Abstract
Uncoupling protein 1 (UCP1) is a mitochondrial protein that is expressed in both brown and beige adipocytes. UCP1 uncouples the mitochondrial electron transport chain from ATP synthesis to produce heat via non-shivering thermogenesis. Due to their ability to dissipate energy as heat and ameliorate metabolic disorders, UCP1-expressing adipocytes are considered as a potential target for anti-obesity treatment. To monitor the expression of UCP1 in live mice in a non-invasive manner, we generated the Ucp1-iRFP720 knock-in (Ucp1-iRFP720 KI) mice, in which the gene encoding a near-infrared fluorescent protein iRFP720 is inserted into the Ucp1 gene locus. Using the heterozygous Ucp1-iRFP720 KI mice, we observed robust iRFP fluorescence in the interscapular region where brown adipose tissue is located. Moreover, the iRFP fluorescence was clearly observable in inguinal white adipose tissues in live mice administered with β3-adrenergic receptor agonist CL316,243. We also found that the homozygous Ucp1-iRFP720 KI mice, which are deficient in UCP1, displayed prominent iRFP fluorescence in the inguinal regions at the standard housing temperature. Consistent with this, the mice exhibited expanded populations of beige-like adipocytes in inguinal white adipose tissue, in which the Ucp1 promoter was dramatically activated. Thus, the Ucp1-iRFP720 KI mice provide a convenient model for non-invasive in vivo imaging of UCP1 expression in both brown and beige adipocytes in live mice.
Introduction
Uncoupling protein 1 (UCP1) is a mitochondrial protein that uncouples respiration from ATP synthesis to produce heat [1]. UCP1 is expressed in brown adipose tissue (BAT) as well as in some white adipose tissues (WATs), in which beige adipocytes are induced upon various stimuli [2]. Recent studies revealed that human adults possess active BAT [3][4][5], which appears to comprise both classical brown adipocytes and beige adipocytes [6][7][8][9]. Human BAT is associated with leanness [3][4][10], and its reduction during aging may accelerate accumulation of body fat [11]. Human BAT may play a protective role against hyperglycemia and related metabolic disorders [12], and, if activated by cold exposure, increases energy dissipation, reduces fat mass, and improves insulin sensitivity [13][14][15][16].
Owing to their anti-obesity potential, brown and beige adipocytes may be manipulated to reduce body weight and ameliorate metabolic disorders [17][18][19]. Beige adipocytes, in particular, are promising targets for treating obesity and its related disorders because of their inducibility in WAT, which is abundant in obese patients. Indeed, when beige adipocytes are ablated by adipocyte-specific knockout of PRDM16, mice develop high-fat diet-induced obesity and insulin resistance [20]. In rodents, beige adipocytes are induced by a number of stimuli, which include cold exposure, thiazolidinediones (peroxisome proliferator-activated receptor gamma [PPAR-γ] agonists) [21], β3-adrenergic receptor agonists [22], or physical exercise [23].
Like classical brown adipocytes, beige adipocytes clearly depend upon UCP1 for thermogenesis in both mice and humans [24][25]. However, studies on UCP1-deficient mice revealed the presence of alternate thermogenesis, which is independent of UCP1 [26][27]. Recent studies on beige adipocytes uncovered mechanisms of alternate thermogenesis, such as creatine-dependent ADP/ATP substrate cycling [28][29][30] and calcium cycling [31], both of which are futile cycles in cellular metabolism that dissipate heat. Thus, beige adipocytes have multiple thermogenic mechanisms that could potentially be targeted and manipulated by drugs. Therefore, in vivo imaging of beige adipocytes could be useful in identifying physiological conditions that induce beige adipocytes, dissecting the molecular mechanisms of beige adipocyte induction, and testing drugs for anti-obesity treatment.
Imaging of biological processes in live mice has been greatly facilitated by the recent development of near-infrared (NIR) fluorescent proteins [32], which are now widely used for in vivo imaging. NIR fluorescent proteins possess red-shifted absorption spectra that range from 670 to 720 nm, and thereby suffer from relatively low absorption by biological components. To emit fluorescence, bacterial phytochrome-based NIR fluorescent proteins require biliverdin, which typically needs to be supplied externally. However, iRFPs, which were engineered to emit fluorescence at the level of endogenous biliverdin in cells, no longer require an external supply of biliverdin [33]. Among the five spectrally distinct iRFPs (iRFP670, iRFP682, iRFP702, iRFP713, and iRFP720), iRFP720 is the most red-shifted NIR fluorescent protein, and presumably best suited for in vivo tissue imaging in live mice [34][35].
Here we used CRISPR/Cas9-based genome editing to generate the Ucp1-iRFP720 knock-in (Ucp1-iRFP720 KI) mice by inserting the iRFP720 gene into the Ucp1 locus and simultaneously inactivating the Ucp1 gene. The mice express UCP1 and iRFP720 under the control of the Ucp1 promoter at its endogenous locus, without any extra amino acids added at their ends. The heterozygous Ucp1-iRFP720 KI mice allowed in vivo imaging of UCP1-expressing brown adipocytes as well as beige adipocytes induced by a β3-adrenergic receptor agonist, CL316,243. The homozygous Ucp1-iRFP720 KI mice showed increased expression of iRFP720 in inguinal WAT (iWAT) when housed at the standard ambient temperature. The Ucp1 promoter activity in the homozygous Ucp1-iRFP720 KI mice is significantly increased in iWAT, which contained clusters of small adipocytes morphologically similar to beige adipocytes. These results indicate that heterozygous and homozygous Ucp1-iRFP720 KI mice may serve as an excellent tool for in vivo imaging of beige adipocyte induction in live mice under various environmental and nutritional conditions.
Materials and methods
Construction of vectors for genome editing
Guide RNA candidates were designed using CRISPR direct software (https://crispr.dbcls.jp) [36]. To construct guide RNA-expression vectors, four pairs of oligonucleotides (5 pmoles each) encoding the guide RNAs were denatured in the buffer containing 10 mM Tris-HCl (pH8.0), 100 mM NaCl and 1 mM ethylenediaminetetraacetic acid (EDTA) at 95°C for 4 minutes and were annealed by gradually cooling to room temperature. The annealed oligonucleotide pairs were individually inserted into the BbsI site of the pX330-U6-Chimeric_BB-CBh-hSp9 vector (Addgene 42230). To construct the donor vector, the mouse Ucp1 gene fragments were prepared by PCR from the BAC clone (pBACe3.6-B6Ng01-162F24, RIKEN RDB07573). Then, the DNA fragment containing the mouse Ucp1 gene (-3.87K to +248 (upstream of ATG)), an artificial intron, the iRFP720 gene, the bovine growth hormone polyadenylation signal (BGHpA) and the mouse Ucp1 gene (+249 to +4.56K) were inserted sequentially into pBluescript II KS (-) vector. The artificial intron, which is a chimera of β-globin and IgG introns, was chemically synthesized. All the constructed plasmids were sequenced entirely to rule out spurious mutations.
