Pharmacogenomics is an important aspect of clinical genomics that is touching an increasingly large number of patients. That is because pharmacogenomic testing is being implemented into clinical care at an increasing pace—initially primarily within academic medical centers, but also across healthcare systems. This Commentary describes the broad implementation of pharmacogenomics within one large, integrated academic medical system, with a focus on challenges that were and are being addressed and lessons that have been learned.
Pharmacogenomics is the aspect of clinical genomics that will eventually have the broadest application—ultimately touching every patient. Pharmacogenomics has evolved from early pharmacogenetic studies of candidate genes, often genes that encoded drug metabolizing enzymes, to become pharmacogenomics with the application of genome‐wide techniques that make it possible to scan across the genome in an unbiased fashion to identify single nucleotide polymorphisms and genes, often unanticipated genes, that contribute to variation in drug response.
There can no longer be any doubt of the clinical utility of certain aspects of pharmacogenomics, e.g., TPMT and DPYD polymorphisms, and the establishment of objective guidelines for the clinical implementation of pharmacogenomics by groups such as the Clinical Pharmacogenetics Implementation Consortium (CPIC) in the United States and the Dutch Working Group on Pharmacogenetics in Europe—among others—has helped to create the necessary peer‐reviewed knowledge infrastructure required to help bring rigor to this important aspect of clinical genomics. As a result, in parallel with the growth of the scientific foundation for pharmacogenomics, a discipline that continues to evolve rapidly, challenges associated with bringing this important aspect of clinical genomics to the bedside have grown in parallel. Former Speaker of the United States House of Representatives “Tip” O'Neil was fond of saying that “all politics are local.” The same is true of pharmacogenomic clinical implementation. Every academic medical center and, eventually, every hospital and clinic will need to address challenges associated with pharmacogenomic implementation, but always within boundaries established by their unique local environments. We have been deeply involved in attempts to bring pharmacogenomics into the practice for every patient seen in our large, highly integrated multiple‐site academic medical center. The perspectives reflected in subsequent paragraphs are based on that experience, but they involve issues faced by every institution attempting to bring this important aspect of clinical genomics to the bedside.
Infrastructure and Reactive Pharmacogenomics
The first requirement for the clinical implementation of pharmacogenomics in any medical institution is commitment and support from organizational leadership. As described in a “Translation Commentary” published in this journal in 2013,1 the Mayo Clinic leadership created a “Center for Individualized Medicine” (CIM) a decade ago that included a Pharmacogenomics Program charged with the clinical implementation of pharmacogenomics. The Mayo Clinic already had a Personalized Genomics Laboratory within the Department of Laboratory Medicine and Pathology capable of performing pharmacogenomic genotyping, so pharmacogenomic laboratory testing was available—although it was not widely ordered at that time.
The CIM Pharmacogenomics Program concluded that electronic health record (EHR) “alerts” at the point of care would be a useful step to help stimulate and assist the medical staff in ordering pharmacogenomic tests. To achieve that goal, a “Pharmacogenomics Task Force” was established within the Formulary Committee to, among other duties, select “Drug–Gene Pairs” that had clinical utility and were appropriate for implementation. Those alerts were designed to “fire” at the point of care the first time that a new drug was prescribed for which clinical utility had been agreed on through an internal peer‐reviewed process, a process that used CPIC and Dutch Working Group information and which always also included internal peer review and approval by appropriate clinical departments and divisions. Those alerts suggested, at the point of care, the possibility of ordering pharmacogenomic testing. Behind each alert, in the EHR in an “AskMayoExpert” database, was scientific and clinical information that provided the provider with data in support of the clinical value and use of the genomic tests. A detailed description of the design and implementation of these Drug–Gene Pair alerts has been published previously.2 Please note that this system of Drug–Gene Pair alerts, which has been in place since 2013, is “reactive”— it requires that the physician order a pharmacogenomic test and await the results before acting. Currently 21 pharmacogenomic alerts of this type are firing in the Mayo Clinic EHR (see Figure 1) across all Mayo sites for all 1.3 million patients seen by the Mayo Clinic annually. It should also be pointed out that pharmacogenomic testing at our institution was and generally remains genotype based. In contrast, the “Mayo‐Baylor RIGHT 10K” pharmacogenomic study described subsequently is preemptive, not reactive, and is DNA sequence based, not genotype based.
