Abstract
Uridine diphosphate (UDP)-glycosyltransferases (UGTs) involved in many metabolic processes are ubiquitous in plants, animals, microorganisms and other organisms and are essential for their growth and development. Upland cotton contains a large number of UGT genes. In this study, we aimed to identify UGT family members in the genome of upland cotton (Gossypium hirsutum L.) and analyze their expression patterns. Bioinformatics methods were used to identify UGT genes from the whole genome of upland cotton (Gossypium hirsutum L. acc. TM-1). Phylogenetic analysis was conducted based on alignment of UGT proteins from upland cotton, and the gene structure, motif and chromosome localization were analyzed for the H subgroup of the UGT family. And the physical and chemical properties and expressions of the genes in the H subgroup of this family were also analyzed. A total of 274 UGT genes were identified from the whole genome of upland cotton and were divided into nine subgroups based on phylogenetic analyses. In subgroup H, 36 genes were distributed on 18 chromosomes. The subfamily genes were simple in the structure, 19 of its members contained two introns, and the others contained only one intron. The qRT-PCR results and transcriptomic data indicated that most of the genes had a wide range of tissue expression characteristics. And the phylogenetic analysis results and expression profiles of these genes revealed tissues and different UGT genes from this crop. Taking RNA-seq, RT-qPCR, and quantitative trait locus (QTL) mapping together, our results suggested that GhUGT6 and GhUGT105 in subgroup H of the GhUGT gene family could be potential candidate genes for cotton yield, and GhUGT16, GhUGT103 might play a vital role in fiber development.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-1984-1) contains supplementary material, which is available to authorized users.
Keywords: Gossypium hirsutum L., GhUGT, Phylogeny, Structure, Expression patterns, Fiber development
Introduction
UDP glycosyltransferases (UGTs) are abundant in plants, which play an indispensable role in plant growth, development, flowering and fruiting. By glycosylation, activated glycosides can develop into more stable and inactive storage forms. The attachment of hydrophilic glucose to hydrophobic glycoside ligands increases water solubility, and glycosylation of UGTs is the last step in the synthesis of natural products in plants (Jones and Vogt 2001; Lim and Bowles 2004; Bowles et al. 2006). Glycosylation is also a key step to detoxify exogenous substances for advanced plants (Jr 1992; Bowles et al. 2006). In addition, glycosylation plays an important biological role in promoting storage and intracellular transport in plants, and it is vital in regulating the balance of growth agents in plants (Gilbert et al. 2013).
Evolutionary analysis of UGTs in plants, animals, fungi, bacteria and viruses showed that UGTs in plants were unique and distinct branch. Plant UGTs were also to be related to synthesis of natural plant products, regulation of plant hormone homeostasis and detoxification of exogenous substances (Jones and Vogt 2001; Paquette et al. 2003; Lim and Bowles 2004). In the study of Mackenzie (Mackenzie et al. 1997) and Paquette (Paquette et al. 2009), a specific UGT sequence in plants was found, which was expressed as a plant secondary product glycosyltransferase (PSPG).
UGTs have been identified in a variety of plants, such as Chlamydomonas in the lowest class and grapes in a higher class (Yonekura-Sakakibara and Hanada 2011). Li identified more than 100 UGTs in Arabidopsis thaliana, which were classified into 14 subgroups according to their amino acid sequences (Li et al. 2001). Various UGTs in plants play an important role in natural metabolites; for example, UGT79B1 and UGT91A1 found in Arabidopsis play a critical role in regulating anthocyanin metabolism (Yonekurasakakibara et al. 2012). UGT genes were found to have similar functions in peaches (Cheng et al. 2014), kiwifruits (Montefiori et al. 2011) and strawberries (Song et al. 2016b). In addition, it was found in Lin’s research that two mutants ko-1 and ko-2, and two restorer lines RE-1 and RE-2, were obtained by inserting T-DNA into UGT72b1 genes in Arabidopsis thaliana (Lin et al. 2016). It was discovered that the mutants aggravated lignification exceptions, thickened secondary cell walls and promoted the binding of glucose to form lignin monomers in flower stems, thus impacting the growth and development of Arabidopsis thaliana. The results demonstrated that the height of the mutant-containing plant was significantly lower than that of the wild plant, the flowering period was later than that of the wild plant and restorer-line-containing plant, and anthocyanin accumulation was also promoted (Lin et al. 2016). It was also found that the diversity of bioactive flavonol glycosides was caused by catalytic modification of UGTs (Ono et al. 2010).
