Abstract
Objective
To determine whether the gut microbiota shows overabundance of commensal bacteria species in patients with anti-NMDA receptor (NMDAR) encephalitis, similar to patients with MS or neuromyelitis optica where they potentially balance pro- and anti-inflammatory immune responses or participate in disease pathogenesis by molecular mimicry.
Methods
Intestinal microbiota was characterized in patients with NMDAR encephalitis (n = 23, mean age: 34 ± 12.7 years; 21 females) and age/sex/environment-matched healthy controls (n = 24, 40 ± 14.2 years; 22 females) using stool bacteria 16S rDNA sequencing and classification in operational taxonomic units (OTUs). Statistical analyses focused on intraindividual and interindividual bacterial diversity and identification of differentially abundant taxa.
Results
Patients with NMDAR encephalitis and controls had similar microbiome profiles of the gut microbiota regarding intraindividual bacterial diversity, OTU distribution, ratio between regional and local species diversity when testing all OTUs, and genera with a relative abundance greater than 0.5%. Similarly, the subgroup of NMDAR encephalitis patients with an ovarian teratoma (n = 3) showed no differences in microbiome variation compared with controls. Patients in the acute encephalitis stage (n = 8) showed significant differences in the numbers of Clostridium XVIII, Clostridium IV, Oscillibacter, Prevotella, and Blautia; however, significance was lost after correction for multiple testing.
Conclusion
Patients with NMDAR encephalitis and controls both had a normal gut microbiome. The lack of overabundance of certain bacterial species in patients suggests that microbiome changes are no major contributors to the pathogenesis, disease course, or prognosis in NMDAR encephalitis. Despite the small sample size and heterogeneous groups, findings indicate differences to other neuroimmunologic diseases.
The role of the gut microbiota has been increasingly recognized in neurologic diseases. Overabundance of commensal bacteria species was seen in Guillain-Barré syndrome, Parkinson disease, MS, and neuromyelitis optica spectrum disorder (NMOSD).1–8 Experimental data in germ-free mice demonstrated increased susceptibility to experimental autoimmune encephalomyelitis after selective bacterial colonization, indicating a role of the gut microbiota for balancing proinflammatory and anti-inflammatory immune responses.9,10 Intestinal microbiota may even be a novel therapeutic target for extraintestinal inflammatory diseases as shown for possible beneficial effects in patients with MS from nutritional administration of a probiotic11 and potentially from fecal microbial transplantation.12 Recently, overabundance of the bacterium Clostridium perfringens, a commensal bacterium of the gut microbiota, was demonstrated in 16 patients with NMOSD, indicating a potential role in the disease pathogenesis.5
Similar to NMOSD, anti-NMDA receptor (NMDAR) encephalitis is an antibody-mediated disease of the CNS, and there are even cases of overlap between the 2 entities.13 Autoantibodies against the NR1 subunit of the NMDAR are directly pathogenic and underlie the clinical disease spectrum ranging from amnesia, psychosis, and epileptic seizures to dyskinesias, coma, and vegetative dysfunction.14,15 The potential role of the gut microbiome for the development of NMDAR encephalitis is particularly interesting as only few disease triggers are known, such as ovarian teratomas,16 viral infections of the brain,17,18 and potentially seasonal factors.19 We therefore aimed to investigate the intestinal microbiome of patients with NMDAR encephalitis and healthy controls.
Methods
Standard protocol approvals, registrations, and patient consents
The study was approved by the Charité ethics committee (EA1/274/16) and Charité data protection (#0626/16/ST3). All study participants gave their written informed consent.
Study population
Twenty-eight patients with NMDAR encephalitis were recruited from the Charité Centre for Autoimmune Encephalitis during spring 2017. Five patients and 3 controls received antibiotic treatment within the last 6 weeks before stool collection (for cystitis, pyelonephritis, tonsillitis, sinusitis, or bacterial vaginosis) and were therefore excluded from the analyses. Of the remaining 23 patients with NMDAR encephalitis, 2 (9%) had acute encephalitis, 6 (26%) were in the recovery phase with persisting clinical deficits, and 15 patients (65%) had recovered. Three patients (13%) had an ovarian teratoma removed during clinical workup. Almost all patients (n = 21; 91%) were female; thus, sex was not considered a covariate. Demographic and clinical characteristics are shown in table e-1 (links.lww.com/NXI/A156).
