Abstract
In order to explore the salt-stress responses of two rice varieties, the physiological responses and biochemical responses were investigated using proteomics and classical biochemical methods. The results showed that the seedling growth was inhibited under salt condition in two rice varieties, the seedling growth in the tolerant variety was better than the sensitive variety. The sensitive variety(L7) appeared obvious salt-injury under 3-day salt stress, the tolerant variety (T07339) keep normal growth under 7-day salt stress except that the shoot length was decreased. Through the growth-parameters analysis, most of them in L7 were restrained by salinity and most in T07339 were unaffected. In T07339, the fresh root weight, the content of chlorophyll and the fresh shoot weight were even increased after 7 days of salt stress. A comparison of two-dimensional gel electrophoresis (2-DGE) protein profiles revealed 8 differently expressed proteins. Four proteins were expressed in different pattern between sensitive and tolerant varieties. These results provide novel insights into the investigations of the salt-response proteins that involved in improved salt tolerance.
Keywords: Physiology, Proteomics, Rice, Salt stress, Morphology
Introduction
Crop production is severely influenced by adverse environmental conditions in agricultural systems. Salt stress is a common abiotic stress worldwide. More than 800 million hectares of land are affected by high salinity throughout the world (Munns and Tester, 2008). A high concentration of salt causes ion imbalance, hyper-osmotic stress and oxidative damage (Zhu 2002), resulting in great losses in productivity.
Rice (Oryza sativa L.) is a staple food source being almost exclusively consumed as grain by the world’s population, and supplying 20% of daily calories (World Rice Statistics, http://www.irri.org, verified 8 October 2010). Soil salinity is a complex and harmful threat to rice, due to disruption of ionic, osmotic, and cell-water homeostasis. Further consequences are disturbance of membrane integrity, nutrient imbalance, altered levels of growth regulators, and disturbances on general metabolic activities. Although higher plants have developed sophisticated strategies to counteract salt stress, including selective ion uptake and exclusion (Shi et al. 2000), compartmentation of Na+ in vacuoles (Apse et al. 1999), detoxification of reactive oxygen species by the antioxidant system (Blokhina et al. 2003; Benavente et al. 2004) and accumulation of osmo-protectants in the cytosol (Waditee et al. 2005; Demiral et al. 2006). The adaptation to saline stress is a complex physiological process accompanied by alterations in the levels of numerous metabolites, proteins and mRNAs (Serrano, 1996) and DNA methylation (Wang et al. 2015). How plants perceive the specific salt stress signal and transmit information internally is an important issue.
Proteomics is one of the high-throughput approaches that is being used to address biological function of plants by studying globally expressed proteins in a given tissue including leaf sheath (Abbasi and Komatsu, 2004), root (Yan et al. 2005; Cheng et al. 2009; Malakshah et al. 2007; Chitteti et al. 2007), leaf (Kim et al. 2005; Nohzadeh et al. 2007; Sengupta and Majumder, 2009; Parker et al. 2006), suspension culture cells (Liu et al. 2013), shoot stem (Song et al. 2011), anther (Sarhadi et al. 2012), young panicle (Dooki et al. 2006). Previous reports have identified various kinds of proteins (Udomchalothorn et al. 2009; Hoai et al. 2003; Filho et al. 2003; Huang et al. 2008; Cheng et al. 2009; Song et al. 2011) including transcription factors (Chen et al. 2013; Liu et al. 2014; Wu et al. 2015) in response to salt stress. Some proteins have been implicated in improving salt tolerance of plants. Udomchalothorn et al. (2009) observed a significant increase in fructose 2,6-bisphosphatase (F26BPas) activity only in the salt-tolerant line LPT123-TC171, which resulted in a decreased F26BP levels leading to an increased partition of carbon to sucrose. This result suggests that increased F26BPas activity may contribute to salt-stress tolerance in rice (Udomchalothorn et al. 2009). Arabidopsis (Col-0) plants over-expressing caseinolytic protease regulatory subunit (ClpD1.3) exhibited higher tolerance to salt and desiccation stresses as compared to wild type plants (Mishra et al. 2016). Hoai et al. (2003) analyzed eight rice cultivars’ nitrogen compounds and found that a high capacity to assimilate ammonium is an important factor in alleviating the consequence of stress. Thus, it is imperative to identify proteins which confer improved salt tolerance, and this strategy can be the basis for genetic engineering to improve salt stress tolerance in plants.
Kim DW (2005) reported that salt stress-responsive proteins expression and leaf morphology has a positive correlation. Ghaffari A (2014) reported the physiology and proteome responses of two contrasting rice mutants and their wild type parent under salt stress conditions at vegetative stage. The standard evaluation score (SES) scores, Na+ and K+ concentrations in shoots and Na+/K+ ratio were significantly different in contrasting mutants, and 34 unique differentially accumulated proteins were identified in various molecular processes. In a comparative complex-omics analysis of leaf whole cell lysate of two rice genotypes with contrasting responses to salt, an interaction was detected for glycolytic enzymes enolase (ENO1) and triosephosphate isomerase (TPI) and also for a chlorophyll a-b binding protein and RuBisCo small subunit elicited by salt stress (Hashemi et al. 2016). Although these studies have investigated the salt stress-related proteins and their relation with physiological and biochemical responses caused by salt stress, the correlation between salt stress-triggered physiological responses in rice plant and changes in root protein profile is still lacking. Since root is the first organ to perceive the salt signal, the investigation of root protein expression under salt stress is critical and important. In this study, we have evaluated the salt stress-triggered damage to two rice varieties in terms of physiological parameters, and report the correlation between identified proteins and salt stress-induced changes in physiological parameters and morphological symptoms.