In vitro assay for evaluating the gRNA/Cas9-mediated homologous recombination
The mouse Ucp1 gene fragment from +1 to +500, which contains the target sequences of the gRNAs, was inserted into the BamHI site of pCAG-EGxxFP vector (Addgene 50716). Human embryonic kidney (HEK293T) cells were seeded in a 24-well plate at 2x105 cells/well one day before transfection and incubated at 37°C in a 5% CO2 incubator. Each gRNA/Cas9-expression vector (0.5 μg), together with the pCAG-EGxxFP-Ucp1(+1 ~ +500) vector (0.5 μg), was transfected into the cells using Lipofectamine LTX (Thermo Fisher Scientific Inc.), and the fluorescence signal was observed two days after transfection.
Generation of the Ucp1-iRFP720 knock-in mice and genomic PCR analyses
The gRNA2/Cas9 expression vector and the donor vector were injected into the pronuclei of 248 fertilized C57BL/6J mouse (Charles River Laboratories Japan, Inc.) eggs, and 236 embryos were transferred into the oviduct of pseudo-pregnant mice. Eighty-two live pups (F0) were obtained, and genome DNA was prepared from the tail of each F0 mouse and analyzed by PCR for the presence of the integrated iRFP720 gene. To detect the iRFP720 gene, 5 ng each of the genome DNA was analyzed by PCR in a 5 μl reactions containing 2.5 μl of AmpliTaq Gold 360 Master Mix (ThermoFisher Scientific Inc.) and 1.5 pmoles each of the primers. To confirm the correct insertion of the iRFP720 gene at the Ucp1 locus, 10 ng each of the genome DNA was analyzed by PCR using 0.25 unit of PrimeStar GXL DNA Polymerase (TaKaRa Bio Inc.) and 3 pmoles each of the primers in a 10 μl reaction. In addition, 20 ng of genome DNA was analyzed by PCR containing 0.1 unit of KOD FX Neo (TOYOBO CO., LTD.) and 1.5 pmoles each of the primers in a 5 μl reaction. To determine the number of the alleles harboring the iRFP720 gene, 20 ng of the genome DNA was analyzed by PCR containing 0.2 unit of KOD FX Neo and 1.5 pmoles each of the primers in a 5 μl reaction. All the PCR reactions were performed according to the manufactures’ instructions, and the sequences of primers are shown in the S1 Table. Genome DNA from a wild-type (WT) mouse was used as a negative control, and a DNA fragment that spans the upstream and downstream regions of the iRFP720-knock-in Ucp1 gene, including all primer target sites, was used as a positive control.
In vivo imaging of the Ucp1-iRFP720 knock-in mice
Mice (male, 16~20 weeks old) were housed at 22–24°C on a 10hr/14hr dark-light cycle, with free access to water and food. Mice were routinely fed standard chow (MF, Oriental Yeast Co., Ltd.), which were replaced with low-fluorescence chow (iVid#2, Oriental Yeast Co., Ltd.) one week before observation of fluorescence by IVIS. Fluorescence images of the Ucp1-iRFP720 knock-in mice were captured and analyzed by IVIS Spectrum imaging system (Summit Pharmaceuticals International Corp.). Quantification of the fluorescence was done by the software Living Image® 4.5.2. (Summit Pharmaceuticals International Corp.). To detect iRFP fluorescence in vivo, the hair was removed in the dorsal and inguinal areas of mice, and the mice were anesthetized by 2.5–3.0% isoflurane during observation of the iRFP720-derived fluorescence (excitation: 675 nm, emission: 720 nm). To detect iRFP fluorescence ex vivo, BAT and WAT were dissected from mice, which were sacrificed by cervical dislocation under isoflurane anesthesia, and observed directly by IVIS Spectrum imaging system. All mouse experiments were prospectively approved by the ethics committee for Animal Experiments of the University of Tsukuba (Approval number: 17–099, 17–397, 18–026).
Western blot analysis
Adipose tissues were minced, and the proteins were extracted by sonication in RIPA buffer [50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 2 mM EDTA (pH 8.0), 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS)] containing protease inhibitor cocktail and 0.5 mM phenylmethylsulfonyl fluoride (PMSF). One microgram (BAT) or 7.5 μg (iWAT) of protein from each cell extract was used for SDS-PAGE, followed by western blotting using the primary antibody against UCP1 (Abcam, ab10983, 1:2000) and the horseradish peroxidase-labeled secondary antibody against rabbit IgG (GE healthcare, NA934V, 1:3000). As a loading control, GAPDH was detected by the primary antibody against GAPDH (Santa Cruz Biotechnology Inc., sc-32233, 1:200) and the horseradish peroxidase-labeled secondary antibody against mouse IgG (GE healthcare, LNA931V/AG, 1:3000). The intensity for UCP1 was normalized by that for GAPDH, and the relative intensity to wild type was calculated.
Hematoxylin and eosin (HE) staining
Adipose tissues were isolated from the mice and soaked in neutral buffered formalin for several days. The tissues were then paraffin-embedded, sectioned, mounted onto slides and stained with hematoxylin and eosin at the Medical Laboratory for Tissue Sample Preparation at the University of Tsukuba according to the standard protocol.
Total RNA preparation and quantitative RT-PCR analysis
Total RNA was prepared from the minced adipose tissues using Sepasol-RNA I Super G (Nacalai Tesque, Inc.), and the complementary DNA (cDNA) was prepared using SuperScript III reverse transcriptase (ThermoFisher Scientific Inc.) according to the manufactures’ instructions. Using the cDNA, quantitative PCR was performed using the primer sets shown in the table S2 Table. The mRNA levels were normalized to the values of Nono (Non-POU domain-containing octamer-binding protein) [37], and the relative amounts of the mRNA were calculated using the the 2−ΔΔCt method.
Results
Generation of Ucp1-iRFP720 Knock-In (Ucp1-iRFP720 KI) mice
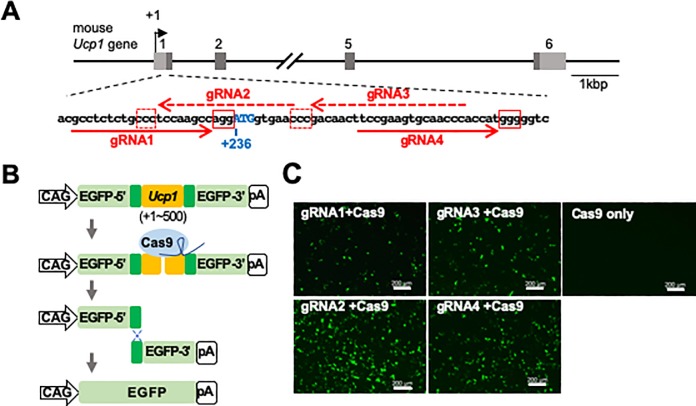
To non-invasively visualize the expression of UCP1 in live mice, we inserted the gene encoding iRFP720, a near-infrared fluorescent protein, into the mouse Ucp1 locus using the CRISPR/Cas9-mediated homologous recombination. Four guide RNAs (gRNAs) were designed around the translation initiation site of the Ucp1 gene (Fig 1A) and tested for the efficiency of double-strand DNA break repair by homologous recombination using the previously reported assay system (Fig 1B) [38]. Among the four tested gRNAs, gRNA2 showed the highest number of EGFP signals in cells when co-expressed with Cas9 (Fig 1C). No EGFP signal was observed without gRNA (Fig 1C, Cas9 only), suggesting that the EGFP signals were derived from the gRNA-induced double-strand DNA break and homologous recombination. These results indicate that gRNA2 induces Cas9-mediated double-strand DNA breaks and homologous recombination with the highest efficiency among tested gRNAs.