So far, there is no report on UGT genes in the UGT gene family in upland cotton. In this study, the whole genome of the UGT gene family in upland cotton was identified, and the prediction of subcellular localization, chromosome distribution, gene structure and expression level of the UGT genes in the H subgroup were analyzed. Our findings provided a foundation for genetic improvement of cotton fiber.
Materials and methods
Identification of UGT genes in Gossypium hirsutum L. and phylogenetic tree construction
To identify members of the UGT gene family in G. hirsutum, G. raimondii, G. arboreum, Therobroma cacao and Arabidopsis, UGT sequences were obtained from the TAIR database (http://www.arabidopsis.org) and used for BLASTP algorithm-based queries against the G. hirsutum genome database (https://www.cottongen.org/species/Gossypium_hirsutum/nbi-AD1_genome_v1.1)(Zhang et al. 2015). The UGT protein domain was analyzed using the Hidden Markov Model (HMM) from the Pfam database (http://pfam.xfam.org/). The PSPG domains were confirmed by Pfam accession number PF00201. And the sequences of all the above proteins were aligned using MAFFT sequence alignment (https://www.ebi.ac.uk/Tools/msa/mafft/) with default parameter settings. The sequence alignment results were obtained using the Gblock (http://molevol.cmima.csic.es/castresana/Gblocks_server.html) to acquire their conservative areas, and the parameters were set to allow smaller final regions, more stringent flank sites and a vacancy in the final area. Finally, the conserved region sequences of the UGT proteins were imported into the online version of PhyML 3.0 (http://phylogeny.lirmm.fr/) provided by the LIRMM laboratory (CNRS–LIRMM) for phylogenetic tree construction. The likelihood ratio test adopted the SH-like method, and the LG model was selected as the substitution model. The result of the phylogenetic tree was visualized by MEGA7.0 and iTOL V4 (http://itol.embl.de/).
Analysis of the Exon/Intron Structure, Motif, Subcellular Localization and Chromosomal Location of the UGT Genes in the H Subfamily in Gossypium hirsutum L.
The exon/intron structures of the H subfamily genes were retrieved according to the GFF annotation file information of Gossypium hirsutum L. using the gene structure display server (GSDS) program (http://gsds.cbi.pku.edu.cn/) (Guo et al. 2007).
The online program of MEME (http://meme-suite.org/) (Bailey et al. 2009) was employed to determine the conserved motifs of GhUGTs with the following optimum parameters: a motif width of 6–50 amino acids and a maximum of 10 motifs. The identified motifs were annotated using the program InterProScan (Quevillon et al. 2005).
The subcellular localization analyzed and predicted localization of large number of proteins by WoLF PSORT: Protein Subcellular Localization Prediction (https://wolfpsort.hgc.jp/).
The chromosomal distribution of the UGT genes was obtained based on the annotation data of the Gossypium hirsutum L. genome. The MapInspect software was used to plot images of their physical locations in G. hirsutum (Ren et al. 2017).
Gene expression analysis
The expressions of the UGT family genes were measured by RNA-sequencing. The raw RNA-sequencing data of CCRI45, and fiber transcriptomic data of excellent fiber quality CCSLs (7747 and 7561) and low-fiber-quality CCSLs 7285 were obtained from the laboratory in seven different periods (5d, 7d, 10d, 15d, 20d, 25d and 28d) (Lu et al. 2017). Raw RNA-sequencing data of six different tissues (root, stem, leave, petal, calycle and ovule) of G. hirsutum TM-1 were downloaded from the NCBI Gene Expression repository under the accession number PRJNA248163 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA248163/).
The relative data were normalized to calculate the expression levels. Hierarchical clustering was performed using Genesis 1.7.7 (Sturn et al. 2002).
RNA Isolation and qRT-PCR analysis
Gossypium hirsutum L. (cv CCRI45) and CCSLs 7747 were cultivated in the field in the experimental farm of the Institute of Cotton Research of Chinese Academy of Agricultural Sciences (ICR–CAAS), Anyang, China. Fibers of seven different periods including 5 days post-anthesis (5DPA), 7DPA, 10DPA, 15DPA, 20DPA, 25DPA and 28DPA and six different tissue parts including the root, stem, leave, petal, calycle and ovule (20DPA) were separately sampled at the full bloom stage of flowers. All samples were immediately frozen in liquid nitrogen and kept at − 80 °C for total RNA extraction.
