Abstract
Li-Fraumeni syndrome (LFS) is an autosomal dominant cancer disorder associated with pathogenic germline variants in TP53, with a high penetrance over an individual`s lifetime. The actual population prevalence of pathogenic germline TP53 mutations is still unclear. The aim of this study was to estimate the prevalence of potentially pathogenic TP53 exonic variants in three data sources, totaling 63,983 unrelated individuals from three sequencing databases. Potential pathogenicity was defined using an original algorithm combining bioinformatic prediction tools, clinical significance evidences, and functional data. We identified 34 different potentially pathogenic TP53 variants in 131/63,983 individuals (0.2%). Twenty-eight (82%) of these variants fell within the DNA-binding domain of TP53, with an enrichment for specific variants which were not previously identified as LFS mutation hotspots, such as the p.R290H and p.N235S variants. Our findings reveal that the population prevalence of potentially pathogenic TP53 variants may be up to 10 times higher than previously estimated from family-based studies. These results point to the need for further studies aimed at evaluating cancer penetrance modifiers as well as the risk associated between cancer and rare TP53 variants.
Keywords: Li-Fraumeni syndrome, TP53, cancer, genetic variation
INTRODUCTION
Li-Fraumeni syndrome (LFS; MIM# 151623) is an autosomal dominant cancer predisposition disorder (Li and Fraumeni, 1969a; Li and Fraumeni, 1969b) associated with germline TP53 (MIM# 191170) mutations (Malkin, et al., 1990). Several clinical definitions based on familial and individual tumor history have been proposed (Li and Fraumeni, 1969a; Li and Fraumeni, 1969b; Birch, et al., 1994; Eeles, 1995; Bougeard, et al., 2015). The core LFS tumor spectrum is dominated by pre-menopausal breast carcinomas and soft-tissue sarcomas in adults, and by brain cancer and adrenocortical carcinomas in children (Bougeard, et al., 2015). The penetrance of cancer is highly variable and associated, at least in part, with the structural and functional effect of the causative TP53 variant (Olivier, et al., 2010). Nevertheless, most studies consistently report that about 50% of the carriers develop at least one malignancy by 30–40 years of age, with a lifetime penetrance of about 90% (Malkin, 2011; Mai, et al., 2016). Population prevalence estimates of germline TP53 mutations range from 1 in 5,000 (Lalloo, et al., 2003) to 1 in 20,000 (Gonzalez, et al., 2009) individuals; however, these observational studies were based on participants selected on personal and familial cancer history.
Large-scale next-generation sequencing (NGS) has generated databases such as the Exome Aggregation Consortium (ExAC) (Lek, et al., 2016) that provide extensive catalogues of genetic variations across different populations unselected for specific disease traits. We have analyzed the prevalence of potentially pathogenic TP53 variants in an aggregated dataset composed of 63,983 individuals unselected for personal or familial cancer history, extracted from 3 different sequencing databases.
METHODS
Populations included
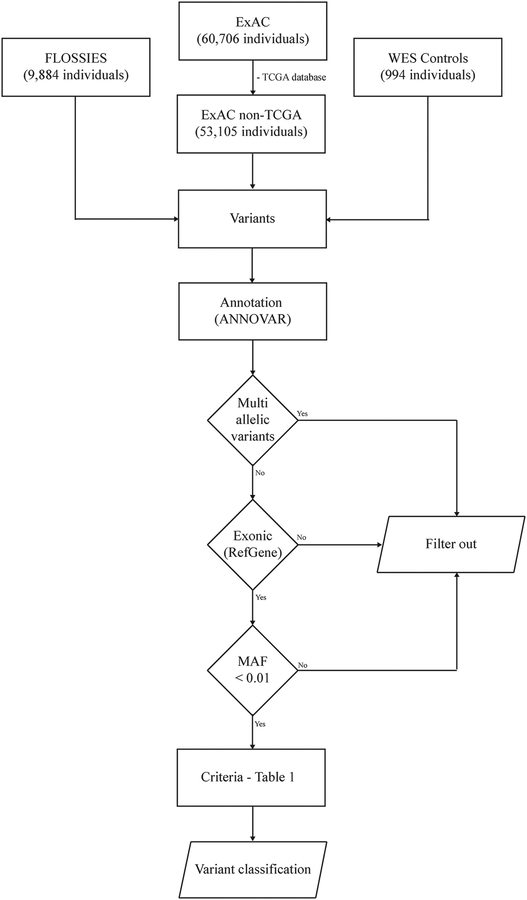
We evaluated TP53 sequencing data from three pooled datasets of unrelated, adult study participants: (1) 53,105 individuals (median age and range not available) from 13 studies included in the Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org/about) (Lek, et al., 2016), excluding individuals selected due to cancer history who were part of The Cancer Genome Atlas Research Network database (TCGA, http://cancergenome.nih.gov/); (2) 9,884 women from the Fabulous Ladies Over Seventy database (FLOSSIES, https://whi.color.com/), over the age of 70 years (median age = 80, range 70–99) without a personal history of cancer; and (3) 994 cancer-free individuals (median age = 68, range 37–88) who underwent exome sequencing as part of three distinct cancer population-based studies: Environment And Genetics in Lung Cancer Etiology – (EAGLE) (Landi, et al., 2008); Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial – (PLCO) (Prorok, et al., 2000) and Cancer Prevention Study II – (CPSII) (Calle, et al., 2002). The combination of these cohorts will be hereafter referenced as Whole-Exome Sequencing Controls (WES Controls). A schematic representation of database selection is shown in Figure 1.