Fig 1. Selection of the optimum gRNA sequence to generate Ucp1-iRFP720 KI mice.
(A) Structure of the mouse Ucp1 gene. The boxes and the numbers above each box indicate the exons of the mouse Ucp1 gene. Light and dark gray regions within the exons indicate untranslated and translated regions, respectively. The position of the transcription start site is indicated by +1. The DNA sequence around the translation start site of the mouse Ucp1 gene is shown. The red arrows indicate the target sequences of the gRNAs (dashed arrows indicate the complementary sequences). The red boxes show the protospacer adjacent motif (PAM) sequences. (B) The assay system for estimating the functional genome editing efficiency of gRNAs. The Ucp1 gene (+1 ~ +500) containing the gRNA target sites (shown in orange) was inserted into pCAG-EGxxFP vector. When recruited by the gRNA (shown in dark blue) to the target site, the Cas9 nuclease (shown in light blue) introduces a double strand break in the target DNA. Because of homologous recombination between the duplicated sequences (shown in dark green), the EGFP-coding gene is recovered. (C) Fluorescence image of HEK293T cells two days after transfection. The Cas9 nuclease was expressed together with one of the four gRNA candidates (gRNA1~gRNA4) or without gRNA (Cas9 only). Scale bar, 200 μm.
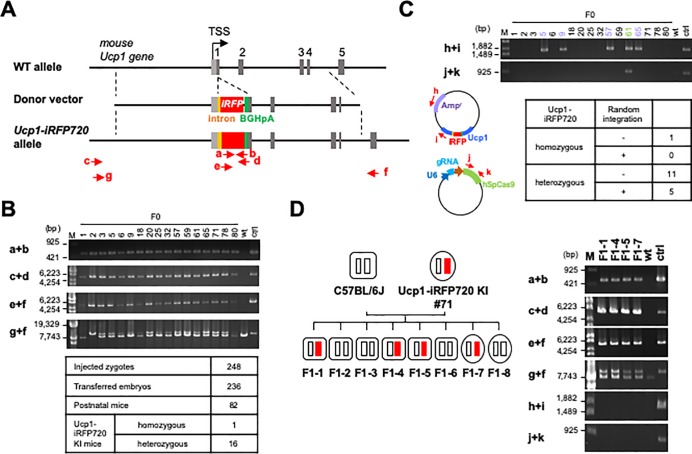
We then constructed the donor vector, that includes the iRFP720 gene and the bovine growth hormone polyadenylation signal (BGHpA) inserted at the translation start site of the Ucp1 gene (Fig 2A). The iRFP720 gene was inserted at the initiation codon of the Ucp1 gene such that the initiation codon of UCP1 is used for translation of iRFP720. Although the Ucp1 gene in the knock-in allele will be disrupted, use of the 2A sequence was avoided to prevent altering the functions of iRFP720 and UCP1. In addition, an intron fragment was inserted into the upstream region of the iRFP720 gene to enhance its expression [39][40]. After injection of the gRNA2/Cas9-expression vector and donor vector into fertilized mouse eggs, 236 embryos were transferred to the oviduct of pseudo-pregnant mice, which delivered 82 postnatal founder mice (F0). Genomic PCR analyses of the F0 mice revealed that 17 mice had the iRFP720 gene inserted at the Ucp1 locus (Fig 2B, a+b, c+d, e+f). Although most iRFP720-positive mice were heterozygous for the knock-in allele, one mouse (F0-2) was homozygous (Fig 2B, g+f). As summarized in the table, the iRFP720 gene was successfully knocked-in at the Ucp1 locus in ~20% (17 out of 82) of the F0 mice. To exclude the presence of randomly integrated vectors, we analyzed the genome of the iRFP720-positive mice using PCR with primer sets specific for the donor vector or the gRNA2/Cas9-expression vector. As shown in Fig 2C, five out of seventeen iRFP720-positive mice (F0-5, 9, 57, 61 and 65) exhibited the donor vector (h+i) integrated in their genome, and one of the mice (F0-61) had the integrated gRNA/Cas9-expression vector as well (j+k). In conclusion, we obtained 12 lines (1 homozygous mouse and 11 heterozygous mice) of the Ucp1-iRFP720 KI F0 mice, C57BL/6J-Ucp1em1(iRFP720)Utgr, which were free of randomly integrated vectors (Fig 2C).
Fig 2. Generation of Ucp1-iRFP720 KI mice.
(A) Targeting of the iRFP720 gene into the Ucp1 locus. The donor vector contains an artificial intron (orange box), iRFP720 gene (red box) and the bovine growth factor gene-derived polyadenylation signal (BGHpA) (green box). Structure of the targeted allele is indicated together with the primers used for genomic PCR (red arrows). (B) Genomic PCR analysis of F0 mice carrying the Ucp1-iRFP720 KI allele. M, marker; wt, wild-type; ctrl, positive control (a DNA fragment that covers the amplicon sequence between the primers c and f in Fig 2A). (C) PCR analysis of the donor vector and the gRNA/Cas9-expression vector randomly integrated into the F0 mouse genome. The table shows the summary of random integration in the F0 mouse genome carrying the Ucp1-iRFP720 allele. (D) Mendelian inheritance of the Ucp1-iRFP720 KI allele. One of the Ucp1-iRFP720 KI mice (#71) was crossed with a WT mouse, and eight F1 mice were born. Their genotypes were analyzed by PCR using the primer sets shown in Fig 2A. Small white and red rectangles show WT Ucp1 and Ucp1-iRFP720 KI alleles, respectively. Rounded rectangles and ellipses indicate male and female mice, respectively.
We then confirmed the inheritance of the iRFP720 transgene to the next generation by crossing F0 mice with WT C57BL/6J mice. Given the numbers of F1 pups in a litter, the iRFP720 gene knocked-in at the Ucp1 locus did not appear to affect fertility. In addition, genomic PCR analysis revealed that the F1 mice inherited the iRFP720 gene at an expected frequency (Fig 2D, a+b, c+d, e+f, g+f), without any random integration of the vectors (Fig 2D, h+i, j+k). These results indicate that the iRFP720 gene of the Ucp1-iRFP720 KI mice is inherited in a Mendelian fashion.