Total RNA was extracted from each sample using the RNA prep Pure Plant Kit (DP441, TIANGEN, Beijing, China). Gel electrophoresis and a Nanodrop2000 nucleic acid analyzer were employed to measure RNA quality. The cDNAs were synthesized using a TranScript All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (Transgen Biotech, Beijing, China). qRT-PCR was carried out following the protocol of the TransStart Top Green qPCR SuperMix kit (Transgen Biotech, Beijing, China) on the ABI 7500 Fast Real-Time PCR system (Applied Biosystems, USA). The specific primers of these differentially expressed genes (DEGs) were designed by Primer-BLAST using the online NCBI database and are listed in Supplementary Table S1. The housekeeping β-Actin gene was used as the reference to normalize the relative expression levels, with its primer sequences of F: 5′-ATCCTCCGTCTTGACCTTG-3′ and R: 5′-TGTCCGTCAGGCAACTCAT-3′. qRT-PCR was carried out on a 20-μL system in the following conditions: one cycle of 94 °C for 30 s; 40 cycles of 94 °C for 5 s and 60 °C for 34 s, and one cycle of 60 °C for 60 s. Three biological and technical replicates were performed to validate the results of the qRT-PCR tests. The relative gene expression levels were quantified by the 2− ΔΔCt method (Livak and Schmittgen 2001).
Mapping subgroup H genes in QTL intervals
Based on the previous studies of QTL mapping and physical locations in the upland cotton genome, the physical locations of related QTLs and the genes in the QTL region were confirmed by comparison, which were compared to the subgroup H genes.
Results
Identification of UGT genes in Gossypium hirsutum L., phylogenetic tree construction and phylogenetic analysis of GOSSYPIUM hirsutum L. UGTs
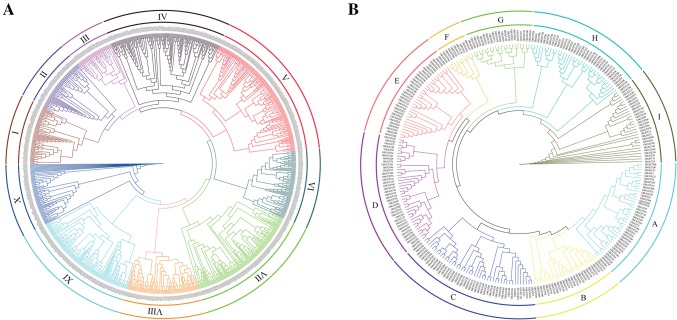
A total of 274 G. hirsutum UGTs in the lengths of 61–1115 amino acids were identified in upland cotton. A phylogenetic tree of upland cotton UGT genes was constructed by aligning full-length amino acid sequences of upland cotton UGTs with functionally characterized plant UGTs, including G. raimondii (151), G. arboreum (150), Arabidopsis (114) and Therobroma cacao (159) (Fig. 1a). The G. hirsutum UGTs were phylogenetically divided into 10 groups (I–X), including almost of all genes that were identified in Arabidopsis (Li et al. 2001). Distribution of the plant UGTs in the phylogenetic groups is summarized in Table 1.
Fig. 1.
a Phylogenetic analysis of the UGT genes in Arabidopsis (AtUGT), G. raimondii (GrUGT), G. arboreum (GaUGT), G. hirsutum (GhUGT) and Therobroma cacao (ThUGT). B. Phylogenetic analysis of the UGTs in G. hirsutum (GhUGT)
Table 1.
Number of the plant UGTs in the different phylogenetic groups
| UGT group | Arabidopsis thaliana | G. hirsutum | G. arboreum | G. raimondii | Therobroma cacao |
|---|---|---|---|---|---|
| I | 7 | 20 | 8 | 11 | 15 |
| II | 10 | 18 | 6 | 12 | 11 |
| III | 6 | 8 | 11 | 6 | 9 |
| IV | 14 | 41 | 22 | 20 | 17 |
| V | 25 | 38 | 24 | 27 | 25 |
| VI | 6 | 24 | 13 | 12 | 17 |
| VII | 13 | 37 | 22 | 21 | 24 |
| VIII | 8 | 27 | 14 | 13 | 12 |
| IX | 16 | 41 | 19 | 18 | 15 |
| X | 9 | 20 | 11 | 11 | 14 |
| Total | 114 | 274 | 150 | 151 | 159 |
To further analyze the evolutionary relationship of the UGT gene family in G. hirsutum, the study built a phylogenetic tree containing only the UGT gene family in upland cotton (Fig. 1b). This tree was phylogenetically divided into nine groups (A–I). Among them, the C subgroup has the largest number of members (44 genes) and the F subgroup contains only ten members. In this research, the study mainly aimed at the H subfamily of the UGT family in upland cotton.