Figure 1 -. Flowchart with a summary of parameters used for variant selection and filtering.
Our analysis is based on a pooled dataset composed by 63,983 individuals, after excluding the TCGA dataset from the ExAC consortium. Variants were annotated using ANNOVAR and selected based on gene region equivalent to exonic and with MAF < 0.01. The canonical transcript NM_00546.5 was used as sequence reference for gene region location based on the RefGene database. Variant classification was established by an original algorithm based on the REVEL score, clinical significance evidences, and the impact on transcriptional activity. Abbreviations: TCGA, The Cancer Genome Atlas; ExAC, Exome Aggregation Consortium; FLOSSIES, Fabulous Ladies Over Seventy; WES, Whole-Exome Sequencing; MAF, Minor Allele Frequency.
Annotations on ethnicity were reported as described in the original ExAC and FLOSSIES databases. Our analysis included all the ancestries reported in these databases. Single nucleotide polymorphism (SNP) array data were previously used to determine that all individuals were of European ancestry in the EAGLE, PLCO, and CPS-II datasets (Savage, et al., 2013).
Variant filtering and classification
Parameters used for variants filtering are shown in Figure 1. Briefly, variants detected and reported in the three databases were annotated using ANNOVAR (Wang, et al., 2010), and selected considering TP53 gene region reported as exonic by the RefGene database, and with minor allele frequency (MAF) less than 0.01 in the ExAC non-TCGA database (Lek, et al., 2016). Variants were filtered and interpreted based on the canonical transcript NM_000546.5 (nucleotide numbering for cDNA uses +1 as the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1) to allow cross referencing with the International Agency for Research on Cancer (IARC) TP53 variant database, which compiles information on variants identified in subjects with LFS or LFS-like profiles described in the literature (Bouaoun, et al., 2016). Multi-allelic, intronic, and UTR variants were not included in this analysis. Variants were further annotated using the VEP dbscSNV plugin (Jian, et al., 2014) to predict splice-site variants. There was no variant predicted to affect TP53 splice donor/acceptor sites in the canonical transcript (NM_000546.5). According to the RefGene database, these variants were annotated as either exonic or intronic and interpreted based on our filtering/classification scheme (Figure 1 and Table 1, respectively).
Table 1 -.
Criteria for TP53 variants classification.
| Variant Category | Bioinformatics | Clinical Significance | Functional assay | |||
|---|---|---|---|---|---|---|
| Variant Classification | Missense or Nonsense | Silent | REVEL > 0.5 | HGMD = DM or ClinVar = P/LP | Non-Functional | Functional |
| Pathogenic (P) | ✓ | ✓ | ✓ | ✓ | ||
| Likely Pathogenic (LP) | ✓ | ✓ | ✓ | ✗ | ||
| Possibly Pathogenic (PP) | ||||||
| ✓ | ✓ | |||||
| Likely Benign (LB) | ||||||
| ✓ | ✗ | ✗ | ||||
| Uncertain Significance (VUS) | ✓ | ✗ | ✗ | ✗ | ||
Variants were classified as pathogenic, likely pathogenic, possibly pathogenic, likely benign, or with uncertain significance based on three main parameters: REVEL score (threshold set to greater than 0.5), clinical significance evidences provided by HGMD and ClinVar (at least one entry supporting pathogenic or likely pathogenic classification), and transcriptional activity. Green check marks represent that the variant must meet the requirement. Red cross marks represent that the variant must not meet the requirement. Abbreviations: REVEL, Rare Exome Variant Ensemble Learner; HGMD, Human Gene Mutation Database; DM, disease causing mutation; P, pathogenic; LP, likely pathogenic; PP, possibly pathogenic; LB, likely benign; VUS, variant of uncertain significance.
For variant classification, we adapted the guidelines proposed by the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (Richards, et al., 2015). Variants were classified as pathogenic (P), likely pathogenic (LP), possibly pathogenic (PP), likely benign (LB), and uncertain significance (VUS) based on the parameters and rules defined in Table 1. Essentially, we used three main criteria: (1) REVEL score with a threshold set to greater than 0.5, which yields a sensitivity of 0.754 and specificity of 0.891 when predicting disease mutations (Ioannidis, et al., 2016); (2) Clinical significance evidences provided by either ClinVar – version 2017013 (Landrum, et al., 2016) or The Human Gene Mutation Database (HGMD) (Stenson, et al., 2014) and; (3) Transcriptional activity based on a yeast-based functional assay (Kato, et al., 2003) compiled in the IARC database of TP53 variants - version R18 April 2016 (Bouaoun, et al., 2016).
For analysis purposes, the combination of pathogenic and likely pathogenic categories will be hereafter denominated as potentially pathogenic.
Statistical analysis
Fisher`s exact tests were performed to compare the proportions of TP53 variants carrier individuals between different ethnic ancestries.
RESULTS
Prevalence of potentially pathogenic TP53 variants in the ExAC non-TCGA database
A total of 142 unique TP53 variants with MAF <1% were detected in the ExAC non-TCGA database (N=53,105), including 84 nonsynonymous, 56 synonymous, 1 nonsense, and 1 nonframeshift deletion. From these, 17 were classified as P, 13 as LP, 22 as PP, 83 as LB, and 7 as VUS. Based on the allele count values for P only or in combination with the allele counts for P and LP, the prevalence of germline TP53 variants in this database ranged from 0.06% to 0.21% (Table 2). Out of 111 individuals carrying potentially pathogenic variants, 46 (41%) had either the p.N235S (27/111, 24%) or the p.R290H (19/111; 17%), both classified as LP (Supp. Table S1).
