Abstract
The stem cell field is hindered by its inability to noninvasively monitor transplanted cells within the target organ in a repeatable, time-sensitive, and condition-specific manner. We hypothesized that quantifying and characterizing transplanted cell–derived exosomes in the recipient plasma would enable reliable, noninvasive surveillance of the conditional activity of the transplanted cells. To test this hypothesis, we used a human-into-rat xenogeneic myocardial infarction model comparing two well-studied progenitor cell types: cardiosphere-derived cells (CDCs) and c-kit+ cardiac progenitor cells (CPCs), both derived from the right atrial appendage of adults undergoing cardio-pulmonary bypass. CPCs outperformed the CDCs in cell-based and in vivo regenerative assays. To noninvasively monitor the activity of transplanted CDCs or CPCs in vivo, we purified progenitor cell–specific exosomes from recipient total plasma exosomes. Seven days after transplantation, the concentration of plasma CPC-specific exosomes increased about twofold compared to CDC-specific exosomes. Computational pathway analysis failed to link CPC or CDC cellular messenger RNA (mRNA) with observed myocardial recovery, although recovery was linked to the microRNA (miRNA) cargo of CPC exosomes purified from recipient plasma. We further identified mechanistic pathways governing specific outcomes related to myocardial recovery associated with transplanted CPCs. Collectively, these findings demonstrate the potential of circulating progenitor cell–specific exosomes as a liquid biopsy that provides a noninvasive window into the conditional state of the transplanted cells. These data implicate the surveillance potential of cell-specific exosomes for allogeneic cell therapies.
INTRODUCTION
Stem/progenitor cell–based therapies have demonstrated varying degrees of efficacy in treating their targeted diseases in preclinical transplantation and tissue injury models. These therapies are now showing promising results in human clinical trials (1–4). Recently, we demonstrated that cardiac progenitor cells (CPCs; c-kit+/Lin−) and cardiosphere-derived cells (CDCs) derived from human neonatal heart tissue reduce cardiac scar size, improve myocardial function, and attenuate adverse myocardial remodeling, secondary to myocardial infarction (MI) in preclinical models when compared to adult-derived CPCs or CDCs (5, 6). Despite encouraging phase 1 clinical trials using either CPC or CDC transplantation in adults with ischemic heart disease, a major limitation to potentiate their clinical efficacy is the inability to noninvasively monitor the transplanted cells and their therapeutic effects during the period of myocardial remodeling (7, 8).
Further, the retention of transplanted cardiac stem/progenitor cells is low; however, the components of the stem cell secretome, including exosomes, promote myocardial recovery through donor age-dependent pathways (5, 9–15). The exponential increase in exosome research related to cardiac repair (16–19) highlights the therapeutic potential of these small vesicles (30 to 150 nm) containing microRNA (miRNA) cargoes, which arise from fusion of multivesicular bodies with plasma membrane (10, 14). Exosomes carry proteins required for immediate repair of the injured heart, as well as miRNAs, which are uniquely capable of facilitating long-term repair by altering the transcriptome of targeted cells (5).
Although intramyocardial transplantation of CPCs or CDCs rescues the infarcted myocardium and improves functional recovery in preclinical studies, the mechanism underlying the beneficial effects of CPCs or CDCs is not well understood. Recently, we performed an in-depth analysis of the CPC secretome, demonstrating that a single intramyocardial injection of the exosomes derived from neonatal CPCs promoted myocardial recovery comparable to that observed after neonatal CPC injection (5). These results, among others, suggest that at least partial therapeutic efficacy of CPCs or CDCs might be attributed to their exosomes (20–25). Circulating tissue-specific exosomes derived from transplanted solid organs have been observed in the transplant recipient’s plasma (26, 27). Exosomes contain specific proteomic and RNA signatures that reflect the conditional and functional status of their cells of origin (23, 28). Recently, we demonstrated that circulating transplant tissue–specific exosome characterization enables noninvasive surveillance of the transplanted solid organ in a time-sensitive, condition-specific manner (25–27). Thus, we hypothesized that transplanted stem/progenitor cells would release an exosome signal into the peripheral circulation, and characterization of stem/progenitor cell–specific exosomes would enable noninvasive surveillance of the functional activity and conditional status of the transplanted cells. However, it was unclear whether transplanted stem/progenitor cell–derived exosomes would have an miRNA cargo profile similar to their in vitro cultured progenitor/stem cells and, subsequently, whether we could detect and use these circulating exosomes for noninvasive surveillance of transplanted progenitor cell residence and activity.
Here, we demonstrate that progenitor cell–specific exosomes are present in the recipient circulation and can be monitored noninvasively. We performed a head-to-head comparison in a xenogeneic rodent MI model to investigate the cardiac regenerative potential of two well-studied progenitor cells, CDCs and CPCs, derived from the same human heart biopsy. We validated the monitoring potential of stem/progenitor cell–specific exosome platform and demonstrated that exosome miRNA cargo reflects the functional myocardial recovery achieved by the transplanted stem cells.
RESULTS
Functional characteristic of CPCs and CDCs in vitro and in vivo
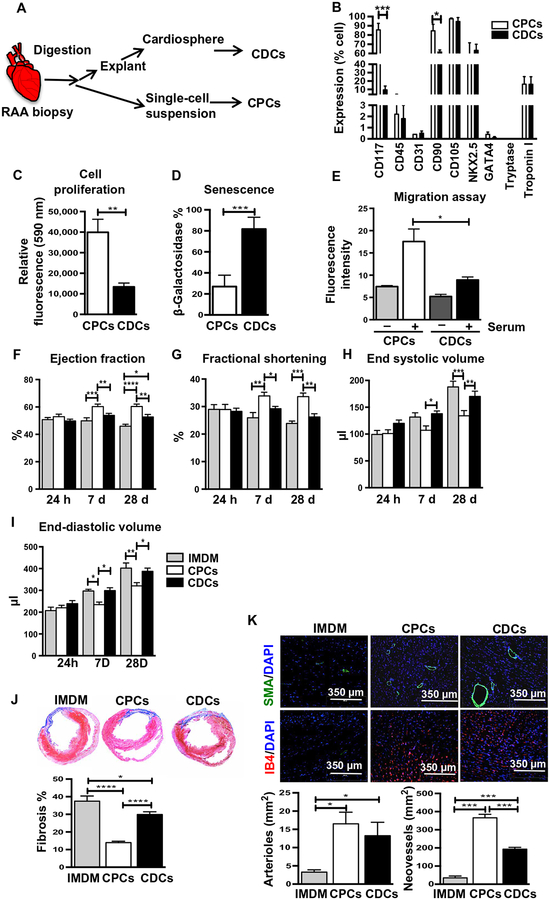
Human myocardial biopsies were obtained from the right atrial appendage (RAA) of adult patients (68 ± 10 years) undergoing coronary artery bypass grafting for severe atherosclerotic disease at the time of cardiac surgery. Adult CPCs and CDCs were isolated from the same RAA biopsy sample using our previously modified protocol involving explant plating, selection processing, and expansion (Fig. 1A and table S1) (5, 6). At passage 3 (P3), CPCs and CDCs were characterized for cell surface markers and had similar cellular morphologies (Fig. 1B and fig. S1), as previously described for each cell type (5, 6). Both cell types expressed mesenchymal stem cell markers (CD105 and CD90) and the cardiomyocyte lineage–specific markers, transcription factor NKX2.5 and troponin I. Both cell types did not express hematopoietic markers (CD34 and CD45), mast cell marker (tryptase), or cardiomyocyte lineage–specific transcription factor (GATA4); however, CPCs were 85% c-kit+ (Fig. 1B).
Fig. 1. Phenotypic characterization and cardiac functional assessment in rat MI model after adult human CPC and CDC transplantation.
(A) Schematic diagram of the isolation of adult CPCs and CDCs. (B) Flow cytometry analysis of CPCs and CDCs for stem cell–specific surface markers (CD117, CD105, CD90, CD45, and CD31), cardiac lineage markers (NKX2.5, troponin I, and GATA4), and mast cell marker tryptase (n = 5); see also fig. S1. (C) Cell proliferation of CPCs and CDCs assessed using alamarBlue assay (n = 8). (D) Senescence assessed using β-galactosidase assay at P3 (n = 4). β-Galactosidase–stained blue cells (see fig. S2A) were quantified as positive. (E) Transwell migration assay with CPCs (n = 4) and CDCs (n = 4) at P3. See also fig. S2B. (F to I) Structural and functional parameters derived from echocardiography measurements at baseline and after MI (n = 10 to 15). Heart sections stained for (J) Masson’s trichrome [fibrosis (blue) and viable mass (pink)] in hearts injected with CPCs, CDCs, or control (IMDM) for n = 7 per group. Sections taken 4 weeks after MI; quantification below. (K) Neovessels stained by IB4 expression, and arterioles marked by α-smooth muscle actin (α-SMA) expression in myocardial sections 4 weeks after MI. Nuclei were labeled with DAPI (4′,6-diamidino-2-phenylindole; blue) in all images (n = 8). All data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Data were analyzed using two-way analysis of variance (ANOVA) followed by Bonferroni post-tests (B, E, and G to J), one-way ANOVA followed by Kruskal-Wallis test, and Student’s t test followed by Mann-Whitney’s analysis (C, D, and K).
Growth properties and functional characteristics of CDCs and CPCs during expansion provide relevant metrics that may reflect their functionality after transplantation. At P3, CPCs were more than four times more proliferative than CDCs derived from the same patient (Fig. 1C). When the two progenitor cell types were allowed to proliferate until P6, most of the adult CDCs developed senescence as evident by increased β-galactosidase activity when compared to CPCs (Fig. 1D and fig. S2A). Another key functional property of the progenitor cells is their intrinsic migration ability after transplantation into the infarcted myocardium. To recapitulate this functional activity in vitro, the migratory potential of both progenitor cell types was measured by a transwell migration assay. Derived from the same RAA of four biological replicates, a significant number (P = 0.0335) of CPCs migrated as compared to CDCs (Fig. 1E and fig. S2B) after 6 hours of incubation in the presence of serum.