In vivo imaging of BAT in Ucp1-iRFP720 KI mice
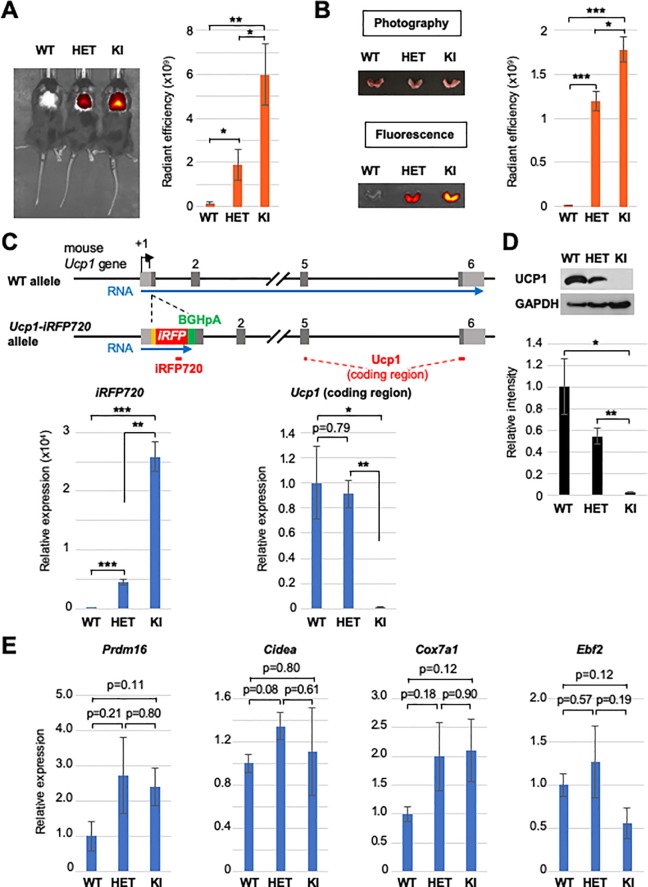
We first analyzed the iRFP720-derived fluorescence from BAT in the dorsal interscapular region of the Ucp1-iRFP720 KI mice. When the heterozygous and homozygous Ucp1-iRFP720 KI mice were observed by In Vivo Imaging System (IVIS), strong fluorescence was detected in the interscapular region of both types of mice, whereas no such fluorescence was detected in WT littermates (Fig 3A). The fluorescence intensity was stronger in homozygous than heterozygous Ucp1-iRFP720 KI mice (Fig 3A). To confirm that the fluorescence in the interscapular region of the Ucp1-iRFP720 KI mice is indeed derived from BAT, we dissected BATs from the interscapular regions of the mice and directly observed fluorescence by IVIS. As shown in Fig 3B, BATs isolated from homozygous and heterozygous Ucp1-iRFP720 KI mice, but not from the WT mice exhibited strong fluorescence ex vivo, and the fluorescence intensity correlated roughly with the number of Ucp1-iRFP720 KI alleles, in a similar manner to the in vivo fluorescence images from the interscapular regions of the live mice (Fig 3A).
Fig 3. In vivo imaging of BAT in Ucp1-iRFP720 KI mice.
(A) In vivo imaging of the BATs of WT, heterozygous Ucp1-iRFP720 KI (HET) and homozygous Ucp1-iRFP720 KI mice (KI). Quantified fluorescence intensity for each mouse is shown in the bar graph (n = 4). ** p<0.01, * p<0.05 (B) BATs isolated from the mice used in Fig 3A. Fluorescence image of the BATs is shown in the lower panel. Quantified fluorescence intensity is shown in the bar graph (n = 4). *** p<0.001, * p<0.05 (C) Quantitative RT-PCR analysis of the iRFP720 and Ucp1 (coding region) in WT mice, heterozygous Ucp1-iRFP720 KI mice and homozygous Ucp1-iRFP720 KI mice (n = 3). The positions of the amplicons are shown as red bars. *** p<0.001, ** p<0.01, * p<0.05 (D) Expression of UCP1 in the Ucp1-iRFP720 KI mice determined by immunoblot. GAPDH was used as a loading control. The bar graph shows the quantified intensities of UCP1 bands in the immunoblot (n = 3). ** p<0.01, * p<0.05 (E) Quantitative RT-PCR analysis of the brown adipocyte marker genes in WT mice, heterozygous Ucp1-iRFP720 KI mice and homozygous Ucp1-iRFP720 KI mice (n = 3).
Analyses of mRNA expression in BAT showed that expression levels of the iRFP720 mRNA displayed a similar pattern to that of the fluorescence levels (Fig 3A and 3C). As previously reported for the Ucp1 knockout mice [41], the Ucp1 mRNA levels were comparable between WT and heterozygous Ucp1-iRFP720 KI mice, whereas homozygous Ucp1-iRFP720 KI mice showed virtually no mRNA expression from the UCP1-coding region (Fig 3C). UCP1 protein expression was reduced to ~50% in heterozygous Ucp1-iRFP720 KI mice, while undetectable in homozygous Ucp1-iRFP720 KI mice (Fig 3D), as was observed for the Ucp1 knockout mice [41]. Thus, due to the bovine growth hormone polyadenylation signal (BGHpA), transcription of the iRFP720 mRNA did not extend into the UCP1-coding region at the Ucp1-iRFP720 KI allele (Fig 3C). Despite the absence of UCP1 (Fig 3C and 3D), homozygous Ucp1-iRFP720 KI mice showed no significant change in expression of other brown marker genes, such as Prdm16, Cidea, Cox7a1, and Ebf2 (Fig 3E), suggesting that UCP1 loss and expression of iRFP720 do not compromise BAT development. Together, these data show that iRFP720 expressed in the BATs of the Ucp1-iRFP720 KI mice can be imaged in live mice in a non-invasive manner.
In vivo imaging of induced beige adipocytes in heterozygous Ucp1-iRFP720 KI mice
In addition to BATs, UCP1 is expressed at a lower level in beige adipocytes, which are induced in WATs by various stimuli, such as cold exposure and β3-adrenergic receptor agonists. To test if UCP1 in the induced beige adipocytes can be imaged in heterozygous Ucp1-iRFP720 KI mice, mice were administered CL316,243, a highly selective β3-adrenergic receptor agonist, for 7 days and observed by IVIS before and after CL316,243 treatment (Fig 4A). As shown in Fig 4B, a low fluorescence signal was detected in both heterozygous Ucp1-iRFP720 KI and WT mice before treatment with CL316,243 (Day 0). However, fluorescence was clearly detected in the inguinal regions of heterozygous Ucp1-iRFP720 KI mice compared with control mice (treated only with PBS) after treatment with CL316,243 for 7 days (Fig 4B, upper panels, Day 7). The WT mice exhibited low levels of fluorescence even after exposure to CL316,243 for 7 days (Fig 4B, lower panels). Fluorescence intensity quantification indicated fluorescence in the inguinal regions of CL316,243-treated heterozygous Ucp1-iRFP720 KI mice was significantly higher than that in PBS-treated mice (Fig 4B, right graph).
Fig 4. In vivo imaging of induced beige adipocytes.