Chromosomal distribution of GhUGT genes
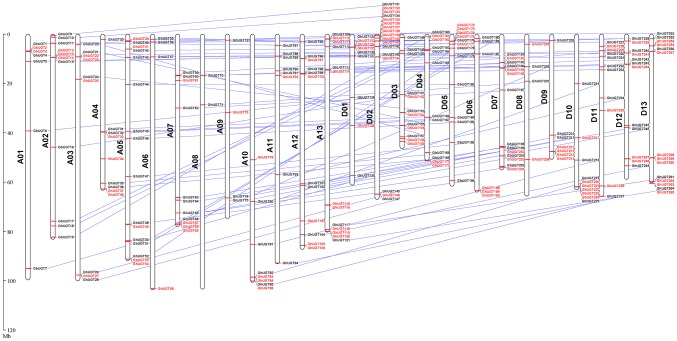
Among all 274 members, 255 UGT genes were physically located on the 13A chromosomes and 13D chromosomes among the 26 G. hirsutum chromosomes (Fig. 2). The remaining 19 were mapped to the scaffold. Most genes were located on D02 (seventeen genes), followed by A05, which had sixteen genes. D13 and D07 had fourteen genes, A06, A09 and D08 had four genes, and only two genes were mapped to A08. The H subfamily of the UGTs had 36 genes, which were distributed on 18 chromosomes. The A and D subgroups had 18 and 17 genes, respectively, and one gene was located on the scaffold. The genes were mostly located on A05, A07 and D04 (four genes on each), followed by A12 and D02 (three genes on each). No genes were located on A03, A04, A09, A11, A13, D02, D09 and D13. Only one gene was mapped to the other chromosomes.
Fig. 2.
Chromosomal locations and collinearity analysis of expansion genes of the UGT gene family in upland cotton. Red and black are both positioning results, and a line indicates that there is a collinearity between two genes
The UGT gene family members were compared by BlastP among the cotton genomes of G. raimondii, G. arboreum and G. hirsutum. We used MCScanX (Kumar et al. 2018) to identify homologous gene pairs and the result was presented by CirCos (Krzywinski et al. 2009). There were 16 UGT homologous pairs between G. arboreum and G. hirsutum. These homologous genes in G. hirsutum were distributed in A02 (two pairs), A04 (one pair), A07 (two pairs), A08 (three pairs), A09 (four pairs), A11 (one pair), and A12 (four pairs). And there were 17 UGT homologous pairs between G. raimondii and G. hirsutum. These homologous genes in G. hirsutum were distributed in D03 (one pair), D04 (one pair), D07 (two pairs), D08 (four pairs), D09 (four pairs), D11 (one pair), and D12 (four pairs). The collinear relationships between G. raimondii, G. arboreum and G. hirsutum are shown in Fig. 3.
Fig. 3.
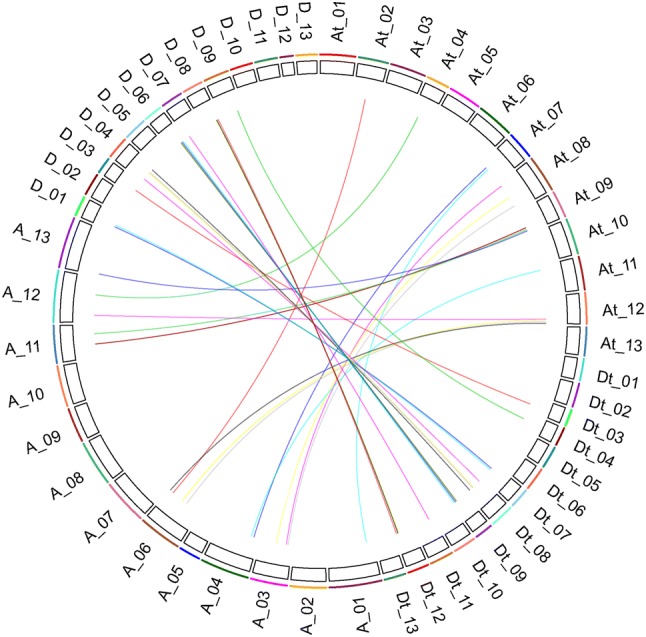
Homology analysis of expansion genes between G. hirsutum, G. raimondii and G. arboreum. a and d Showed the G. hirsutum’s chromosome, At was on behalf of G. arboretum’s chromosomes, and Dt represented G. raimondii’s chromosomes
Information of the H subfamily of the UGT family in upland cotton and its protein physico-chemical and biochemical characteristics
In the H subfamily, the coding sequence lengths of these genes were between 303 bp (GhUGT272) and 1509 bp (GhUGT105), and the average length was 1300 bp; they coded 109 (GhUGT272) to 502 (GhUGT105) amino acid molecules, with an average value of 432.5; their relative molecular masses were between 11823.34 (GhUGT272) and 55,345.47 (GhUGT105), and the average was 48,005.98; the isoelectric points of these proteins ranged between 5.49 (GhUGT53) and 8.96 (GhUGT272), and the mean was 6.52. Subcellular localization is a key characteristic for protein function research. The subcellular localization prediction of the H subfamily showed that most of the proteins were located on the cell membrane and chloroplast, four were located in the cytoplasm and three proteins were in the nucleus. All proteins in this subfamily did not have signal peptides, and only GhUGT6 and GhUGT103 had trans-membrane domains. They are demonstrated in Table 2 and the physico-chemical properties of all UGT proteins in this family in upland cotton are shown in Supplementary Table S2.