To examine whether the growth and migratory properties of CPCs are associated with an improved ability to recover myocardial function after infarction, CPCs and CDCs from the same heart biopsy were transplanted in the rodent MI model. After ligation of the left anterior descending (LAD) artery, 1 million CPCs, 1 million CDCs, or cell-free Iscove’s modified Dulbecco’s medium (IMDM) was injected into the injured myocardium of the left ventricle (LV), as described previously (5, 6). The consistency of the MI model was verified for each treatment group by performing echocardiography 24 hours after MI, which demonstrated similar ejection fraction (EF) among the three groups at this time point (Fig. 1F). Groups were evaluated for cardiac function and LV remodeling by echocardiogram and post-mortem histology. Compared to medium control, transplanted CPCs and CDCs significantly improved cardiac function and structure as indicated by increased EF (PCDCs = 0.0299, PCPCs < 0.0001) and fractional shortening (FS) (PCPCs < 0.0001, PCDCs = 0.4978) and decreased end-diastolic volume (EDV) (PCPCs < 0.0026, PCDCs = 0.7909,) and end-systolic volume (ESV) (PCPCs < 0.0002, PCDCs = 0.3544). The LV functional improvement was significantly greater with CPCs as compared to CDCs (Fig. 1, F to I; EFCPCs vs. CDCs, P = 0.0045; FSCPCs vs. CDCs, P = 0.0006; EDVCPCs vs. CDCs, P = 0.0034; ESVCPCs vs. CDCs, P = 0.0034). The functional improvement was apparent 1 week after MI and was sustained over the 4 weeks of follow-up. Structural changes in the LV were further evaluated by histologic analysis at 28 days after MI, focusing on fibrosis (Masson trichrome), arteriolar density (smooth muscle actin), and total vascular density (isolectin IB4). Representative images of myocardial fibrosis and quantification of the three different treatment groups are shown in Fig. 1J. Four weeks after MI, infarct size (the area of fibrosis relative to total stained myocardial area) was significantly smaller in hearts treated with either CPCs or CDCs relative to IMDM controls, and fibrosis was significantly (CPCs versus CDCs, P < 0.0001) reduced in CPC-treated hearts versus CDC-treated hearts (Fig. 1J). Although both CPC and CDC treatments significantly increased arteriolar (SMA) and total neovascular density (IB4) compared to IMDM control (SMAIMDM vs. CPCs, P = 0.0359; SMAIMDM vs. CDCs, P = 0.044) (IB4IMDM vs. CDCs, P = 0.0006; IB4IMDM vs. CPCs, P = 0.0002; Fig. 1K), CPCs outperformed CDCs in regard to neovascular density.
Increased c-kit+ stoichiometry in CDC population improved therapeutic efficacy
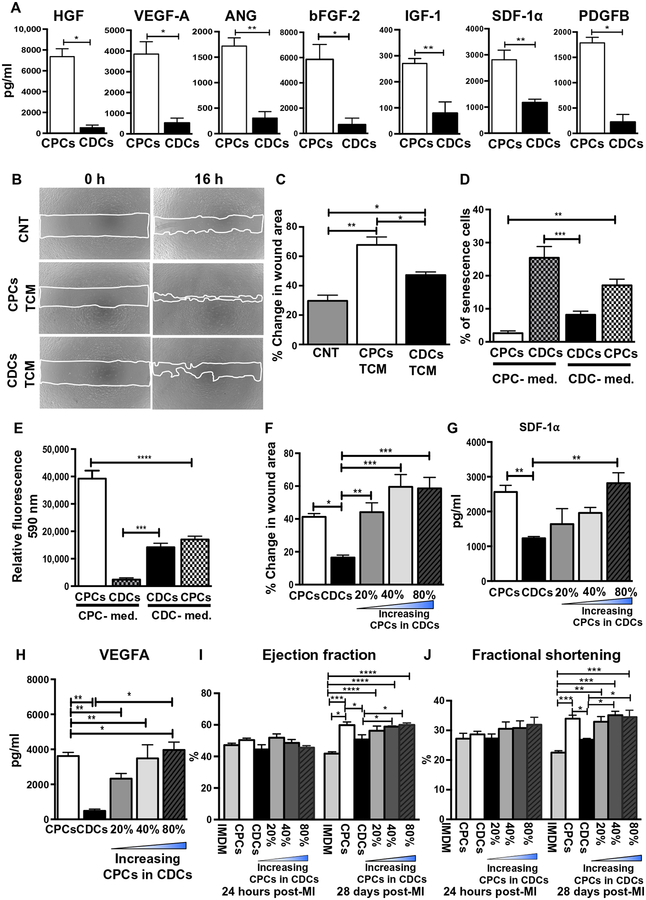
Recent reports have demonstrated that the functional unit of transplanted progenitor cells is their secretome, containing independently secreted proteins and exosomes (5, 29). Initially, we compared the concentrations of seven well-studied paracrine factors [hepatocyte growth factor (HGF), insulin-like growth factor–1 (IGF-1), stromal cell–derived growth factor 1α (SDF-1α), angiopoietin-1 (ANG-1), vascular endothelial growth factor A (VEGFA), platelet-derived growth factor B (PDGFB), and basal fibroblast growth factor (bFGF)] released by CPCs and CDCs into the total conditioned medium (TCM). These factors have previously been suggested to play a role in stem/progenitor cell–mediated repair (29–34). Enzyme-linked immunosorbent assay (ELISA)–based quantitative analysis of these seven paracrine factors in the TCM of CPCs and CDCs revealed that HGF (P = 0.035), IGF-1 (P = 0.0079), SDF-1α (P = 0.0159), ANG-1 (P = 0.0286), VEGFA (P = 0.0285), PDGFB (P = 0.0357), and bFGF (P = 0.0286) are secreted at higher concentrations by CPCs compared to CDCs (Fig. 2A). To assess the angiogenic potential of the TCM derived from CDCs and CPCs, wound-healing assays were performed on human mammary epithelial cells (HMECs), keeping TCM protein concentration constant (protein concentration, 50 ng/μl). Wound area was significantly (P = 0.003) reduced in the presence of TCM derived from CPCs compared to CDCs TCM (Fig. 2, B and C). These data suggested that the concentrations of independently secreted molecules in the secretome may be directly correlated with the c-kit+ cell proportions within CDCs. To test this hypothesis, we increased the c-kit+ cell stoichiometry by varying the c-kit+ cell concentration from 20% to 80% within the CDC population while maintaining a constant total cell number. Because growth medium composition can affect the biochemical cellular and functional properties of CPCs and CDCs, including the c-kit+ cell concentration, we attempted to grow CDCs in CPC culture medium and vice versa using P3 cells. Changing to the nondesignated growth medium formulation of CDCs and CPCs resulted in early onset of senescence, reduced cellular proliferation, and loss of mesenchymal cell markers (CD90 and CD105) (Fig. 2, D and E, and fig. S3, A to E). CPCs and CDCs could not be derived from RAA explant when cultured in the nondesignated growth medium due to limited cell outgrowth (fig. S3F). Consequently, we cultured CPCs and CDCs in their prescribed medium and modulated the c-kit+ cell proportion within the CDC population just before in vitro assays or intramyocardial injection into the rat MI model. A scratch migration assay demonstrated that increasing the c-kit+ cell proportion in CDCs progressively enhanced wound healing (Fig. 2F and fig. S4).
Fig. 2. Functional assessment of medium (TCM) derived from CPCs, CDCs, and mixed cell populations in vitro and in vivo.
(A) ELISA-based quantitative analysis of seven paracrine factors secreted by CPCs and CDCs (n = 4 to 6) in vitro. (B) Representative images and (C) quantification of in vitro wound-healing assay using HMECs (n = 4). Treatments included basal medium [serum/growth factor free, control (CNT)] and the secretome of CPCs and CDCs (TCM). (D) Effect of growth medium on growth properties of CPCs and CDCs as assessed by onset of senescence and (E) cellular proliferation by alamarBlue (see also fig. S3). (F) Wound-healing assay composed of groups with increasing c-kit+ stoichiometry (20, 40, and 80%) in CDC populations including unaltered CPCs and CDCs (n = 4 to 6; see also fig. S4). (G and H) Quantification of secreted SDF-1α and VEGF-A with increasing percentage of c-kit+ cells in CDCs (n = 4) by ELISA (n = 4). (I and J). Cardiac parameters of EF and FS determined by echocardiographs at 24 hours and 28 days after injection of mixed cell populations or unaltered CPCs or CDCs in a rat MI model (n = 5 to 6). All data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Grouped data are analyzed using one-way ANOVA followed by Bonferroni post-tests, two-way ANOVA followed by Tukey post-test (C to J), and t test followed by Mann-Whitney’s analysis (A).
Next, TCM was harvested from CDCs with varying proportions of c-kit+ cells. Two well-established paracrine factors, SDF-1a and VEGFA, were shown to increase with increasing c-kit+ cell concentration by ELISA (Fig. 2, G and H) (35–37). Last, we tested the myocardial recovery potential of these combination groups in our established rodent MI model while maintaining a constant total number of transplanted cells (1 million) (5, 6). EF and FS were significantly (PEF, FS < 0.0001) increased with increasing concentrations of c-kit+ cells (Fig. 2, I and J). Together, these data suggest that the higher concentration (pg/ml) of cardioprotective factors in the secretome of c-kit+ cells may contribute to their enhanced functional abilities when compared to CDCs.
CPCs and CDCs release progenitor cell–specific human major histocompatibility complex–containing exosomes into the recipient plasma
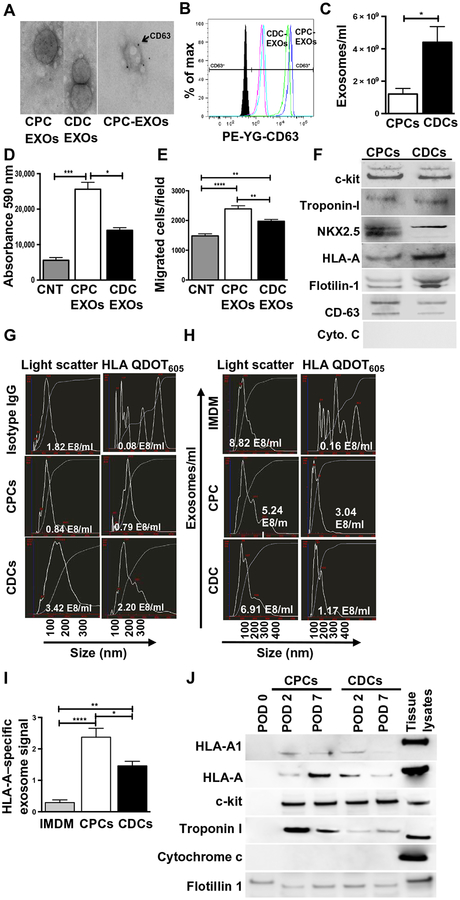
Although the roles of CDC- or CPC-derived exosomes in cardiac repair are being actively investigated (5, 10, 11), in vivo validation of exosome production and functional potential was lacking. We hypothesized that transplanted human CDCs or CPCs would release exosomes composed of their parent cell–specific constituents into the recipient circulation during myocardial recovery in the xenogeneic rodent MI model. To first test this hypothesis in vitro, >85% confluent CPCs or CDCs derived from the same heart biopsy were conditioned in serum-free medium for 48 hours and their exosome output was analyzed. Subsequent to conditioning in serum-free basal medium, both cell types retained viability for at least 48 hours. Exosomes were purified from the conditioned medium using size exclusion chromatography (5, 38). Transmission electron microscopy (TEM) confirmed that the isolated extracellular vesicles were in the size range of exosomes and expressed the canonical exosome marker CD63 (Fig. 3A). Flow cytometry using CD63-conjugated magnetic beads demonstrated higher CD63 expression on CDC-derived exosomes as compared to CPC-derived exosomes (Fig. 3B). Further characterization of these exosomes by nanoparticle tracking analysis (NTA) revealed that CDC-derived exosomes not only were larger in size (average size, 165 nm) than CPC-derived exosomes (average size, 124 nm) but also existed at a higher concentration as compared to CPC-derived exosomes (Fig. 3C and fig. S5A). However, CPC-derived exosomes induced more proliferation in HMECs and exhibited more angiogenic potential compared to CDC-derived exosomes, with equal and constant numbers of exosomes (Fig. 3, D and E, and fig. S5, B to D). Together, we concluded that despite being fewer in number, CPC-derived exosomes may be more potent than CDC-derived exosomes for myocardial repair.