All patients had received immunotherapy according to current guidelines. These included the following:
Steroids in 18 patients (78%), 1,000 mg IV methylprednisolone for 5 days, and mean 2.0 cycles;
IV immunoglobulins in 11 (48%), 2 g/kg body weight, and mean 1.9 cycles;
Plasmapheresis in 15 (65%) and mean 3.6 series of 5–10 sessions each;
Rituximab in 13 (57%), 1,000 mg per cycle every 6 months, and mean 2.7 cycles (3 patients ongoing);
Azathioprin in 3 (13%), 250 mg/d for 4 months, 75 mg/d for 1.5 years, and 100 mg/d for 6 months, respectively;
Cyclophosphamide in 2 (9%), 750 mg/kg once in 1 patient, and 5 times in the second;
Methotrexate in 1 (4%), 10–15 mg/wk over 3.5 years;
Bortezomib in 1 patient (4%) and 2 cycles of 4 subcutaneous injections of 1.3 mg/m2.
Five patients (22%) still received immunosuppression at the time of stool collection. In the others, the mean immunotherapy-free interval was 4.4 years.
The control group consisted of 24 case-matched healthy individuals. Matching was prioritized for similar living environment, lifestyle, and dietary habits. Most controls were friends of the same sex and age or family members (mostly siblings) who shared the apartment. Controls had no history of autoimmune or gastrointestinal disease or cancer.
Study protocol and clinical assessment
Patients and controls collected fecal samples at home in standard stool collection tubes. The samples were shipped immediately (within 24 hours) at room temperature and were stored at −80°C until processing.
A standardized survey was completed by all patients and controls including demographic data (birth, sex, body size and weight, and size of city of residence [metropolis, city, center city, small town, and village]), information on smoking history (never smoked, current smoker, and the number of pack-years), general activity level (type and extent of physical activity), defecation history (general character of feces using Bristol stool scales, incidence of constipation or diarrhea within the last 3 months [never, sometimes 25%, often 50%, very often 75%, and always 100%], number of average defecation per week, rectal tenesmus, and presence of hematochezia and abdominal pain), dietary habits (nutritional style [omnivore, ovo-lacto-vegetarian, pesco-vegetarian, vegan diet, and others] and regular intake of probiotics), preexisting illnesses (including fever in the last 7 days), regular medication (beginning of intake, cause of medication, dosage, and antibiotics within the last 6 months), current general state of health (very good, impaired, bad, very bad, and unacceptable), need for current antibiotic treatment, and travel history (longer than 4 weeks within the last 12 months).
DNA extraction, 16S rDNA sequencing, and quality control
DNA was extracted using the QIAcube and the QIAamp DNA stool kit (Qiagen) and a prior beat-beating step. Variable regions v1-v2 of the 16S rRNA gene were amplified using the primer pair 27F/338R. PCR products were normalized using the SequalPrep Normalization Plate Kit (Life Technologies) and sequenced on an Illumina MiSeq (2 × 300 bp).
Raw data were obtained with exact agreement of the index sequences, demultiplexed with a dual-indexing approach, and subjected to quality control. This consisted of trimming low-quality sequence ends,20 combining forward and backward amplicon reads into a single sequence (VSEARCH), quality control based on estimated error (VSEARCH), and sequence quality (FastX toolkit). Reference and DeNovo-based identification of chimeric sequences (VSEARCH) took place,21 as well as a filtering step to exclude nonbacterial (and thus unspecific or erroneous) sequence data (Simple non-Bayesian taxonomy classifier [SINTAX]). The clean sequences were classified in operational taxonomic units (OTUs) (VSEARCH), normalized to 10,000 sequences per sample, and taxonomically annotated (SINTAX).22 Based on this information OTU, an operational definition used to classify groups of closely related individuals, and taxon abundance tables were created for subsequent analysis.
Statistical analysis
Statistical analysis was performed on alpha- (intraindividual) and beta- (inter-individual) diversity measures (Bray-Curtis dissimilarity and UniFrac distance). For differences in alpha-diversity, the Wilcoxon rank-sum test was used when value distribution deviated from normality, otherwise the 2-sample t test. Alpha-diversity indices contained the Shannon diversity index (accounts for both abundance and evenness of the species present) and the Chao1 index (reflecting species richness).
Unconstrained multidimensional scaling (MDS) plots of beta-diversity measures were generated using the cmdscale function in R.23 MDS plots generally visualize the level of similarity of individual cases of a data set. To test for differences in beta-diversity, permutational multivariate analysis of variance was performed using the adonis function of the vegan software package with the option sqrt.dist = T when using abundance tables, but not when using UniFrac distances, and 10,000 permutations. Tests for differential abundance were performed using zero-truncated generalized linear models (GLMs) with negative binomial distribution on taxonomic groups present in at least 2 samples per group and 10 samples in total.