Materials and Methods
Plant Material and Salt Treatment
Rice (Oryza sativa L. japonica) cvs T07339 and L7 were used in this study. They were sterilized and germinated by the same method. The rice seeds were sterilized by 70% ethanol and 3% bleach, then washed with water for three times. After that the seeds were soaked in water for one day and then germinated on wet filter paper for one day. After germination, 24 rice seedlings for each variety were cultured in two plastic containers containing YS (Yoshida’s nutrition solution) (Yoshida et al. 1976). Rice seedlings grown in a greenhouse at 30°C during the day and 22°C at night under natural light conditions. When rice seedlings were at the second leaf fully unfolded stage, the uniformly grown seedlings were selected and transferred to nutrition solution containing 60 mM NaCl. The rice seedlings transferred to the nutrition solution without NaCl served as the control. After 7 days, they were evaluated under salt stress and control condition.
Plant Physiological Analysis
Rice seedlings were harvested randomly to measure growth parameters. Lengths of shoot (the whole seedling above the seed), leaf (3rd), leaf sheath (the area from above the seed to the joint at the base of the 3rd leaf), and root, and the fresh weight (FW) of shoot and root were measured. The shoot of seedlings were oven-dried at 80°C for 48 hours, and the resulting dry weight (DW) was measured. The values are presented as means from ten seedlings. Statistical analysis was performed using the Student’s t-test. P< 0.05 and P< 0.01 was considered statistically significant.
Chlorophyll content was measured on the 2nd leaf of control and salt-treated rice seedlings using a portable chlorophyll meter (SPAD-502 Chlorophyll Meter Model SPAD-502). Values are presented as means from ten seedlings. Statistical analysis was performed using the Student’s t-test and p<0.05 was considered statistically significant.
Measurement of ion concentration
Following 3 days of oven-drying of seedling shoots at 60°C, approximately 0.1 g of each dried powdered sample (see above) was weighed and extracted in 100mM nitric acid for 2 h at 90°C (Deng et al. 2015). The concentrations of Na+ and K+ in the extracts were determined using inductively coupled plasma-optical emission spectrometry (Perkin-Elmer, Norwalk, CT, USA).
Two-Dimensional Gel Electrophoresis (2-DGE) of Rice Root Proteins
Total protein was extracted from rice roots using the method of Wang et al. (2006). For the iso-electro-focusing (IEF), the precast immobilized pH gradient (IPG) strips (Bio-Rad) were rehydrated for 12 hours in rehydration buffer containing 300 μg of proteins. After isoelectric focusing, the IPG strips were equilibrated in equilibration buffers according to the manufactural instruction and then placed directly onto 12% polyacrylamide–SDS slab gels. The gels were run in parallel at 32 mA per gel using a Protean II electrophoresis cell (Bio-Rad). Gels were stained with silver. Two-D gel electrophoresis of rice root proteins was conducted with three replications.
Image Analysis
For each treatment, three 2-DE gels were used for data analysis. Protein spots showing reproducible change in staining intensity were subjected to mass spectrometric analysis. Those interest spots of interest were excised from the stained gels using a scalpel and then stored at 4°C for subsequent mass spectrometry analysis. Protein spots from the 2-D gels were digested by trypsin as described previously (He and Li, 2008).
Protein Identification by Liquid Chromatography-tandem Mass Spectrometry (LC-MS/MS)
The dried digests were re-suspended in 10 μl of a solution of 2.5% acetonitrile and 2.5% formic acid in water and 5 μl of the digest were analyzed by LC-MS/MS on a linear ion trap (LTQ) mass spectrometer (Thermos Fisher Scientific). Peptides were loaded onto a 100 μm x 120 mm capillary fused silica column packed with MAGIC C18 (5 μm particle size, 20 nm pore size, Michrom Bioresources, CA) at a flow rate of 500 nL/min. Peptides were separated by a gradient of 5–35% acetonitrile / 0.1% formic acid over 50 min, 35–100% acetonitrile /0.1% formic acid in 1 min, and 100% acetonitrile /0.1% formic acid for 9 min, followed by an immediate return to 2.5% acetonitrile /0.1% formic acid and a isocratic hold at 2.5% acetonitrile /0.1% formic acid until the next injection. Peptides were introduced into the linear ion trap via a nanospray ionization source and a laser pulled ~3 μm orifice with a spray voltage of 1.8 kV. Mass spectrometry data were acquired in a data-dependent “Top 10” acquisition mode, in which a survey scan from m/z 360–2000 is followed by 10 collision-induced dissociation (CID) tandem mass spectrometry MS/MS scans of the most abundant ions. MS/MS scans were acquired with the following parameters: isolation width: 2 m/z, normalized collision energy: 35%, Activation Q: 0.250 and activation time = 30 ms. Dynamic exclusion was enabled (repeat count: 2; repeat duration: 30 sec; exclusion list size: 500; exclusion duration: 60 sec). The minimum threshold was 500. Product ion spectra were searched using the SEQUEST HT engine implemented on the Proteome Discoverer 1.4 (Thermo Fisher Scientific, Waltham, MA, USA) against a curated Uniprot database of Oryza-sativa (subsp. japonica and indica combined; downloaded on January 26, 2015). Search parameters were as follows: (1) full trypsin enzymatic activity, (2) two missed cleavages, (3) min. peptides length = 6, (4) mass tolerance at 2 Da for precursor ions and 0.8 Da for fragment ions, (5) Dynamic modifications on methionine (+15.9949 Da: oxidation), (6) 4 maximum dynamic modifications allowed per peptide; and (7) static modification on cysteine (+57.0215 Da: carbamidomethylation). Peptides were filtered according to XCorr criteria of 2.0, 2.5, 3.0 for singly, doubly and triply charged peptides and minimal number of peptides =2. The identifications in each spot were ranked by sequence coverage and the top hits were reported, provided that its theoretical MW and pI information match those obtained from the 2-D gels..