Fig. 3. Characterization of exosomes obtained from CPCs and CDCs both in vitro and from rat plasma after injection of CPCs or CDCs.
(A). TEM visualization of exosomes obtained in vitro from CPCs and CDCs, labeled with exosome marker CD63 using immunogold. (B) Fluorescence-activated cell sorting (FACS) verification of exosomes using anti-CD63–phycoerythrin (PE) antibody (n = 4). (C) Measurement of exosome concentration and size by NanoSight in the secretome of CDCs and CPCs (n = 4; see also fig. S5A). (D) HMEC proliferation assay performed in basal medium or CPC- or CDC-derived exosomes (n = 6 to 10), as indicated. (E) Transwell cell migration assay using basal medium or CPC- or CDC-derived exosomes (n = 6), as indicated (see also fig. S5B). Wound-healing assay with basal medium or CPC- or CDC-derived exosomes (n = 5) and quantification were depicted in fig. S5C. (F) Immunoblot for stem cell markers and cardiac-associated proteins in exosomes isolated from CDCs and CPCs (n = 4; see also fig. S6). (G) CPC- and CDC-derived exosomes analyzed for surface expression of HLA-A on light scatter (total exosomes) and fluorescence modes (HLA-A, goat secondary Qdot 605) by NTA (NanoSight NS300) (n = 7). (H) Exosomes from rat plasma analyzed on NanoSight nanoparticle detector on light scatter (total exosomes) and fluorescence modes (HLA Qdot 605) for transplanted CPC- and CDC-derived exosomes using anti–HLA-A. Human exosomes were isolated from rat plasma obtained after transplanted myocardial injections in rat MI model (n = 8; see also schematic diagram in fig. S7). (I) Quantification of human exosomes retained in rat plasma 7 days after MI (n = 8). (J) Immunoblot characterization of the human exosomes isolated from rat plasma on PODs 0, 2, and 7 using human HLA-A1, HLA-A, c-kit, troponin I, cytochrome c, and flotillin 1 (n = 4; see also fig. S8). All data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Data are analyzed using t test followed by Mann-Whitney’s analysis (C) and two-way ANOVA followed by Tukey’s post-tests (D, E, and I).
Next, using immunoblot analyses, we tested whether progenitor cells would release exosomes containing progenitor-specific cellular markers (Fig. 1B). The exosomes derived from the CPCs and CDCs contained homeobox protein NKX2.5, human leukocyte antigen A (HLA-A), troponin I, c-kit+, and exosome markers flotillin-1 and CD63 (Fig. 3F and fig. S6). Cytochrome c, a marker for apoptotic bodies, could not be detected. Similar to its cellular counterpart, HLA-A is selectively present on the surface of human stem/progenitor cell exosomes. Therefore, we used major histocompatibility complex (MHC) class I specificity to quantify and purify the human exosome subpopulation (fig. S7) from the rat plasma after cellular transplantations (35, 39). This platform was tested in vitro on exosomes derived from CDCs or CPCs in culture, and HLA-A–specific signal was detected on the exosomes (Fig. 3G). To validate this concept in vivo, the progenitor cell–specific exosome signal within the total exosomes was quantified in the recipient rat plasma 2 and 7 days after progenitor cell transplantation. These time points were chosen because our previous analysis demonstrated that despite the loss of most of the transplanted progenitor cells, improved cardiac function of the injured myocardium was observed by 7 days after transplantation (5, 36). At day 7, the total number of plasma exosomes was similar among the IMDM (negative control), CDC, and CPC groups, but progenitor cell–specific exosome signal was detected only in the CDC and CPC groups (Fig. 3H). Further, about twofold higher progenitor cell–specific HLA-A exosome signal was seen in CPC-derived exosomes compared to CDC-derived exosomes (Fig. 3I).
To validate our in vitro findings that the MHC mismatch enabled purification and characterization of progenitor cell–derived exosomes and their cargo, we assessed whether circulating progenitor cell exosomes coexpressed MHC and other surface markers and proteins reflecting the cellular constituents of CDCs and CPCs. Immunoblot analysis of human progenitor cell–specific exosomes purified from rat plasma on post-operative days (PODs) 2 and 7 demonstrated that at both selected time points, transplanted cell–specific exosome subpopulations expressed HLA molecules (HLA-A and HLA-A1), flotillin 1 (exosome marker), c-kit (progenitor cell marker), and cardiomyocyte marker troponin I (Fig. 3J and fig. S8). These subpopulations of exosomes were negative for cytochrome c (cellular/apoptotic body contamination). Pretransplant samples (POD 0) did not express the markers seen in the CPC and CDC exosome populations.
Together, these data demonstrate that human progenitor cells transplanted into ischemic myocardium release distinct, quantifiable exosomes into the peripheral circulation, and that CPC-derived exosomes contain a larger HLA-A exosome signal than CDC-derived exosomes in this model. Progenitor cell exosomes carry protein markers specific to the cytoplasmic and membrane constituents of their cellular counterparts. Collectively, these findings support the concept that circulating progenitor cell–specific exosome subpopulation may reflect the conditional state of the transplanted progenitor cells, and its quantitative and exosome cargo characterization may enable noninvasive monitoring of the functional transplanted progenitor cell mass.
Computational modeling of miRNAs and systems biology of the genes involved in myocardial recovery
To understand the mechanisms by which CPCs or CDCs potentiate myocardial recovery, we performed computational analyses on (i) cellular mRNA from CDC and CPC cultures, (ii) miRNA cargoes of exosomes from CDC and CPC culture supernatants, and (iii) miRNA cargoes of circulating progenitor cell–specific exosomes purified from recipient total plasma exosome pools 7 days after CPC or CDC transplant. Analyses of cellular mRNA profiles of in vitro cultured CPCs and CDCs could not predict the outcomes involved with improved cardiac function purely based on the differential expression patterns of mRNA (table S2). Previously, we have used computational tools such as principal components analysis (PCA) and partial least squares regression (PLSR) modeling of pediatric CPC cellular mRNA content and CPC-derived exosome miRNA cargo harvested from in vitro cultures to understand potential mechanisms and predictors of myocardial remodeling (37, 40). We had identified the top 300 mRNA signals and used them to generate a predictive model to quantitatively link signals to functional outcomes triggered by the transplanted CPCs and CDCs (40). Therefore, in this study, we selected the same top 300 genes and matched them with CDC- and CPC-specific mRNA expression patterns using PCA (fig. S9A). Individuals in each age/cell type groups were clustered together and were localized in close proximity. Despite clustering uniquely from previous studies and matching signals, because of low correlation between the observed and expected ratios, the model was unable to predict most endpoints for this study, including angiogenesis (fig. S9B). We retrained a new PLSR prediction model by combining the adult CDC and CPC mRNA signals (37) to create a new model to understand potential shared mechanisms. We then used PLSR analysis to establish a relationship between the covarying mRNA signals and responses of angiogenesis, fibrosis reduction, migration, and proliferation that were shared among the studies (fig. S9C). Ingenuity Pathway Analysis (IPA) revealed top genes, which have a prominent role for inflammatory and cardiac development in the reparative process. Last, the loading plot from PLSR analysis demonstrated specific shared cardiac and immune system–related genes from the past (37) and present studies that clustered with cardiac functions (fig. S9D).
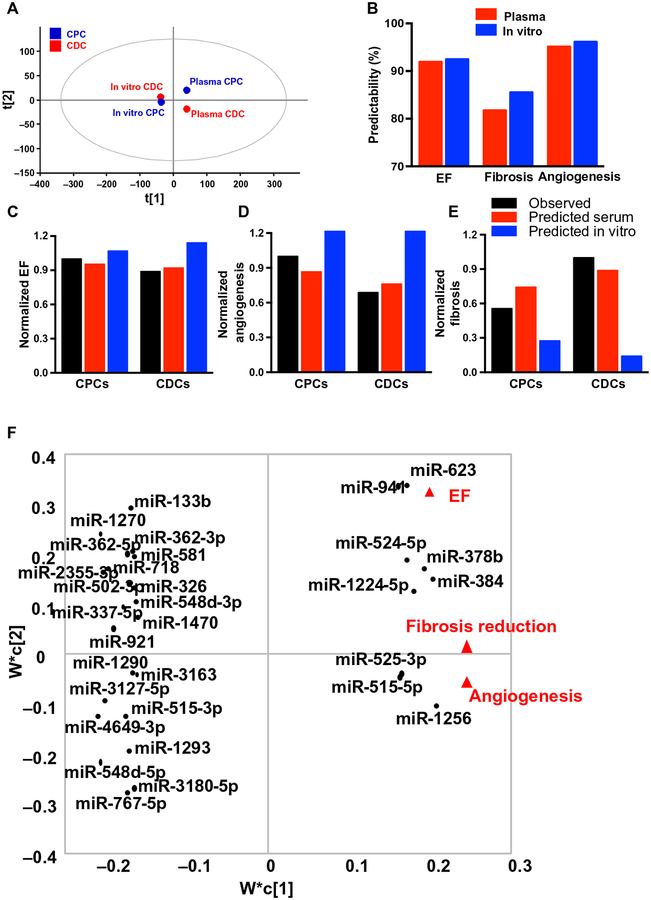
We also developed a model to predict functional effects based on exosome miRNA content, using both CPC- and CDC-derived exosomes isolated in vitro from TCM, as well as circulating CPC- and CDC-specific exosome subpopulations purified from rat plasma after transplantation (37). PCA showed that exosomes derived in vitro, whether from CPCs or CDCs, clustered very closely together, whereas progenitor cell exosomes purified from recipient plasma had divergent expression patterns (Fig. 4A). Both datasets fit our previously published computational model well, with high predictability ranging from 82 to 97% (Fig. 4B). When our computational model was used to predict outcomes of the current study (improvement in EF and angiogenesis, reduction in fibrosis), the model using plasma CPC- and CDC-derived exosome subsets more closely matched the observed results than the model using in vitro CDC- and CPC-derived exosomes (Fig. 4, C to E, and table S3). Because of the high predictability, we then retrained the model on all datasets to determine miRNAs likely involved in the response using PLSR and plotted them in PC space along with the cellular mRNA signals (Fig. 4F and table S4). We identified miRNAs associated with improvement in EF [miRNA 378b (miR-378b), miR-623, and miR-941], reduction in fibrosis (miR-1256 and miR-384), and induction of angiogenesis (miR-525–3p, miR-5155p, and miR-1224). Functional analyses of these miRNAs using IPA identified 45 cardioprotective pathways that were up-regulated (table S5), favoring cell growth and proliferation [phosphatidylinositol 3-kinase (PI3K)/AKT, mammalian target of rapamycin (mTOR), hypoxia-inducible factor-1α (HIF-1α), Janus kinase (JAK)/signal transducer and activator of transcription (STAT), high-mobility group box protein 1 (HMGB1), platelet-derived growth factor (PDGF), IGF-1, FGF, transforming growth factor–β (TGF-β)], resistance to stress (oxidative stress response, nitric oxide signaling, PXR/RXR), anti-fibrosis and anti-inflammation [inhibition of matrix metalloproteinases (MMPs) and interleukin-6 (IL-6)], cardiomyocyte regeneration (NOTCH and HIPPO), angiogenesis (FAK/PAK, HIF-1α, VEGF), and cardiomyocyte proliferation (NOTCH and HIPPO; fig. S10). Collectively, these data demonstrate that the miRNA cargo of CPC-derived exosomes isolated from recipient plasma was different than the cargo isolated from in vitro collected exosomes, and that the in vivo exosome cargo was more predictive of mechanistic cardioprotective potency than the in vitro exosome profiles.