Data availability
Anonymized data will be shared by request from any qualified investigator.
Results
Study participants
Subject characteristics of the 23 patients with NMDAR encephalitis and 24 unaffected controls are given in table e-1 (links.lww.com/NXI/A156). Forty-three of 47 participants were female, and the mean age was 34 years in patients and 40 years in controls. There were no differences between groups regarding smoking, obesity, place of residence, defecation at the time of stool collection with general stool consistency and stool frequency per week, occurrence of hematochezia, occasional abdominal pain, nutritional intake, diet, travel history, and general medication, such as for hypertension, contraception, or hypothyroidism (table e-1). Five (22%) patients with NMDAR encephalitis with persisting symptoms still received immunosuppression or neuroleptic medication (rituximab [n = 3], cortisone [n = 1], bortezomib [n = 1], risperidone [n = 2], and lamotrigine [n = 1]), which could theoretically alter the gut microbiome. Two patients and 3 controls had probiotic intake.
Similar microbiome profiles in patients with NMDAR encephalitis and controls
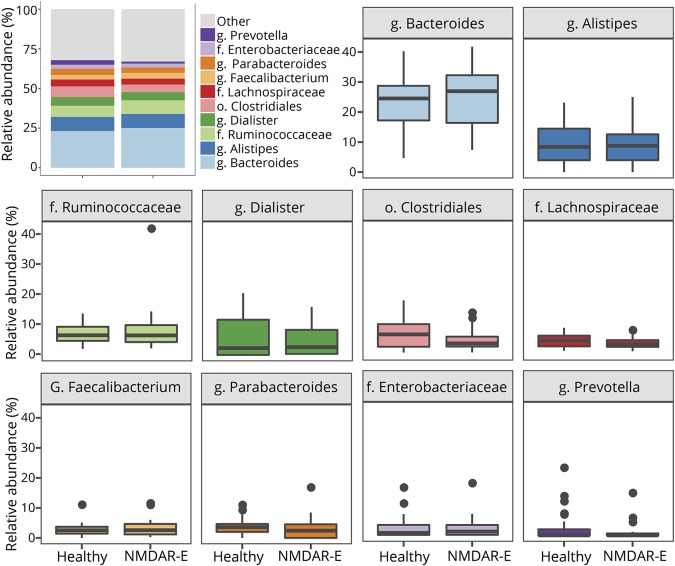
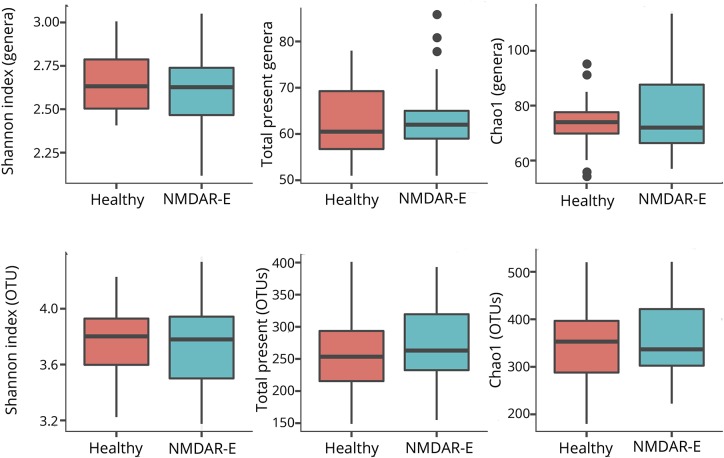
The average microbiome profile of patients with encephalitis and healthy controls did not reveal any differences between groups (figure 1). Regarding alpha-diversity (reflecting intraindividual bacterial diversity), no difference in species diversity within the gut microbiota was observed. The Shannon diversity index based on OTU distribution (groups of closely related individuals) did not reveal any significant difference between both groups. Similarly, the Chao1 index was not different (Mann-Whitney U test, figure 2).
Figure 1. Average gut flora profiles of patients with NMDAR encephalitis and healthy controls.
Shown are all families and genera with a mean abundance of more than 2.5%. Remaining genera are summarized in the group “other.”
Figure 2. Boxplots of the alpha-diversity indices.
There were no significant differences in diversity (Shannon Index) and number or richness (Chao1) of gut microbiome genera and species within genera (OTUs) between patients with NMDAR encephalitis and healthy controls.