Proteomic Data Analysis
Functional categorization of proteins was carried out according to the GO rules using the gene ontology tools at http://www.agbase.msstate.edu. The ontologies contained three parts: (1) the biological process in which the gene product participates; (2) the molecular function that describes the gene product activities; and (3) the cellular compartment where the gene product can be found.
Results and Discussion
Morphological Responses at Seedling Stage
Several studies indicated that rice becomes very sensitive during early seedling stage (Gregorio et al. 1997), thus the effect of salinity on morphological characteristics was evaluated for the two rice varieties during early seedling stage. After 7 days of salt treatments, there was a clear distinction between the two entries. The two rice varieties had different visual symptoms of salt toxicity (Fig. 1), most of T07339 plants had normal growth and no salt injury leaf symptoms, except the length of seedlings is shorter than the control. L7 plants were more sensitive to salinity, e.g. the first leaf was whitish and rolled, half of the second leaves were rolled, and the growth of the third leaf was restrained.
Fig.1.
Morphological changes associated with salt stress exposure in rice seedling. Rice seedlings at the second leaf fully unfolded stage were grown in nutrient solutions containing 60 mM NaCl (salt stress) or without NaCl (control) for 7 days. 1: T07339 control plants; 2: salt-stressed T07339 plants; 3: L7 control plants; 4: salt-stressed L7 plants.
Salinity stress at early seedling stage manifests on the first leaf, followed by the second, and finally on the growing leaf. After 7 days of salt treatment, the seedlings’ leaves appeared salt injury symptoms gradually from the first leaf to the third leaf. Seedling survival was not affected by salinity stress, probably because the salt treatment was mild salt stress condition (Negrao et al., 2011). We classified the leaves symptoms of salt injury, and the statistics (Table 1) showed that the percentage of salt-harmed leaves on the second leaf was ninety-four in the variety of L7, otherwise the percentage is forty-seven in T07339; over one half of plant seedlings showed that only the first leaf was rolled in T07339. The results strongly demonstrated that T07339 plants have higher tolerance than L7 plants to salt stress.
Table 1.
The comparison of visual salt injury at seedling stage
| Visual salt stress symptoms | Percentage of yellowed leaves in two test entries | |
|---|---|---|
| L7 | T07339 | |
| Only the first leaf yellowed | 0 | 0.53 |
| The second leaf yellowed | 0.94 | 0.47 |
| The third leaf yellowed | 0.60 | 0 |
Physiological Responses to Salt Stress
K+/Na+ ratios in roots and shoots
The K+/Na+ ratios of rice seedlings were decreased among all the two rice cultivars either under 3 day or 7 day of salt stress (Table 2). The difference of K+/Na+ ratios in root were not statistically significant between L7 and T07339. However the K+/Na+ ratios in T07339 shoots were doubles of those in L7 shoots, especially the K+/Na+ ratio in T07339 shoots at 3 days salt treatment was 2.67 times that in L7 shoots. The difference of K+/Na+ ratios in shoots were statistically significant (Table 2). The higher K+/Na+ ratio might suggest that T07339 have more ability of stimulating the ion equilibrium under salt stress.
Table 2.
Comparison of K+/Na+ ratio in two rice cultivars exposed to 60 mM NaCl
Salt tolerant variety—T07339
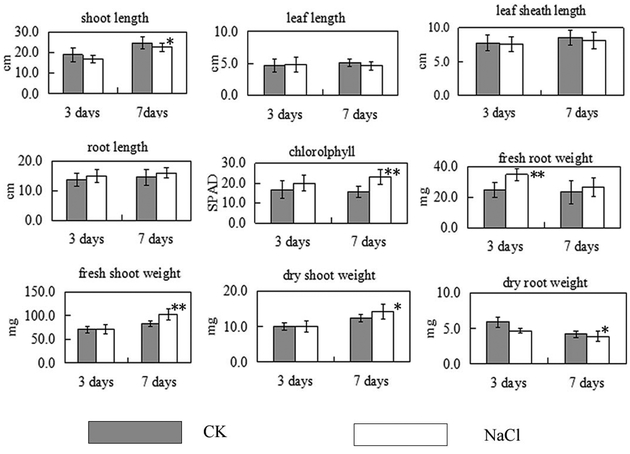
Most growth parameters (Fig. 2) was not significantly affected after 3 days of salt stress except that the fresh root weight dramatically increased, about 30% increase under salinity, and the dry root weight decreased. That is a very interesting phenomenon. This results might suggest that the root absorbed much water to dilute the large amounts of Na+ in its tissues, which is another effective approach to cope with salinity stress. In previous research, rice seedlings were shown to be able to dilute the absorbed Na+ through its higher growth rate (Nakhoda et al. 2012). After 7 days of salt stress the shoot length and dry root weight of T07339 plants were reduced, and the content of chlorophyll, fresh shoot weight and dry shoot weight increased significantly (Fig. 2). The content of chlorophyll increased dramatically which contributes to the increase of the fresh shoot weight and dry shoot weight.