(A) Experimental scheme for beige adipocyte induction by CL316,243, which was injected intraperitoneally (1 μg/g body weight) every day for one week. Fluorescence image of the mice was captured on day 0 and day 7 by IVIS. (B) Fluorescence image of the heterozygous Ucp1-iRFP720 KI mice and WT mice before (day 0) and after (day 7) CL316,243 treatment. Quantified fluorescence intensity is shown in the bar graph (n = 4). (C) Fluorescence image of the iWATs isolated from the mice (day 7) used in Fig 4B. The graphs show the quantification of the fluorescence (n = 4) and expression of the iRFP720 mRNA (n = 5). ** p<0.01 (D) The expression of Ucp1 (coding region) mRNA as well as the UCP1 protein in the iWATs used in Fig 4C (n = 3). * p<0.05 (E) Expression of the browning marker genes in the iWATs used in Fig 4C (n = 3). ** p<0.01, * p<0.05 (F) Hematoxylin and eosin staining of the iWAT sections isolated from the heterozygous Ucp1-iRFP720 KI mice and WT mice after one-week treatment with PBS or CL316,243. Scale bar, 50 μm.
To confirm the source of fluorescence, iWATs were isolated and observed directly by IVIS. As shown in Fig 4C, robust fluorescence was detected in iWATs derived from heterozygous Ucp1-iRFP720 KI mice that were treated with CL316,243. However, low fluorescence levels were observed in iWATs derived from heterozygous Ucp1-iRFP720 KI mice treated with PBS alone, or from WT mice (Fig 4C). Consistent with greater fluorescence, the iRFP720 mRNA was induced in iWATs from CL316,243-treated heterozygous Ucp1-iRFP720 KI mice (Fig 4C). Moreover, the Ucp1 mRNA and protein were induced from the intact Ucp1 allele in heterozygous Ucp1-iRFP720 KI mice, in a similar manner to those in WT mice (Fig 4D). In addition to Ucp1 induction, iWAT from CL316,243-treated heterozygous Ucp1-iRFP720 KI mice showed induction of other beige adipocyte marker genes, such as Cox7a1, Cox8b, and Elovl3 (Fig 4E). In support of beige adipocyte induction in iWAT, histological analysis revealed clusters of smaller adipocytes with multilocular lipid droplets in HE-stained sections of iWAT from CL316,243-treated mice (Fig 4F). These data show that heterozygous Ucp1-iRFP720 KI mice enable in vivo imaging of beige adipocyte induction in iWAT via iRFP720 fluorescence.
Induction of beige-like adipocytes in homozygous Ucp1-iRFP720 KI mice
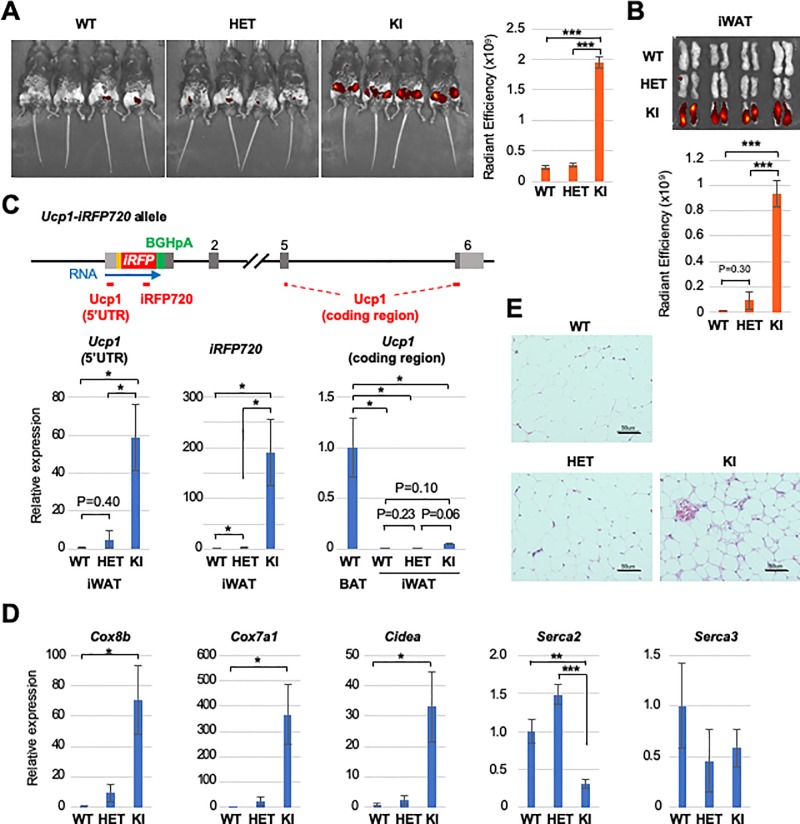
Previous studies showed that adipocytes in the inguinal region of UCP1-deficient mice adopt a brown adipocyte morphology when the mice are housed at an ambient temperature of 4~20°C [27][42][43]. To address if beige adipocytes are induced in WAT from Ucp1 knockout mice, we generated homozygous Ucp1-iRFP720 KI mice. When housed at the standard ambient temperature (22–24°C), which provides a mild cold stress that induces UCP1-mediated thermogenesis [44][45], homozygous Ucp1-iRFP720 KI mice exhibited strong fluorescence in the inguinal regions, even without CL316,243 treatment (Fig 5A). Inguinal fluorescence intensity in homozygous Ucp1-iRFP720 KI mice was comparable to that observed in CL316,243-treated heterozygous Ucp1-iRFP720 KI mice (Figs 4B and 5A), and was significantly higher than that in heterozygous Ucp1-iRFP720 KI or WT mice, both of which exhibited lower levels of fluorescence (Fig 5A). Indeed, iWATs isolated from homozygous Ucp1-iRFP720 KI mice exhibited significantly stronger fluorescence than those from heterozygous Ucp1-iRFP720 KI or WT mice (Fig 5B), indicating that fluorescence derives from inguinal iWAT. Quantitative RT-PCR analysis revealed that iRFP720 mRNA was expressed at a high level in iWAT from homozygous Ucp1-iRFP720 KI mice (Fig 5C). PCR analysis of the 5’ UTR region of iRFP720 mRNA, which detects pre-mRNA as well as mRNA, also showed increased expression (Fig 5C), suggesting that the Ucp1 gene promoter is activated in homozygous Ucp1-iRFP720 KI mice. Due to the polyadenylation signal, the UCP1-coding region was not transcribed (Fig 5C) as observed for BAT (Fig 3C). Moreover, beige adipocyte markers, Cox8b, Cox7a1, and Cidea, were also induced in iWAT from homozygous Ucp1-iRFP720 KI mice. Further, expression of Serca2, which is implicated in energy metabolism via calcium cycling-dependent non-canonical thermogenesis [31], was altered in homozygous Ucp1-iRFP720 KI mice (Fig 5D). HE-stained tissue sections of iWAT showed clusters of small adipocytes with multilocular lipid droplets in homozygous Ucp1-iRFP720 KI mice (Fig 5E). These results indicate that iWAT adipocytes from homozygous Ucp1-iRFP720 KI mice show elevated Ucp1 promoter activity, displaying gene expression patterns and morphology characteristics for beige adipocytes.