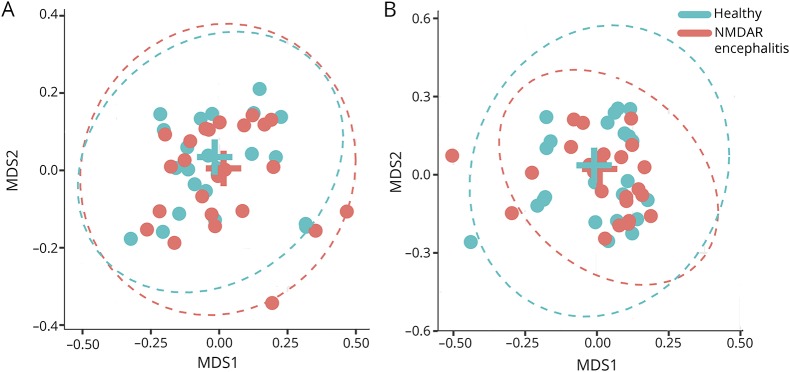
Beta-diversity (interindividual dissimilarity) also did not differ significantly between patients and healthy controls. The lack of differences for genus and OTU is visualized using MDS plots (figure 3), i.e., showing the level of similarity of individual cases (adonis analysis: genera: p > 0.3; OTU: p > 0.8). All OTUs and genera with a mean abundance greater than 50 reads per sample (equivalent to 0.5% relative abundance) were tested in a GLM or hurdle model. None of the results were significantly different (all q > 0.05) after correction for multiple testing.
Figure 3. MDS plots of Bray-Curtis distances at the genera and OTU levels.
Data show a high level of similarity regarding interindividual differences between patients with NMDAR encephalitis and healthy controls for genera (A) and OTU level (B). The ellipses represent the 99% CIs of the SDs within the groups around the mean values marked by the position of the group names.
Alpha- and beta-diversity in patients with NMDAR encephalitis with a teratoma
Three of the 23 patients with NMDAR encephalitis had a teratoma, thus representing a subgroup with potentially different immunologic mechanisms. Patients were compared with age- and sex-matched controls living in the same household. None of the 3 patients received immunosuppressive medication at the time of stool collection, and 2 had l-thyroxine for hypothyroidism. None of the controls required medication including antibiotics. All subjects were on omnivore diet, and the stool was inconspicuous.
No differences in microbiome variation were seen between patients with NMDAR encephalitis after teratoma removal and controls. Alpha-diversity was equal between both groups regarding Shannon diversity (genera/OTUs), the number of genera/OTUs, or the Chao1 estimator at the generic and OTU levels. Also, beta-diversity was not significantly different at the genus (p = 0.62) and OTU levels (p = 0.32, data not shown).
Alpha- and beta-diversity in patients with NMDAR encephalitis with acute illness
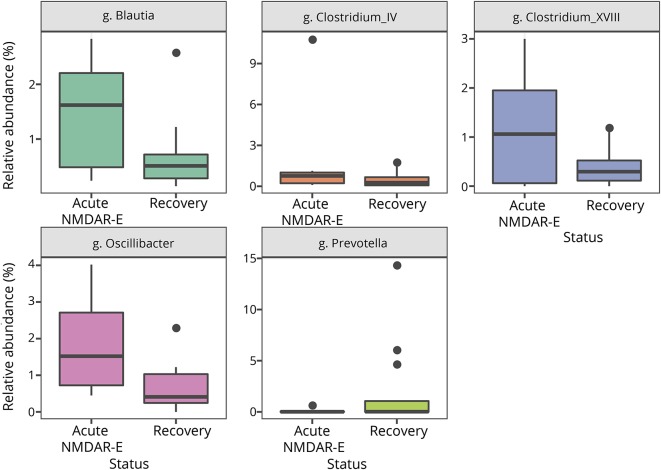
Eight of the 23 patients still had clinical symptoms of NMDAR encephalitis at the time of examination. Seven (88%) were female, and 5 (63%) received immunosuppressive therapy at the time of stool collection. These patients were compared with the 15 NMDAR encephalitis after disease recovery (14 [93%] females, 2 [13%]) on immunosuppressive therapy. All participants had normal stool habits and stool conditions at the time of examination. Comparing the intraindividual bacterial diversity between 2 patient groups showed no significant differences in the Shannon diversity index. However, significant differences in the number of genera were detected for Clostridium XVIII, Clostridium IV, Oscillibacter, Prevotella, and Blautia (figure 4), although significance was lost after Bonferroni correction (table e-2, links.lww.com/NXI/A156). Regarding beta-diversity, there were again no differences at the genus (p = 0.082) and OTU levels (p = 0.15).