Fig.2.
Salt stress-triggered changes in physiological parameters of T07339 rice seedlings. Values are means from ten seedlings. Error bars indicate SDs. * indicate significant difference at P<0.05 and ** indicate significant difference at P<0.01 when compared to control seedlings.
Salt sensitive variety—L7
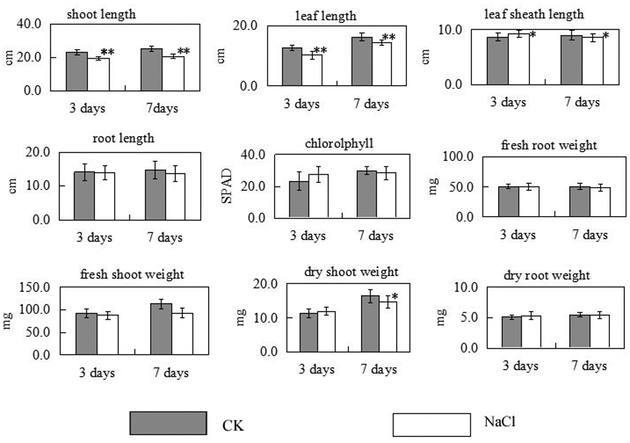
The growth of rice seedlings was significantly inhibited by salt stress. The shoot length of rice seedlings and the 3rd leaf length were dramatically reduced compared with the control seedlings after 3 or 7 days of salt stress (Fig. 3). The 2nd leaf sheath length decreased after 7 days of salt stress. The dry shoot weight was significantly reduced after 7 day salt stress. All these results shown in Fig. 3 indicate that L7 is more sensitive to salinity than T07339.
Fig.3.
Salt stress-triggered changes in physiological parameters of L7 rice seedlings. Values are means from ten seedlings. Error bars indicate SDs. * indicate significant difference at P<0.05 and ** indicate significant difference at P<0.01 when compared to control seedlings.
Shoot biomass
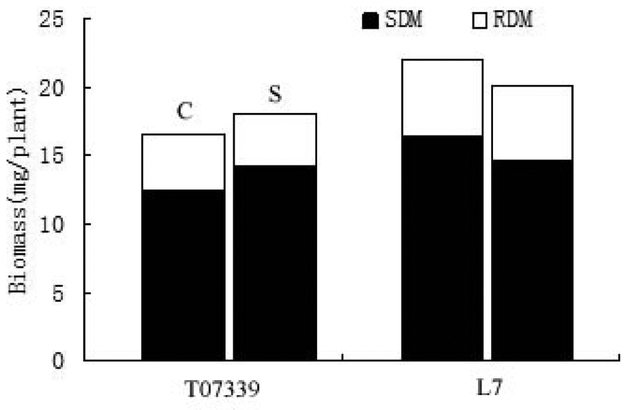
The shoot biomass production was significant different in the two rice varieties. As shown in Fig. 4, the shoot biomass of salt-treated T07339 seedlings was higher than control condition. On the other hand, the shoot biomass of L7 under salt stress was reduced compared with control seedlings. This abnormal performance appeared in T07339 might be owing to the low Na+ concentration stress (Nakhoda et al. 2012). The result confirmed that T07339 has an effective way to resist mild salinity stress. “Which” protocol and “how” to modulate the salinity for rice normal growth is still not clear.
Fig.4.
Root and shoot dry matter and total biomass of T07339 and L7 under control and salt stress conditions. Data are mean values of ten replicates. Vertical bar shows LSD value for total biomass. C: Control plants; S: Salt-stressed plants; SDM: Shoot Dry Matter, RDM: Root Dry Matter.
Proteome Analysis of Roots from Two Cultivars in Response to Salt Stress
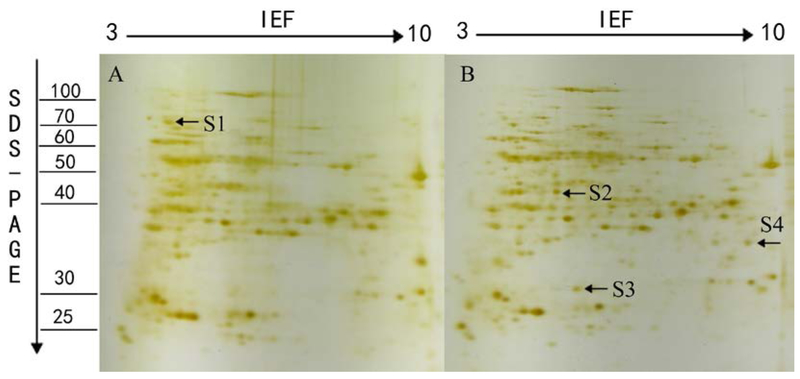
Most of the previous studies focus on leaves’ proteome, but root is the initial organ to confront the salinity stress and the K+/Na+ ratio serves as a reliable indicator of salt stress tolerance in rice (Chunthaburee et al. 2016), the mechanism how the root transmits the stress signal and keeps ionic balance is essentially important. Therefor the root was chosen for the current proteomic analysis. After 7 days of salt treatment, the root proteins from L7 and T07339 plants were analyzed separately using 2-DGE in three independent biological replicates. The seedlings roots are harvested separately, then the total proteins in roots from three independent biological replicates were extracted and separated by 2-DGE using pH 3–10 IPG strips for IEF. The protein profiles are revealed by silver staining. Differently regulated protein spots showing reproducible change in abundance were excised and subjected to tandem mass spectrometric analysis. Proteins that were identified with the highest sequence coverage and more than 2 sequenced distinct peptides were deemed as top hits (Table 3). Sequence information of the proteins that were identified with more than 5 distinct peptides except for protein S4 and S7, where EIF3 and salt stress-induced protein were identified as top hits, respectively, with equal to or more than 2 distinct peptides. In L7 protein profile (Fig. 5), 4 protein spots showed significant changes in abundance. Out of them, 1 spot showed significant decrease while 3 spots showed significant increase under salt stress. While, 4 protein spots’ abundance changed in T07339 protein profile (Fig. 6), 3 spots were down regulated and 1 spot up-regulated under stress.
