plasma levels of miR-194 and miR-210 in patients with stage I were not significantly different from control group (P= 0.174 and P= 0.095, respectively). Furthermore, the plasma levels of the three miRNAs were evaluated based on the depth of invasion, haematogenous metastasis, and lymph node metastasis. The results showed that the level of the three miRNAs were not significantly different (Figure 2).
Figure 2.
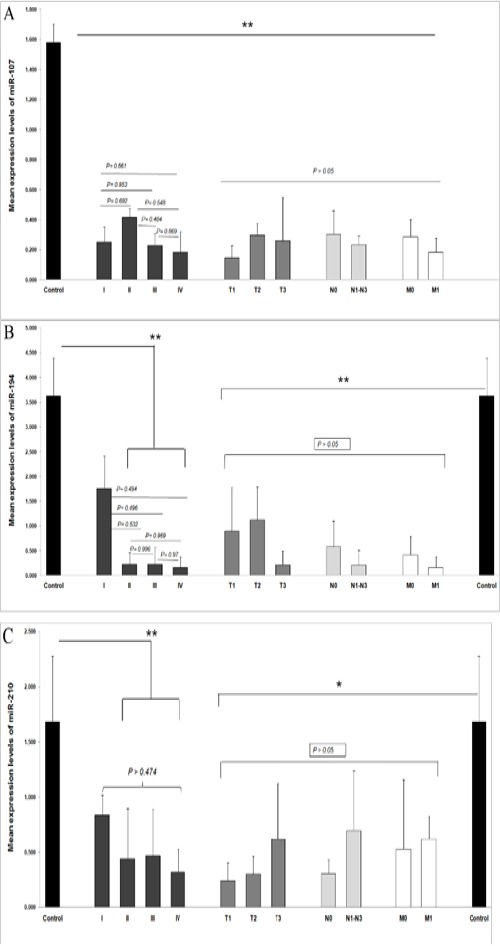
Plasma Concentrations of miR-107, miR-194, and miR-210 in Different Clinicopathological Status. The Plasma Levels of miR-107, miR-194, and miR-210 Are Shown in Figure a-c, Respectively. Error bars indicate standard deviation. Statistically significant results are shown by asterisks (*P < 0.05, **P < 0.01). The P-values of insignificant comparisons are shown above the horizontal bars.
Determination of expression level cutoff
ROC curve analyses were performed to determine diagnostic values for miR-107, miR-194, and miR-210 to differentiate IGC patients from healthy individuals. The cutoff levels were chosen as the point with the maximum sum of sensitivity and specificity on the ROC curves using the Youden`s index ([sensitivity + specificity] -1) (Khansarinejad et al., 2015). Based on the results shown in Figure 3, at the cutoff level of 0.504, miR-107 exhibited 93.8% sensitivity and 78.8% specificity with an AUC of 0.947. For miR-194, the cutoff level of 0.266 showed 87.5% sensitivity and 75.8% specificity, with an AUC of 0.862. Finally, at the cutoff level of 0.394, miR-210 exhibited 100% sensitivity and 72.7% specificity with an AUC of 0.82 (Figure 3).
Figure 3.
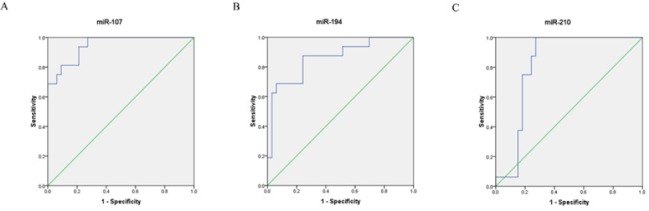
Receiver Operating Characteristic (ROC) Curve Analyses Corresponds to miR-107, miR-194, and miR-210 Expression Levels to Discriminate IGC Patients from Healthy Ones Individuals. a, The area under the curve of miR-107; b, The area under the curve of miR-194; c, The area under the curve of miR-210.