Table 2.
Physico-chemical and biochemical characteristics of the H subfamily
| H group number | Gene ID | Chromosome | Chromosome location | CDS length/bp | Protein | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Length/aa | pI | MW/Mr | SP | TM | Subcellular locatin | |||||
| GhUGT6 | Gh_A01G1073 | A01(−) | 39,143,032–39,144,431 | 1377 | 458 | 8.22 | 52,308.4 | × | √ | Cell membrane, Chloroplast |
| GhUGT7 | Gh_A01G1652 | A01(−) | 94,938,092–94,939,474 | 1383 | 460 | 6.13 | 51,544.93 | × | × | Cell membrane |
| GhUGT16 | Gh_A02G1034 | A02(−) | 45,732,158–45,733,447 | 1290 | 429 | 6.02 | 47,612.01 | × | × | Chloroplast |
| GhUGT18 | Gh_A02G1341 | A02(−) | 77,646,919–77,648,591 | 1362 | 453 | 5.97 | 49,431.93 | × | × | Cell membrane, Chloroplast |
| GhUGT50 | Gh_A05G3195 | A05(−) | 83,670,360–83,671,790 | 1431 | 476 | 6.24 | 51,711.21 | × | × | Chloroplast |
| GhUGT52 | Gh_A05G3535 | A05(+) | 91,070,553–91,072,167 | 1341 | 446 | 7.18 | 49,297.84 | × | × | Cell membrane, Chloroplast |
| GhUGT53 | Gh_A05G3536 | A05(+) | 91,074,815–91,076,896 | 1035 | 344 | 5.49 | 37,972.31 | × | × | Chloroplast, Cytoplasm, Nucleus |
| GhUGT54 | Gh_A05G3537 | A05(+) | 91,084,484–91,086,802 | 1362 | 453 | 6.02 | 49,915.26 | × | × | Cell membrane, Chloroplast |
| GhUGT57 | Gh_A06G0476 | A06(−) | 9,142,093–9,143,635 | 1350 | 449 | 5.85 | 49,587.85 | × | × | Chloroplast, Cytoplasm |
| GhUGT61 | Gh_A07G0980 | A07(+) | 18,622,020–18,623,152 | 915 | 304 | 6.45 | 33,850.28 | × | × | Chloroplast |
| GhUGT66 | Gh_A07G2017 | A07(+) | 76,443,883–76,445,300 | 1200 | 399 | 5.88 | 44,687.12 | × | × | Chloroplast, Cytoplasm |
| GhUGT67 | Gh_A07G2018 | A07(+) | 76,456,977–76,458,844 | 1449 | 482 | 6.18 | 53,853.94 | × | × | Cell membrane, Chloroplast |
| GhUGT68 | Gh_A07G2019 | A07(+) | 76,482,343–76,483,761 | 1419 | 472 | 6.18 | 52,607.6 | × | × | Cell membrane, Chloroplast, Nucleus |
| GhUGT71 | Gh_A08G0769 | A08(−) | 28,533,116–28,535,751 | 1368 | 455 | 6.93 | 50,476.79 | × | × | Cell membrane, Chloroplast |
| GhUGT86 | Gh_A10G2112 | A10(−) | 99,949,243–99,950,630 | 1365 | 454 | 8.36 | 51,538.65 | × | × | Cell membrane |
| GhUGT103 | Gh_A12G1553 | A12(+) | 75,599,992–75,601,334 | 1290 | 429 | 5.71 | 48,080.43 | × | √ | Chloroplast |
| GhUGT105 | Gh_A12G2287 | A12(−) | 85,570,104–85,571,612 | 1509 | 502 | 6.09 | 55,345.47 | × | × | Cell membrane, Chloroplast |
| GhUGT106 | Gh_A12G2288 | A12(−) | 85,574,775–85,576,145 | 1371 | 456 | 5.53 | 50,372.39 | × | × | Cell membrane, Chloroplast |
| GhUGT128 | Gh_D01G1155 | D01(−) | 25,559,353–25,560,753 | 1401 | 466 | 6.85 | 52,956.21 | × | × | Cell membrane, Chloroplast |