Fig. 4. Computational model of covariant miRNAs using the cue-signal-response paradigm.
Computational model and the prediction of cardiac functions of exosomal miRNA cargo. (A) PCA. In vitro derived exosomes from CPCs/CDCs and rat plasma–purified exosomes after CPC/CDC transplantation were analyzed on the basis of their matching miRNA expression. (B) Predictability measurements of cardiac functions. Predictive model was created using exosome microarray data from this study and to identify the predictability of EF, fibrosis, and angiogenesis functions. The top 100 VIP (variable importance for projection) miRNAs were selected and matched with plasma (blue bars) as well as in vitro (red bars) exosome CPC and CDC miRNAs individually (42). (C to E) The predicted plasma (red bars) and in vitro (blue bars) CPC and CDC functional outcomes were compared with the observed functional data (green bars) for EF (C) and angiogenesis (D) in comparison with fibrosis function (E). (F). PLSR and miRNA target analysis. Top miRNAs with known validated targets were identified among the 60 matched miRNAs using miRTarBase and plotted in PC space. Thirty-one miRNAs with validated targets were identified by miRTarBase. Clusters of miRNAs are formed on the basis of the functional outcome.
Experimental validation of miRNA function predicted by computational analyses
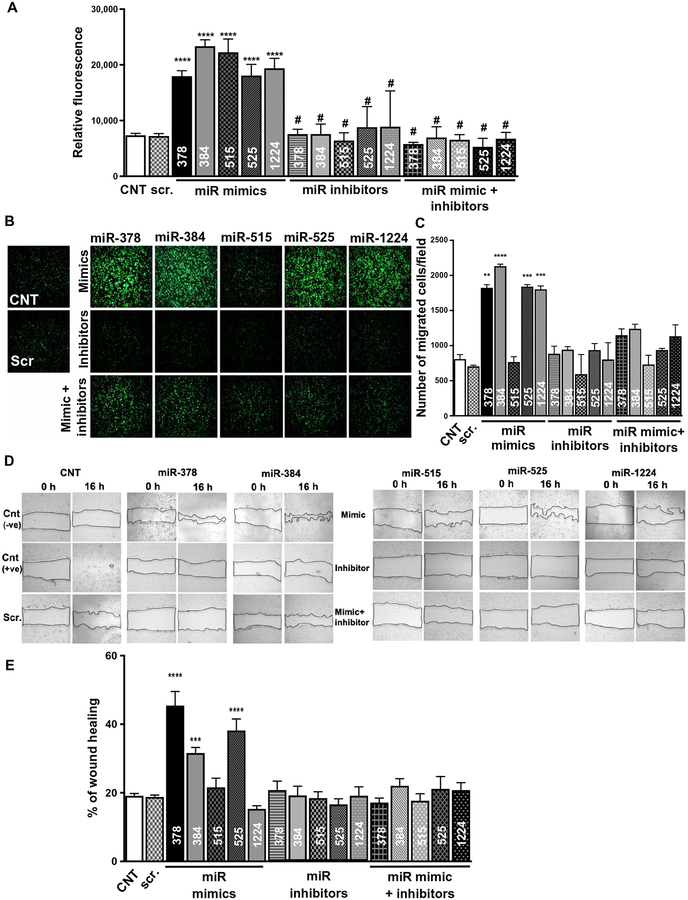
Bioinformatics tools, based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, facilitate the study of miRNAs by providing a list of their potential functions. However, because of poorly conserved specificity of miRNAs’ targets across species, it is important to validate the critical predicted pathways by miRNAs in humans (41). On the basis of our computational analyses, we identified five miRNAs (fig. S11A) predicted to improve cardiac function after MI by enhancing angiogenesis. miR-378, miR-384, miR-515, miR-525, and miR-1224 were not detected in in vitro cultured CPC-derived exosomes but were enriched in the transplanted CPC-derived circulating exosome subset purified from rat plasma in the MI model. Literature-based software analyses (TargetScan and DIANA Tools) and previous reports also suggested that forced overexpression of these synthetic miRNAs may promote angiogenesis in the injured myocardium (fig. S11A) (42). To validate our predictions, we transfected HMECs with mimics (gain of function) of these miRNAs and assessed their angiogenic potential by three well-accepted in vitro angiogenic assays: (i) endothelial cell proliferation, (ii) transwell migration, and (iii) wound-healing assay (43, 44). Because the shortlisted miRNAs could not be detected by reverse transcription polymerase chain reaction (RT-PCR) analysis in cultured cells (fig. S11B), we transfected HMECs with the mimics and then transfected with inhibitors of particular mimics 72 hours later. Transfection with a specific miRNA mimic resulted in multifold enrichment of the specific miRNAs (fig. S11, C and D). All miRNAs, as predicted, significantly (P < 0.0001) induced cellular proliferation (Fig. 5A) in comparison to nonspecific miRNA transfection controls. miRNA mimic–induced increases in cell proliferation were neutralized by their respective inhibitors. The transwell migration assay (Fig. 5, B and C) demonstrated that miR-378, miR-384, miR-525, and miR-1224 significantly (P ≤ 0.01) enhanced the migration of HMECs, whereas no significant (P > 0.9999) migration was observed with miRNA 515–transfected HMECs. In the wound-healing assay, HMECs transfected with mimics of miR-378, miR-384, and miR-525 significantly (P < 0.001) reduced the wound area, whereas miRNA 515–transfected HMECs had no significant (P > 0.9999) effect on the healing of wound area (Fig. 5, D and E). Together, these results showed that our computational model could predict the functional potential of miRNAs.
Fig. 5. Verification of functional role of miRNAs as identified by computational analysis.
(A) Assessment of miRNAs on cell proliferation using alamarBlue assay. HMECs were transfected with the mimics, inhibitors, or mimics + inhibitors of miR-378, miR-384, miR-515, miR-525, and miR-1224, as indicated. (B and C) Transwell migration assay images and quantification assessing the migration potential of HMECs transfected with mimics, inhibitors, or mimics + inhibitors of miR-378, miR-384, miR-515, miR-525, and miR-1224. (D and E) Wound-healing assay images and quantification of HMECs treated with mimics, inhibitors, or mimics + inhibitors of miR-378, miR-384, miR-515, miR-525, and miR-1224. All data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Data are analyzed using one-way ANOVA followed by Tukey’s analyses (A, C, and E).
DISCUSSION
Although multiple cell-based therapies have been proposed for the treatment of heart failure, few clinical trials have demonstrated efficacy using the strategy of ex vivo expansion and reintroduction of cells into the injured myocardium (4, 7, 8, 45, 46). To monitor cell transplantation and retention, attempts have been made to quantify exogenous cells in the recipient myocardium using PCR, but this technique cannot be applied to humans (9). The field therefore lacks a method to monitor the presence and remodeling abilities of therapeutic stem/progenitor cells in real time. A noninvasive platform enabling surveillance of the cellular footprint and identification of the pathways triggered by stem/progenitor cells would facilitate the development of new therapeutic approaches for cardiac regeneration in humans. Because of the condition-specific and dynamic nature of the quantitative and cargo profiles, tissue-specific exosomes are being investigated for their potential as diagnostic biomarkers and as therapeutic modulators in cardiovascular disorders (47–49). Here, we investigated the potential of transplanted progenitor cell–derived exosome characterization as a noninvasive tool to monitor their cellular counterpart’s presence and function in vivo.
We report that (i) transplanted human CPCs release exosomes reflecting parent cell–specific surface markers into the recipient circulation; (ii) transplanted progenitor cell–derived exosomes can be isolated, quantified, and profiled noninvasively from the recipient plasma; (iii) progenitor/stem cell–based remodeling of the myocardium may lead to changes in their numbers and miRNA cargo of circulating exosomes; and (iv) miRNA signatures of progenitor cell–derived exosomes may reflect the mechanistic pathways in the tissue microenvironment during myocardial recovery. For these reasons, we believe that progenitor/stem cell–specific exosomes have potential to serve as a liquid biopsy of the functional progenitor cell mass.
Our study demonstrates that miRNA profiling of transplanted progenitor cell–derived exosomes isolated from recipient plasma more accurately predicts the clinical outcomes seen with stem cell therapy than the RNA profiles of cultured progenitor cells or their exosomes. In support of this idea, in the xenogeneic rat MI model, the total plasma exosome numbers and exosome particle size were similar between the CDC and CPC groups, but the transplanted CPC-derived exosome signal significantly increased during the early post-operative period when compared to the CDC exosome signal. This higher CPC-specific exosome signal was associated with better functional myocardial recovery and biochemical properties of the transplanted CPCs. The diagnostic potential of this platform is further validated by the ability to noninvasively enrich for the progenitor cell–specific exosomes containing the cellular constituents of the transplanted CPCs and CDCs. Future studies investigating quantitative and cargo changes in the progenitor cell exosome subpopulation during the first 30 PODs will help to further understand its diagnostic accuracy. In addition, changes in the transplant exosome cargoes may reflect different responses by the recipient to the transplanted cells or different responses by the transplanted cells to the environmental cues triggered by the injured myocardium. Last, the kinetics of the exosome presence in the circulation after progenitor/stem cell transplantation needs further clarification to determine the diagnostic window of this platform.
