Abstract
Background:
The use of targeted specific genes in therapeutic and treatment decisions has been considered for lung cancer. The epidermal growth factor receptor (EGFR) gene, which is over expressed in non-small cell lung cancer (NSCLC), was considered as one of the targeted specific genes. EGFR mutations in exons 18–21, which encode a portion of the EGFR kinase domain, were found in NSCLC patients and were associated with the response of EGFR- tyrosine kinase inhibitors (EGFR-TKIs). Therefore, a molecular technique for EGFR mutation detection has important benefits for therapy in NSCLC patients. This study aims to determine the EGFR mutations in patients with NSCLC using polymerase chain reaction-single strand conformational polymorphism (PCR-SSCP) in exons 18-21.
Methods:
DNA samples were extracted from formalin fixed paraffin embedded tissues of NSCLC patients who attended hospital. The extracted DNA was used as a template for the EGFR gene amplification.
Results:
Occurrence of EGFR mutations were found in 29 out of 50 cases (58%).The frequency of EGFR mutations by first PCR at exon 18, 19, 20 and 21 were 6 (12%), 19 (38%) 20 (40%) and at 21 (42%), respectively. By PCR-SSCP, the frequencies of EGFR mutations at exon 18, 19, 20 and 21 were 3(6%), 18(36%), 23(46%) and 13(26%), respectively. All of the mutations found were in agreement with DNA sequencings.
Conclusion:
The high frequency of EGFR mutations in NSCLC suggests that PCR-SSCP is a efficient screening method and useful for treatment plan.
Keywords: Epidermal growth factor receptor, polymerase chain reaction-single strand conformational polymorphism
Introduction
Lung cancer is a crucial health problem and one of the most prevalent causes of death worldwide. There were estimated to be 1.8 million new cases in 2012 (12.9%), and 58% occurred in the less developed regions (Ferlay et al., 2015). The disease is the most common cancer in men worldwide (1.2 million, 16.7% of total), and it has the highest estimated age-standardized incidence rates in Central and Eastern Europe (53.5 per 100,000) and Eastern Asia (50.4 per 100,000) (Chan and Hughes, 2015; Zhou, 2014). In women, the incidence rates were lower in general and the geographical pattern was a little different, mainly reflecting different historical exposures to tobacco smoking. Thus, the highest estimated rates were in Northern America (33.8 per 100,000) and Northern Europe (23.7 per 100,000) with a relatively high rate in Eastern Asia (19.2 per 100,000) and the lowest rates also in Western and Middle Africa (1.1 and 0.8 per 100,000 respectively) (Ferlay et al., 2015).
In South East Asia, lung cancer incidence rates were ranked as first and fifth for males and females, respectively. Lung cancer can be divided into two types depending on the histology: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC is the most commonly found lung cancer (85%) (Midha et al., 2015). There are four pathologies of NSCLC: adenocarcinoma (ADC), squamous cell carcinoma (SCC), large cell carcinoma (LCD) and adenosquamous cell carcinoma (ASC); ADC is the most commonly found in Asia (Dearden et al., 2013; Midha et al., 2015). Beside genetics, environmental factors, such as smoking, air pollution, occupation and infection, could coordinate to induce the pathogenesis of lung cancer, leading to genetic mutations (Taga et al., 2013; Zhou, 2014). The use of targeted specific genes has increased in therapeutic and treatment decisions for lung cancer. The epidermal growth factor receptor (EGFR) gene, which is overexpressed in NSCLC patients, is also found in exons (Rootthumnong et al., 2010). The engagement between EGFR and epidermal growth factor (EGF) promotes higher phosphorylation of intracellular signaling protein levels than in normal cells (Seshacharyulu et al., 2012). Previous studies showed that genetic mutations in EGFR were associated with lung cancer, especially NSCLC type. The most common mutations in EGFR were deletions in exon 19 affecting the amino acid motif LREA (delE746-750) or the substitution of arginine for leucine at position 858 (L858R) in exon 21(Boch et al., 2013; Lee et al., 2016).
Indeed EGFR might be over activated mainly as a consequence of mutations or due to gene amplification, and its inhibition could be exploited either through anti-EGFR monoclonal antibodies (mAbs) or through small tyrosine kinase inhibitors (TKIs). Therefore, the detection of EGFR mutations using a molecular technique may be an alternative procedure for the evaluation of medication in NSCLC patients. This study aimed to investigate EGFR mutations in patients with NSCLC by polymerase chain reaction-single strand conformational polymorphism (PCR-SSCP) in exons 18 to 21. Moreover, we determined the banding patterns, including normal, mutation and deletion patterns.