Table 3.
Differentially accumulated proteins identified by tandem mass spectrometry
| Spot No. | Protein name | # peptides | # unique peptides | Coverage (%) | Theo. Mr |
Theo. pI |
Accession number |
|---|---|---|---|---|---|---|---|
| S1 | 70 kDa heat shock protein [japonica]/ | 27 | 8 | 44 | 71.1 | 5.21 | Q10NA9/ |
| DnaK-type molecular chaperone hsp70 [japonica]/ | 26 | 4 | 43.91 | 71.1 | 5.21 | Q53NM9/ | |
| Heat shock cognate 70 kDa protein 2 [japonica]/ | 27 | 3 | 43.76 | 71.3 | 5.21 | Q84TA1/ | |
| 70 kDa heat shock protein [japonica]/ | 23 | 4 | 40.59 | 70.9 | 5.21 | Q943K7/ | |
| Os05g0460000 protein [japonica] | 24 | 4 | 39.47 | 70.8 | 5.21 | Q6L509 | |
| S2 | S-adenosylmethionine synthase 2 [japonica]/ | 17 | 6 | 56.6 | 42.9 | 6.05 | P93438/ |
| S-adenosylmethionine synthase 1 [japonica]/ | 12 | 1 | 45.71 | 43.2 | 6.14 | Q0DKY4 | |
| S3 | DIP3 [japonica]/ | 5 | 0 | 20.20 | 32.5 | 6.54 | Q5WMX0/ |
| Putative uncharacterized protein [indica] | 5 | 0 | 20.20 | 32.5 | 6.30 | A2Y2C4 | |
| S4 | Eukaryotic translation initiation factor 3 subunit [japonica] | 2 | 1 | 9 | 32.0 | 8.50 | Q6K4P1 |
| S5 | DnaK-type molecular chaperone hsp70 [japonica] / | 23 | 3 | 38.98 | 71.1 | 5.21 | Q53NM9/ |
| Os05g0460000 protein [japonica] / | 20 | 3 | 36.22 | 70.8 | 5.21 | Q6L509/ | |
| 70 kDa heat shock protein [japonica] / | 23 | 9 | 36.00 | 71.1 | 5.21 | Q10NA9/ | |
| 70 kDa heat shock protein [japonica] / | 18 | 2 | 32.87 | 70.9 | 5.21 | Q943K7/ | |
| Heat shock cognate 70 kDa protein 2, putative [japonica] | 18 | 3 | 31.28 | 71.3 | 5.21 | Q84TA1/ | |
| S6 | Putative uncharacterized protein [indica]/ | 10 | 0 | 34.7 | 46.4 | 5.03 | A2Y9V7/ |
| Os06g0173100 protein [japonica] / | 10 | 0 | 31 | 47.8 | 5.07 | P46465/ | |
| Putative uncharacterized protein [indica]/ | 9 | 0 | 33.57 | 47.7 | 5.07 | Q5SNC0/ | |
| 26S protease regulatory subunit 6A homolog [japonica]/ | 9 | 0 | 31.82 | 46.8 | 5.07 | A2XAQ0 | |
| Protein SGT1 homolog [japonica] | 9 | 6 | 29.7 | 40.9 | 5.06 | Q0JL44 | |
| S7 | Salt stress root protein RS1 [japonica] | 4 | 2 | 31.86 | 21.8 | 4.92 | Q0JPA6 |
| Salt stress-induced protein [indica]/ | 2 | 0 | 20.69 | 15.2 | 5.40 | A2WPN7/ | |
| Salt stress-induced protein [japonica] | 2 | 0 | 20.69 | 15.2 | 5.15 | Q0JMY8 | |
| DIP3 [japonica]/ | 5 | 0 | 20.54 | 32.5 | 6.54 | Q0JMY8 | |
| Putative uncharacterized protein [indica] | 5 | 0 | 20.54 | 32.5 | 6.30 | A2Y2C4 | |
| 26S proteasome regulatory particle non-ATPase subunit12 [japonica]/ | 4 | 0 | 16.85 | 30.8 | 5.07 | Q8W423/ | |
| Putative uncharacterized protein [indica] | 4 | 0 | 16.85 | 30.8 | 5.07 | B8B5M9 | |
| S8 | DIP3 [japonica]/ | 7 | 0 | 27.61 | 32.5 | 6.54 | Q5WMX0/ |
| DIP3 [indica] | 7 | 0 | 27.61 | 32.5 | 6.30 | A2Y2C4 |
Top hits with the highest sequence coverage are listed. Proteins with identified peptides that were matched to multiple protein accessions in the database (e.g. isoforms, truncated versions of the protein sequence, different annotations of the same protein, etc) are listed together and separated by /. If a different protein was identified in the spot with distinct sequences from the top hits, the protein is listed separately in another row.