Fig 5. Beige-like adipocyte induction in homozygous Ucp1-iRFP720 KI mice.
(A) Fluorescence image of the inguinal regions of WT, heterozygous Ucp1-iRFP720 KI and homozygous Ucp1-iRFP720 KI mice. Quantified fluorescence intensity is shown in the bar graph below (n = 4). *** p<0.001 (B) Fluorescence image of the iWATs isolated from the mice shown in Fig 5A. The bar graph shows the quantified fluorescence intensity (n = 4). (C) Quantitative RT-PCR analysis of iRFP720, Ucp1 5’ untranslated region (5’ UTR) and Ucp1 coding region in the iWATs used in Fig 5B (n = 4). The positions of the amplicons for quantitative PCR are shown as red lines below the Ucp1-iRFP720 allele. (D) Expression of the browning marker genes (Cox8b, Cox7a1, Cidea) (n = 3) and UCP1-independent thermogenesis genes (Serca2, Serca3) (n = 4) in the iWATs used in Fig 5B. *** p<0.001, ** p<0.01, * p<0.05 (E) Hematoxylin and eosin staining of the iWAT sections isolated from WT, heterozygous Ucp1-iRFP720 KI and homozygous Ucp1-iRFP720 KI mice. Scale bar, 50 μm.
Discussion
In this study we generated the Ucp1-iRFP720 KI mice that allow in vivo imaging of UCP1 expression via iRFP720 fluorescence. iRFPs are near-infrared fluorescent proteins recently engineered from bacterial phytochrome photoreceptors [33]. iRFPs bind to chromophore biliverdin, which is naturally present in mammalian tissues, and exhibit bright fluorescence without an exogenous supply of the chromophore [33]. As the near-infrared light from 650 nm to 900 nm is minimally absorbed by various body components, such as hemoglobin and melanin, iRFPs are suitable for in vivo imaging [35]. We show here that iRFP720 expressed from the Ucp1 locus is easily visualized in BAT of live mice. Moreover, the iRFP fluorescence is robust enough to detect lower levels of UCP1 in iWAT of live mice treated with CL316,243.
Previous studies reported bioluminescence-based in vivo imaging systems for UCP1 using the luciferase gene as a reporter [46][47][48]. ThermoMouse carries the Ucp1 genomic region containing the Ucp1 promoter-driven luciferase-T2A-tdTomato cassette on the Y chromosome. Luciferase expression faithfully mimics that of UCP1 in BAT and iWAT, which can be imaged in live mice upon injection of the natural substrate of luciferase, D-luciferin [46]. ThermoMouse in vivo imaging is restricted to males only, since the transgene is inserted randomly on the Y chromosome. Similarly, Ucp1-2A-luciferase knock-in mice, which carry the luciferase gene at the Ucp1 locus, also allow in vivo imaging of UCP1 expression in BAT and iWAT by the luciferase reporter gene [47]. In the mice, insertion of a 2A peptide sequence in place of the stop codon of Ucp1 inevitably attaches a 20 amino acid tag to the C-terminus of UCP1 after 2A-peptide-mediated cleavage. Although the homozygous Ucp1-2A-luciferase knock-in mice appear to behave normally, the effect of the added tag on UCP1-mediated thermogenesis remains unknown. Moreover, both ThermoMouse and Ucp1-2A-luciferase knock-in mice utilize the bioluminescence method, which requires injection of D-luciferin for in vivo imaging. The intensity of the luciferin signal is known to change dynamically over ~30 min, and the time course of the signal intensity can vary depending on injection methods and observed tissues [49][50]. Thus, accurate measurements of the luciferase signal in live mice is somewhat cumbersome and inconvenient.
Recently, Wang et al reported Ucp1-T2A-luciferase-T2A-iRFP713 knock-in mice, which carry a dual Ucp1 reporter, consisting of luciferase and iRFP713, inserted after the UCP1-coding region [48]. The mice supposedly enable in vivo imaging of UCP1 expression via both luciferase and iRFP713. Indeed, UCP1 expression is visualized in vivo by bioluminescence from the luciferase reporter in these mice. However, the fluorescence from iRFP713 appears to be weak and only observable in interscapular BAT of cold acclimated mice, and its use is largely restricted to ex vivo imaging of isolated tissues. Weak fluorescence from iRFP713 may be due to the position of the iRFP gene in the tri-cistronic construct, because expression of the fluorescent protein at the third position in tri-cistronic vectors with two 2A sequences tends to be weak [51]. In the Ucp1-iRFP720 KI mice, the iRFP720-coding region is inserted directly at the translation initiation site of the Ucp1 gene and expressed as a single cistron without the use of a 2A sequence. The knock-in iRFP720 gene mimics UCP1 expression and enables in vivo imaging of not only BAT, but also beige adipocytes in iWAT in live, CL316,243-treated mice. The iRFP fluorescence in iWAT is almost comparable with that observed in BAT, and is significantly stronger than background autofluorescence of the liver, where bilirubin is abundant [52].
Despite its robust fluorescence, knock-in of the iRFP gene at the initiation codon of the Ucp1 gene inevitably ablates Ucp1 expression from the cognate allele. The UCP1 level in heterozygous Ucp1-iRFP720 KI mice is reduced to ~50% of that observed in WT mice, and UCP1 was undetectable in homozygous Ucp1-iRFP720 KI mice (Fig 3D). However, given that heterozygous Ucp1 knockout mice exhibit minimal phenotypic characteristics [41], heterozygous Ucp1-iRFP720 KI mice are presumed to be phenotypically normal. Since heterozygous Ucp1-iRFP720 KI mice express iRFP720 from the knock-in allele, and UCP1 from the other allele, it is possible to image UCP1 expression in vivo via iRFP720 fluorescence, and to identify brown and beige adipocytes in isolated tissue samples via UCP1 immunostaining.
In contrast, homozygous KI mice serve as a UCP1-deficient mouse model, with the added advantage of imaging and identifying cells that would otherwise express UCP1. Previous studies showed that UCP1-deficient mice are sensitive to cold, but paradoxically more resistant to diet-induced obesity [42]. This resistance to obesity occurs during cold stress (4~20°C), but is absent at thermoneutrality (27°C), suggesting the presence of alternative UCP1-independent non-shivering thermogenesis that is less efficient than UCP1-dependent thermogenesis. iWAT from UCP1-deficient mice housed at 4~20°C exhibit the appearance of beige-like adipocytes, suggesting a probable location for alternative thermogenesis [27][42][43], which are also observed in Ucp1-iRFP720 KI mice (Fig 5). Given the recent identification of futile cycles in beige adipocytes, such as creatine-dependent ADP/ATP substrate cycling [28][29][30] and calcium cycling [31], it would be interesting to investigate the expansion of beige adipocytes in iWAT and the rise of alternative thermogenesis in these adipocytes. Elevated activation of the Ucp1 gene promoter in homozygous Ucp1-iRFP720 KI mice housed under standard conditions indicates possible auto-regulatory pathways, which are constitutively activated by UCP1 loss. The mice described in this study should be particularly useful for identifying such pathways that lead to beige adipocyte induction and alternative non-shivering thermogenesis in iWAT in vivo. A recent study using single-cell RNA sequencing revealed a developmental hierarchy of adipocyte progenitors [53]. A similar method can be applied to stromal vascular fraction cells in iWAT from Ucp1-iRFP720 KI mice to investigate how iRFP-positive and–negative adipocytes arise from mesenchymal progenitors and where their developmental pathways diverge. In addition to stromal vascular fraction cells, mature adipocytes may be sorted by using iRFP720 as a reporter in a flow cytometry method [54]. The isolated iRFP-positive and–negative mature adipocytes may allow analysis of distinct metabolic and thermogenic properties of these two types of adipocytes. In conclusion, Ucp1-iRFP720 KI mice described here serve as a convenient mouse model for in vivo imaging and in vitro analyses of UCP1-expressing adipocytes in both BAT and iWAT.