In addition to its diagnostic potential, the ability to enrich progenitor cell–derived circulating exosomes will provide an unparalleled window into the unique cross-talk between the recipient myocardium and the transplanted progenitor cell mass. Limited studies have been able to determine how the transplanted cells remodel the myocardium, and the mechanistic pathways underlying this process have been performed in preclinical studies (not in humans). The ability to profile circulating transplant cell–specific exosomes gives a unique opportunity into this process and may have clinical translational potential. In support of this idea, we have recently shown that in clinical allogeneic islet cell transplantation, donor islet cell–specific exosomes can be reliably profiled from recipient blood (25, 26). In addition to its diagnostic value, the clinical translation of this platform may provide a better understanding of the underlying reasons why certain patients are more responsive to stem/progenitor cell–based therapies. Another major advantage of this noninvasive surveillance platform might be the facilitation of clinically optimal progenitor cell dose/multiple doses in humans. Moreover, once targeted miRNAs are identified, these molecules may be titrated within exosomes to elicit the optimal effect of the stem/progenitor cells for organ recovery/regeneration. In the MI model, the miRNA cargo of progenitor cell exosomes in recipient plasma most accurately represented how transplanted cells may be remodeling the myocardium. This platform may facilitate the local application of key miRNAs using bioengineered delivery approaches to improve ischemic heart regeneration. Thus, progenitor cell–specific exosomes have diagnostic as well as therapeutic implications for allogeneic cell–based therapies.
To identify the molecular mechanisms responsible for cardiac repair, we used an unbiased computational biology approach and found that the in vitro cellular mRNA model was unable to predict functional outcomes of the current study (40). This is most likely due to the previous study’s focus on pediatric CPCs and use of a different injury model focusing on right ventricular dysfunction (37). Despite this, we were able to retrain the model to gain insights into potential paracrine mediators, which supported the idea that cell therapy modulates the inflammatory cascade. When we trained the model on either data from exosomes derived from cultured cells or progenitor cell–specific exosomes from rat plasma, predictability values were high regardless of exosome source, indicating the validity of the model. However, only progenitor cell exosomes derived from the plasma predicted the outcomes seen in the current study. When limited to the in vitro exosome data, our results would predict that exosomes derived from CDCs would be much more reparative than CPCs, which suggests that, to better understand the clinical potential of CPCs versus CDCs, additional head-to-head comparisons of purified exosomes are warranted. These findings support that progenitor cells release different exosomes in vivo versus in vitro, and that the microenvironmental cues in vivo may play a role in triggering release of different exosome quantities with different cargoes.
Our previous studies showed that physiological stresses on the CPCs, for instance, hypoxia, improved the functional abilities of these treated CPCs to recover the injured myocardium by directly changing their exosome miRNA cargo (37, 50). Analysis of miRNAs enriched in the transplanted CPC-derived circulating exosomes identified various signaling pathways, including cellular metabolism, cell survival and proliferation, and HIPPO and FoxO signaling, that potentially contribute to functional recovery of injured myocardium (42). We have identified miRNAs within CPC-derived exosomes that may have roles in cardiac repair (specifically miR-384, miR-378, miR-1224–5p, miR-525–3p, and miR-515). Because exosomal miRNAs exist in low copy numbers, the unbiased selection of pools of signals will be critical in determining potential mechanistic contributions.
Computational modeling provides an avenue for determining the mechanistic pathways driving progenitor cell–mediated remodeling of the MI myocardium. Similar to our previous validation study for our computational modeling of transplanted pediatric progenitor cells (37, 51), we performed the initial validation of our model by examining miRNAs involved in angiogenesis. Transplanted stem/progenitor cells induce recovery of injured myocardium by several cellular pathways, among which angiogenesis is one of the most important pathways. Therefore, we choose to assess the specificity of miRNAs predicted by our computational model to induce angiogenesis using well-established in vitro assays. Although miR-378 and miR-525 failed to promote cell migration, all miRNAs induced cellular proliferation and transwell migration, suggesting that the methodology adopted by our computational analyses is accurate and the exosomes secreted after cellular transplantation are more effective in predicting miRNAs involved in angiogenesis. The complete mechanism and verification of direct miRNA targets during myocardial remodeling remain the focus of future work.
Plasma-based exosome profiling could be very useful for monitoring the efficacy of allogeneic cell–based therapies in future clinical trials. Pathways driven by miRNA cargo of transplanted CPC-derived circulating exosomes give an unparalleled insight into the mechanisms of cellular recovery after cellular transplantation. By direct head-to-head comparison of miRNA function in vitro, we demonstrated that CPC-derived exosomes had a stronger ability to induce angiogenic response compared to CDC-derived exosomes. Because both cell types were derived from the same heart biopsy sample, our approach eliminated patient variability, strengthening the observed comparative data. CPCs were superior in terms of paracrine factor secretion, angiogenesis, myocardial tissue preservation, and cardiac functional improvement. As expected, increasing the c-kit+ cell proportion in the CDC population increased myocardial recovery, supporting the importance of the c-kit+ cell population within CDCs. Despite increased exosome secretion by CDCs in vitro, CPCs demonstrated increased exosome secretion in vivo and improved myocardial functional recovery in this model.
There are limitations to our study. One limitation is that although we have verified the effect of miRNAs predicted to promote angiogenesis, we will need to complete the characterization of all mechanism(s) necessary for myocardial recovery by determining other miRNA cargo involved in these processes. Additional studies will verify whether the predicted miRNAs enriched in circulating exosomes will offer cardioprotection. Despite our studies involving two time points, a more detailed study will determine the correlation between the number of transplanted cells’ circulating exosomes and myocardial functional recovery. Although HLA mismatch between the donor and recipient provides an excellent marker for donor exosome detection, specificity of HLA antibodies may limit their clinical application. Another approach to detect circulating exosomes may involve transplanting technetium-99m (99mTc)–labeled cells and thus their exosomes (52). We anticipate that the exosomes generated from these cells will also be labeled with 99mTc and can be detected even with smaller quantities of recipient plasma.
In summary, effective use of stem/progenitor cell therapy for human cardiac recovery will require a noninvasive surveillance method that can measure exogenous cell retention and interrogate cardioprotective pathways. Quantitative and cargo profiles of circulating progenitor/stem cell–derived exosomes may serve as a diagnostic platform. Our study suggests that miRNA cargoes of circulating exosomes are different than those secreted by cells in vitro. Here, we implicated the potential role of progenitor cell–derived exosome cargoes in the enhanced cardioprotective effects of CPCs relative to CDCs in the rat MI model. Progenitor cell–derived exosomes could facilitate development of next-generation allogeneic stem/progenitor cell–based therapies. Nevertheless, our results provide strong evidence that the transplanted cell–specific HLA can be used as a hallmark to identify and purify their exosomes from recipient serum and may be potentially applied to other allogeneic therapeutics.
MATERIALS AND METHODS
Study design
The research objective was to establish a circulating exosome-based diagnostic biomarker platform to noninvasively monitor the status of transplanted cardiac progenitor/stem cells using rodent MI model. We used two well-studied and clinically tested cardiac progenitor/stem cells derived from the same human cardiac biopsies of both male and female patients as mentioned in table S1. We verified their cell surface markers using FACS and tested them in our in vitro cell-based assays and in vivo rodent MI model studies. Our initial analyses identified CPCs to have more potential of cardiac function recovery and attenuation of LV remodeling than CDCs as assessed by echocardiography and immunohistochemistry. These results were verified by in vivo experiments with increasing CPC stoichiometry in CDCs. Concurrently, we performed computational analyses coupled with systems biology to determine miRNA profile of exosomes derived from these two cells types either from culture conditions or purified from recipient plasma after transplantation. For purification of transplanted cell–derived exosomes from the rodent serum, we used HLA-A surface marker. Our studies determined that, in response to the microenvironment of injured myocardium, transplanted cells modulate miRNA profile of their exosomes and the changes in miRNA profile of transplanted CPCs were predicted to be more cardioprotective. In silico predictions were further verified by various cell-based angiogenesis assays. These studies were conducted with rigorous experimental design (randomization, analysis by investigators blinded to group assignment). All data and detailed methods are made available in the Supplementary Materials with hyperlinks to original files to enable verification by other research groups. To achieve robust and unbiased results from our experiments, we adopted the following criteria: (i) For MI experiments, animals were assigned to treatment groups randomly. (ii) Myocardial function and histological analysis were assessed by performers blinded to the treatment group. (iii) Analysis of 7 to 10 animals per treatment group and (iv) data was analyzed using nonparametric t test (comparison of two groups), one-way ANOVA (comparison of more than two groups), and two-way ANOVA (comparison of more than two groups with multiple time points) followed by appropriate post hoc test. Group numbers of rodents used in in vivo experiments are provided below and in the figure legends. Data points from in vivo experiments were excluded if the animal died before the completion of the experiment. Chemicals, molecular reagents, and chemically synthesized miRNAs of highest-grade purity were purchased from reputable vendors. To account for variability between sexes, all in vivo models were performed on animals of both sexes. Data were disaggregated to enable assessment of possible sex differences.
Human tissue samples and cell culture
The Institutional Review Board and the Institute of Animal Care and Use Committee at the University of Maryland School of Medicine approved this study. After patient consent was given, specimens (70 ± 80 mg) from the RAA were obtained from 14 adult patients during coronary artery bypass grafting (table S1). These tissue samples were cut into two equal halves: One half was processed for isolating c-kit+ CPCs, and the other half was used for generation of human CDCs via the cardiosphere development. We randomly chose paired CPC and CDC cell lines derived from the same heart biopsies for this study, with each biological paired sample treated as n = 1.
Generation of c-kit+/CD45− cardiac stem cell- and cardiosphere-derived cells
c-kit+/CD45− cells were isolated from RAA biopsies of adult myocardium using a previously described protocol (6, 53). Briefly, samples were minced and digested in Ham’s F12 (Lonza, 12–615F) basal medium containing collagenase type II (1 to 2 mg/ml; Worthington, 4177) on an orbital shaker for 45 min at 37°C. After the collagenase treatment, cells were washed twice with complete growth medium [Ham’s F12, 10% fetal bovine serum (FBS), recombinant human FGF-basic (10 ng/ml), 0.2 mM l-glutathione, and human erythropoietin 250 (5 U/ml)] before being plated. At subconfluency, cells were trypsinized and sorted for c-kit+ cell surface antigen with Miltenyi MicroBeads (CD117 MicroBead Kit human, 130-091-332) as per the manufacturer’s instructions. Consequently, c-kit+ cells were collected and cultured in growth medium (5). Human CDCs were generated according to the protocol with modifications (6, 53). Briefly, RAA biopsies are diced into small pieces (1 to 2 mm in diameter) and digested with digestion cocktail (dispase, 0.85 mg/ml), collagenase type II (1 mg/ml), and trypsin (0.05%) for 15 min at 37°C in orbital shaker for three times. Explants were collected in CDC complete growth medium [IMDM, 20% FBS (heat-inactivated), and 100 μM β-mercaptoethanol] and plated in fibronectin-coated flasks. After 2 to 3 weeks, the phase-bright cells originating from explants were removed by mild trypsinization and plated on fibronectin-coated flasks at low density (1.5 × 104 to 3 × 104 cells/ml) in cardiosphere-growing medium [3.5% FBS, 35% IMDM, 1% penicillin-streptomycin, 1% glutamine, 2% B27 serum substitute, cardiotroponin I (4 ng/ml), epidermal growth factor (25 ng/ml), human basic FGF (80 ng/ml), thrombin (1 U/ml), 100 μM β-mercaptoethanol, and 65% Dulbecco’s modified Eagle’s medium (DMEM)/Ham’s F12 (1:1)]. After a week of culture, the phase-bright cells formed cardiospheres. After 24 hours, floating cardiospheres were collected by centrifugation and expanded on fibronectin-coated flasks in CDC complete growth medium to obtain CDCs.