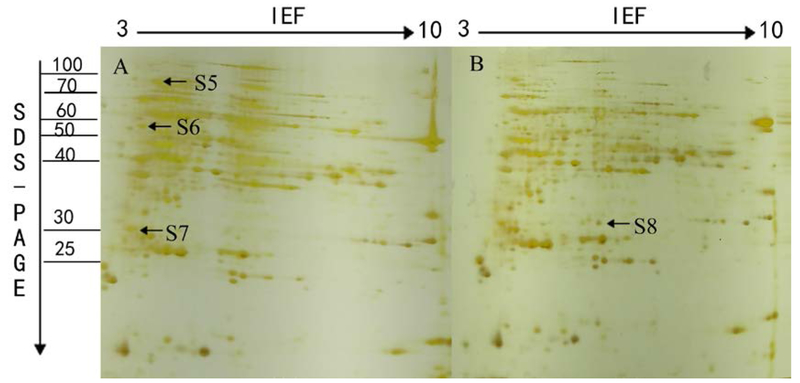
Fig.5.
A representative 2-DE gel image of proteins extracted from seedlings of L7 under control (A) and salt stress (B) condition. Total soluble proteins were loaded in a 11 cm IPG strip with a pH gradient of 3–10. IEF was performed for a total 70 KVh. In the second dimension, 12% SDS–PAGE gel was used. Proteins were visualized by silver staining. The arrows show the position of the identified protein spots that showed significant changes during salt stress treatment along with numbers which represent spot IDs as shown in Table 2.
Fig.6.
A representative 2-DE gel image of proteins extracted from seedlings of T07339 under control (A) and salt stress (B) condition. Total soluble proteins were loaded in a 11 cm IPG strip with a pH gradient of 3–10. IEF was performed for a total 70 KVh. In the second dimension, 12% SDS–PAGE gel was used. Proteins were visualized by silver staining. The arrows show the position of the identified protein spots that showed significant changes during salt stress treatment along with numbers which represent spot IDs as shown in Table 2.
Among the eight identified proteins (Table 3), DIP3 (S3, S8) acts as hydrolase activity in carbohydrate metabolic process. 70 kDa heat shock protein (S1, S5), acting as intracellular molecular chaperones, was reported to assist in repair or degradation of mis-folded proteins within the cell under stress conditions (Liu et al. 2013; Song et al. 2011; Sarhadi et al. 2012). They have the same expression patterns in the two rice varieties. S2 and S4 were expressed only in L7. S-adenosylmethionine synthase (S2) catalyzes the formation of S-adenosylmethionine (SAM) from methionine and ATP, involving Amino-acid biosynthesis, one-carbon metabolic process, and S-adenosylmethionine biosynthetic process. SAM is an important methyl group donor utilized in most trans-methylation reactions (Parker et al. 2006). Several studies showed that the increased induction of SAMS genes might be for the biosynthesis of lignin and the polyamine glycine betaine during water- or salt- stress (Sanchez-Aguayo et al. 2004; Tabuchi et al. 2004). S4 (Eukaryotic translation initiation factor 3 subunit) is a component of the eukaryotic translation initiation factor 3 (eIF-3) complex, which is involved in protein synthesis and, together with other initiation factors, stimulates binding of mRNA and methionyl-tRNAi to the 40S ribosome. The result suggests that the two proteins’ over-expression was to resist the salt stress. While, S6 and S7 were expressed only in T07339. S6 (Os06g0173100 protein) takes part in hydrolase activity of protein catabolic process, which might take part in the ubiquitin 26S proteasome system. S7 was identified as salt stress root protein/salt stress induced protein/26S proteasome regulatory particle non-ATPase subunit 12. Salt stress root protein is embedded in the membrane as the component of the plasma membrane. Salt stress induced protein is in response to salt and related osmotic stresses. 26S proteasome regulatory particle non-ATPase subunit 12 is involved in protein catabolic process. It is known to catalyze the ATP-dependent degradation of ubiquitinated proteins and degrade the proteins that were unnecessary or damaged. These results implied that this protein might be the donor that probably contributes to the salt tolerance of T07339.
Conclusion
The physiological characteristics of two varieties with contrasting responses to salt stress were evaluated by morphological and physiological analysis and a proteomic approach. The tolerant variety has better performance than sensitive variety. In tolerant variety (T07339), the fresh root weight was increased after 3-days salt-stress, the content of chlorophyll, the fresh shoot weight and the dry shoot weight were increased after 7-days salt-stress. In sensitive variety (L7), the shoot weight, leaf length, leaf sheath length and the dry shoot weight were decreased compared with the control. Through proteomic analysis, we identified four proteins expressed in different patterns between tolerant and sensitive variety. Two proteins, involved in salt stress response and ubiquitin 26S proteasome system, might contribute to the salt tolerance of T07339. Those results provide a new insight into rice seedlings’ tolerance mechanisms.