| GhUGT130 | Gh_D01G1899 | D01(−) | 57,203,897–57,205,279 | 1383 | 460 | 6.55 | 51,676.09 | × | × | Cell membrane |
| GhUGT150 | Gh_D03G0476 | D03(−) | 7,798,230–7,800,199 | 1362 | 453 | 5.97 | 49,345.84 | × | × | Chloroplast |
| GhUGT151 | Gh_D03G0695 | D03(+) | 24,354,545–24,355,987 | 1443 | 480 | 6.51 | 53,214.82 | × | × | Cell membrane, Chloroplast |
| GhUGT152 | Gh_D03G0696 | D03(+) | 24,357,294–24,357,929 | 636 | 211 | 7.7 | 23,765.57 | × | × | Chloroplast |
| GhUGT160 | Gh_D04G0070 | D04(−) | 974,261–976,657 | 1353 | 450 | 6.12 | 49,373.67 | × | × | Chloroplast |
| GhUGT161 | Gh_D04G0071 | D04(−) | 984,331–986,797 | 1365 | 454 | 7.67 | 50,208.68 | × | × | Cell membrane, Chloroplast |
| GhUGT162 | Gh_D04G0072 | D04(−) | 992,432–994,273 | 1365 | 454 | 7.66 | 50,035.55 | × | × | Cell membrane, Chloroplast |
| GhUGT164 | Gh_D04G0410 | D04(+) | 6,473,721–6,475,151 | 1431 | 476 | 6.68 | 51,665.17 | × | × | Chloroplast |
| GhUGT177 | Gh_D05G0494 | D05(−) | 3,975,820–3,977,211 | 1392 | 463 | 6.35 | 50,856.63 | × | × | Cell membrane, Chloroplast |
| GhUGT187 | Gh_D06G0522 | D06(−) | 7,937,638–7,939,188 | 1350 | 449 | 6.09 | 49,512.87 | × | × | Cytoplasm |
| GhUGT203 | Gh_D07G2238 | D07(+) | 53,452,297–53,453,715 | 1308 | 435 | 5.83 | 48,470.88 | × | × | Chloroplast |
| GhUGT207 | Gh_D08G0915 | D08(−) | 18,968,845–18,971,453 | 1368 | 455 | 7.72 | 50,563.96 | × | × | Cell membrane, Chloroplast |
| GhUGT226 | Gh_D10G2487 | scaffold4399_D10(+) | 18,141–19,430 | 1290 | 429 | 6.35 | 48,787.25 | × | × | Cell membrane, Chloroplast |
| GhUGT236 | Gh_D11G3004 | D11(+) | 61,450,780–61,452,198 | 1419 | 472 | 5.75 | 52,711.69 | × | × | Chloroplast |
| GhUGT250 | Gh_D12G2681 | scaffold4576_D12(−) | 159,855–161,288 | 1434 | 477 | 5.55 | 52,596.12 | × | × | Cell membrane, Chloroplast |
| GhUGT251 | Gh_D12G2682 | scaffold4576_D12(−) | 164,564–165,934 | 1371 | 456 | 5.97 | 50,456.43 | × | × | Cell membrane, Chloroplast |
| GhUGT272 | Gh_Sca144839G01 | scaffold144839(+) | 24–353 | 330 | 109 | 8.96 | 11,823.34 | × | × | Cell membrane, Nucleus |
pI isoelectric point, MW molecular weight (mass), SP signal peptide, TM trans-membrane
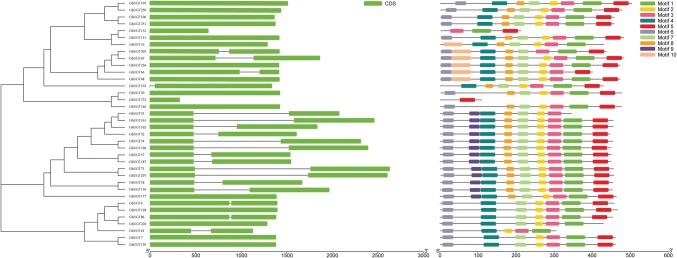
Gene structure and protein motif analysis of subgroup H
The UGT gene structure helps to determine phylogenetic relationships. Within the same subfamily, most members have similar exon and intron quantities, and they have important sequence characteristics, indicating that they have very close evolutionary relationships. The most important differences lay in the exon–intron lengths (Fig. 4). In subgroup H, there were 36 members, among which 17 genes contained only one intron and 19 genes had two introns.