Flow cytometric analysis
c-kit+/CD45− cells derived from adult patients at P4 for both CPCs and CDCs were labeled with fluorochrome-conjugated primary antibodies: mesenchymal stem cell marker CD105 or CD90, cardiac-specific transcription factors NKX2.5 and GATA4, cardiac stem cell marker c-kit+, endothelial cell marker CD31, mast cell marker tryptase, and hematopoietic cell lineage markers CD45 and CD34. Conjugated isotype antibodies were used as negative controls. The labeled cells were evaluated by flow cytometry with FACSCalibur (Becton Dickinson), with 25,000 events per sample collected.
Cell transplantation and echocardiography
MI was induced by permanent ligation of the LAD coronary artery in athymic nude male rats (weight, 250 to 300 g). The heart was exposed via a left thoracotomy, and the proximal LAD was ligated. Subsequently, 1 million adult CPCs or CDCs suspended in 100 μl of vehicle (IMDM) were injected into the myocardium at four sites adjacent to the infarct. Transthoracic echocardiograms were acquired 1, 7, and 28 days after MI surgery. Cardiac function was evaluated on the basis of echocardiographic parameters (5). M-mode images of the LV in the parasternal short-axis view were obtained at the level of the papillary muscles using high-resolution ultrasound (Vevo 2100: VisualSonics) equipped with a 20-MHz scan head for calculation/estimation of LV fraction shortening and LV EF. Data were calculated from five cardiac cycles according to the generally accepted formula (54).
Myocardial histology
Tissues were processed as previously described (5, 6, 53). Briefly, rat hearts were excised under anesthesia after collection of echocardiographic data and perfused with 4% paraformaldehyde. Tissues were cryopreserved using 30% sucrose and embedded in OCT (optimal cutting temperature) (Tissue-Tek). Sections were cut to 7 μm using a cryostat and immunostained for isolectin B4 (Invitrogen) and α-SMA (Sigma). Cells and tissue sections were counterstained with DAPI nuclear stain (Sigma).
Myocardial viability
To calculate infarct size, Masson trichrome–stained sections at various levels along the long axis were analyzed for collagen deposition, and the infarct size was determined by midline technique (55). The stained sections were analyzed using Image-Pro software (5). To calculate the amounts of viable and nonviable tissues, the number of pink pixels (viable tissue) and blue pixels (nonviable tissue) was measured and the ratio of nonviable tissue/overall number of the pixels was presented. Six sections per animal and at least 15 animals per group were analyzed.
Preparation of cell mixtures with increasing stoichiometry of CPCs in CDC populations
CPCs and CDCs are mixed together in the proportion of 20, 40, and 80% of CPCs in the CDC population for in vivo and in vitro studies. CDC populations inherently contain 10% c-kit+ cells, which were factored into the proportions presented below, and the artificial mixtures started from 20% CPCs in CDCs. One million adult CPCs and CDCs suspended in 100 μl of vehicle (IMDM) were injected into the rat myocardium as a control. In case of 20% CPC group (2 × 105 CPCs + 8 × 105 CDCs), 40% CPC group (4 × 105 cells from CPCs + 6 × 105 cells from CDCs), and 80% CPC group (8 × 105 cells from CPCs + 2 × 105 cells from CDCs), a total of 1 million cells (in 100 μl) were injected in the rat myocardium.
Paracrine factor quantification
Adult CPCs and CDCs at P3 were grown in complete medium until they reached 85% confluence (~1 × 106 cells) in a T-25 flask. The conditioned medium was collected and filtered through 0.22-μm filters and concentrated using 3-kDa filters (Millipore Inc.). Protein content was quantified using the Pierce bicinchoninic assay (BCA) method (Thermo Fisher Scientific). To normalize the protein content, we used the following formula: (concentration factor) × (total volume of medium)/(total protein content) of conditioned medium (5). The conditioned medium was quantified using BCA method and normalized to a total of 1 mg of protein. ELISA was performed for human VEGF-1A, SDF-1α, PDGFB, IGF-1, ANG-1, bFGF, and HGF in the core facility at the University of Maryland School of Medicine using human-specific ELISA kits (Millipore and R&D Systems), according to the manufacturers’ protocols.
Transwell migration assay
CPCs and CDCs were placed on the upper layer of a cell culture insert with permeable fluorescence block (8.0-μm pore size, catalog no. 351152) membrane, and the media with serum and without serum are placed below the cell-permeable membrane in a 24-well cell culture plate (catalog no. 353504). After an incubation period (6 to 7 hours) at 37°C, the cells that migrated through the membrane were stained with calcein (Calcein AM, Thermo Fisher Scientific, C3100MP), imaged as whole wells by an automated EVOS microscope, and quantified using Image-Pro software. In case of endothelial migration assay by HMECs, 50 ng of exosomes isolated from CPCs and CDCs was incubated along with the HMECs on the cell culture insert (upper chamber). After 3 hours of the incubation, migrated cells were imaged by EVOS Systems (Thermo Fisher Scientific Inc.) and quantified using Image-Pro software.
alamarBlue cell proliferation assay
Cell proliferation was assessed using alamarBlue as per the manufacturer’s instructions (10% of the total volume of the medium). Briefly, 5000 cells per well were seeded in 96-well plates in their respective medium. After overnight incubation at 37°C, 10 μl of alamarBlue cell viability reagent (Invitrogen, catalog no. 1933424) was added per well, and absorbance was taken immediately (basal absorbance) and after 3 hours (proliferation absorbance) of incubation at 37°C. To obtain the actual absorbance, basal absorbance was subtracted from proliferation absorbance.
Senescence-associated β-galactosidase staining
Cellular senescence was identified as described previously (5). Briefly, adult CPCs and CDCs (5.0 × 104) were plated in a 24-well plate at P6. A β-galactosidase staining kit (Cell Signaling Technology, catalog no. 9860) was used to stain the enzyme using the manufacturer’s protocol. Cell growth medium was removed from the cells; cells were rinsed with phosphate-buffered saline (PBS) and fixed with fixative solution (4% paraformaldehyde) for 15 min at room temperature. Cells were incubated with 1 ml of β-galactosidase staining solution. The β-galactosidase–positive (blue) cells were imaged in EVOS microscope (20×). The number of positive cells was quantified using Image-Pro software.
In vitro wound-healing assay
An in vitro scratch assay was performed to assess the relative migratory potential of CPCs, CDCs, and several mixtures of CPCs in CDCs. Cells were seeded in a 12-well plate to create a confluent monolayer. After 12 hours of serum starvation with basal medium, a 1-ml pipette tip was used to create linear scratches along the cell monolayer to simulate a wound. Cell debris was removed by washing the cells once with basal medium. Images of each wound were taken at specific reference points along the scratch at times 0 hours and 6 to 7 hours after treatment. Image-Pro software was used to measure the total wound area before and after treatment to calculate percentage change in wound closure. For endothelial wound-healing assay, HMEC was seeded in 24-well plates to create confluent monolayer. HMEC scratches were treated with basal medium (MCDB 131) or basal medium containing exosomes derived from CPCs or CDCs. Cells were fixed in their wells, and after 16 hours, migrated distances were calculated using image-Pro software.
Immunoblotting
Exosome proteins were separated using NuPAGE 4 to 12% bis-tris gels and transferred onto nitrocellulose membranes (Life Technologies). The blots were blocked with 5% nonfat dry milk at room temperature for 1 hour and incubated overnight at 4°C with desired primary antibodies at concentration per the manufacturer’s protocol, followed by incubation with horseradish peroxidase–conjugated secondary antibodies (Santa Cruz Biotechnologies Inc.) at room temperature for 1 hour. The membrane blots were developed with enhanced chemiluminescence detection reagent (Millipore Corporation) per the manufacturer’s protocol and detected through chemiluminescence using ImageQuant LAS 400 Phospho-Imager (GE Healthcare). As a standard protein marker, we used Thermo Fisher Scientific PageRuler Plus Prestained Protein Ladder (no. 26619). Antibodies specific to c-kit (catalog no. 18696–1-AP), troponin-I (ab56357), HLA-A (ab52922), flotilin-1 (CST 3253), HLA-A1 (One Lambda BIH0331), NKX2.5 (SC-376565) (according to the company’s data sheet, this antibody recognizes two bands in some cell lines), CD-63 (Sc-7080), and cytochrome c (Sc-13156) were purchased from Santa Cruz Biotechnology.
Affinity antibody coupled purification of human-specific exosomes
HLA-A–specific antibodies were covalently conjugated to N-hydroxysuccinamide magnetic beads (Pierce Inc.) as per the manufacturer’s suggestions. Protein equivalent (50 μg) of exosomes was incubated with antibody–magnetic beads complex overnight at 4°C on a rocker platform. The bead-bound exosomes were washed using PBS, eluted using the manufacturer’s protocol, and used for downstream analysis.
Isolation procedure of exosomes and exosomal miRNA
Exosomes were isolated from CPC- and CDC-conditioned medium (48 hours) by size exclusion chromatography, and miRNA was immediately isolated from the exosomes using an exoRNeasy kit (catalog no. 77023) as per the manufacturer instruction. Total RNA was quantified on a NanoDrop ND-1000 spectrophotometer followed by RNA quality assessment on Agilent TapeStation. miRNA labeling was performed using the FlashTag Biotin HSR RNA Labeling Kit (Applied Biosystems). GeneChip miRNA 4.0 Arrays were hybridized with FlashTag Biotin-Labeled total RNA (100 ng) from experimental and control samples in 100 μl of hybridization cocktail. Target denaturation was performed at 99°C for 5 min and then at 45°C for 5 min followed by hybridization for 18 hours at 48°C. Arrays were washed and stained using GeneChip Fluidics Station 450 according to protocol. Chips were scanned on Affymetrix GeneChip Scanner 3000, using Command Console Software. These studies were performed at Cancer Genomics Laboratory of the Thomas Jefferson University.