Figure 1.
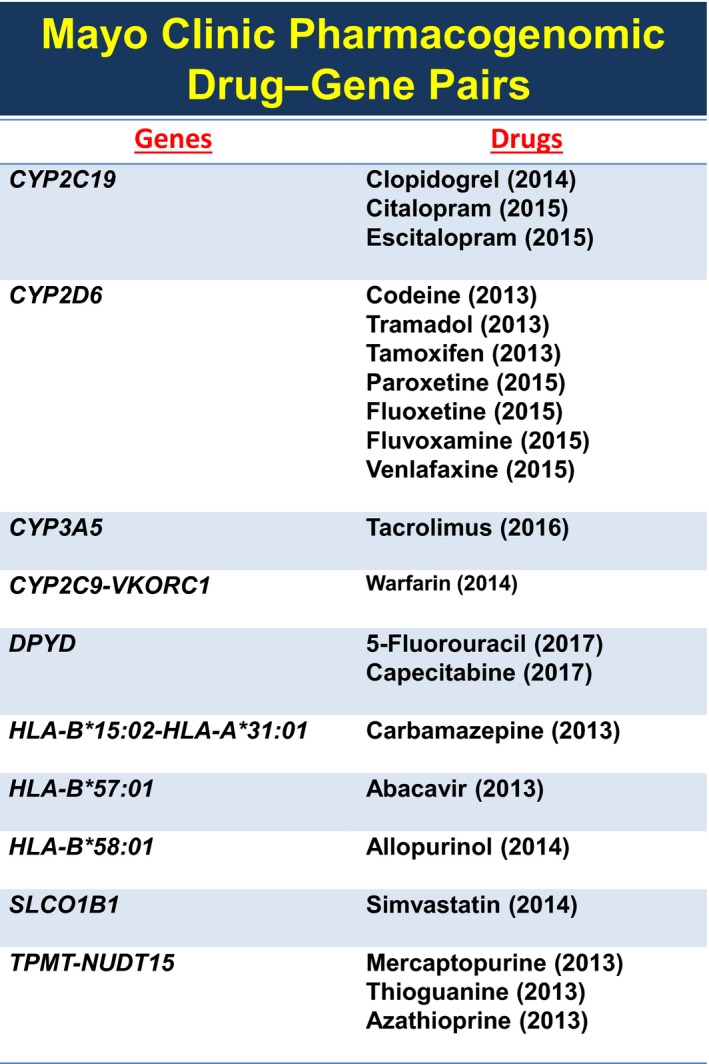
The figure lists the 21 drug–gene pairs for which “alerts” currently fire in the Mayo Clinic electronic health record across all Mayo Clinic clinical sites as well as the year when each alert was approved for inclusion in the Mayo Clinic electronic health record.
Preemptive Sequence‐Based Pharmacogenomics
After having created the infrastructure required to implement pharmacogenomic EHR alerts across the geographically dispersed Mayo Clinic Health System, a system that extends from Minnesota to Florida and Arizona, it became clear that, eventually, it would be necessary to move from the present “reactive” approach to a “preemptive” approach. In a preemptive alert pharmacogenomic information would not be acquired after the initial patient–physician encounter—requiring a waiting period for genomic testing to be completed—but rather that information would have already been deposited in the EHR so alerts could be “individualized” instantaneously for that particular patient at the point of care. As described subsequently, “preemptive” alerts could use either genotype or sequence‐based data, but—for the Mayo‐Baylor 10K Project described subsequently—we chose sequencing because that is clearly where pharmacogenomics is moving. Irrespective of whether the alert might be reactive or preemptive, a critical issue involves helping physicians and other providers understand and use the pharmacogenomic information. Furthermore, when an alert fires, the physician might not have time to review the detailed information in “AskMayoExpert,” so they would pick up the telephone and call the pharmacy to ask for advice. It quickly became clear that we needed to develop educational modules for pharmacists before each new alert “went live.” Therefore, one consequence of our clinical implementation effort has been the training and/or hiring of pharmacists with pharmacogenomic subspecialty expertise. Finally, the nature of clinical decision‐support tools needed by members of the healthcare team has also presented significant challenges. As we considered these facts and the broader question of how pharmacogenomic testing might evolve in the future, it became increasingly clear that we should be planning for preemptive, DNA sequence‐based pharmacogenomics for a panel of genes with known or potential clinical utility—leading to the “Mayo‐Baylor RIGHT 10K Pharmacogenomic Study.”