Figure 4. Overabundance of bacterial species in active NMDAR encephalitis.
Five genera showed significant difference in overall abundance in patients with acute NMDAR encephalitis compared with recovered patients. The differences were not present after correction for multiple testing.
Discussion
In this first study on the gut microbiome in patients with NMDAR encephalitis, data showed no overabundance of certain bacterial taxa compared with controls. Both patients and healthy participants had a “normal” microbiome consistent with previous reports of the healthy gut where gram-negative Bacteroides and gram-positive Firmicutes, including Clostridiales and Lactobacillaceae, dominate.24,25 Similarly, no differences were seen in subgroups of patients with ovarian teratomas or in patients during the active disease phase.
The study therefore provides no support for the hypothesis that microbiome changes are major contributors to pathogenesis, disease course, or prognosis in NMDAR encephalitis, although it is still possible that higher patient numbers could reveal differences. It is also possible that the microbiome is different only during the initial phase of the disease compared with remission. Indeed, our subgroup of patients with acute encephalitis had overabundance of 5 genera including Clostridium XVIII and IV, which, however, did not withstand statistical correction for multiple testing.
In the somewhat related neurologic disease NMOSD with autoantibodies targeting the water channel aquaporin-4 (AQP4), C perfringens was enriched in the gut microbiota of 16 patients vs 16 controls.5 As T cells from NMOSD cross-react with a homologous sequence of a C perfringens adenosine triphosphate-binding cassette transporter,26 the authors concluded that the microbiota might be involved in NMOSD pathogenesis.
It is therefore likely that different mechanisms initiate and drive disease in NMOSD and NMDAR encephalitis. For example, approximately one-third of female patients with NMDAR encephalitis have an ovarian teratoma, which contains neuronal elements expressing NMDAR and is thought to trigger the encephalitis.16,27 In contrast, tumors are rare in patients with NMOSD. Another potential difference is the role of T cells for disease. Although T cells in patients with NMOSD proliferate in response to AQP4 protein,26,28 no such role has yet been established for T cells from patients with NMDAR encephalitis. In fact, neuropathologic findings including the absence of relevant numbers of T cells in the brain suggested that NMDAR encephalitis is a predominantly humoral autoimmune disease.27,29 Furthermore, molecular mimicry of NMOSD T cells between AQP4 and a Clostridium protein might contribute to NMOSD pathogenesis, whereas no cross-reaction of T cells and antibodies were so far observed in NMDAR encephalitis. The absence of detection of a consistent infectious agent makes an immune response by molecular mimicry unlikely.14
Differences in immunosuppression between groups had no effect on the composition of the gut microbiota, although conclusions about a clear effect would likely require higher patient numbers or stool samples from untreated patients with NMDAR encephalitis. These might be difficult to obtain as immunotherapy is usually started immediately after confirmation of the diagnosis with positive serum and CSF antibodies. In NMOSD, rituximab treatment did not account for changes in the microbiome.5
Despite the limitations of relatively small sample sizes, heterogeneity of the groups, and the majority of patients being in the recovery phase, the present work argues against a major influence of the gut microbiome in NMDAR encephalitis. It thereby reminds us of how little we still know about etiology and disease mechanisms in NMDAR encephalitis, in particular about the role of T cells and further immune cells. It is finally possible that microbiota-driven effects are still in place—not reflected by mere overrepresentation of certain bacterial species, but rather by dysfunctional immune responses to numerically normal microbiota, potentially shaped by genetic, metabolic, or psychosocial factors.
Acknowledgment
The authors gratefully acknowledge the technical help of Doreen Brandl, Ilona Urbach, Ines Wulff, and Tonio Hauptmann. They acknowledge the contributions of the patients and their families to this project.
Glossary
- AQP
aquaporin-4
- EAE
experimental autoimmune encephalomyelitis
- GLM
generalized linear model
- MDS
multidimensional scaling
- NMDAR
NMDA receptor
- NMOSD
neuromyelitis optica spectrum disorder
- OTU
operational taxonomic unit
Appendix. Authors

Study funding
No targeted funding reported.
Disclosure
The authors report no conflicts of interest. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NN.
