Abstract
Introduction
The use of optical coherence tomography (OCT) images is increasing in the medical treatment of age-related macular degeneration (AMD), and thus, the amount of data requiring analysis is increasing. Advances in machine-learning techniques may facilitate processing of large amounts of medical image data. Among deep-learning methods, convolution neural networks (CNNs) show superior image recognition ability. This study aimed to build deep-learning models that could distinguish AMD from healthy OCT scans and to distinguish AMD with and without exudative changes without using a segmentation algorithm.
Methods
This was a cross-sectional observational clinical study. A total of 1621 spectral domain (SD)-OCT images of patients with AMD and a healthy control group were studied. The first CNN model was trained and validated using 1382 AMD images and 239 normal images. The second transfer-learning model was trained and validated with 721 AMD images with exudative changes and 661 AMD images without any exudate. The attention area of the CNN was described as a heat map by class activation mapping (CAM). In the second model, which classified images into AMD with or without exudative changes, we compared the learning stabilization of models using or not using transfer learning.
Results
Using the first CNN model, we could classify AMD and normal OCT images with 100% sensitivity, 91.8% specificity, and 99.0% accuracy. In the second, transfer-learning model, we could classify AMD as having or not having exudative changes, with 98.4% sensitivity, 88.3% specificity, and 93.9% accuracy. CAM successfully described the heat-map area on the OCT images. Including the transfer-learning model in the second model resulted in faster stabilization than when the transfer-learning model was not included.
Conclusion
Two computational deep-learning models were developed and evaluated here; both models showed good performance. Automation of the interpretation process by using deep-learning models can save time and improve efficiency.
Trial Registration
No15073.
Keywords: Age-related macular degeneration, Artificial intelligence, Class activation mapping, Convolution neural network, Deep learning, Machine learning, Optical coherence tomography, Transfer learning
Introduction
Age-related macular degeneration (AMD) is the leading cause of severe visual loss, and the number of patients with this condition is increasing with the rapid aging of the population in developed countries [1]. The 10-year cumulative incidence of AMD was reported to be 12.1% in the Beaver Dam Study in the United States and 14.1% in the Blue Mountains Eye Study in Australia [2, 3]. AMD is categorized into dry and wet AMD, based on the absence or presence of neovascularization [4]. In wet AMD, fluid leakage or bleeding from the permeable capillary network in the sub-retinal pigment epithelium (RPE) and fibrotic scars in the subretinal areas result in severe photoreceptor degeneration [5]. Several recent large clinical trials have shown the effectiveness of anti-vascular endothelial growth factor (VEGF) therapy (ranibizumab, bevacizumab, and aflibercept) for neovascular AMD [6–10]. However, follow-up studies of these trials have also reported that retaining good visual acuity for a long period of time in real practice was difficult, indicating that monthly monitoring of patients receiving anti-VEGF treatment requires much effort from patients and medical staff, and increases medical costs [11].
Spectral-domain optical coherence tomography (SD-OCT) is a commercially available device that clearly describes particular findings of AMD, such as drusen, intra-retinal fluid (IRF), sub-retinal fluid (SRF), sub-retinal hyper-reflective material, including hemorrhage and retinal pigment epithelium detachment, etc. [12]. Among these, exudative changes (intra- and sub-retinal fluid and hemorrhage) are the key indication for most physicians to initiate anti-VEGF therapy and evaluate the therapeutic effect [13]. Zero tolerance has been applied for the large clinical trial mentioned above [14]. Therefore, the amount of OCT data requiring analysis is increasing, beyond clinical capacity [15].
Advances in machine-learning techniques provide a solution for meaningful interpretation of large amounts of medical image data arising from the frequent treatment and follow-up monitoring of patients [16]. In particular, convolution neural networks (CNNs) have greatly advanced the classification of medical images using multi-layer neural networks and deep-learning algorithms [17]. In ophthalmology, the excellent accuracy of CNNs has already been reported in the classification of diabetic retinopathy from fundus photographs, visual field examination of glaucoma patients, grading of pediatric nuclear cataracts, etc. [18–20]. The impressive performance of neural networks in the classification of AMD images has also been reported for the automated detection of AMD features in OCT and fundus photographs, for guidance of anti-VEGF therapy, and monitoring disease progression [18, 21–28]. Although some studies have classified the exudative feature of AMD for automated segmentation [21, 29], to our knowledge, there are no reports about the classification of exudative change with AMD in deep learning models without segmentation.
The purpose of the present study was to build two deep-learning models and to evaluate their performance without using a segmentation algorithm. The first CNN model was built to distinguish AMD eyes from healthy eyes on OCT scans, and the second transfer-learning model was used to distinguish between AMD with and without exudative changes.
Methods
This research was conducted in line with the Helsinki Declaration of 1964, and the research protocols and their implementation were approved by the Ethics Committee of Kobe City Medical Center General Hospital (approval date; Sept. 17, 2015 and approval number: No15073). The committee waived the requirement for obtaining informed consent given that this was a retrospective observational study of medical records, and retrospectively registered.