It should be emphasized that the evolution of pharmacogenomics as a scientific discipline and its clinical implementation have depended on a partnership between academic medical centers like the Mayo Clinic and biomedical funding agencies such as the National Institutes of Health (NIH). The RIGHT 10K Study rests on a foundation created by two major NIH grants, the National Institute of General Medical Sciences (NIGMS)‐funded Pharmacogenomics Research Network (PGRN) grants and the National Human Genome Research Institute (NHGRI)‐funded Electronic Medical Records and Genomics (eMERGE) grants. Specifically, the eMERGE network supported a series of pilot projects in which 84 “pharmacogenes” were sequenced using a DNA capture reagent designed by the Baylor Human Genome Sequencing Center through the PGRN which, at our center, was applied to 1,013 DNA samples from the Mayo Clinic Biobank. The results were so striking, 99.1% of those 1,013 subjects had a clinically actionable pharmacogenomic variant in just the top five genes tested,3 that the Mayo Clinic and Baylor subsequently collaborated to design a study during which 10,000 adult patients who had deposited DNA in the Mayo Clinic Biobank consented to have 77 pharmacogenes sequenced, with information for the genes for which we currently fire pharmacogenomic alerts (see Figure 1) being deposited in the EHR for clinical use. Sequencing of DNA from those 10,000 subjects is now complete, and their pharmacogenomic data, obtained in a College of American Pathologists, Clinical Laboratory Improvement Amendments (CAP‐CLIA) setting, are already in the Mayo EHR for all 10K participants—making it possible to perform both retrospective and prospective pharmacogenomic studies using data from these 10,000 subjects.
Conclusions and Future Directions
Pharmacogenomics is advancing rapidly at the discovery, translational, and implementation levels. It is now clear that pharmacogenomics, as a clinical discipline, is being applied at the bedside to enhance the care of patients who are suffering from a variety of diseases and are being treated with a variety of drugs. However, it is also clear that we are only at the beginning of the process of using data from genomics and other omic testing to individualize drug therapy—to optimize both drug selection and dosing. It is also clear that the “Mayo‐Baylor RIGHT 10K Study,” large and ambitious though it might seem, is itself only one step pointing the way to the future. It is also obvious that significant challenges remain for both pharmacogenomic discovery and implementation. The role of the pharmacist was mentioned in previous paragraphs, and it will continue to be critical. The way in which we deliver pharmacogenomic information to physicians and other members of the healthcare team also cries out for novel approaches—many of which may come back to the EHR. Finally, we have not mentioned rapidly growing and very promising attempts to apply artificial intelligence and machine learning to create predictive models for drug response, models that incorporate genomics and other “omics” data,4 not merely to avoid adverse drug reactions and to optimize dosage, but ultimately to help the provider select the right drug at the right dose for the right patient—the goal of pharmacogenomics since its origins over half a century ago.5
Funding
Supported by National Institutes of Health Grants R01 GM2815, R01 GM125633, R01 AA27486, and U19 GM61388 and by the Mayo Clinic Center for Individualized Medicine.
Conflict of Interest
L.W. and R.W. are cofounders of and stockholders in OneOme, LLC.
References
- 1. Farrugia, G. & Weinshilboum, R.M. Challenges in implementing genomic medicine: the Mayo Clinic Center for Individualized Medicine. Clin. Pharmacol. Ther. 94, 204–206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Caraballa, P.J. et al Multidisciplinary model to implement pharmacogenomics at the point of care. Genet. Med. 19, 421–429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ji, Y. et al Preemptive pharmacogenomic testing for precision medicine: a comprehensive analysis of five actionable pharmacogenomic genes using next‐generation DNA sequencing and a customized CYP2D6 genotype cascade. J. Mol. Diagn. 18, 438–445 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Athreya, A.P. et al Pharmacogenomics‐driven prediction of antidepressant treatment outcomes: a machine learning approach with multi‐trial replication. Clin. Pharmacol. Ther. 106, 855–865 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weinshilboum, R.M. & Wang, L. Pharmacogenomics: precision medicine and drug response. Mayo Clin. Proc. 92, 1711–1722 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