Supplementary Material
Acknowledgements
This research was supported by the national the national Science & Technology Support Program (CN, (2015BAD01B01) and Science & Technology Support Program of Jiangsu Province (CN, BE2016370). Proteomic research was conducted at Mississippi State University by Dr. Yan Liu as a visiting scholar. The research was partially supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture, Hatch/Multi-State Project (Multistate No. NC 1200; Project No. MIS-153180) under Accession No. 232044. The Vermont Genetics Network Proteomics Facility is supported through NIH grant P20GM103449 from the INBRE Program of the National Institute of General Medical Sciences.
References
- Abbasi FM, Komatsu S (2004) A proteomic approach to analyze salt-responsive proteins in rice leaf sheath. Proteomics 4:2072–2081 [DOI] [PubMed] [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by over-expression of a vacuolar Na 1/H1 antiport in Arabidopsis. Science 285:1256–1258 [DOI] [PubMed] [Google Scholar]
- Benavente LM, Teixeira FK, Kamei CLA, Pinheiro MM (2004) Salt stress induces altered expression of genes encoding antioxidant enzymes in seedlings of a Brazilian indica rice (Oryza sativa L.). Plant Science 16:6323–331 [Google Scholar]
- Blokhina O, Virolainen E, Fagerstedt KV (2003) Antioxidants oxidative damage and oxygen deprivation stress. Ann. Bot 91:179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Li F, Ma Y, Chong K, Xu YY (2013) Overexpression of OrbHLH001, a putative helix–loop–helix transcription factor, causes increased expression of AKT1 and maintains ionic balance under salt stress in rice. Journal of Plant Physiology 170:93–100 [DOI] [PubMed] [Google Scholar]
- Cheng YW, Qi YC, Zhu Q, Chen X, Wang N, Zhao X, Chen HY, Cui XJ, Xu LL, Zhang W (2009) New changes in the plasma-membrane-associated proteome of rice roots under salt stress. Proteomics 9:3100–3114. [DOI] [PubMed] [Google Scholar]
- Chitteti BR, Peng ZH (2007) Proteome and phosphor-proteome differential expression under salinity stress in rice (Oryza sativa) roots. Journal of Proteome Research 6:1718–1727 [DOI] [PubMed] [Google Scholar]
- Chunthaburee S, Dongsansuk A, Sanitchon J, Pattanagul W, Theerakulpisut P (2016) Physiological and biochemical parameters for evaluation and clustering of rice cultivars differing in salt tolerance at seedling stage. Saudi Journal of Biological Sciences 23:467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demiral T, T¨urkan I (2006) Exogenous glycinebetaine affects growth and proline accumulation and retards senescence in two rice cultivars under NaCl stress. Environ. Exp. Bot 56:72–79 [Google Scholar]
- Deng P, Jiang D, Dong Y, Shi X, Jing W, Zhang W (2015) Physiological characterization and fine mapping of a salt-tolerant mutant in rice (Oryza sativa). Functional Plant Biology 42:1026–1035 [DOI] [PubMed] [Google Scholar]
- Dooki AD, Mayer-Posner FJ, Askari H, Zaiee A, Salekdeh GH (2006) Proteomic responses of rice young panicles to salinity. Proteomics 6: 6498–6507 [DOI] [PubMed] [Google Scholar]
- Filho GAS, Ferreira BS, Dias JM, Queiroz KS, Branco AT, Bressan-Smith RE, Oliveira JG, Garcia AB (2003) Accumulation of SALT protein in rice plants as a response to environmental stresses. Plant Science 164:623–628 [Google Scholar]
- Ghaffaria A, Gharechahib J, Nakhodaa B, Salekdehc GH (2014) Physiology and proteome responses of two contrasting rice mutants and their wild type parent under salt stress conditions at the vegetative stage. Journal of Plant Physiology 171:31–44 [DOI] [PubMed] [Google Scholar]
- Gregorio GB, Senadhira D, Mendoza RD (1997) Screening Rice for Salinity Tolerance. IRRl Discussion paper series NO. 22 [Google Scholar]
- Hashemi A, Gharechahi J, Nematzadeh G, Shekari F, Hosseini SA, Salekdeh GH (2016) Two-dimensional blue native/SDS-PAGE analysis of whole cell lysateprotein complexes of rice in response to salt stress. Journal of Plant Physiology 200:90–101 [DOI] [PubMed] [Google Scholar]
- He HQ, Li JX (2008) Proteomic analysis of phosphoproteins regulated by abscisic acid in rice leaves. Biochemical and Biophysical Research Communications 371: 883–888 [DOI] [PubMed] [Google Scholar]
- Hoai NTT, Shim IS, Kobayashi K, Kenji U (2003) Accumulation of some nitrogen compounds in response to salt stress and their relationships with salt tolerance in rice (Oryza sativa L.) seedlings. Plant Growth Regulation 41:159–164 [Google Scholar]
- Huang YW, Tsay WS, Chen CC, Lin CW, Huang HJ (2008) Increased expression of the rice C-type cyclin-dependent protein kinase gene, Orysa;CDKC;1, in response to salt stress. Plant Physiology and Biochemistry 46:71–81 [DOI] [PubMed] [Google Scholar]