Materials and Methods
Specimen collection
NSCLC was diagnosed and confirmed by a pathologist. Specimens were obtained from the Department of Pathology, Faculty of Medicine, Khon Kaen University, Thailand. This study was approved by the Human Research Ethics Committee of Khon Kaen University, Thailand (HE.561262). Lung cancer tissue (10 μm) was collected from 50 patients (30 males and 20 females) who attended Srinagarind Hospital.
DNA extraction
The protocol for the DNA extraction was as follows: firstly, tissues were deparafined by heating at 100°C in alkaline solution. After cooling, the tissue was centrifuged at room temperature for 5 min (Shi et al., 2004). The mixture was transferred into a new micro-centrifuge tube and digestion buffer was added. Then, 100 μl of lysis solution, 100 μl of lysis buffer and 200 μl of enzyme reaction solution were added for cellular and nuclear membrane breaking. The reaction was incubated at 37 °C for 20 min before adding 5 μl of 20 mg/ml Proteinase K, and then incubated at 37 °C overnight (Wang et al., 1994). Finally the mixture was precipitated using the phenol:chloroform:isoamyl alcohol method. The quantity and quality of DNA were determined by a NanoVue™ spectrophotometer (GE Healthcare UK Limited, UK) and 1% agarose gel electrophoresis under UV light by a gel document system Syngene, USA.
PCR for sample preparation
The EGFR gene was amplified using the PCR technique employing specific primers (Table 1). The appropriate temperature for each exon was optimized. The gradient Tm was calculated by a G-Strom GS482 Thermo Cycler (Gene Technology Ltd, UK). PCR under optimal conditions was performed in 25 μl of reaction solution, including 10× PCR buffer, 2.5 μM of each specific primer, 25 mM MgCl2, 2.5 mM dNTP, five units of Taq DNA polymerase and 20 ng/μl of DNA products. Amplifications were performed with the following cycling profile: pre-denaturation at 95 °C for 5 min followed by 40 cycles of denaturing 95 °C for 30 s; annealing for exon 18 at 58.5 °C, exon 19 at 57.5 °C, exon 20 at 55.5 °C and exon 21 at 59.5 °C; extension at 72°C for 1 min and the last cycle was followed by a final extension at 72 °C for 10 min. Amplified products were analyzed by agarose gel electrophoresis and ethidium bromide staining (Figure 1).
Table 1.
Specific Primer for Conventional PCR Detected EGFR Mutation
| Gene | Product Size (bp.) | Chromosome/Locus | Sequence |
|---|---|---|---|
| Exon18 | 277 | 7p11.2 | F:5’-AGGGCTGAGGTGACCCTTGT-3’ |
| R:5’-TCCCCACCAGACCATGAGAG-3’ | |||
| Exon19 | 192 | F:5’-ACCATCTCACAATTGCCAGTTAAC-3’ | |
| R:5’ -ACCATCTCACAATTGCCAGTTAAC-3’ | |||
| Exon20 | 123 | F:5’-ATGGCCAGCGTGGACAAC-3’ | |
| R:5’-TTTGTG TTCCCGGACATAGTC-3’ | |||
| Exon21 | 212 | F: 5’ -TCACAGCAGGGTCTTCTCTGTTT- 3 | |
| R:5’ -ATGCTGGCTGACCTAAAGCC-3’ |
Figure 1.

Optimized Condition for PCR of Control and Exons 18 - 21. Lane M for marker, lanes 1, 3, 5, 7 and 9 for blank and lane 2 for control (β-actin, 295 bp.). Lanes 4, 6, 8 and 10 were exon 18 (277 bp.), 19 (192 bp.), 20 (123 bp.) and 21 (212 bp.) respectively. The annealing temperature was different for each exon, for exon 18 (58.5 °C), 19 (57.5 °C), 20 (55.5 °C) and 21 (59.5 °C)
PCR-SSCP for EGFR gene mutation
Single-strand conformation polymorphism (SSCP) analysis was first described by Orita and coworkers (Orita et al., 1989), and in this technique, double-stranded (ds) DNA is denatured to single-stranded (ss) DNA. PCR-SSCP is associated with the following steps: PCR amplification of interesting DNA sequences and denaturation of double-stranded DNA (dsDNA) to single-stranded DNA (ssDNA) by heating in loading buffer; then cooling of the ssDNA to maximize self-annealing; and finally detection of mobility differences in the single-stranded DNAs by electrophoresis on a non-denaturing polyacrylamide gel. A single base change in the sequence can cause single-stranded DNA to migrate differently. This study investigated suitable conditions for detecting EGFR mutant genes in four exons of NSCLC patients using PCR products from Veriti® Thermal Cycler, Serial No; 2990216160, GenePlus Co., Ltd., USA. The reaction was mixed, spun down and denatured at 97°C for 10 min. Then the reaction was immediately cooled on ice at -20 °C for 10 min. The ratio of acrylamide:bis for gel preparation was 50:1. The optimum PAGs exon 18 to exon 21 were determined. The voltage used was 150 V on ice for 150 min. The gel was separated from the gel cassette carefully in water due to the polyacrylamide gel being quite thin. After that, the gel was stained with ethidium bromide and photographed under ultraviolet light.