NanoSight particle analysis for exosomes
Exosomes from CDCs and CPCs were isolated by size exclusion chromatography using a Sepharose 2B column (Sigma-Aldrich), and eluted fractions were analyzed using NanoSight NS300 (405-nm laser diode) for the presence of 40- to 120-nm-diameter vesicles (26). For cell-based in vitro assays, exosomes from CDCs and CPCs were used at the constant number 0.5 × 109/100 μl, equivalent to 10 ng/100 μl proteins. The surface expression of HLA class I was analyzed using exosomes (2 × 108) incubated with 0.5 μg of anti–HLA class I (BioLegend, catalog no. 311402) for 2 hours. Thereafter, goat anti-mouse Qdot 605 (1:50 dilution; Q-11001MP, Thermo Fisher Scientific) was added as fluorescent secondary antibody and incubated for 2 hours. The unbound primary and secondary antibodies were removed using ExoQuick PLUS (EQPL10A-1, System Biosciences) exosome isolation kit according to the manufacturer’s protocol. Total exosomes were counted in bright-field emission, and the HLA class I–labeled exosomes were counted using fluorescent emission in NanoSight. HLA-specific exosome signal was quantified using the following formula: (HLA fluorescence/HLA light scatter) − (POD 0 fluorescence/POD 0 light scatter) – (IgG isotype fluorescence/IgG isotype light scatter).
miRNA mimic transfections
To identify the functional role of miRNAs, human miRIDIAN mimics (miR-378, miR-384, miR-515–5p, miR-525–3p, and miR-1224), along with the transfection control-Dy547 (catalog no. CP-004500-01-05), positive control (catalog no. CP-001000-02-05), and scrambled (non-targeting) miRNAs, were procured from Dharmacon. Corresponding miRNA inhibitors were procured from Thermo Fisher Scientific Inc. Cells were transfected with 25 nM of each miRNA mimic or inhibitor using the reverse transfection protocol of Lipofectamine RNAiMAX (catalog no. P/N 100014472). In the miRNA mimic + inhibitor group, after 72 hours of miRNA mimic transfection, cells were transfected with 25 nM of corresponding miRNA inhibitors. After 72 hours of final transfection, HMECs were trypsinized and plated for different cell-based assays: (i) transwell migration assay, (ii) cell proliferation assay, and (iii) wound-healing assay.
Partial least squares regression modeling
PCA and PLSR were performed using SIMCA-P software (UMetrics, now part of Sartorius Stedim Biotech; https://umetrics.com) that solves the PLSR problem with the nonlinear partial least squares algorithm (37). Gene and miRNA data were analyzed by IPA. miRTarBase was used to identify miRNAs with known targets (validated by at least three assays, http://mirtarbase.mbc.nctu.edu.tw).
Transmission electron microscopy
Exosomes were negatively stained after absorption onto carbon-coated copper grids for 2 min. Grids were washed twice for 1 min each in dH2O and stained for 1 min with 1% aqueous uranyl acetate (Ted Pella). Samples were viewed on a JEOL 1200EX TEM (JEOL) equipped with an AMT 8-megapixel digital camera (Advanced Microscopy Techniques). For immunogold labeling of exosomes, grids were incubated with mouse anti-CD63 antibody (AB193349, Abcam) for 30 min, followed by secondary goat anti-mouse IgG antibody conjugated to colloidal gold (Jackson ImmunoResearch Laboratories) for 30 min. Grids were washed and stained with uranyl acetate and viewed by TEM as described above.
Statistical analyses
Data were analyzed using GraphPad Prism 7 software. When comparing two conditions, we used Student’s t tests with Mann-Whitney’s test. More than two comparisons were made using one-way ANOVA followed by Dunn’s or Tukey’s post hoc test. Two-way ANOVA with Bonferroni correction was used for grouped analysis of echocardiographic data. P values of less than 0.05 were considered significant, and two-sided tests were performed. Data are represented as mean ± SEM. Individual subject-level data are reported in data file S1.
Supplementary Material
Fig. S1. Representative histograms of flow cytometry analysis for antigenic phenotype of CPCs and CDCs at P3.
Fig. S2. Comparative analysis of senescence and transwell migration potential of CPCs and CDCs.
Fig. S3. Growth potential of CPCs and CDCs in CDC and CPC growth medium.
Fig. S4. Effect of increasing number (percentage) of c-kit+ cells in CDCs on their migration potential.
Fig. S5. Quantification and functionality of exosomes derived from CPCs and CDCs.
Fig. S6. Complete immunoblots related to Fig. 3F.
Fig. S7. Schematic of donor human exosome purification and identification from recipient rat serum after cellular transplantation.
Fig. S8. Complete immunoblots related to Fig. 3J.
Fig. S9. Computational modeling of exosome miRNA cargo.
Fig. S10. PLSR analysis of gene array.
Fig. S11. miRNAs predicted to improve cardiac function by enhancing angiogenesis.
Table S1. Source of heart biopsies for CPC and CDC generation.
Table S2. Top 50 mRNA changes (CPC compared to CDC).
Table S3. Exosome miRNA VIP.
Table S4. miRNAs with their roles in cardioprotective processes.
Table S5. List of canonical signaling pathways affected by VIP miRNAs.
Data file S1. Individual subject-level data.
Funding:
S.K. was supported by NIH grants 1R01HL118491, 1R01HL139060-02, and R01HL141922-02 and the Maryland Stem Research Fund. S.S. is supported by the Maryland Stem Cell Research Fund and AHA-CDA-18CDA34110282-2. S.R.D. was supported by the Maryland Stem Cell Research Fund. P.V and L.K. are supported by internal funds from the University of Pennsylvania.
Footnotes
SUPPLEMENTARY MATERIALS
Competing interests: A patent was filed on the circulating exosomal biomarker platform for allogeneic therapeutics.
Data and materials availability: All data associated with this study are presented in the manuscript or the Supplementary Materials.
REFERENCES AND NOTES
- 1.Zwetsloot PP, Vegh AM, Jansen of Lorkeers SJ, van Hout GPJ, Currie GL, Sena ES, Gremmels H, Buikema JW, Goumans M-J, Macleod M-R, Doevendans PA, Chamuleau SAJ, Sluijter JPG, Cardiac stem cell treatment in myocardial infarction: A systematic review and meta-analysis of preclinical studies. Circ. Res 118, 1223–1232 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Gyöngyösi M, Wojakowski W, Lemarchand P, Lunde K, Tendera M, Bartunek J, Marban E, Assmus B, Henry TD, Traverse JH, Moyé LA, Sürder D, Corti R, Huikuri H, Miettinen J, Wöhrle J, Obradovic S, Roncalli J, Malliaras K, Pokushalov E, Romanov A, Kastrup J, Bergmann MW, Atsma DE, Diederichsen A, Edes I, Benedek I, Benedek T, Pejkov H, Nyolczas N, Pavo N, Bergler-Klein J, Pavo IJ, Sylven C, Berti S, Navarese EP, Maurer G, Meta-Analysis of Cell-based CaRdiac stUdiEs (ACCRUE) in patients with acute myocardial infarction based on individual patient data. Circ. Res 116, 1346–1360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cambria E, Pasqualini FS, Wolint P, Günter J, Steiger J, Bopp A, Hoerstrup SP, Emmert MY, Translational cardiac stem cell therapy: Advancing from first-generation to next-generation cell types. NPJ Regen. Med 2, 17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu R, Hu X, Wang J, Concise review: Optimized strategies for stem cell-based therapy in myocardial repair: Clinical translatability and potential limitation. Stem Cells 36, 482–500 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Sharma S, Mishra R, Bigham GE, Wehman B, Khan MM, Xu H, Saha P, Goo YA, Datla SR, Chen L, Tulapurkar ME, Taylor BS, Yang P, Karathanasis S, Goodlett DR, Kaushal S, A deep proteome analysis identifies the complete secretome as the functional unit of human cardiac progenitor cells. Circ. Res 120, 816–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simpson DL, Mishra R, Sharma S, Goh SK, Deshmukh S, Kaushal S, A strong regenerative ability of cardiac stem cells derived from neonatal hearts. Circulation 126, S46–S53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolli R, Chugh AR, D’Amario D, Loughran JH, Stoddard MF, Ikram S, Beache GM, Wagner SG, Leri A, Hosoda T, Sanada F, Elmore JB, Goichberg P, Cappetta D, Solankhi NK, Fahsah I, Rokosh DG, Slaughter MS, Kajstura J, Anversa P, Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): Initial results of a randomised phase 1 trial. Lancet 378, 1847–1857 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Makkar RR, Smith RR, Cheng K, Malliaras K, Thomson LEJ, Berman D, Czer LSC, Marbán L, Mendizabal A, Johnston PV, Russell SD, Schuleri KH, Lardo AC, Gerstenblith G, Marbán E, Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): A prospective, randomised phase 1 trial. Lancet 379, 895–904 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong KU, Guo Y, Li QH, Cao P, Al-Maqtari T, Vajravelu BN, Du J, Book MJ, Zhu X, Nong Y, Bhatnagar A, Bolli R, c-kit+ Cardiac stem cells alleviate post-myocardial infarction left ventricular dysfunction despite poor engraftment and negligible retention in the recipient heart. PLOS ONE 9, e96725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibrahim AG-E, Cheng K, Marban E, Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Rep. 2, 606–619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallet R, Dawkins J, Valle J, Simsolo E, de Couto G, Middleton R, Tseliou E, Luthringer D, Kreke M, Smith RR, Marbán L, Ghaleh B, Marbán E, Exosomes secreted by cardiosphere-derived cells reduce scarring, attenuate adverse remodelling, and improve function in acute and chronic porcine myocardial infarction. Eur. Heart J 38, 201–211 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibrahim A, Marban E, Exosomes: Fundamental biology and roles in cardiovascular physiology. Annu. Rev. Physiol 78, 67–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L, Wang Y, Pan Y, Zhang L, Shen C, Qin G, Ashraf M, Weintraub N, Ma G, Tang Y, Cardiac progenitor-derived exosomes protect ischemic myocardium from acute ischemia/reperfusion injury. Biochem. Biophys. Res. Commun 431, 566–571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arslan F, Lai RC, Smeets MB, Akeroyd L, Choo A, Aguor ENE, Timmers L, van Rijen HV, Doevendans PA, Pasterkamp G, Lim SK, de Kleijn DP, Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res 10, 301–312 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Bian S, Zhang L, Duan L, Wang X, Min Y, Yu H, Extracellular vesicles derived from human bone marrow mesenchymal stem cells promote angiogenesis in a rat myocardial infarction model. J. Mol. Med 92, 387–397 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Singla DK, Stem cells and exosomes in cardiac repair. Curr. Opin. Pharmacol 27, 19–23 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Kishore R, Khan M, More than tiny sacks: Stem cell exosomes as cell-free modality for cardiac repair. Circ. Res 118, 330–343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahoo S, Losordo DW, Exosomes and cardiac repair after myocardial infarction. Circ. Res 114, 333–344 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Suzuki E, Fujita D, Takahashi M, Oba S, Nishimatsu H, Stem cell-derived exosomes as a therapeutic tool for cardiovascular disease. World J. Stem Cells 8, 297–305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barile L, Lionetti V, Cervio E, Matteucci M, Gherghiceanu M, Popescu LM, Torre T, Siclari