- Kim DW, Rakwal R, Agrawal GK, Jung YH, Shibato J, Jwa NS, Iwahashi Y, Iwahashi H, Kim DH, Shim IS, Usui K (2005) A hydroponic rice seedling culture model system for investigating proteome of salt stress in rice leaf. Electrophoresis 26:4521–4539 [DOI] [PubMed] [Google Scholar]
- Liu CT, Mao BG, Ou SJ, Wang W, Liu LC, Wu YB, Chu CC, XP Wang(2014) OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol Biol 84:19–36 [DOI] [PubMed] [Google Scholar]
- Liu DW, Ford KL, Roessner U, Natera S, Cassin AM, Patterson JH, Bacic A (2013) Rice suspension cultured cells are evaluated as a model system to study salt responsive networks in plants using a combined proteomic and metabolomic profiling approach. Proteomics 13:2046–2062 [DOI] [PubMed] [Google Scholar]
- Mishra RC, Richa, Grover A (2016) Constitutive over-expression of rice ClpD1 protein enhances tolerance to salt and desiccation stresses in transgenic Arabidopsis plants. Plant Science 250: 69–78 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–81 [DOI] [PubMed] [Google Scholar]
- Nakhoda B, Leung H, Mendioro MS, Mohammadi-nejad G, Ismail AM (2012) Isolation, characterization, and field evaluation of rice (Oryza sativa L., Var. IR64) mutants with altered responses to salt stress. Field Crops Research 127:191–202 [Google Scholar]
- Negrao S, Courtois B, Ahmadi N, Abreu I, Saibo N, Oliveira MM (2011). Recent updates on salinity stress in rice: from physiological to molecular responses. Critical Reviews in Plant Sciences 30:329–377. [Google Scholar]
- Nohzadeh MS, Habibi RM, Heidari M, Salekde GH (2007) Proteomics reveals new salt responsive proteins associated with rice plasma membrane. biosci. Biotechnol. Biochem 71:2144–2154 [DOI] [PubMed] [Google Scholar]
- Parker R, Flowers TJ, Moore AL, Harpham NVJ (2006) An accurate and reproducible method for proteome profiling of the effects of salt stress in the rice leaf lamina. Journal of Experimental Botany 57:1109–1118 [DOI] [PubMed] [Google Scholar]
- Sanchez-Aguayo I, Rodriguez-Galan JM, Garcia R, Torreblanca J, Pardo JM (2004) Salt stress enhances xylem development and expression of S-adenosyl-L-methionine synthase in lignifying tissues of tomato plants. Planta 220:278–285 [DOI] [PubMed] [Google Scholar]
- Sarhadi E, Bazargani MM, Sajise AG, Abdolahi S, Vispo NA, Arceta M, Nejad GM, Singh RK, Salekdeh(2012) GH Proteomic analysis of rice anthers under salt stress. Plant Physiology and Biochemistry 58:280–287 [DOI] [PubMed] [Google Scholar]
- Sengupta S, Majumder AL (2009) Insight into the salt tolerance factors of a wild halophytic rice, Porteresia coarctata: a physiological and proteomic approach. Planta 229:911–929 [DOI] [PubMed] [Google Scholar]
- Serrano R (1996) Salt tolerance in plants and microorganisms: toxicity targets and defense responses. Int. Rev. Cytol 165:1–52 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na1/H1 antiporter Proc. Natl. Acad. Sci. USA 97:6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Zhang C, Ge WN, Zhang YF, Burlingame AL, Guo Y (2011) Identification of NaCl stress-responsive apoplastic proteins in rice shoot stems by 2D-DIGE. Journal of proteomics 74:1045–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi T, Kawaguchi Y, Azuma T, Nanmori T, Yasuda T (2004) Salt induction of S-adenosylmethionine synthetase in leaves of halophyte Atriplex nummularia L. Plant and Cell Physiology 45:S56–S56 [DOI] [PubMed] [Google Scholar]
- Udomchalothorn T, Maneeprasobsuk S, Bangyeekhun E, Boon-Long P, Chadchawan S (2009) The role of the bifunctional enzyme, ructose-6-phosphate-2-kinase/fructose-2,6-bisphosphatase, in carbon partitioning during salt stress and salt tolerance in Rice (Oryza sativa L.). Plant Science 17:6334–341 [Google Scholar]
- Waditee R, Bhuiyan MN, Rai V, Aoki K, Tanaka Y, Hibino T, Suzuki S, Takano J, Jagendorf AT, Takabe T, Takabe T (2005) Genes for direct methylation of glycine provide high levels of glycinebetaine and abiotic-stress tolerance in Synechococcus and Arabidopsis. Proc. Natl. Acad. Sci. USA 102:1318–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Vignani R, Scali M, Cresti M (2006) A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 27:2782–2786 [DOI] [PubMed] [Google Scholar]
- Wang WS, Huang F, Qin Q, Zhao XQ, Li ZK, Fu BY (2015) Comparative analysis of DNA methylation changes in two rice genotypes under salt stress and subsequent recovery. Biochemical and Biophysical Research Communications 465:790–796 [DOI] [PubMed] [Google Scholar]
- Wu H, Ye HY, YaO RF, Zhang T, Xiong LZ (2015) OsJAZ9 acts as a transcriptional regulator in jasmonate signaling andmodulates salt stress tolerance in rice. Plant Science 232:1–12 [DOI] [PubMed] [Google Scholar]
- Yan SP, Tang ZC, Su W, Sun WN (2005) Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics 5:235–244 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol 53:247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.