Table 2 -.
Prevalence of TP53 variants across the three databases.
| ExAC non-TCGA | FLOSSIES | WES Controls | Total | |
|---|---|---|---|---|
| Total Individuals (n) | 53,105 | 9,884 | 994 | 63,983 |
| Median age (range), years | N/A | 80 (70–99) | 68 (37–88) | |
| Total Variants MAF < 1% | 142 | 45 | 7 | 194 |
| Nonsynonymous | 84 | 26 | 6 | 116 |
| Synonymous | 56 | 19 | 1 | 76 |
| Nonsense | 1 | 0 | 0 | 1 |
| Nonframeshift deletion | 1 | 0 | 0 | 1 |
| Variant Classification (n) | ||||
| Pathogenic (P) | 17 | 1 | 1 | 19 |
| Likely Pathogenic (LP) | 13 | 7 | 2 | 22 |
| Possibly Pathogenic (PP) | 22 | 7 | 1 | 30 |
| Likely Benign (LB) | 83 | 27 | 3 | 113 |
| Uncertain Significance (VUS) | 7 | 3 | 0 | 10 |
| Prevalence | ||||
| Pathogenic (P) only | ||||
| Variants (n) | 17 | 1 | 1 | 19 |
| llele Count (n) | 34 | 1 | 1 | 36 |
| Prevalence | 0.0006 | 0.0001 | 0.0010 | 0.0006 |
| Pathogenic (P) + Likely Pathogenic (LP) | ||||
| Variants (n) | 30 | 8 | 3 | 41 |
| Allele Count (n) | 111 | 17 | 3 | 131 |
| Prevalence | 0.0021 | 0.0017 | 0.0030 | 0.0020 |
Distribution of all the 194 variants (including duplicates) with MAF less than 1% identified in a total of 63,983 individuals. Prevalence distribution separated by pathogenic only, and in combination with likely pathogenic variants. Abbreviations: MAF, Minor Allele Frequency; WES, Whole-Exome-Sequencing; N/A, not available; P, pathogenic; LP, likely pathogenic; PP, possibly pathogenic; LB, likely benign; VUS, variant of uncertain significance.
Prevalence of potentially pathogenic TP53 variants in the FLOSSIES database
A total of 45 unique TP53 variants with MAF <1% were detected in the FLOSSIES database (N=9,884), including 26 nonsynonymous and 19 synonymous. From these, 1 was classified as P, 7 as LP, 7 as PP, 27 as LB, and 3 as VUS. The frequency of P variants only, and in combination with LP, ranged from 0.01% to 0.17% (Table 2). Seventeen individuals were detected with potentially pathogenic variants. Among them, 7 carried the p.R290H (7/17; 41%) and 4 carried the p.N235S (4/17, 23.5%) variants, both classified as LP (Supp. Table S1).
Prevalence of potentially pathogenic TP53 variants in the WES Controls database
We also queried our in-house WES Controls data of cancer-free individuals (N=994). A total of 7 unique variants with MAF < 1% were detected, including 6 nonsynonymous and 1 synonymous. One variant was classified as P, 2 as LP, 1 as PP, 3 as LB, and none was VUS. Based on the respective allele count values for P variants only, and in combination with LP, the prevalence of germline TP53 variants ranged from 0.1% to 0.3% (Table 2). There were no allele count enrichments for specific variants within this cohort (Supp. Table S1).
Distribution of TP53 variants across p53 protein domains
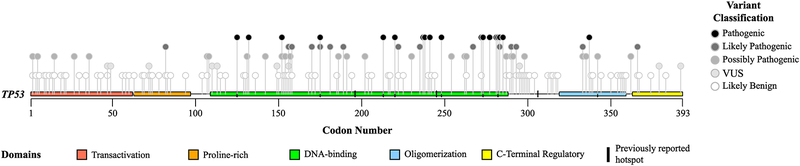
Figure 2 and Supp. Table S2 show the distribution of the variants detected in all three databases across TP53 protein domains. Thirteen nonsynonymous and 13 synonymous variants were detected in more than one database, including three that were observed in all three databases; therefore, after excluding duplicates, we evaluated a total of 165 unique variants. For potentially pathogenic variants, 28 out of 34 (82%) fall within the DNA binding domain, 2 in the oligomerization domain, 2 in DNA-binding and oligomerization domains transition region, 1 in the proline-rich domain, and 1 in the C-terminal regulatory domain.
Figure 2 -. Distribution of TP53 variants by protein domains.
According to the canonical transcript NM_000546.5, domains of TP53 were divided in: transcriptional activation (residues 1 to 62), proline-rich region (residues 63–97), DNA-binding domain (residues 109–288), oligomerization domain (residues 319–359), and C-terminal regulatory domain (residues 363–393). Point markers do not include duplicate values for variants detected in more than one database. Supp. Table S2 demonstrates exact number of variants in each region.