We included the records of patients who visited Kobe City Medical Center General Hospital from March 2016 to November 2017. We included SD-OCT (Heidelberg Spectralis, Heidelberg Engineering, Heidelberg, Germany) images of normal subjects and of patients with AMD.
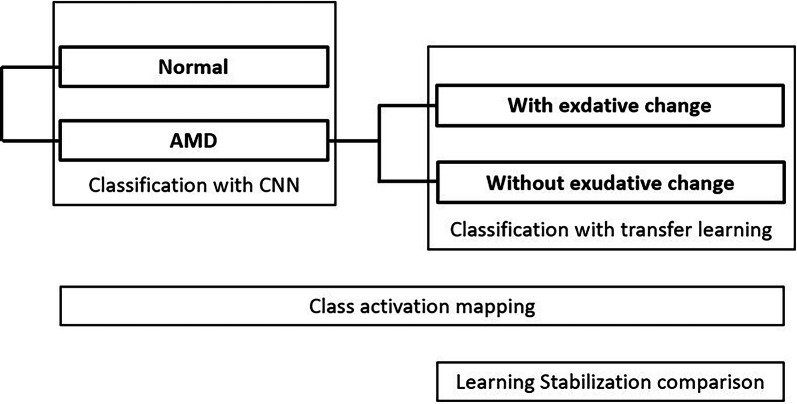
The outline we have researched is as follows (Fig. 1). We built two classification models. In the first CNN model, we classified images into normal and AMD images. In the second transfer-learning model, we classified AMD images into those with and those without exudative changes. We used class activation mapping (CAM) as a heat map to show the location of the images that the CNN models emphasized in the classification. Additionally, in the second model, we compared the speed of learning stability with the model using transfer learning and the single CNN model.
Fig. 1.
We built two classification models. In the first CNN model, we classified images into normal and AMD images. In the second transfer-learning model, we classified AMD images into those with and those without exudative changes. We used CAM to show the location of the image that CNN models emphasized in the classification as the heat map. Additionally, in the second model, we compared the speed of learning stability with the model using transfer learning and the single CNN model. AMD age-related macular degeneration, CNN convolution neural network, CAM class activation mapping
Patients diagnosed with AMD and normal control subjects were enrolled at the outpatients’ clinic during the same period. AMD was diagnosed by means of fundus examinations, fluorescein angiography/indocyanine green angiography, and OCT images by independent retinal specialists. Only one eye per patient was selected for analysis. The exclusion criteria were poor image quality and the presence of other potentially confounding retinal pathologies.
We included 120 eyes of 120 AMD patients, and 49 eyes from 49 normal subjects, as the training data group, and 77 eyes of 77 AMD patients, and 25 eyes from 25 normal subjects, as the test data group. We used OCT images for training and validating deep-learning models. In the first model, 185 normal images and 1049 AMD images were used for training data, and the CNN model was subsequently evaluated using another 49 normal images and 333 AMD images. In the second model, 535 AMD images with exudative changes and 514 AMD images without exudative changes were selected as training data, and the second model was subsequently evaluated using another 188 images with exudative changes and 154 images without exudative changes.
Building the Models
We built two classification models. In the first CNN model, we classified images into normal and AMD images. In the second transfer-learning model, we classified AMD images into those with and those without exudative changes (Fig. 1).
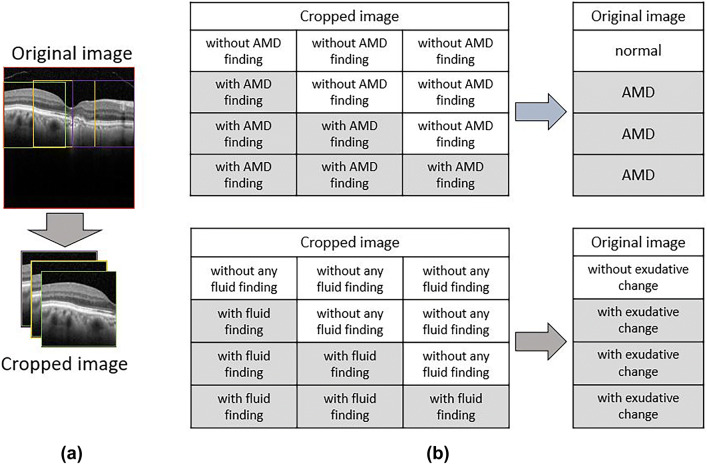
The process of image classification is summarized in Fig. 2. The CNN classification models were constructed using a cropped image obtained by dividing the original image into three in order not to degrade the image quality. After classifying the cropped images with the CNN models, three cropped images were reassembled into the original image to determine the original image classification.