Discussion
Several studies have shown alterations in microRNA expression in patients with gastric cancers and suggested circulating microRNAs as novel biomarkers (Calin and Croce, 2006; Komatsu et al., 2013). It has been shown that the expression levels of miR-21, miR-221, miR-223, miR-106, miR-17-5p, miR-19b, miR-194, miR-744 and miR-376 were significantly upregulated in these patients (Zhou et al., 2010; Li et al., 2012; Song et al., 2012; Wang et al., 2014; Tsujiura et al., 2015), whereas miR-218, miR-143, miR-195, miR-141, miR-1233, and let-7a were downregulated (Li et al., 2012; Wang et al., 2013; Huang and Yu, 2015).
In the present study, the plasma levels of miR-103, miR-107, miR-194 and miR-210 were compared in 50 preoperative IGC patients and 50 healthy individuals. The result showed that the expression levels of miR-107, miR-194, and miR-210 were significantly lower in patients with IGC, whereas, the level of miR-103 exhibited no significant difference with control group. These findings are in contrast to those found by Rotkrua et al., who reported that the expression of all four miRNAs were higher in the mice model of DGC (Rotkrua et al., 2013). The main reason for this inconsistency might be largely due to the difference in the host species. Recently, the same group reported that, in contrast to the animal model, the plasma levels of miR-103, miR-107, and miR-194 are not significantly changed in human plasma and are not suitable for human DGC diagnosis (Oze et al., 2017). However, the results of the current study showed downregulations of miR-107, miR-194, and miR-210. The type of gastric cancer (diffuse-type versus intestinal-type) and the methods of relative quantification calculations could be other possible reasons for the disagreement. The earlier studies used “comparative CT (or ΔΔCT) method, while in the current study, the calculations were performed using Pfaffl method with the Relative Expression Software tool, which is more reliable (Pfaffl et al., 2002). On the other hand, the main limitation of the current study was difficulty in finding more plasma specimens from IGC patients who had not been received therapy and the sample size of more than 50 IGC specimen would have been more appropriate.
ROC curve analysis showed that IGC patients could be distinguished from healthy individuals at the cutoff levels of 0.504, 0.266, and 0.394 of miR-107 (sensitivity=93.8% and specificity=78.8%) , miR-194 (sensitivity=87.5% and specificity=75.8%), and miR-210 (sensitivity=100% and specificity=72.7%), respectively.
Furthermore, comparison of the plasma levels of these three miRNAs in different clinicopathological stages showed that the expression levels of the miRNAs were not significantly different among four stages. However, each stage showed lower plasma levels when compared with the control group. The results indicate that the differential expression of miR-107, miR-194, and miR-210 could not be used for staging intestinal-type gastric cancer. The finding is in accordance with some previous studies that evaluated expression levels of microRNAs in different stages of gastric cancer (Li et al., 2012). Nevertheless, some studies reported differential expression of some microRNAs in different stages of gastric cancer. Song et al. showed that the serum expression levels of miR-221, miR-744 and miR-376c were higher in poorly differentiated GC patients than in patients with well or moderately differentiated gastric cancer (Song et al., 2012). Mir-143 and miR-195 were also associated with clinicopathological parameters, including depth of invasion and lymph node metastasis (Guo et al., 2013).
On the other hand, some of these miRNAs were studied in other types of malignant disease, including colorectal cancer (Luo et al., 2011), breast cancer (Stuckrath et al., 2015; Xiong et al., 2017), lung cancer (Zhu et al., 2016; Qu et al., 2017), colorectal cancer (Nonaka et al., 2015; Basati et al., 2016; Wang et al., 2016b), glioma (Ji et al., 2015), hepatocellular carcinoma (Zhao et al., 2015; Zhang et al., 2017), and clear cell renal cell carcinoma (Li et al., 2017). However, combined expression profile of miR-107, miR-194 and miR-210 were only evaluated in gastric cancer.
In Conclusion, the results of this study indicate that miR-107, miR-194 and miR-210 were downregulated in the plasma samples of patients with intestinal-type of gastric cancer and suggest them as potential non-invasive biomarkers for detection of IGC.
Funding Statement
This study was supported by the Research Deputy of Arak University of Medical Sciences.
Statement conflict of Interest
The authors declare no competing interests.