Distribution of TP53 variants across different ethnic groups
The distribution of variants across different population ancestries are shown in Supp. Table S3. In the ExAC dataset, the prevalence of potentially pathogenic variants was higher among individuals of non-Finnish European ancestry (75/27,173; 0.28%), followed by individuals of Finnish ancestry (8/3,307; 0.24%). Populations of Latino, East-Asian, and South-Asian ancestries had similar prevalence rates of potentially pathogenic variants. A pooled comparison revealed statistically significant difference between the prevalence of potentially pathogenic variants in individuals of European ancestries (both subgroups combined: 83/30,480) compared with the other five ancestries (28/22,625; p-value = 0.00016, Fisher`s exact test). Specific variants were enriched in certain ethnic groups. Two specific potentially pathogenic variants were mostly observed in individuals of European ancestry; among the 83 carriers, 27 (32.5%) harbored the p.N235S variant, and 16 (19%) harbored the p.R290H variant. The variant p.N263D, classified as PP, was found only among individuals of South-Asian ancestry (10/10). The variant p.Y107H, classified as PP, was detected only among individuals of African/African American ancestry (6/6). Among 6 individuals of East-Asian ancestry identified with a potentially pathogenic variant, 4 (66.6%) carried the p.A189V variant (Supp. Table S1).
In the FLOSSIES database, no statistically difference was found between ancestry subgroups (p=0.2685, Fisher`s exact test), although there was a higher prevalence of potentially pathogenic variants among individuals of European ancestry (15/7,325; 0.2%) in comparison with those of African American ancestry (2/2,559; 0.08%), (Supp. Table S3). This difference was mainly driven by the variants p.R290H and p.N235S detected in 6 and 3 individuals of European ancestry, respectively (Supp. Table S1). In addition, the population of African American ancestry had a higher proportion of PP variants (Supp. Table S3), primarily due to the presence of the p.Y107H variant in 6 individuals (Supp. Table S1).
No ancestry comparison was performed among individuals from our in-house WES Controls database since all of them had European ancestry (Supp. Table S3).
DISCUSSION
LFS has been considered a rare highly penetrant multi-cancer predisposition syndrome (Bougeard, et al., 2015). The criteria for identifying LFS are summarized in the “Revised Chompret criteria” developed by the French LFS working group (Bougeard, et al., 2015) which considers four typical familial or individual presentations suggestive of LFS. Germline variants in the TP53 gene are the only unequivocally identified genetic alterations underlying LFS. These variants may be detected in about 30% of the subjects meeting the Revised Chompret criteria and in about 80% of the families with the most severe clinical patterns of LFS (Mai, et al., 2012). To date, estimates of the prevalence of germline TP53 variants in the general population have been based on small-scale studies and our current understanding of LFS may be biased due to preferential ascertainment of highly affected LFS families, and/or individuals with multiple early cancers. The first published population frequency estimate of 1:5,000 was based on a cohort selected for personal early-breast cancer history (Lalloo, et al., 2003), whereas the second estimate of 1:20,000 consisted of 341 clinically categorized families, of which 296 (87%) met any LFS or Li-Fraumeni-like (LFL) criteria established at the time of the study (Gonzalez, et al., 2009). Based on our analysis of three aggregated sequencing databases of unrelated individuals unselected for either personal or familial history of cancer, we estimate that the prevalence of potentially pathogenic TP53 exonic variants may be as high as 0.2%. Our results suggest that even with a conservative variant categorization, the actual prevalence of potentially pathogenic TP53 variants may be substantially higher than previous projections. Similar observations have also recently been identified for the DICER1 syndrome, another rare autosomal dominant disease (Kim, et al., 2017).
The presence of undiagnosed adult carriers with potentially pathogenic TP53 variants mostly in the DNA binding and oligomerization domains, the TP53 hotspot regions (Olivier, et al., 2010; Hainaut and Pfeifer, 2016), raises the question of whether all germline potentially pathogenic TP53 variants confer a diagnosis of LFS. This is further illustrated by the consistent prevalence of potentially pathogenic variants across the three databases, especially with regard to the FLOSSIES and WES Controls which are composed of older cancer-free individuals who presumably should not have pathogenic TP53 variants. This suggests that the penetrance of cancer associated with potentially pathogenic TP53 variants may be lower than originally estimated, indicating that only a fraction of carriers will actually develop a typical LFS phenotype. This also implies that our current knowledge of TP53 variant classification does not account for the diversity of individual risk. Future population-based and clinical studies should consider the extent to which pathogenic TP53 variants confer a true high risk for early cancer development. Moreover, our results have potential implications for the interpretation of results among individuals without any clinical presentation of LFS, in those undergoing gene panel testing and/or in individuals in whom a germline TP53 mutation was detected in the course of somatic exome analysis of cancer tissue for molecular diagnostic purposes.
Another lesson from this large-scale analysis of sequencing database is that potentially pathogenic variants might be present in subjects and families who may not carry an increased risk of developing cancer as compared with non-carriers. This situation has been well demonstrated in the case of the TP53 p.R337H variant, which is common in the Brazilian population (Achatz, et al., 2007) due to a founder effect (Pinto, et al., 2004; Garritano, et al., 2010). This variant carries an individual cancer risk which is extremely variable, from fully penetrant LFS to cancer-free over a lifetime (Achatz and Zambetti, 2016), suggesting that additional components may regulate its penetrance. Among these factors, we and others have provided evidences that intragenic TP53 polymorphisms or MDM2 could modulate penetrance and age at cancer diagnosis in LFS (Bougeard, et al., 2006; Marcel, et al., 2009). Accordingly, it is likely that other non-genetic, genetic, or epigenetic events may play a role in cancer penetrance in LFS, modulating the effect of different TP53 variants.