Supporting information
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank other members of the laboratory for advice on designing guide RNAs for CRISPR/Cas9 system and helpful discussion.
Abbreviations
- UCP1
Uncoupling protein 1
- iRFP
near-infrared fluorescent protein
- BAT
brown adipose tissue
- WAT
white adipose tissue
- NIR
near-infrared
- BGHpA
bovine growth hormone polyadenylation signal
- WT
wild-type
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by JSPS KAKENHI Grant Numbers JP16K08610 and JP19K07343 (AF), JP16K07244 (KN) and JP17H04036 (KH) as well as by Nakanishi Foundation for Advancement of Measuring in Biomedical Engineering (KH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kozak LP, Anunciado-Koza R. UCP1: Its involvement and utility in obesity. International Journal of Obesity. 2008. 10.1038/ijo.2008.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikeda K, Maretich P, Kajimura S. The Common and Distinct Features of Brown and Beige Adipocytes. Trends in Endocrinology and Metabolism. 2018. 10.1016/j.tem.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cypess AM, Lehman S, Williams G, Tal I, Goldfine AB, Kuo FC, et al. Identification and Importance of Brown Adipose Tissue in Adult Humans A BS TR AC T. N Engl J Med. 2009. 10.1056/NEJMoa0810780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JMAFL, Kemerink GJ, Bouvy ND, et al. Cold-Activated Brown Adipose Tissue in Healthy Men. N Engl J Med. 2009; 10.1056/nejmoa0808718 [DOI] [PubMed] [Google Scholar]
- 5.Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, et al. Functional Brown Adipose Tissue in Healthy Adults. N Engl J Med. 2009; 10.1056/nejmoa0808949 [DOI] [PubMed] [Google Scholar]
- 6.Lee P, Swarbrick MM, Zhao JT, Ho KKY. Inducible brown adipogenesis of supraclavicular fat in adult humans. Endocrinology. 2011; 10.1210/en.2011-1349 [DOI] [PubMed] [Google Scholar]
- 7.Cypess AM, White AP, Vernochet C, Schulz TJ, Xue R, Sass CA, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat Med. 2013; 10.1038/nm.3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jespersen NZ, Larsen TJ, Peijs L, Daugaard S, Homøe P, Loft A, et al. A classical brown adipose tissue mrna signature partly overlaps with brite in the supraclavicular region of adult humans. Cell Metab. 2013; 10.1016/j.cmet.2013.04.011 [DOI] [PubMed] [Google Scholar]
- 9.Shinoda K, Luijten IHN, Hasegawa Y, Hong H, Sonne SB, Kim M, et al. Genetic and functional characterization of clonally derived adult human brown adipocytes. Nat Med. 2015; 10.1038/nm.3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: Effects of cold exposure and adiposity. Diabetes. 2009; 10.2337/db09-0530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoneshiro T, Aita S, Matsushita M, Okamatsu-Ogura Y, Kameya T, Kawai Y, et al. Age-related decrease in cold-activated brown adipose tissue and accumulation of body fat in healthy humans. Obesity. 2011; 10.1038/oby.2011.125 [DOI] [PubMed] [Google Scholar]
- 12.Matsushita M, Yoneshiro T, Aita S, Kameya T, Sugie H, Saito M. Impact of brown adipose tissue on body fatness and glucose metabolism in healthy humans. Int J Obes. 2014; 10.1038/ijo.2013.206 [DOI] [PubMed] [Google Scholar]
- 13.Chondronikola M, Volpi E, Børsheim E, Porter C, Annamalai P, Enerbäck S, et al. Brown adipose tissue improves whole-body glucose homeostasis and insulin sensitivity in humans. Diabetes. 2014; 10.2337/db14-0746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoneshiro T, Aita S, Matsushita M, Kayahara T, Kameya T, Kawai Y, et al. Recruited brown adipose tissue as an antiobesity agent in humans. J Clin Invest. 2013; 10.1172/JCI67803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orava J, Nuutila P, Lidell ME, Oikonen V, Noponen T, Viljanen T, et al. Different metabolic responses of human brown adipose tissue to activation by cold and insulin. Cell Metab. 2011; 10.1016/j.cmet.2011.06.012 [DOI] [PubMed] [Google Scholar]
- 16.Hanssen MJW, Hoeks J, Brans B, Van Der Lans AAJJ, Schaart G, Van Den Driessche JJ, et al. Short-term cold acclimation improves insulin sensitivity in patients with type 2 diabetes mellitus. Nat Med. 2015; 10.1038/nm.3891 [DOI] [PubMed] [Google Scholar]
- 17.Townsend K, Tseng Y-H. Brown adipose tissue: Recent insights into development, metabolic function and therapeutic potential. Adipocyte. 2012; 10.4161/adip.18951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lshibashi J, Seale P. Beige can be slimming. Science. 2010. 10.1126/science.1190816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiefer FW. The significance of beige and brown fat in humans. Endocr Connect. 2017; 10.1530/ec-17-0037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen P, Levy JD, Zhang Y, Frontini A, Kolodin DP, Svensson KJ, et al. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell. 2014; 10.1016/j.cell.2013.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Chronic peroxisome proliferator-activated receptor γ (PPARγ) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem. 2010; 10.1074/jbc.M109.053942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Contreras GA, Lee Y-H, Mottillo EP, Granneman JG. Inducible brown adipocytes in subcutaneous inguinal white fat: the role of continuous sympathetic stimulation. Am J Physiol Metab. 2014; 10.1152/ajpendo.00033.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aldiss P, Betts J, Sale C, Pope M, Budge H, Symonds ME. Exercise-induced ‘browning’ of adipose tissues. Metabolism: Clinical and Experimental. 2018. 10.1016/j.metabol.2017.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shabalina IG, Petrovic N, deJong JMA, Kalinovich A V., Cannon B, Nedergaard J. UCP1 in Brite/Beige adipose tissue mitochondria is functionally thermogenic. Cell Rep. 2013; 10.1016/j.celrep.2013.10.044 [DOI] [PubMed] [Google Scholar]
- 25.Nyman E, Bartesaghi S, Melin Rydfalk R, Eng S, Pollard C, Gennemark P, et al. Systems biology reveals uncoupling beyond UCP1 in human white fat-derived beige adipocytes. npj Syst Biol Appl. 2017; 10.1038/s41540-017-0027-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Granneman JG, Burnazi M, Zhu Z, Schwamb LA. White adipose tissue contributes to UCP1-independent thermogenesis. Am J Physiol Metab. 2003; 10.1152/ajpendo.00197.2003 [DOI] [PubMed] [Google Scholar]