Acknowledgements
The authors thank Dr. Amir Nader Emami Razavi and Dr. Hamid Abtahi for their assistance in this research.
References
- Basati G, Razavi AE, Pakzad I, et al. Circulating levels of the miRNAs, miR-194, and miR-29b, as clinically useful biomarkers for colorectal cancer. Tumour Biol. 2016;37:1781–8. doi: 10.1007/s13277-015-3967-0. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez MA, Bueso-Ramos C, Ferdin J, et al. MicroRNAs in body fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–77. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs:are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fitzmaurice C, Dicker D, Pain A, et al. The global burden of cancer 2013. JAMA Oncol. 2015;1:505–27. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Li J, Liu L, et al. Dysregulation of miRNAs and their potential as biomarkers for the diagnosis of gastric cancer. Biomed Rep. 2013;1:907–12. doi: 10.3892/br.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YK, Yu JC. Circulating microRNAs and long non-coding RNAs in gastric cancer diagnosis:An update and review. World J Gastroenterol. 2015;21:9863–86. doi: 10.3748/wjg.v21.i34.9863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Wei Y, Wang J, et al. Decreased expression of microRNA-107 predicts poorer prognosis in glioma. Tumour Biol. 2015;36:4461–6. doi: 10.1007/s13277-015-3086-y. [DOI] [PubMed] [Google Scholar]
- Khansarinejad B, Soleimanjahi H, Mirab Samiee S, et al. Monitoring human cytomegalovirus infection in pediatric hematopoietic stem cell transplant recipients:using an affordable in-house qPCR assay for management of HCMV infection under limited resources. Transpl Int. 2015;28:594–603. doi: 10.1111/tri.12545. [DOI] [PubMed] [Google Scholar]
- Komatsu S, Ichikawa D, Tsujiura M, et al. Prognostic impact of circulating miR-21 in the plasma of patients with gastric carcinoma. Anticancer Res. 2013;33:271–6. [PubMed] [Google Scholar]
- Li BS, Zhao YL, Guo G, et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS One. 2012;7:e41629. doi: 10.1371/journal.pone.0041629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Zhao A, Peoch M, et al. Detection of urinary cell-free miR-210 as a potential tool of liquid biopsy for clear cell renal cell carcinoma. Urol Oncol. 2017;35:294–9. doi: 10.1016/j.urolonc.2016.12.007. [DOI] [PubMed] [Google Scholar]
- Liu R, Zhang C, Hu Z, et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47:784–91. doi: 10.1016/j.ejca.2010.10.025. [DOI] [PubMed] [Google Scholar]
- Luo X, Burwinkel B, Tao S, et al. MicroRNA signatures:novel biomarker for colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2011;20:1272–86. doi: 10.1158/1055-9965.EPI-11-0035. [DOI] [PubMed] [Google Scholar]
- Mondanizadeh M, Arefian E, Mosayebi G, et al. MicroRNA-124 regulates neuronal differentiation of mesenchymal stem cells by targeting Sp1 mRNA. J Cell Biochem. 2015;116:943–53. doi: 10.1002/jcb.25045. [DOI] [PubMed] [Google Scholar]
- Nonaka R, Miyake Y, Hata T, et al. Circulating miR-103 and miR-720 as novel serum biomarkers for patients with colorectal cancer. Int J Oncol. 2015;47:1097–102. doi: 10.3892/ijo.2015.3064. [DOI] [PubMed] [Google Scholar]
- Oze I, Shimada S, Nagasaki H, et al. Plasma microRNA-103, microRNA-107, and microRNA-194 levels are not biomarkers for human diffuse gastric cancer. J Cancer Res Clin Oncol. 2017;143:551–4. doi: 10.1007/s00432-016-2316-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L, Li L, Zheng X, et al. Circulating plasma microRNAs as potential markers to identify EGFR mutation status and to monitor epidermal growth factor receptor-tyrosine kinase inhibitor treatment in patients with advanced non-small cell lung cancer. Oncotarget. 2017;8:45807–24. doi: 10.18632/oncotarget.17416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotkrua P, Shimada S, Mogushi K, et al. Circulating microRNAs as biomarkers for early detection of diffuse-type gastric cancer using a mouse model. Br J Cancer. 