Fig. 2.
a CNN classification models were constructed using a cropped image obtained by dividing the original image into three in order not to degrade the image quality. b After classifying the cropped images with CNN models, the three cropped OCT images were returned to the original image to determine the original image classification. If at least one of the three cropped OCT images showed AMD findings, the original image was judged as AMD, and it was judged as normal only if all three cropped images were without AMD findings. The second judgement of the presence of exudative fluid was similarly performed. CNN convolution neural network, OCT optical coherence tomography, AMD age-related macular degeneration
SD-OCT images obtained with either a radial-scan (scan length: 6.0 mm) or cross-scan (scan length: 9.0 mm) protocol were included. In the cross-scan image, the central 6.0-mm area was cropped to obtain the same scan length as radial scans and resized to 496 × 496 pixels.
Considering (0,0) as the bottom-left corner of an OCT image, and the left-to-right as the x-axis direction and top-to-bottom as the y-axis direction, 3 points for which y was on the RPE line were determined automatically, with a fixed value of x as 112, 247, or 399. With these points as centers, we cropped three images from each OCT image to a size of 224 × 224 pixels to increase the numbers in the data set without resizing images.
Three ophthalmologists (N. M., M. M, Y. H.) who have extensive experience with macular outpatients independently labeled the three cropped images. In the first model, labelling the images as “normal” or “AMD” was conducted in two steps. First, each cropped image was labeled as “without AMD finding” and “with AMD finding,” depending on whether the image contained any AMD findings, such as drusen, pseudo-drusen, pigmented epithelial detachment (PED), drusenoid PED, geographic atrophy, hyperreflective foci, or sub-retinal hyper-reflective material. Only images in which the three physician’s diagnoses matched were included in the next step. Original images were judged as “normal” when all three images were labeled as “without AMD finding” and judged as “AMD” only when at least one of the three images was labeled as “with AMD finding.”
In the second model, we cropped three images from the AMD OCT images, as in the first model. We then labeled these images as “exudative change = fluid” or “without exudative change = no fluid” in two steps. First, each cropped image was labeled as “with fluid finding” and “without any fluid finding,” depending on whether the image contained any fluid, such as exudative changes (intra- and sub-retinal fluid and hemorrhage), as judged independently by the same three well-trained ophthalmologists. Only the images for which the three physicians reached agreement were included in the next step. Original images were then categorized as “without exudative change = no fluid” when all three images were labeled as “without any fluid finding,” and were categorized as “exudative change = fluid” only when there was at least one of the three images that was labeled as “with fluid finding.”
The number of images was expanded about 1000 times with flips, translations, or rotations. A batch size of 32 images and an epoch number of 1000 times was used during the training phase, using an optimization function of the Adam algorithm. Finally, from 1000 CNN models, we selected the one with the highest area under the receiver operating characteristic curve (AUROC). In this study, a system running Ubuntu 16.04 OS, and with a single GTX 1080 TI graphics card (Nvidia, Santa Clara, CA, USA), was used.
We examined the AUROC to assess the performance of the CNN models for classification of the cropped images. In order to classify the original image, the cropped images were recombined, and the sensitivity, specificity, and accuracy of the classification models were determined.
Furthermore, we compared the necessary number of epochs for the training loss to converge, and classification performance, between the transfer-learning model and the CNN of the same architecture without transfer learning. To build a robust classification model, the data augmentation and dropout technique were also applied in the training phase for both models.
CNN Model Architecture
We applied a deep-learning model using CNNs for use in our classification system (Table 1). Two classification models were built. We arranged convolutional layers as layer 1, 2, 4, 5, 7, 8, 10, 12, and 14 for the activation functions of rectified linear units (ReLU) and batch normalization next to convolutional layers [29, 30]. We arranged max. pooling layers as layers 3, 6, 9, 11, and 13 after convolutional layers. The global average pooling layer was layer 15. We set the dropout layer (drop rate: 0.2) as layer 16 and the fully connected layers as layers 16 and 17. Finally, in the final output layer, we arranged layer 18 as the softmax layer. This CNN architecture was applied to both models. Transfer learning was used to retrain a CNN that had previously been constructed, and most unit weights were pretrained.
Table 1.