NGS data from different regions worldwide has the potential to provide novel insights on cancer predisposition syndromes in underrepresented populations, such as those of African and Asian ancestries (see Supp. Information for further details). Our findings suggest the need for developing studies on LFS in these regions, especially given the high prevalence of some TP53 variants that may be associated with low penetrance effects, and/or with specific cancer patterns which may not fully match current clinical definitions of LFS. Larger databases comprised of underrepresented populations could provide more accurate prevalence estimates as our results may be inflated by specific ethnic genetic diversity.
Our study identified several variants that have been extensively described as strongly associated with LFS, including well-characterized hotspots TP53 mutations such as p.R175H (the most frequent somatic or germline cancer variant), p.R248Q, p.R273H, or p.Y220C. Together, these 4 cancer hotspot variants were present in 8.4% of the unrelated individuals carrying potentially pathogenic variants in our dataset (11/131), whereas they account for 17.8% of the LFS families documented in the IARC TP53 database (Bouaoun, et al., 2016). However, the two most common variants in our analysis, p.R290H and p.N235S, have less certain pathogenic significance (see Supp. Information for further details). The high representation of these two variants in sequencing databases of individuals unselected for cancer risk calls for caution in interpreting them as causal for LFS when detected in a familial setting.
Although TP53 is one of the most widely studied genes, the prediction of deleteriousness by the current annotation tools still needs to be refined. Variants in TP53 should be interpreted not solely based on structure/function prediction algorithms but also considering the cumulative knowledge of clinical outcomes and functional assays in experimental systems. Variants may affect p53 functionality in many ways and a number of variants predicted to be likely benign, or with uncertain significance, may exert a detrimental effect through other mechanisms based on the nature of the amino-acid substitution, such as aberrant splicing. Furthermore, even some synonymous variants such as p.T125T have been shown to activate aberrant cryptic splicing sites (Varley, et al., 2001) and are now considered as potentially pathogenic variants. One of the limitations of this analysis is that we were unable to analyze insertions, deletions, or indels, UTR, and intronic variants, for which current annotation tools are still very imprecise. However, we acknowledge that some variants in these regions may also have a detrimental effect, such as the recently published germline variant (rs78378222) in the TP53 3` UTR region (Macedo, et al., 2016). Therefore, due to conflicting results and limited available data, the variant classification presented herein may change in the near future as collaborative efforts such as the Clinical Genome Resource (ClinGen) consortium (Rehm, et al., 2015) has aimed to provide more comprehensive variant curation guidelines specific for TP53 variants.
While the prevalence of TP53 pathogenic mutations was consistent across the data sources in our study, limitations of the data might affect our ability to infer our results to the general population as we relied predominantly on public genomic datasets with limited phenotype data. We excluded individuals with a known prior history of cancer included in the dataset from the TCGA consortium to avoid variant overrepresentation either due to individuals that were recruited based on cancer history or even blood samples contaminated by circulating tumor cells. Nevertheless, the ExAC database includes both cases and controls for other clinical outcomes that might enrich for variants that are also associated with cancer risk. Also, although the ExAC database restricted inclusion to unrelated adults without severe pediatric diseases, it may still contain cancer-affected individuals or some clinical characteristics that should be excluded depending upon the aim of the analyses (Lek, et al., 2016). Another potential bias that may lead to an overestimation of the true prevalence is the presence of somatic TP53 mutations due to clonal hematopoiesis. In this context, the FLOSSIES and the WES Controls databases would be particularly prone as the prevalence of clonal hematopoiesis has a significant increase after the sixth decade of life (Genovese, et al., 2014; Jaiswal, et al., 2014). The FLOSSIES database used an allele fraction cut-off of 0.25; therefore, there may be residual somatic variants whose allele fractions were between 0.25 and the desirable cut-off of 0.3. This 0.3 threshold has been proposed to reduce the proportion of somatic TP53 variants that could be wrongly ascertained as of germline origin (Coffee, et al., 2017). However, prevalence estimates were comparable for the FLOSSIES database and our WES Cohort, which used an allele fraction cut-off of 0.3 and is also mostly comprised by older people, thus results for the FLOSSIES database may not have been substantially impacted (allele fraction cut-off for the ExAC database was not available). On the other hand, the available sequencing data were based primarily on studies that recruited participants in adulthood, excluding all those who may have developed cancer in childhood, a common occurrence in TP53 mutation carriers (Bougeard, et al., 2015). This is a potential bias that may lead to an underestimation of the true frequency of TP53 pathogenic carriers in the general population. Ultimately, we acknowledge that our findings raise more questions than provide definitive answers. Altogether, additional family studies, functional data, analysis of genetic co-factors, and improved variant curation guidelines are still required to better stratify clinical management of LFS patients.
In summary, our results suggest that the prevalence of pathogenic germline TP53 variants in the population may be substantially higher than previous estimates. Consequently, the actual cancer penetrance of LFS may be lower than expected, illustrating that sequencing data and variant curation should be carefully interpreted. We encourage further studies of penetrance modifiers and of individuals with TP53 variants to better classify variants with regards to their pathogenicity and enrich knowledge of variant curation for clinical purposes.
Supplementary Material
Supp. Table S1 - List of all germline TP53 variants identified and respective classification, allele count, and reported ancestry. Abbreviations: REF, reference allele; VAR, variant allele; AC, allele count; HET, heterozygous; AFR, African/African American ancestry; AMR, Latino ancestry; EAS, East-Asian ancestry; FIN, Finnish ancestry; NFE, non-Finnish European ancestry; OTH, other ancestry; SAS, South-Asian ancestry; WES, Whole-Exome-Sequencing. According to the ExAC consortium group, individuals were classified as “OTH” if they did not cluster with any of the major populations in a principal component analysis. Columns for FIN and NFE allele counts were merged for the FLOSSIES and WES Controls databases as there was no separated data for these populations. Variants reported based on the canonical transcript NM_000546.5 (nucleotide numbering for cDNA uses +1 as the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1).