Statistical Analysis
The variables measured in the study were investigated for associations using the SPSS statistics program, v. 17.0, for data analysis. The association of EGFR mutational status, as the independent variable, with age, sex and histology was determined.
Results
A total of 50 FFPE NSCLC tissue samples were screened for the mutant EGFR gene using the PCR technique. The samples were divided in 30 case (60%) males and 20 case (40%) females and included the ages from 37-75 years. There were two cases found from patients with an age lower than 40 year (4%), 12 cases from 41 – 50 year (24%), 20 cases from 51 – 60 year (40%), 11 cases from 61 – 70 year (22%) and five cases from over 71 year (10%) (Figure 2). In addition, pathologies of NSCLC patients were divided in ADC 47(94%), SCC 1(2%) and BAC 2(4%), respectively. The results showed that exons 18, 19, 20 and 21 had 6(12%), 24(48%), 36 (72%) and 24 (48%) cases, respectively (Figure 2).
Figure 2.

Age Ranges of NSCLC Patients.
In addition, the PCR-SSCP showed that exons 18, 19, 20 and 21 had 3(6%), 8(16%), 13(26%) and 13 (26%) cases, respectively (Figures 3-4). There were no mutations in exons 18 and 21. Only one case (12.5%) found a deletion at 2235_2249 del ggaattaagagaagc or Del E746-A750 was the most common mutation in exon 19 (Figures 5-6). An insertion at 2313_2314insCAC or 771_772insHis one case (7.6%) of exon 20 was also found in this study. The deletion or insertion of a mutation was found with different patterns when compared to the control, which was confirmed by DNA sequencing. The detections and insertions were shown with different patterns when compared to the control cases, and they were correlated with the DNA sequencing result (Figures 7-8).
Figure 3.

PCR-SSCP Patterns on 11% PAG for Exon 19 at 150 v for 150 mins. Lane C, control and NSCLC cases 32, 33, 35, 37 and 38 in lanes 1 - 5 respectively
Figure 4.

PCR-SSCP Pattern on 10% PAG for Exon 20 at 150 v for 150 mins. Lane C: control and NSCLC cases 4, 6, 7 and 12 in lanes 1- 4 respectively
Figure 5.

DNA Sequencing of Control Normal EGFR Gene Exon 19, 100% Identity
Figure 6.
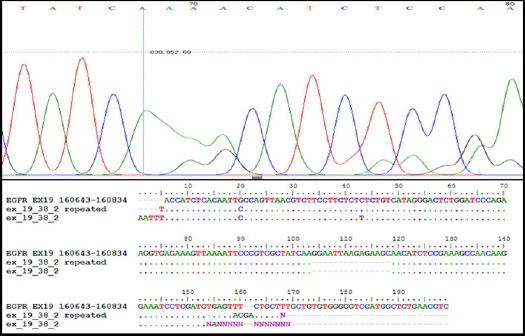
Histogram and Data Base of EGFR Exon 19 (Case No. 38) That had 15 bp Deletion at 2235_2249 Delggaattaagagaagc, Amino Acid Change (E746-A750del.), Detected by DNA Sequencing
Figure 7.
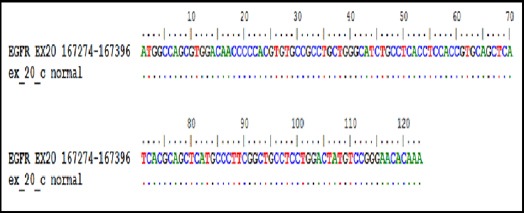
DNA Sequencing of Control Normal EGFR Gene Exon 20,100% Identity
Figure 8.