2013;108:932–40. doi: 10.1038/bjc.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Ward E, Brawley O, et al. Cancer statistics, 2011:the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology. 2012;143:35–47.e2. doi: 10.1053/j.gastro.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Song MY, Pan KF, Su HJ, et al. Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS One. 2012;7:e33608. doi: 10.1371/journal.pone.0033608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckrath I, Rack B, Janni W, et al. Aberrant plasma levels of circulating miR-16, miR-107, miR-130a and miR-146a are associated with lymph node metastasis and receptor status of breast cancer patients. Oncotarget. 2015;6:13387–401. doi: 10.18632/oncotarget.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujiura M, Ichikawa D, Komatsu S, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–9. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujiura M, Komatsu S, Ichikawa D, et al. Circulating miR-18a in plasma contributes to cancer detection and monitoring in patients with gastric cancer. Gastric Cancer. 2015;18:271–9. doi: 10.1007/s10120-014-0363-1. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Wu Z, et al. Three dysregulated microRNAs in serum as novel biomarkers for gastric cancer screening. Med Oncol. 2014;31:298. doi: 10.1007/s12032-014-0298-8. [DOI] [PubMed] [Google Scholar]
- Wang J, Chen J, Sen S. MicroRNA as biomarkers and diagnostics. J Cell Physiol. 2016a;231:25–30. doi: 10.1002/jcp.25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Qu A, Liu W, et al. Circulating miR-210 as a diagnostic and prognostic biomarker for colorectal cancer. Eur J Cancer Care (Engl) 2016b;26:49–57. doi: 10.1111/ecc.12448. [DOI] [PubMed] [Google Scholar]
- Wang WN, Chen Y, Zhang YD, et al. The regulatory mechanism of CCR7 gene expression and its involvement in the metastasis and progression of gastric cancer. Tumour Biol. 2013;34:1865–71. doi: 10.1007/s13277-013-0728-9. [DOI] [PubMed] [Google Scholar]
- Wittekind C, Compton CC, Greene FL, et al. TNM residual tumor classification revisited. Cancer. 2002;94:2511–6. doi: 10.1002/cncr.10492. [DOI] [PubMed] [Google Scholar]
- Xie M, Dart DA, Owen S, et al. Insights into roles of the miR-1, -133 and -206 family in gastric cancer (Review) Oncol Rep. 2016;36:1191–8. doi: 10.3892/or.2016.4908. [DOI] [PubMed] [Google Scholar]
- Xiong B, Lei X, Zhang L, et al. miR-103 regulates triple negative breast cancer cells migration and invasion through targeting olfactomedin 4. Biomed Pharmacother. 2017;89:1401–8. doi: 10.1016/j.biopha.2017.02.028. [DOI] [PubMed] [Google Scholar]
- Yin Y, Li J, Chen S, et al. MicroRNAs as diagnostic biomarkers in gastric cancer. Int J Mol Sci. 2012;13:12544–55. doi: 10.3390/ijms131012544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li T, Qiu Y, et al. Serum microRNA panel for early diagnosis of the onset of hepatocellular carcinoma. Medicine (Baltimore) 2017;96:e5642. doi: 10.1097/MD.0000000000005642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li Z, Gao C, et al. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358–66. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Li F, Zhang X, et al. MicroRNA-194 acts as a prognostic marker and inhibits proliferation in hepatocellular carcinoma by targeting MAP4K4. Int J Clin Exp Pathol. 2015;8:12446–54. [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Guo JM, Lou YR, et al. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using microRNA as a marker. J Mol Med (Berl) 2010;88:709–17. doi: 10.1007/s00109-010-0617-2. [DOI] [PubMed] [Google Scholar]
- Zhu W, Zhou K, Zha Y, et al. Diagnostic value of serum miR-182, miR-183, miR-210, and miR-126 levels in patients with early-stage non-small cell lung cancer. PLoS One. 2016;11:e0153046. doi: 10.1371/journal.pone.0153046. [DOI] [PMC free article] [PubMed] [Google Scholar]