The CNN configurations used in this study
| Layer | Type | Kernel numbers | Kernel size | Stride | Activation |
|---|---|---|---|---|---|
| 0 | Input | 3 | 224 × 224 | – | – |
| 1 | Convolution | 32 | 3 × 3 | – | – |
| 2 | Convolution | 32 | 3 × 3 | – | ReLU |
| 3 | Max pooling | – | – | 2 | – |
| 4 | Convolution | 64 | 3 × 3 | – | – |
| 5 | Convolution | 64 | 3 × 3 | – | ReLU |
| 6 | Max pooling | – | – | 2 | – |
| 7 | Convolution | 64 | 3 × 3 | – | – |
| 8 | Convolution | 64 | 3 × 3 | – | ReLU |
| 9 | Max pooling | – | – | 2 | – |
| 10 | Convolution | 128 | 3 × 3 | – | ReLU |
| 11 | Max pooling | – | – | 2 | – |
| 12 | Convolution | 128 | 3 × 3 | – | ReLU |
| 13 | Max pooling | – | – | 2 | – |
| 14 | Convolution | 256 | 3 × 3 | – | ReLU |
| 15 | Global average pooling | ||||
| 16 | Fully connected | 256 | – | – | ReLU, dropout |
| 17 | Fully connected | 2 | – | – | ReLU |
| 18 | Softmax | – | – | – |
With each OCT image in the training data labeled, we trained a CNN classification model to distinguish AMD OCT images from normal images. To build a robust classification model, data augmentation and the dropout technique were applied in the training phase
CNN convolution neural network, AMD age-related macular degeneration, ReLu rectified linear unit
Heatmap Creation
One method that represents the area of the CNN classification model on a heatmap is class activation mapping (CAM); this technique was applicable in our case, as a global average pooling layer was used in our proposed CNN architecture. Using this technique, heat maps were generated for each model. For the first model, we generated a heat map for the AMD category, indicating the effective region for the model to identify AMD, while for the second model, a heat map was created for the discriminative region used to identifying AMD with or without the presence of fluid [31].
Results
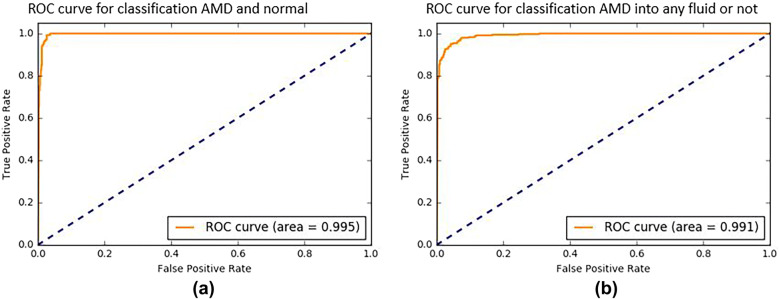
We evaluated the performance of the deep-learning models using cropped OCT images. In the first model, we obtained an AUROC of 99.5% for each individual cropped OCT image (Fig. 3). From the constructed ROC curve, we calculated the optimal threshold for distinguishing AMD from normal eyes, and the peak sensitivity and specificity using this threshold were 97.33% and 98.51%, respectively. In the second model, we obtained an AUROC of 99.1% for each individual cropped OCT image (Fig. 3). From the ROC curve constructed, the optimal threshold was calculated for distinguishing AMD with and without exudative changes, and the peak sensitivity and specificity with this threshold were 93.78% and 96.54%, respectively.
Fig. 3.
a The ROC curve for classification of AMD and healthy eyes from cropped OCT images. The ROC curve for the first model yielded an AUROC of 99.5%. b The ROC curve for classification of AMD cropped images into the presence or absence of fluid. The ROC curve for the second model yielded an AUROC of 99.1%. ROC receiver operating characteristic curve, AUROC area under the ROC curve
To evaluate the classification of the original image, the cropped images were then recombined. In the first model, correct AMD diagnosis (true positives) was made in 333 cases. There were no cases of non-detection of AMD (false negatives). In the normal control group, correct diagnosis (true negative) was made in 4 cases, while AMD was diagnosed (false positives) in 45 cases. Therefore, the sensitivity of the classifier was 100%, the specificity was 91.8%, and the accuracy was 99.0% (Tables 2, 3).
Table 2.
The sensitivity, specificity, and accuracy of the model for classification of AMD and normal OCT images
| Based on images | AMD by doctors | Normal by doctors | Specificity |
|---|---|---|---|
| AMD by the model | 333 | 4 | 98.8% |
| Normal by the model | 0 | 45 | 100.0% |
| Sensitivity | 100.0% | 91.8% |
Accuracy 99.0% |
The sensitivity of the classifier was 100%, the specificity was 91.8%, and the accuracy was 99.0%
Table 3.
The sensitivity, specificity, and accuracy of the model for classification of the presence or absence of exudative changes
| Based on images | Fluid by doctors | No fluid by doctors | Specificity |
|---|---|---|---|
| Fluid by the model | 185 | 18 | 91.1% |
| No fluid by the model | 3 | 136 | 97.8% |
| Sensitivity | 98.4% | 88.3% |
Accuracy 93.9% |
The sensitivity of the classifier was 98.4%, the specificity was 88.3%, and the accuracy was 93.9%
In the second model, correct diagnosis of the presence of fluid (true positives) was made in 185 cases, while in 3 cases with exudative changes, fluid was not detected (false negatives). In the no-fluid group, correct diagnosis (true negatives) was made in 18 cases, and an incorrect diagnosis (false positives) was made in 136 cases. Therefore, the sensitivity of the classifier was 98.4%, the specificity was 88.3%, and the accuracy was 93.9% (Tables 2, 3).