Supp. Table S2 - Distribution of TP53 variants by protein domains. Tabular representation of Figure 2. According to the NM_000546.5 reference, domains of TP53 were divided in: transcriptional activation (residues 1 to 62), proline-rich region (residues 63–97), DNA-binding domain (residues 109–288), oligomerization domain (residues 319–359), and C-terminal regulatory domain (residues 363–393). Numbers do not include duplicate values for variants detected in more than one database. Nonsynonymous variants include 1 nonframeshift deletion and 1 nonsense variant. Abbreviations: P, pathogenic; LP, likely pathogenic; PP, possibly pathogenic; LB, likely benign; VUS, variant of uncertain significance.
Supp. Table S3 – Distribution of TP53 variants across different population ancestries. Variant distribution across ancestries comprised in each database. Prevalence calculated by the number of individuals within a certain category divided by the total number of individuals of a given ancestry. Abbreviations: WES, Whole-Exome-Sequencing; P, pathogenic; LP, likely pathogenic; PP, possibly pathogenic; LB, likely benign; VUS, variant of uncertain significance.
Acknowledgements
The authors would like to acknowledge the contribution provided by the Exome Working group from the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health. We are also thankful for the funding and personnel responsible for creating and maintaining sequencing databases and original studies used in this analysis. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Funding
This study was funded by the intramural research program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health.
Footnotes
Conflict of Interest
The authors hereby declare that they have no conflict of interest.
REFERENCES
- Achatz MI, Olivier M, Le Calvez F, Martel-Planche G, Lopes A, Rossi BM, Ashton-Prolla P, Giugliani R, Palmero EI, Vargas FR, Da Rocha JC, Vettore AL, Hainaut P. 2007. The TP53 mutation, R337H, is associated with Li-Fraumeni and Li-Fraumeni-like syndromes in Brazilian families. Cancer Lett 245(1–2):96–102. [DOI] [PubMed] [Google Scholar]
- Achatz MI, Zambetti GP. 2016. The Inherited p53 Mutation in the Brazilian Population. Cold Spring Harb Perspect Med 6(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcand SL, Maugard CM, Ghadirian P, Robidoux A, Perret C, Zhang P, Fafard E, Mes-Masson AM, Foulkes WD, Provencher D, Narod SA, Tonin PN. 2008. Germline TP53 mutations in BRCA1 and BRCA2 mutation-negative French Canadian breast cancer families. Breast Cancer Res Treat 108(3):399–408. [DOI] [PubMed] [Google Scholar]
- Birch JM, Hartley AL, Tricker KJ, Prosser J, Condie A, Kelsey AM, Harris M, Jones PH, Binchy A, Crowther D. 1994. Prevalence and diversity of constitutional mutations in the p53 gene among 21 Li-Fraumeni families. Cancer Res 54(5):1298–304. [PubMed] [Google Scholar]
- Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, Olivier M. 2016. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum Mutat 37(9):865–76. [DOI] [PubMed] [Google Scholar]
- Bougeard G, Baert-Desurmont S, Tournier I, Vasseur S, Martin C, Brugieres L, Chompret A, Bressac-de Paillerets B, Stoppa-Lyonnet D, Bonaiti-Pellie C, Frebourg T. 2006. Impact of the MDM2 SNP309 and p53 Arg72Pro polymorphism on age of tumour onset in Li-Fraumeni syndrome. J Med Genet 43(6):531–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougeard G, Renaux-Petel M, Flaman JM, Charbonnier C, Fermey P, Belotti M, Gauthier-Villars M, Stoppa-Lyonnet D, Consolino E, Brugières L, Caron O, Benusiglio PR, Bressac-de Paillerets B, Bonadona V, Bonaïti-Pellié C, Tinat J, Baert-Desurmont S, Frebourg T. 2015. Revisiting Li-Fraumeni Syndrome From TP53 Mutation Carriers. J Clin Oncol 33(21):2345–52. [DOI] [PubMed] [Google Scholar]
- Calle EE, Rodriguez C, Jacobs EJ, Almon ML, Chao A, McCullough ML, Feigelson HS, Thun MJ. 2002. The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer 94(9):2490–501. [DOI] [PubMed] [Google Scholar]
- Coffee B, Cox HC, Kidd J, Sizemore S, Brown K, Manley S, Mancini-DiNardo D. 2017. Detection of somatic variants in peripheral blood lymphocytes using a next generation sequencing multigene pan cancer panel. Cancer Genet 211:5–8. [DOI] [PubMed] [Google Scholar]
- Eeles RA. 1995. Germline mutations in the TP53 gene. Cancer Surv 25:101–24. [PubMed] [Google Scholar]
- Garritano S, Gemignani F, Palmero EI, Olivier M, Martel-Planche G, Le Calvez-Kelm F, Brugiéres L, Vargas FR, Brentani RR, Ashton-Prolla P, Landi S, Tavtigian SV, Hainaut P, Achatz MI. 2010. Detailed haplotype analysis at the TP53 locus in p.R337H mutation carriers in the population of Southern Brazil: evidence for a founder effect. Hum Mutat 31(2):143–50. [DOI] [PubMed] [Google Scholar]
- Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, Purcell SM, Svantesson O, Landén M, Höglund M, Lehmann S, Gabriel SB, Moran JL, Lander ES, Sullivan PF, Sklar P, Grönberg H, Hultman CM, McCarroll SA. 2014. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371(26):2477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez KD, Noltner KA, Buzin CH, Gu D, Wen-Fong CY, Nguyen VQ, Han JH, Lowstuter K, Longmate J, Sommer SS, Weitzel JN. 2009. Beyond Li Fraumeni Syndrome: clinical characteristics of families with p53 germline mutations. J Clin Oncol 27(8):1250–6. [DOI] [PubMed] [Google Scholar]