Fig. 4.
Phylogenetic relationships and structures of the genes in the H subfamily of the UGT gene family in Gossypium hirsutum L.
Protein motif analysis of the H subfamily in the UGT gene family in G. hirsutum L. showed that almost each subfamily member possessed the same or similar number of motifs and order of arrangement (Fig. 4). Almost all members had eight motifs and the same arrangement order. Among them, 12 members had nine identical and same-order motifs, five had eight identical and same-order motifs, five had seven identical and same-order motifs, and only one member had one motif, demonstrating that they had a very close evolutionary relationship. The gene structure and motif analysis of all members of the UGT gene family in upland cotton was shown in Supplementary Fig. S1.
Expression pattern analyses of the genes in subgroup H
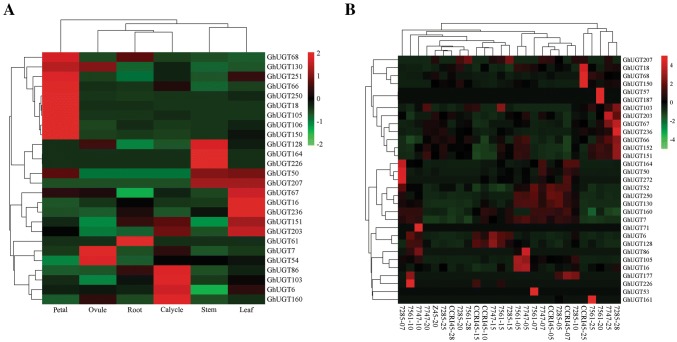
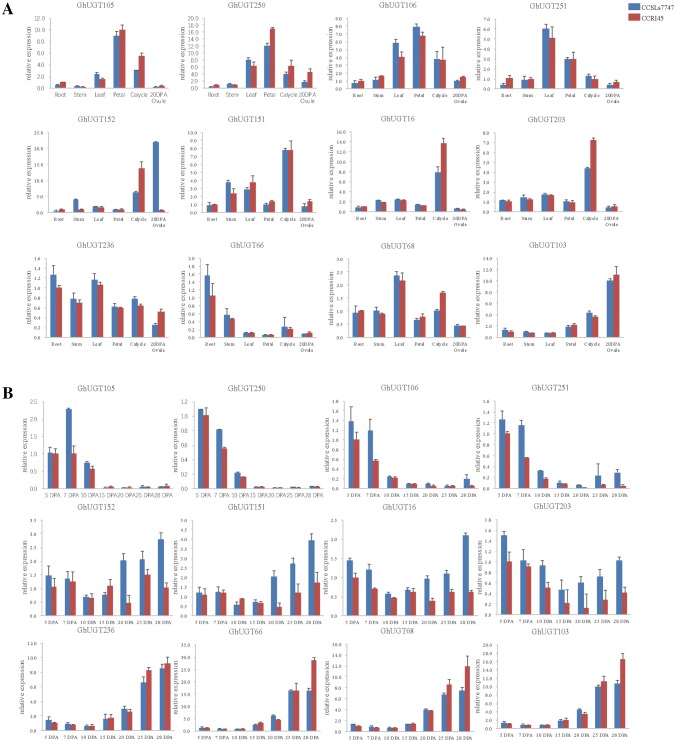
To explore the expression patterns of the genes in subgroup H of the UGT family in G. hirsutum-specific developmental processes, the expression profiles of 36 genes were detected in six different tissues (root, stem, leaf, petal, calycle, and ovule) by transcriptome sequencing (Fig. 5a) and the expression patterns of fibers in seven different periods (5d, 7d, 10d, 15d, 20d, 25d, and 28d) were obtained from the transcriptomic data of fibers in the laboratory (Fig. 5b) (Lu et al. 2017).The heat map revealed similar expression patterns of different genes within subfamily H.