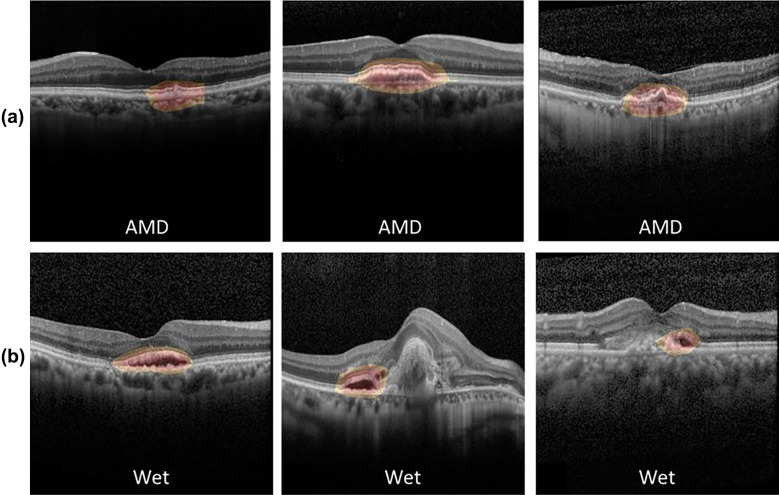
In both models, CAM was able to identify the pathologic region on OCT successfully, based on the generated heat map (Fig. 4). Additionally, in the second model, the learning stabilized faster with transfer learning than with CNN (Fig. 5).
Fig. 4.
Heat maps for two CNN classification models. a Heat map for the first model for classifying normal and AMD OCT images, b heat map for the second model for classifying the presence or absence of any fluid. CAM was able to identify characteristic areas on the OCT, and it is presented as a heat map. CAM class activation mapping, AMD age-related macular degeneration, CNN convolution neural network, OCT optical coherence tomography
Fig. 5.
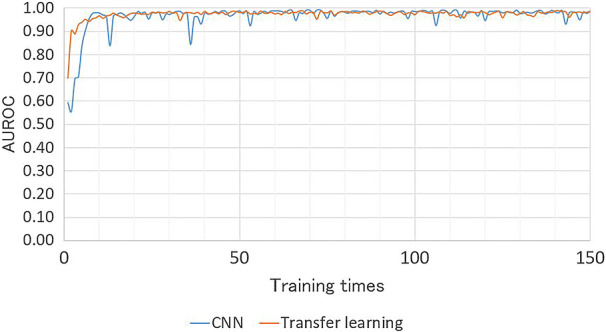
Comparison of the necessary number of epochs for convergence of the training loss, and classification performance, between the transfer-learning model and the CNN of the same architecture without transfer learning. In the second model, for classification of AMD images into with or without exudative changes, learning stabilized faster when using transfer learning. CNN convolution neural network, AMD age-related macular degeneration, AUROC area under the receiver operating characteristic curve
Discussion
In this study, we built two deep-learning models and evaluated their performance in either distinguishing normal and AMD eyes on OCT images, or classifying AMD into the presence or absence of exudative changes.
Using the first model, we could classify AMD and normal OCT images with 100% sensitivity, 91.8% specificity, and 99.0% accuracy. Recent studies have also reported that deep-learning techniques have achieved a high accuracy in identifying AMD from normal eyes using fundus photographs and OCT images. Our results are consistent with those of recent reports [24, 26]. Lee et al. reported that a CNN trained on about 80,000 OCT images could classify normal and AMD images with 92.64% sensitivity, 93.69% specificity, with an AUROC of 97.46% [24]. Treder et al. reported building a CNN model in which all layers except the last layer were pretrained with ImageNet data consisting of more than 1.2 million non-OCT images; the last layer was trained with 1012 cross-sectional SD-OCT scans. This model was able to distinguish normal from AMD images with 100% sensitivity, 92% specificity, and 96% accuracy [26]. As the sensitivity was as high as 100%, it was considered to be useful for a screening test. Classifying AMD-affected eyes from normal eyes in screening tests can be useful for preventive healthcare and ensuring appropriate treatment [32, 33]. Our results add to the evidence that CNNs are able to distinguish between disease images and normal images with high accuracy.