- Hainaut P, Pfeifer GP. 2016. Somatic TP53 Mutations in the Era of Genome Sequencing. Cold Spring Harb Perspect Med 6(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, Musolf A, Li Q, Holzinger E, Karyadi D, Cannon-Albright LA, Teerlink CC, Stanford JL, Isaacs WB, Xu J, Cooney KA, Lange EM, Schleutker J, Carpten JD, Powell IJ, Cussenot O, Cancel-Tassin G, Giles GG, MacInnis RJ, Maier C, Hsieh CL, Wiklund F, Catalona WJ, Foulkes WD, Mandal D, Eeles RA, Kote-Jarai Z, Bustamante CD, Schaid DJ, Hastie T, Ostrander EA, Bailey-Wilson JE, Radivojac P, Thibodeau SN, Whittemore AS, Sieh W. 2016. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am J Hum Genet 99(4):877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, Higgins JM, Moltchanov V, Kuo FC, Kluk MJ, Henderson B, Kinnunen L, Koistinen HA, Ladenvall C, Getz G, Correa A, Banahan BF, Gabriel S, Kathiresan S, Stringham HM, McCarthy MI, Boehnke M, Tuomilehto J, Haiman C, Groop L, Atzmon G, Wilson JG, Neuberg D, Altshuler D, Ebert BL. 2014. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371(26):2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian X, Boerwinkle E, Liu X. 2014. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res 42(22):13534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, Ishioka C. 2003. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci U S A 100(14):8424–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Field A, Schultz KAP, Hill DA, Stewart DR. 2017. The prevalence of DICER1 pathogenic variation in population databases. Int J Cancer 10.1002/ijc.30907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalloo F, Varley J, Ellis D, Moran A, O’Dair L, Pharoah P, Evans DG, Group EOBCS. 2003. Prediction of pathogenic mutations in patients with early-onset breast cancer by family history. Lancet 361(9363):1101–2. [DOI] [PubMed] [Google Scholar]
- Landi MT, Consonni D, Rotunno M, Bergen AW, Goldstein AM, Lubin JH, Goldin L, Alavanja M, Morgan G, Subar AF, Linnoila I, Previdi F, Corno M, Rubagotti M, Marinelli B, Albetti B, Colombi A, Tucker M, Wacholder S, Pesatori AC, Caporaso NE, Bertazzi PA. 2008. Environment And Genetics in Lung cancer Etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health 8:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, Jang W, Katz K, Ovetsky M, Riley G, Sethi A, Tully R, Villamarin-Salomon R, Rubinstein W, Maglott DR. 2016. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44(D1):D862–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG, Consortium EA. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536(7616):285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FP, Fraumeni JF. 1969a. Rhabdomyosarcoma in children: epidemiologic study and identification of a familial cancer syndrome. J Natl Cancer Inst 43(6):1365–73. [PubMed] [Google Scholar]
- Li FP, Fraumeni JF. 1969b. Soft-tissue sarcomas, breast cancer, and other neoplasms. A familial syndrome? Ann Intern Med 71(4):747–52. [DOI] [PubMed] [Google Scholar]
- Macedo GS, Araujo Vieira I, Brandalize AP, Giacomazzi J, Inez Palmero E, Volc S, Rodrigues Paixão-Côrtes V, Caleffi M, Silva Alves M, Achatz MI, Hainaut P, Ashton-Prolla P. 2016. Rare germline variant (rs78378222) in the TP53 3’ UTR: Evidence for a new mechanism of cancer predisposition in Li-Fraumeni syndrome. Cancer Genet 209(3):97–106. [DOI] [PubMed] [Google Scholar]
- Mai PL, Best AF, Peters JA, DeCastro RM, Khincha PP, Loud JT, Bremer RC, Rosenberg PS, Savage SA. 2016. Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute Li-Fraumeni syndrome cohort. Cancer 122(23):3673–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai PL, Malkin D, Garber JE, Schiffman JD, Weitzel JN, Strong LC, Wyss O, Locke L, Means V, Achatz MI, Hainaut P, Frebourg T, Evans DG, Bleiker E, Patenaude A, Schneider K, Wilfond B, Peters JA, Hwang PM, Ford J, Tabori U, Ognjanovic S, Dennis PA, Wentzensen IM, Greene MH, Fraumeni JF Jr., Savage SA. 2012. Li-Fraumeni syndrome: report of a clinical research workshop and creation of a research consortium. Cancer Genet 205(10):479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin D. 2011. Li-fraumeni syndrome. Genes Cancer 2(4):475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin D, Li FP, Strong LC, Fraumeni JF Jr., Nelson CE, Kim DH, Kassel J, Gryka MA, Bischoff FZ, Tainsky MA, et al. 1990. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 250(4985):1233–8. [DOI] [PubMed] [Google Scholar]