F, Moccetti T, Vassalli G, Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc. Res 103, 530–541 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Lawson C, Vicencio JM, Yellon DM, Davidson SM, Microvesicles and exosomes: New players in metabolic and cardiovascular disease. J. Endocrinol 228, R57–R71 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Colombo M, Raposo G, Théry C, Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol 30, 255–289 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Julich H, Willms A, Lukacs-Kornek V, Kornek M, Extracellular vesicle profiling and their use as potential disease specific biomarker. Front. Immunol 5, 413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo DJ, Kaplan B, Kirk AD, Biomarkers for kidney transplant rejection. Nat. Rev. Nephrol 10, 215–225 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Vallabhajosyula P, Korutla L, Habertheuer A, Reddy S, Schaufler C, Lasky J, Diamond J, Cantu III E, Ex vivo lung perfusion model to study pulmonary tissue extracellular microvesicle profiles. Ann. Thorac. Surg 103, 1758–1766 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Vallabhajosyula P, Korutla L, Habertheuer A, Yu M, Rostami S, Yuan C-X, Reddy S, Liu C, Korutla V, Koeberlein B, Trofe-Clark J, Rickels MR, Naji A, Tissue-specific exosome biomarkers for noninvasively monitoring immunologic rejection of transplanted tissue. J. Clin. Invest 127, 1375–1391 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Habertheuer A, Korutla L, Rostami S, Reddy S, Lal P, Naji A, Vallabhajosyula P, Donor tissue-specific exosome profiling enables noninvasive monitoring of acute rejection in mouse allogeneic heart transplantation. J. Thorac. Cardiovasc. Surg 155, 2479–2489 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Valadi H, Ekstrom K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO, Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol 9, 654–659 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Hodgkinson CP, Bareja A, Gomez JA, Dzau VJ, Emerging concepts in paracrine mechanisms in regenerative cardiovascular medicine and biology. Circ. Res 118, 95–107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rota M, Padin-Iruegas ME, Misao Y, De Angelis A, Maestroni S, Ferreira-Martins J, Fiumana E, Rastaldo R, Arcarese ML, Mitchell TS, Boni A, Bolli R, Urbanek K, Hosoda T, Anversa P, Leri A, Kajstura J, Local activation or implantation of cardiac progenitor cells rescues scarred infarcted myocardium improving cardiac function. Circ. Res 103, 107–116 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams AR, Hare JM, Mesenchymal stem Cells. Circ. Res 109, 923–940 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellison GM, Torella D, Dellegrottaglie S, Perez-Martinez C, Perez de Prado A, Vicinanza C, Purushothaman S, Galuppo V, Iaconetti C, Waring CD, Smith A, Torella M, Cuellas Ramon C, Gonzalo-Orden JM, Agosti V, Indolfi C, Galinanes M, Fernandez-Vazquez F, Nadal-Ginard B, Endogenous cardiac stem cell activation by insulin-like growth factor-1/hepatocyte growth factor intracoronary injection fosters survival and regeneration of the infarcted pig heart. J. Am. Coll. Cardiol 58, 977–986 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Gnecchi M, He H, Liang OD, Melo LG, Morello F, Mu H, Noiseux N, Zhang L, Pratt RE, Ingwall JS, Dzau VJ, Paracrine action accounts for marked protection of ischemic heart by Akt-modified mesenchymal stem cells. Nat. Med 11, 367–368 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Mirotsou M, Jayawardena TM, Schmeckpeper J, Gnecchi M, Dzau VJ, Paracrine mechanisms of stem cell reparative and regenerative actions in the heart. J. Mol. Cell. Cardiol 50, 280–289 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gardiner C, Ferreira YJ, Dragovic RA, Redman CW, Sargent IL, Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell. Vesicles 2, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnston PV, Sasano T, Mills K, Evers R, Lee S-T, Smith RR, Lardo AC, Lai S, Steenbergen C, Gerstenblith G, Lange R, Marban E, Engraftment, differentiation, and functional benefits of autologous cardiosphere-derived cells in porcine ischemic cardiomyopathy. Circulation 120, 1075–1083 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agarwal U, George A, Bhutani S, Ghosh-Choudhary S, Maxwell JT, Brown ME, Mehta Y, Platt MO, Liang Y, Sahoo S, Davis ME, Experimental, systems, and computational approaches to understanding the MicroRNA-mediated reparative potential of cardiac progenitor cell-derived exosomes from pediatric patients. Circ. Res 120, 701–712 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lobb RJ, Becker M, Wen Wen S, Wong CSF, Wiegmans AP, Leimgruber A, Möller A, Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Exracell. Vesicles 4, 27031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJ, Hole P, Carr B, Redman CW, Harris AL, Dobson PJ, Harrison P, Sargent IL, Sizing and phenotyping of cellular vesicles using nanoparticle tracking analysis. Nanomedicine 7, 780–788 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agarwal U, Smith AW, French KM, Boopathy AV, George A, Trac D, Brown ME, Shen M, Jiang R, Fernandez JD, Kogon BE, Kanter KR, Alsoufi B, Wagner MB, Platt MO, Davis ME, Age-dependent effect of pediatric cardiac progenitor cells after juvenile heart failure. Stem Cells Transl. Med 5, 883–892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, Giacca M, Functional screening identifies miRNAs inducing cardiac regeneration. Nature 492, 376–381 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG, DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 43, W460–W466 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathiyalagan P, Sahoo S, Exosomes-based gene therapy for MicroRNA Delivery. Methods Mol. Biol 1521, 139–152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nowak-Sliwinska P, Alitalo K, Allen E, Anisimov A, Aplin AC, Auerbach R, Augustin HG, Bates DO, van Beijnum JR, Bender RHF, Bergers G, Bikfalvi A, Bischoff J, Böck BC, Brooks PC, Bussolino F, Cakir B, Carmeliet P, Castranova D, Cimpean AM, Cleaver O, Coukos G, Davis GE, De Palma M, Dimberg A, Dings RPM, Djonov V, Dudley AC, Dufton NP, Fendt S-M, Ferrara N, Fruttiger M, Fukumura D, Ghesquiere B, Gong Y, Griffin RJ, Harris AL, Hughes CCW, Hultgren NW, Iruela-Arispe ML, Irving M, Jain RK, Kalluri R, Kalucka J, Kerbel RS, Kitajewski J, Klaassen I, Kleinmann HK, Koolwijk P, Kuczynski E, Kwak BR, Marien K, Melero-Martin JM, Munn LL, Nicosia RF, Noel A, Nurro J, Olsson AK, Petrova TV, Pietras K, Pili R, Pollard JW, Post MJ, Quax PHA, Rabinovich GA, Raica M, Randi AM, Ribatti D, Ruegg C, Schlingemann RO, Schulte-Merker S, Smith LEH, Song JW, Stacker SA, Stalin J, Stratman AN, Van de Velde M, van Hinsbergh VWM, Vermeulen PB, Waltenberger J, Weinstein BM, Xin H, Yetkin-Arik B, Yla-Herttuala S, Yoder MC, Griffioen AW, Consensus guidelines for the use and interpretation of angiogenesis assays. Angiogenesis 21, 425–532 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costantino S, Paneni F, Stem cell therapy in heart failure: Is the best yet to come? Int. J. Cardiol 260, 135–136 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Jeong H, Yim HW, Park H-J, Cho Y, Hong H, Kim NJ, Oh I-H, Mesenchymal stem cell therapy for ischemic heart disease: Systematic review and meta-analysis. Int. J. Stem Cells 11, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Broughton KM, Wang BJ, Firouzi F, Khalafalla F, Dimmeler S, Fernandez-Aviles F, Sussman MA, Mechanisms of cardiac repair and regeneration. Circ. Res 122, 1151–1163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phinney DG, Pittenger MF, Concise review: MSC-derived exosomes for cell-free therapy. Stem Cells 35, 851–858 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Boon RA, Dimmeler S, MicroRNAs in myocardial infarction. Nat. Rev. Cardiol 12, 135–142 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Gray WD, French KM, Ghosh-Choudhary S, Maxwell JT, Brown ME, Platt MO, Searles CD, Davis ME, Identification of therapeutic covariant microRNA clusters in hypoxia-treated cardiac progenitor cell exosomes using systems biology. Circ. Res 116, 255–263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garikipati VNS, Shoja-Taheri F, Davis ME, Kishore R, Extracellular vesicles and the application of system biology and computational modeling in cardiac repair. Circ. Res 123, 188–204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Musialek P, Tekieli L, Kostkiewicz M, Miszalski-Jamka T, Klimeczek P, Mazur W, Szot W, Majka M, Banys RP, Jarocha D, Walter Z, Krupinski M, Pieniazek P, Olszowska M, Zmudka K, Pasowicz M, Kereiakes DJ, Tracz W, Podolec P, Wojakowski W, Infarct size determines myocardial uptake of CD34+ cells in the peri-infarct zone: Results from a study of (99m)Tc-extametazime-labeled cell visualization integrated with cardiac magnetic resonance infarct imaging. Circ. Cardiovasc. Imaging 6, 320–328 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Mishra R, Vijayan K, Colletti EJ, Harrington DA, Matthiesen TS, Simpson D, Goh SK, Walker BL, Almeida-Porada G, Wang D, Backer CL, Dudley SC Jr., L. E. Wold, S. Kaushal, Characterization and functionality of cardiac progenitor cells in congenital heart patients. Circulation 123, 364–373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen L, Zhang J, Hu X, Philipson KD, Scharf SM, The Na+/Ca2+ exchanger-1 mediates left ventricular dysfunction in mice with chronic intermittent hypoxia. J. Appl. Physiol 109, 1675–1685 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharma S, Mishra R, Simpson D, Wehman B, Colletti EJ, Deshmukh S, Datla SR, Balachandran K, Guo Y, Chen L, Siddiqui OT, Kaushal S, Kaushal S, Cardiosphere-derived cells from pediatric end-stage heart failure patients have enhanced functional activity due to the heat shock response regulating the secretome. Stem Cells 33, 1213–1229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Representative histograms of flow cytometry analysis for antigenic phenotype of CPCs and CDCs at P3.
Fig. S2. Comparative analysis of senescence and transwell migration potential of CPCs and CDCs.
Fig. S3. Growth potential of CPCs and CDCs in CDC and CPC growth medium.
Fig. S4. Effect of increasing number (percentage) of c-kit+ cells in CDCs on their migration potential.
Fig. S5. Quantification and functionality of exosomes derived from CPCs and CDCs.
Fig. S6. Complete immunoblots related to Fig. 3F.
Fig. S7. Schematic of donor human exosome purification and identification from recipient rat serum after cellular transplantation.
Fig. S8. Complete immunoblots related to Fig. 3J.
Fig. S9. Computational modeling of exosome miRNA cargo.
Fig. S10. PLSR analysis of gene array.
Fig. S11. miRNAs predicted to improve cardiac function by enhancing angiogenesis.
Table S1. Source of heart biopsies for CPC and CDC generation.
Table S2. Top 50 mRNA changes (CPC compared to CDC).
Table S3. Exosome miRNA VIP.
Table S4. miRNAs with their roles in cardioprotective processes.
Table S5. List of canonical signaling pathways affected by VIP miRNAs.
Data file S1. Individual subject-level data.