DNA Sequencing of Insertion at 2313-ACC-2314; Overlapping Peaks of Normal and Mutant, Insertion of Three Base Nucleotides -GGT- at 2313-ACC-2314, Producing Amino Acid Histamine Insertion at Amino Acid Position 772
Table 2.
| Parameters | NSCLC patients (n = 50) | Mean age ± SEM |
|---|---|---|
| Age range | N (%) | |
| < 40 | 2 (4) | 37.00± 3.00 |
| 41-50 | 12 (24) | 46.08±0.76 |
| 51-60 | 20 (40) | 55.65±0.16 |
| 61-70 | 11 (22) | 66.00±1.00 |
| >70 | 5 (10) | 75.00±1.26 |
| Total | 50 (100) | |
| Sex | ||
| Female | 20(40) | |
| Male | 30(60) | |
| Histological type | ||
| ADC | 47 (94) | |
| SCC | 1 (2) | |
| BAC | 2 (4) | |
| BAC | ||
NS, not significant; ADC, Adenocarcinoma; SCC, Squamous cell carcinoma; BAC, bronchioloalveolar carcinoma
Discussion
From the analyses of FFPE samples of NSCLC patients in Srinagarind hospital, Khon Kaen University, we have detected up to 58% of mutant EGFR genes by PCR-SSCP. The PCR-SSCP technique yielded different banding patterns according to types of mutation which is a rapid and reliable method for screening mutant EGFR genes. The method is also costs less than DNA sequencing because only one sample from each pattern is selected for DNA sequencing while the results are still correlated. The resulting gene mutations should be beneficial for choosing effective and appropriated chemotherapy.
The detection of the mutant EGFR gene in FFPE of NSCLC in lung cancer patients is difficult and challenging. High quality DNA is required for molecular techniques (Khoo et al., 2015). This study selected a new block of tissue to decrease DNA degradation due to formalin exposure, prevent a major crosslink problem in the DNA and the denaturation of the DNA to short fragments that are difficult to amplify. Heat induced retrieval in an alkaline solution, which is a simple technique, was conducted, and samples were deparaffined by boiling at 100 °C for 30 min. The NaOH alkaline solutions play a role in the disruption of cell membranes for solubilization of proteins through the ionization of aspartine (A), glutamin (G), cysteine (C) and tyrosine (T) residues, while the DNA structure is relatively stable under an alkaline condition. These events promote highly efficient DNA extraction and results in a higher yield (Rudbeck and Dissing, 1998; Shi et al., 2004; Taga et al., 2013; Turashvili et al., 2012).There are two molecular techniques for EGFR gene study, firstly long preserved biopsies that lead to interference in the PCR amplification and secondly the contamination of normal and tumor cells in FFPE. However, if the DNA is a good quality, it can be amplified even if there is a low yield from the DNA extraction. SSCP analysis was first described by Orita et al., (1989). SSCP is a rapid method for the detection of minor sequence changes in polymerase chain reaction-amplified DNA. This technique has been used widely to detect mutations in oncogenes, tumor suppressor genes and genes responsible for genetic diseases. However, many factors can influence the sensitivity of SSCP, and its optimization is highly empirical (Hayashi, 1992; Hayashi and Yandell, 1993). The PCR-SSCP technique that was used in this study could screen EGFR gene mutations in lung cancer patients by combined SSCP that finds minor sequence changes first and then uses PCR to amplify the DNA sequences of interest. Thus, a change of only 1 bp in the DNA could be detected.
From previous studies, a mutation in the EGFR at 2235_2249 delggaattaagagaagc (Del E746-A750) in exon 19 (Marchetti et al., 2005; Jorge et al., 2014) and a new insertion at 2313_2314insCAC position (771_772insHis) in exon 20 have been found. The database showed a correlation between the pattern from the PCR-SSCP and that from the control that was confirmed by DNA sequencing, which previous studies have not reported (Arcila et al., 2013; Improta et al., 2016; Zupa et al., 2012).
In conclusion, we demonstrated the detection of a mutant EGFR gene from FFPE in NSCLC patients by PCR-SSCP. This technique could screen mutant EGFR genes via the different patterns in the bands. PCR-SSCP is a rapid and reliable method for screening mutant EGFR genes, and it costs less than DNA sequencing because only one sample from each pattern is selected for DNA sequencing while the results are still correlated. This technique may help clinicians in rapidly choosing the optimal therapy for patients.
Acknowledgements
We would like to thank all the staff in the Department of Pathology, Faculty of Medicine, Khon Kaen University for providing the FFPE of NSCLC patients. Many thanks to all staff who supported the work and gave advise. The authors appreciated the Faculty of Associated Medical Sciences and the Faculty of Graduated School, Khon Kean University for research grant support.