Using the second model, we were able to classify AMD according to the presence or absence of any fluid, with 98.4% sensitivity, 88.3% specificity, and 93.9% accuracy. Identification of IRF and SRF is a key indication for commencing anti-VEGF treatment administration and re-dosing [12]. Moreover, IRF and SRF have been reported as prognostic factors for visual impairment [34]. Recent reports have shown that deep-learning methods can accurately quantify the amount of fluid and can clearly distinguish between IRF and SRF [35]. Previous reports about automatic classification of exudative changes used a segmentation algorithm [21, 29]. Chakravarthy et al. reported that their automated segmentation algorithm could distinguish between IRF and SRF in AMD OCT scans with 92% sensitivity, 91% specificity, and 93% accuracy [21]. Our results are consistent with their report. We built deep-learning models without a segmentation algorithm using transfer learning and a method of dividing an original image into three cropped images without reducing the image pixels. In general, reducing the image resolution makes the classification inaccurate. There have been other reports of deep learning that do not use a segmentation algorithm of AMD, but they included classifications of AMD, drusen, and normal, and the classification purpose was different [27]. It is increasingly being reported that deep-learning models are as accurate as ophthalmology experts [18]. Meta-analyses have shown that active treatment can typically maintain visual function, although in some cases, there is a risk of loss of vision, despite long-term treatment. Starting treatment while eyesight is still good leads to a good prognosis; thus, early detection of exudative changes and early treatment are important for maximizing visual acuity, and deep-learning models to distinguish medical images are likely to be used more in the future. Telemedicine is becoming a more frequently used method of increasing the opportunities to see patients who have difficulty in attending clinical institutions, and thereby can prevent severe chronic diseases, such as AMD. Advances in OCT technology have yielded several scan-pattern OCT images, increasing the amount of OCT data that needs to be analyzed to the extent that it exceeds clinical capacity. Therefore, image analysis by artificial intelligence (AI) using deep learning is likely to contribute markedly to the field of medicine in the future.
CNNs are neural networks with many layers that demonstrate particularly excellent performance in the field of image recognition [17]. A CNN contains a repeated structure of convolution layers, which is responsible for local feature extraction of the image, and a pooling layer (subsampling layer) which summarizes the features of each area. The number of parameters is significantly reduced by CNNs, as compared to a simple all-coupled network, because the parameters of the convolution filter are shared in all locations in the image. Weight is adjusted so as to eliminate the difference calculated from the supervised data and the input image. In our second model, we applied transfer learning, which is an efficient technique for retraining a previously constructed CNN; and most unit weights are pretrained. The efficiency of training a CNN with transfer learning is high, and in most cases, stable classification performance can be achieved more rapidly when including transfer learning than when training a CNN without transfer learning. In our second model using transfer learning, we could obtain a highly accurate result with a smaller dataset. This is therefore particularly useful when it is difficult to amass image data, such as in cases of rare diseases.
We found that CAM was able to identify the characteristic area on the OCT image and represent it as a heat map. In essence, the purpose of CAM is to specify the part of the image with a large effect on the probability score for each class by averaging the differential coefficients. It quantifies changes in a certainty degree, as a heat map, and thereby highlights important areas. Therefore, it is possible to confirm the area of interest on the image by using CAM. When many OCT examination images obtained by frequent examinations and long-term follow-up need to be analyzed, AI judgment making use of a heat map can be used to both shorten the analysis time and prevent overlooking, and as a training mechanism for non-experts. In the future, AMD clinics may make use of real-time automated detection of many AMD findings.
Our study had several limitations. We used only images from patients who met our inclusion criteria, and the neural network was trained only on these images. We excluded low-quality images and patients who had other concomitant diseases. It is necessary to implement image classification by adding a process of determining good image quality [36] to ensure utility in future clinical practice, such as telemedicine. In this case, only OCT images passing through the fovea of the cross scan and radial scan were used. However, to ensure utility for classification of diseases in clinical practice, it is necessary to develop machine learning, including OCT volume scanning, more widely We also excluded images on which the three ophthalmologists did not agree regarding the presence or absence of SRF or IRF. Therefore, it is unclear whether our results can be applied in general. In addition, our models were trained using images from a single OCT device at a single academic center. In the future, images from various OCT devices at multiple facilities should be included, and it is desirable to develop deep learning from more images to ensure reliability.
Conclusion
In this study, we developed and evaluated reliable computational deep-learning models for distinguishing AMD from normal OCT images, and for classifying AMD according to the presence or absence of exudative changes without a segmentation algorithm. In this era, where treatment of AMD is decided based on interpretation of OCT images, it is important to maximize the capacity to process the increasing number of images from the growing number of patients. Automation of the interpretation process by using deep-learning models can improve cost and time efficiency.
Acknowledgements
We thank the participants of the study.
Funding
No funding or sponsorship was received for this study or publication of this article. The Department of Ophthalmology of Kobe City Eye Hospital are funding the Rapid Service Fees.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Author Contributions
Concept and design: Naohiro Motozawa, Guangzhou An, Seiji Takagi and Michiko Mandai; data acquisition: Naohiro Motozawa, Guangzhou An and Seiji Takagi; data analysis/interpretation: Naohiro Motozawa, Guangzhou An, Michiko Mandai and Yasuhiko Hirami; drafting manuscript: Naohiro Motozawa, Guangzhuo An and Seiji Takagi; supervision: Naohiro Motozawa, Guangzhou An, Seiji Takagi, Shohei Kitahata, Michiko Mandai, Yasuhiko Hirami, Hideo Yokota, Masahiro Akiba and Yasuo Kurimoto; All authors read and approved the final manuscript.