References
- 1.Miyake S, Kim S, Suda W, et al. . Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to clostridia XIVa and IV clusters. PLoS One 2015;10:e0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berer K, Mues M, Koutrolos M, et al. . Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011;479:538–541. [DOI] [PubMed] [Google Scholar]
- 3.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med 2016;375:2369–2379. [DOI] [PubMed] [Google Scholar]
- 4.Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, et al. . Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol 2010;185:4101–4108. [DOI] [PubMed] [Google Scholar]
- 5.Cree BA, Spencer CM, Varrin-Doyer M, et al. . Gut microbiome analysis in neuromyelitis optica reveals overabundance of Clostridium perfringens. Ann Neurol 2016;80:443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez Y, Rojas M, Pacheco Y, et al. . Guillain-Barre syndrome, transverse myelitis and infectious diseases. Cell Mol Immunol 2018;15:547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van den Berg B, Walgaard C, Drenthen J, et al. . Guillain-Barre syndrome: pathogenesis, diagnosis, treatment and prognosis. Nat Rev Neurol 2014;10:469–482. [DOI] [PubMed] [Google Scholar]
- 8.Pröbstel AK, Baranzini SE. The role of the gut microbiome in multiple sclerosis risk and progression: towards characterization of the “MS microbiome”. Neurotherapeutics 2018;15:126–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berer K, Gerdes LA, Cekanaviciute E, et al. . Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci U S A 2017;114:10719–10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A 2011;108(suppl 1):4615–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tankou SK, Regev K, Healy BC, et al. . A probiotic modulates the microbiome and immunity in multiple sclerosis. Ann Neurol 2018;83:1147–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wing AC, Kremenchutzky M. Fecal microbial transplantation in multiple sclerosis: trial design. Neurology 2018;90:P2.356. [Google Scholar]
- 13.Titulaer MJ, Höftberger R, Iizuka T, et al. . Overlapping demyelinating syndromes and anti–N-methyl-D-aspartate receptor encephalitis. Ann Neurol 2014;75:411–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalmau J, Lancaster E, Martinez-Hernandez E, et al. . Clinical experience and laboratory investigations in patients with anti-NMDAR encephalitis. Lancet Neurol 2011;10:63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kreye J, Wenke NK, Chayka M, et al. . Human cerebrospinal fluid monoclonal N-methyl-D-aspartate receptor autoantibodies are sufficient for encephalitis pathogenesis. Brain 2016;139:2641–2652. [DOI] [PubMed] [Google Scholar]
- 16.Day GS, Laiq S, Tang-Wai DF, Munoz DG. Abnormal neurons in teratomas in NMDAR encephalitis. JAMA Neurol 2014;71:717–724. [DOI] [PubMed] [Google Scholar]
- 17.Armangue T, Leypoldt F, Malaga I, et al. . Herpes simplex virus encephalitis is a trigger of brain autoimmunity. Ann Neurol 2014;75:317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prüss H, Finke C, Höltje M, et al. . N-methyl-D-aspartate receptor antibodies in herpes simplex encephalitis. Ann Neurol 2012;72:902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adang LA, Lynch DR, Panzer JA. Pediatric anti-NMDA receptor encephalitis is seasonal. Ann Clin Transl Neurol 2014;1:921–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sickle [computer program]. Version 1.33. San Francisco, CA: GitHub; 2011. [Google Scholar]
- 21.Edgar RC, Haas BJ, Clemente JC, et al. . UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC. SINTAX: a simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv 2016. 10.1101/074161. [DOI] [Google Scholar]
- 23.R [computer program]. Vienna, Austria: The R Foundation; 2013. [Google Scholar]
- 24.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falony G, Joossens M, Vieira-Silva S, et al. . Population-level analysis of gut microbiome variation. Science 2016;352:560–564. [DOI] [PubMed] [Google Scholar]
- 26.Varrin-Doyer M, Spencer CM, Schulze-Topphoff U, et al. . Aquaporin 4-specific T cells in neuromyelitis optica exhibit a Th17 bias and recognize Clostridium ABC transporter. Ann Neurol 2012;72:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tüzün E, Zhou L, Baehring JM, et al. . Evidence for antibody-mediated pathogenesis in anti-NMDAR encephalitis associated with ovarian teratoma. Acta Neuropathol 2009;118:737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zamvil SS, Spencer CM, Baranzini SE, Cree BAC. The gut microbiome in neuromyelitis optica. Neurotherapeutics 2018;15:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bien CG, Vincent A, Barnett MH, et al. . Immunopathology of autoantibody-associated encephalitides: clues for pathogenesis. Brain 2012;135:1622–1638. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Anonymized data will be shared by request from any qualified investigator.