References
- Arcila ME, Nafa K, Chaft JE, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas:prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12:220–9. doi: 10.1158/1535-7163.MCT-12-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch C, Kollmeier J, Roth A, et al. The frequency of EGFR and KRAS mutations in non-small cell lung cancer (NSCLC):routine screening data for central Europe from a cohort study. BMJ Open. 2013;3:e002560. doi: 10.1136/bmjopen-2013-002560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan BA, Hughes BG. Targeted therapy for non-small cell lung cancer:current standards and the promise of the future. Transl Lung Cancer Res. 2015;4:36–54. doi: 10.3978/j.issn.2218-6751.2014.05.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearden S, Stevens J, Wu YL, Blowers D. Mutation incidence and coincidence in non small-cell lung cancer:meta-analyses by ethnicity and histology (mutMap) Ann Oncol. 2013;24:2371–6. doi: 10.1093/annonc/mdt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide:sources, methods and major patterns in GLOBOCAN. Int J Cancer 2012. 2012;136:359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Hayashi K. PCR-SSCP:a method for detection of mutations. Genet Anal Tech Apple. 1992;9:73–9. doi: 10.1016/1050-3862(92)90001-l. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yandell DW. How sensitive is PCR-SSCP. Hum Mutat. 1993;2:338–46. doi: 10.1002/humu.1380020503. [DOI] [PubMed] [Google Scholar]
- Improta G, Pettinato A, Gieri S, et al. Epidermal growth factor receptor exon 20 p.S768I mutation in non-small cell lung carcinoma:A case report combined with a review of the literature and investigation of clinical significance. Oncol Lett. 2016;11:393–8. doi: 10.3892/ol.2015.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo C, Rogers TM, Fellowes A, Bell A, Fox S. Molecular methods for somatic mutation testing in lung adenocarcinoma:EGFR and beyond. Transl Lung Cancer Res. 2015;4:126–41. doi: 10.3978/j.issn.2218-6751.2015.01.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Lee B, Choi YL, et al. Non-small cell lung cancer with concomitant EGFR, KRAS, and ALK mutation:clinicopathologic features of 12 cases. J Pathol Transl Med. 2016;50:197–203. doi: 10.4132/jptm.2016.03.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology:a systematic review and global map by ethnicity (mutMapII) Am J Cancer Res. 2015;5:2892–911. [PMC free article] [PubMed] [Google Scholar]
- Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A. 1989;86:2766–70. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rootthumnong E, Lertprasersuke N, Saeteng S, et al. Mutation analysis at Tyrosine kinase domain of EGFR of lung cancer patient in the upper Northern Thailand. KKU Res J. 2013;10:55–65. [Google Scholar]
- Rudbeck L, Dissing J. Rapid, simple alkaline extraction of human genomic DNA from whole blood, buccal epithelial cells, semen and forensic stains for PCR. BioTechniques. 1998;25:588–92. doi: 10.2144/98254bm09. [DOI] [PubMed] [Google Scholar]
- Seshacharyulu P, Ponnusamy MP, Haridas D, et al. Targeting the EGFR signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16:15–31. doi: 10.1517/14728222.2011.648617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SR, Datar R, Liu C, et al. DNA extraction from archival formalin-fixed, paraffin-embedded tissues:heat-induced retrieval in alkaline solution. Histochem Cell Biol. 2004;122:211–8. doi: 10.1007/s00418-004-0693-x. [DOI] [PubMed] [Google Scholar]
- Taga M, Eguchi H, Shinohara T, et al. Improved PCR amplification for molecular analysis using DNA from long-term preserved formalin-fixed, paraffin-embedded lung cancer tissue specimens. Int J Clin Exp Pathol. 2013;6:76–9. [PMC free article] [PubMed] [Google Scholar]
- Turashvili G, Yang W, McKinney S, et al. Nucleic acid quantity and quality from paraffin blocks:defining optimal fixation, processing and DNA/RNA extraction techniques. Exp Mol Pathol. 2012;92:33–43. doi: 10.1016/j.yexmp.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Wang L, Hirayasu K, Ishizawa M, Kobayashi Y. Purification of genomic DNA from human whole blood by isopropanol-fractionation with concentrated Nal and SDS. Nucleic Acids Res. 1994;22:1774–5. doi: 10.1093/nar/22.9.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C. Lung cancer molecular epidemiology in China:recent trends. Transl Lung Cancer Res. 2014;3:270–9. doi: 10.3978/j.issn.2218-6751.2014.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupa A, Vita G, Landriscina M, et al. Identification of a new insertion in exon 20 of EGFR in a woman with NSCLC. Med Oncol. 2012;29:3198–201. doi: 10.1007/s12032-012-0304-y. [DOI] [PubMed] [Google Scholar]