Disclosures
Naohiro Motozawa, Guangzhou An, Seiji Takagi, Shohei Kitahata, Michiko Mandai, Yasuhiko Hirami, Hideo Yokota, Masahiro Akiba, Akitaka Tsujikawa, Masayo Takahashi, and Yasuo Kurimoto have nothing to declare.
Compliance with Ethics Guidelines
This research was conducted in line with the Helsinki Declaration of 1964, and the research protocols and their implementation were approved by the ethics committee of Kobe City Medical Center General Hospital (approval date; Sep 17th, 2015 and approval number: No15073). The committee waived the requirement for obtaining informed consent given that this was a retrospective observational study of medical records, and retrospectively registered.
Data Availability
The datasets during collected and/or analyzed during the current study are available from the corresponding author on reasonable request.
Footnotes
Enhanced Digital Features
To view enhanced digital features for this article, go to 10.6084/m9.figshare.8977010.
References
- 1.Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY. Age-related macular degeneration. Lancet. 2012;379:1728–1738. doi: 10.1016/S0140-6736(12)60282-7. [DOI] [PubMed] [Google Scholar]
- 2.Klein R, Klein BE, Tomany SC, Meuer SM, Huang GH. Ten-year incidence and progression of age-related maculopathy: the Beaver Dam Eye study. Ophthalmology. 2002;109:1767–1779. doi: 10.1016/S0161-6420(02)01146-6. [DOI] [PubMed] [Google Scholar]
- 3.Wang JJ, Rochtchina E, Lee AJ, et al. Ten-year incidence and progression of age-related maculopathy. Ophthalmology. 2007;114:92–98. doi: 10.1016/j.ophtha.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Bird AC, Bressler NM, Bressler SB, et al. An international classification and grading system for age-related maculopathy and age-related macular degeneration. Surv Ophthalmol. 1995;39:367–374. doi: 10.1016/S0039-6257(05)80092-X. [DOI] [PubMed] [Google Scholar]
- 5.Bhutto I, Lutty G. Understanding age-related macular degeneration (AMD): relationships between the photoreceptor/retinal pigment epithelium/Bruch’s membrane/choriocapillaris complex. Mol Aspects Med. 2012;33:295–317. doi: 10.1016/j.mam.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown DM, Soubrane G, Schneider S. Ranibizumab versus verteporfin for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1432–1444. doi: 10.1056/NEJMoa062655. [DOI] [PubMed] [Google Scholar]
- 7.Oishi A, Kojima H, Mandai M, et al. Comparison of the effect of ranibizumab and verteporfin for polypoidal choroidal vasculopathy: 12-month LAPTOP study results. Am J Ophthalmol. 2013;156:644–651.e1. doi: 10.1016/j.ajo.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 8.Heier JS, Brown DM, Chong V, et al. Intravitreal aflibercept (VEGF trap-eye) in wet age-related macular degeneration. Ophthalmology. 2012;119:2537–2548. doi: 10.1016/j.ophtha.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Tufail A, Patel PJ, Egan C, et al. Bevacizumab for neovascular age related macular degeneration (ABC Trial): multicentre randomised double masked study. BMJ. 2010;340:c2459. doi: 10.1136/bmj.c2459. [DOI] [PubMed] [Google Scholar]
- 10.Agarwal A, Aggarwal K, Gupta V. Management of neovascular age-related macular degeneration: a review on landmark randomized controlled trials. Middle East Afr J Ophthalmol. 2016;23:27–37. doi: 10.4103/0974-9233.173133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rofagha S, Bhisitkul RB, Boyer DS, Sadda SR, Zhang K. Seven-year outcomes in ranibizumab-treated patients in ANCHOR, MARINA, and HORIZON: a multicenter cohort study (SEVEN-UP) Ophthalmology. 2013;120:2292–2299. doi: 10.1016/j.ophtha.2013.03.046. [DOI] [PubMed] [Google Scholar]
- 12.Regatieri CV, Branchini L, Duker JS. The role of spectral-domain OCT in the diagnosis and management of neovascular age-related macular degeneration. Ophthalm Surg Lasers Imaging. 2011;42:S56–S66. doi: 10.3928/15428877-20110627-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fung AE, Lalwani GA, Rosenfeld PJ, et al. An optical coherence tomography-guided, variable dosing regimen with intravitreal ranibizumab (Lucentis) for neovascular age-related macular degeneration. Am J Ophthalmol. 2007;143(566–583):e2. doi: 10.1016/j.ajo.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 14.Freund K, Korobelnik J-F, Devenyi R, et al. Treat-and-extend regimens with anti-VEGF agents in retinal diseases. Retina. 2015;35:1489–1506. doi: 10.1097/IAE.0000000000000627. [DOI] [PubMed] [Google Scholar]