- 27.Ukropec J, Anunciado RP, Ravussin Y, Hulver MW, Kozak LP. UCP1-independent thermogenesis in white adipose tissue of cold-acclimated Ucp1-/- mice. J Biol Chem. 2006; 10.1074/jbc.M606114200 [DOI] [PubMed] [Google Scholar]
- 28.Kazak L, Chouchani ET, Jedrychowski MP, Erickson BK, Shinoda K, Cohen P, et al. A Creatine-Driven Substrate Cycle Enhances Energy Expenditure and Thermogenesis in Beige Fat. Cell. 2015; 10.1016/j.cell.2015.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazak L, Chouchani ET, Lu GZ, Jedrychowski MP, Bare CJ, Mina AI, et al. Genetic Depletion of Adipocyte Creatine Metabolism Inhibits Diet-Induced Thermogenesis and Drives Obesity. Cell Metab. 2017; 10.1016/j.cmet.2017.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertholet AM, Kazak L, Chouchani ET, Bogaczyńska MG, Paranjpe I, Wainwright GL, et al. Mitochondrial Patch Clamp of Beige Adipocytes Reveals UCP1-Positive and UCP1-Negative Cells Both Exhibiting Futile Creatine Cycling. Cell Metab. 2017; 10.1016/j.cmet.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ikeda K, Kang Q, Yoneshiro T, Camporez JP, Maki H, Homma M, et al. UCP1-independent signaling involving SERCA2bmediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat Med. 2017; 10.1038/nm.4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K, Verkhusha V V. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011; 10.1038/nbt.1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shemetov AA, Oliinyk OS, Verkhusha V V. How to Increase Brightness of Near-Infrared Fluorescent Proteins in Mammalian Cells. Cell Chem Biol. 2017; 10.1016/j.chembiol.2017.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shcherbakova DM, Baloban M, Verkhusha V V. Near-infrared fluorescent proteins engineered from bacterial phytochromes. Current Opinion in Chemical Biology. 2015. 10.1016/j.cbpa.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shcherbakova DM, Stepanenko O V., Turoverov KK, Verkhusha V V. Near-Infrared Fluorescent Proteins: Multiplexing and Optogenetics across Scales. Trends in Biotechnology. 2018. 10.1016/j.tibtech.2018.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naito Y, Hino K, Bono H, Ui-Tei K. CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015; 10.1093/bioinformatics/btu743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arsenijevic T, Grégoire F, Delforge V, Delporte C, Perret J. Murine 3T3-L1 Adipocyte cell differentiation model: Validated reference genes for qPCR gene expression analysis. PLoS One. 2012; 10.1371/journal.pone.0037517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A, Ikawa M. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep. 2013; 10.1038/srep03355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang MTF, Gorman CM. Intervening sequences increase efficiency of RNA 3’ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990; 10.1093/nar/18.4.937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmiter RD, Sandgren EP, Avarbock MR, Brinster RL. Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci U S A. 1991; 10.1073/pnas.88.2.478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enerbäck S, Jacobsson A, Simpson EM, Guerra C, Yamashita H, Harper ME, et al. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature. 1997; 10.1038/387090a0 [DOI] [PubMed] [Google Scholar]
- 42.Liu X, Rossmeisl M, McClaine J, Kozak LP. Paradoxical resistance to diet-induced obesity in UCP1-deficient mice. J Clin Invest. 2003; 10.1172/JCI200315737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keipert S, Kutschke M, Lamp D, Brachthäuser L, Neff F, Meyer CW, et al. Genetic disruption of uncoupling protein 1 in mice renders brown adipose tissue a significant source of FGF21 secretion. Mol Metab. 2015; 10.1016/j.molmet.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. UCP1 Ablation Induces Obesity and Abolishes Diet-Induced Thermogenesis in Mice Exempt from Thermal Stress by Living at Thermoneutrality. Cell Metab. 2009; 10.1016/j.cmet.2008.12.014 [DOI] [PubMed] [Google Scholar]
- 45.Maloney SK, Fuller A, Mitchell D, Gordon C, Overton JM. Translating Animal Model Research: Does It Matter That Our Rodents Are Cold? Physiology. 2014; 10.1152/physiol.00029.2014 [DOI] [PubMed] [Google Scholar]
- 46.Galmozzi A, Sonne SB, Altshuler-Keylin S, Hasegawa Y, Shinoda K, Luijten IHN, et al. ThermoMouse: An In Vivo Model to Identify Modulators of UCP1 Expression in Brown Adipose Tissue. Cell Rep. 2014; 10.1016/j.celrep.2014.10.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mao L, Nie B, Nie T, Hui X, Gao X, Lin X, et al. Visualization and quantification of browning using a Ucp1-2A-luciferase knock-in mouse model. Diabetes. 2017; 10.2337/db16-0343 [DOI] [PubMed] [Google Scholar]
- 48.Wang H, Willershäuser M, Karlas A, Gorpas D, Reber J, Ntziachristos V, et al. A dual Ucp1 reporter mouse model for imaging and quantitation of brown and brite fat recruitment. Mol Metab. 2019; 10.1016/j.molmet.2018.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Close DM, Xu T, Sayler GS, Ripp S. In vivo bioluminescent imaging (BLI): Noninvasive visualization and interrogation of biological processes in living animals. Sensors. 2011. 10.3390/s110100180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inoue Y, Kiryu S, Izawa K, Watanabe M, Tojo A, Ohtomo K. Comparison of subcutaneous and intraperitoneal injection of d-luciferin for in vivo bioluminescence imaging. Eur J Nucl Med Mol Imaging. 2009; 10.1007/s00259-008-1022-8 [DOI] [PubMed] [Google Scholar]
- 51.Liu Z, Chen O, Wall JBJ, Zheng M, Zhou Y, Wang L, et al. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci Rep. 2017; 10.1038/s41598-017-02460-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Croce AC, Ferrigno A, Santin G, Vairetti M, Bottiroli G. Bilirubin: An autofluorescence bile biomarker for liver functionality monitoring. J Biophotonics. 2014; 10.1002/jbio.201300039 [DOI] [PubMed] [Google Scholar]
- 53.Merrick D, Sakers A, Irgebay Z, Okada C, Calvert C, Morley MP, et al. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science (80-). 2019; 10.1126/science.aav2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hagberg CE, Li Q, Kutschke M, Bhowmick D, Kiss E, Shabalina IG, et al. Flow Cytometry of Mouse and Human Adipocytes for the Analysis of Browning and Cellular Heterogeneity. Cell Rep. 2018; 10.1016/j.celrep.2018.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.