- Marcel V, Palmero EI, Falagan-Lotsch P, Martel-Planche G, Ashton-Prolla P, Olivier M, Brentani RR, Hainaut P, Achatz MI. 2009. TP53 PIN3 and MDM2 SNP309 polymorphisms as genetic modifiers in the Li-Fraumeni syndrome: impact on age at first diagnosis. J Med Genet 46(11):766–72. [DOI] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. 2010. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2(1):a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto EM, Billerbeck AE, Villares MC, Domenice S, Mendonça BB, Latronico AC. 2004. Founder effect for the highly prevalent R337H mutation of tumor suppressor p53 in Brazilian patients with adrenocortical tumors. Arq Bras Endocrinol Metabol 48(5):647–50. [DOI] [PubMed] [Google Scholar]
- Prorok PC, Andriole GL, Bresalier RS, Buys SS, Chia D, Crawford ED, Fogel R, Gelmann EP, Gilbert F, Hasson MA, Hayes RB, Johnson CC, Mandel JS, Oberman A, O’Brien B, Oken MM, Rafla S, Reding D, Rutt W, Weissfeld JL, Yokochi L, Gohagan JK, Prostate L, C.lorectal and Ovarian Cancer Screening Trial Project Team. 2000. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin Trials 21(6 Suppl):273S–309S. [DOI] [PubMed] [Google Scholar]
- Quesnel S, Verselis S, Portwine C, Garber J, White M, Feunteun J, Malkin D, Li FP. 1999. p53 compound heterozygosity in a severely affected child with Li-Fraumeni syndrome. Oncogene 18(27):3970–8. [DOI] [PubMed] [Google Scholar]
- Rajkumar T, Meenakumari B, Mani S, Sridevi V, Sundersingh S. 2015. Targeted Resequencing of 30 Genes Improves the Detection of Deleterious Mutations in South Indian Women with Breast and/or Ovarian Cancers. Asian Pac J Cancer Prev 16(13):5211–7. [DOI] [PubMed] [Google Scholar]
- Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, Ledbetter DH, Maglott DR, Martin CL, Nussbaum RL, Plon SE, Ramos EM, Sherry ST, Watson MS, ClinGen. 2015. ClinGen--the Clinical Genome Resource. N Engl J Med 372(23):2235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage SA, Mirabello L, Wang Z, Gastier-Foster JM, Gorlick R, Khanna C, Flanagan AM, Tirabosco R, Andrulis IL, Wunder JS, Gokgoz N, Patiño-Garcia A, Sierrasesúmaga L, Lecanda F, Kurucu N, Ilhan IE, Sari N, Serra M, Hattinger C, Picci P, Spector LG, Barkauskas DA, Marina N, de Toledo SR, Petrilli AS, Amary MF, Halai D, Thomas DM, Douglass C, Meltzer PS, Jacobs K, Chung CC, Berndt SI, Purdue MP, Caporaso NE, Tucker M, Rothman N, Landi MT, Silverman DT, Kraft P, Hunter DJ, Malats N, Kogevinas M, Wacholder S, Troisi R, Helman L, Fraumeni JF, Yeager M, Hoover RN, Chanock SJ. 2013. Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat Genet 45(7):799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. 2014. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 133(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hest LP, Ruijs MW, Wagner A, van der Meer CA, Verhoef S, van’t Veer LJ, Meijers-Heijboer H. 2007. Two TP53 germline mutations in a classical Li-Fraumeni syndrome family. Fam Cancer 6(3):311–6. [DOI] [PubMed] [Google Scholar]
- Varley JM, Attwooll C, White G, McGown G, Thorncroft M, Kelsey AM, Greaves M, Boyle J, Birch JM. 2001. Characterization of germline TP53 splicing mutations and their genetic and functional analysis. Oncogene 20(21):2647–54. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp. Table S1 - List of all germline TP53 variants identified and respective classification, allele count, and reported ancestry. Abbreviations: REF, reference allele; VAR, variant allele; AC, allele count; HET, heterozygous; AFR, African/African American ancestry; AMR, Latino ancestry; EAS, East-Asian ancestry; FIN, Finnish ancestry; NFE, non-Finnish European ancestry; OTH, other ancestry; SAS, South-Asian ancestry; WES, Whole-Exome-Sequencing. According to the ExAC consortium group, individuals were classified as “OTH” if they did not cluster with any of the major populations in a principal component analysis. Columns for FIN and NFE allele counts were merged for the FLOSSIES and WES Controls databases as there was no separated data for these populations. Variants reported based on the canonical transcript NM_000546.5 (nucleotide numbering for cDNA uses +1 as the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1).
Supp. Table S2 - Distribution of TP53 variants by protein domains. Tabular representation of Figure 2. According to the NM_000546.5 reference, domains of TP53 were divided in: transcriptional activation (residues 1 to 62), proline-rich region (residues 63–97), DNA-binding domain (residues 109–288), oligomerization domain (residues 319–359), and C-terminal regulatory domain (residues 363–393). Numbers do not include duplicate values for variants detected in more than one database. Nonsynonymous variants include 1 nonframeshift deletion and 1 nonsense variant. Abbreviations: P, pathogenic; LP, likely pathogenic; PP, possibly pathogenic; LB, likely benign; VUS, variant of uncertain significance.
Supp. Table S3 – Distribution of TP53 variants across different population ancestries. Variant distribution across ancestries comprised in each database. Prevalence calculated by the number of individuals within a certain category divided by the total number of individuals of a given ancestry. Abbreviations: WES, Whole-Exome-Sequencing; P, pathogenic; LP, likely pathogenic; PP, possibly pathogenic; LB, likely benign; VUS, variant of uncertain significance.