- 15.Alexandru MR, Alexandra NM. Wet age-related macular degeneration management and follow-up. Rom J Ophthalmol. 2016;60:9–13. [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang F, Jiang Y, Zhi H, et al. Artificial intelligence in healthcare: past, present and future. Stroke Vasc Neurol. 2017;2:230–243. doi: 10.1136/svn-2017-000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeCun Y, Bengio Y, Hinton G. Deep-learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt-Erfurth U, Sadeghipour A, Gerendas BS, Waldstein SM, Bogunović H. Artificial intelligence in retina. Prog Retinal Eye Res. 2018;67:1–29. doi: 10.1016/j.preteyeres.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Liu X, Jiang J, Zhang K, et al. Localization and diagnosis framework for pediatric cataracts based on slit-lamp images using deep features of a convolutional neural network. PLoS One. 2017;12:e0168606. doi: 10.1371/journal.pone.0168606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yousefi S, Kiwaki T, Zheng Y, et al. Detection of longitudinal visual field progression in glaucoma using machine learning. Am J Ophthalmol. 2018;193:71–79. doi: 10.1016/j.ajo.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Chakravarthy U, Goldenberg D, Young G, et al. Automated identification of lesion activity in neovascular age-related macular degeneration. Ophthalmology. 2016;123:1731–1736. doi: 10.1016/j.ophtha.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Venhuizen FG, van Ginneken B, van Asten F, et al. Automated staging of age-related macular degeneration using optical coherence tomography. Invest Ophthalmol Vis Sci. 2017;58:2318–2328. doi: 10.1167/iovs.16-20541. [DOI] [PubMed] [Google Scholar]
- 23.Burlina PM, Joshi N, Pekala M, Pacheco KD, Freund DE, Bressler NM. Automated grading of age-related macular degeneration from color fundus images using deep convolutional neural networks. JAMA Ophthalmol. 2017;135:1170–1176. doi: 10.1001/jamaophthalmol.2017.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee CS, Baughman DM, Lee AY. Deep-learning is effective for classifying normal versus age-related macular degeneration OCT images. Ophthalmol Retina. 2017;1:322–327. doi: 10.1016/j.oret.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burlina P, Pacheco KD, Joshi N, Freund DE, Bressler NM. Comparing humans and deep-learning performance for grading AMD: a study in using universal deep features and transfer learning for automated AMD analysis. Comput Biol Med. 2017;82:80–86. doi: 10.1016/j.compbiomed.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Treder M, Lauermann JL, Eter N. Automated detection of exudative age-related macular degeneration in spectral domain optical coherence tomography using deep-learning. Graefes Arch Clin Exp Ophthalmol. 2018;256:259–265. doi: 10.1007/s00417-017-3850-3. [DOI] [PubMed] [Google Scholar]
- 27.Kermany DS, Goldbaum M, Cai W, et al. Identifying medical diagnoses and treatable diseases by image-based deep-learning. Cell. 2018;172(1122–31):e9. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Fauw JD, Ledsam JR, Romera-Paredes B, et al. Clinically applicable deep-learning for diagnosis and referral in retinal disease. Nat Med. 2018;24:1342–1350. doi: 10.1038/s41591-018-0107-6. [DOI] [PubMed] [Google Scholar]
- 29.Lee H, Kang KE, Chung H, Kim HC. Automated segmentation of lesions including subretinal hyperreflective material in neovascular age-related macular degeneration. Am J Ophthalmol. 2018;191:64–75. doi: 10.1016/j.ajo.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Glorot X, Bordes A, Bengio Y. Deep sparse rectifier neural networks. Proc AISTATS. 2010;9:249–256. [Google Scholar]
- 31.Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv:150203167 [cs] [Internet]. 2015 Feb 10 [cited 2019 Feb 11]; http://arxiv.org/abs/1502.03167.
- 32.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning deep features for discriminative localization. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) [Internet]. Las Vegas, NV, USA: IEEE; 2016 [cited 2019 Feb 11]. p. 2921–9. http://ieeexplore.ieee.org/document/7780688/.
- 33.Age-Related Eye Disease Study Research Group A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss. Arch Ophthalmol. 2001;119:1417–1436. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oliver-Fernandez A, Bakal J, Segal S, Shah GK, Dugar A, Sharma S. Progression of visual loss and time between initial assessment and treatment of wet age-related macular degeneration. Can J Ophthalmol. 2005;40:313–319. doi: 10.1016/S0008-4182(05)80074-2. [DOI] [PubMed] [Google Scholar]
- 35.Waldstein SM, Philip A-M, Leitner R, et al. Correlation of 3-dimensionally quantified intraretinal and subretinal fluid with visual acuity in neovascular age-related macular degeneration. JAMA Ophthalmol. 2016;134:182–190. doi: 10.1001/jamaophthalmol.2015.4948. [DOI] [PubMed] [Google Scholar]
- 36.Schlegl T, Waldstein SM, Bogunovic H, et al. Fully automated detection and quantification of macular fluid in OCT using deep-learning. Ophthalmology. 2018;125:549–558. doi: 10.1016/j.ophtha.2017.10.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets during collected and/or analyzed during the current study are available from the corresponding author on reasonable request.