Fig. 5.
Expression levels of flax UGT genes in various tissues in transcriptomic data. The robust multi-array average (RMA)-normalized, average log2 signal values of upland cotton UGTs in various tissues and fiber developmental stages (listed at the top of the heat map) were used for the construction of the heat map. The hierarchical clustering results based on Pearson correlation matrix were shown on the left and top of the heat maps
Currently, qRT-PCR has been proven to be the most accurate method for detecting differential gene expressions. To validate the participation of UGT genes in regulating flowering, we selected 16 genes from a small cluster of orthologous genes in subgroup H to test their expressions in six different tissues (root, stem, leaf, petal, calycle, and 20 DPA ovule) by qRT-PCR, which covered almost all plant parts (Fig. 6a) and seven different periods (5d, 7d, 10d, 15d, 20d, 25d and 28d) in the entire fiber development stage in G. hirsutum L. cv CCRI45 and CCSLs 7747 (Fig. 6b).
Fig. 6.
Validation of RNA-seq data by qRT-PCR. a Relative transcript abundance of each gene in comparison with the root. b Relative transcript abundance of each gene in comparison with the gene of CCRI45′s 5 DPA fiber
A total of 12 genes could be detected by qRT-PCR, and the results showed that they had similar expression patterns. These genes had a wide range of tissue expression characteristics. GhUGT105, GhUGT250 and GhUGT106 had a higher expression level in petal than those in other different tissues, they showed preferential expression in flowers and were likely to involve in the regulation of flower development. The GhUGT152 and GhUGT103 had a higher expression level in 20 day ovule than those in other different tissues.
The expression levels of GhUGT105, GhUGT250, GhUGT106 and GhUGT251 showed a continuous decrease trend from 5 DPA to 15 DPA, and the expression levels of these genes in CSSLs 7747 were higher than those in CCRI 45, In addition, these genes had low expression levels in the other periods, which suggested these four genes may be related to fiber elongation. The expression levels of the other four genes of GhUGT152, GhUGT151, GhUGT16 and GhUGT203 showed a decrease trend until 15 DPA, and then gradually increased, and the expression of these genes in CCSLs7747 was higher than that in CCRI45, which suggested that the four genes may be involved in the development of cotton fibers. Yet the other four genes of GhUGT236, GhUGT66, GhUGT68 and GhUGT103 had higher expression in the later fiber development stages of secondary wall biosynthesis. The expression levels of most genes among these 12 genes from 5 DPA to 28 DPA implied these genes might have specific relationships with fiber development. Their expression was also supported by the expression results of transcriptome sequencing.
The other four genes of GhUGT67, GhUGT50, GhUGT164 and GhUGT272 were not detected in different tissues and fibers, and their expressions were very low in transcriptome sequencing. In short, RNA-seq analysis results were basically consistent with the qRT-PCR results for the 12 genes, which indicated the reliability and accuracy of the two analytical methods, and the potential functions of GhUGT genes in fiber development.
Mapping subgroup H genes in QTL intervals
A total of nine subgroup H genes were located in the previous QTL interval, of which seven genes were located in the QTL interval associated with fiber quality, three genes were located in the QTL interval associated with the yield, and one gene was located in the QTL region for both fiber quality and yield.
Discussion
Recent studies involving functional characterization of plant UGTs suggested their important roles in growth, development and interaction with the environment. They played an essential role in regulating glucose metabolism and homeostasis as well as participating in detoxification of xenobiotics, and they were also important in biosynthesis, storage and transport properties of secondary metabolites (Wu et al. 2017). The UGT multigene family had been identified in plant species including Arabidopsis (Caputi et al. 2012), flax (Barvkar 2012), rice (Moon et al. 2013) and fruit species such as grapes (Bönisch et al. 2014), kiwifruits (Yauk et al. 2015), strawberries (Song et al. 2016a) and peaches (Wu et al. 2017).
Although the roles of many UGTs still remained unknown, a thorough and detailed analysis of multigene families was conducted for the whole genome sequences of many plants. For example, a whole genome survey of six plant species helped to identify 56 (Carica papaya) to 242 (Glycine max) UGTs (Yonekura-Sakakibara and Hanada 2011). The study identified 274 upland cotton UGTs, which were more than those identified in Arabidopsis, rice, flax and fruit species.
A phylogenetic tree provided a framework to compare the properties of gene family members and identify their similarities and differences (Jung et al. 2008). In this present study, G. raimondii (151), G. arboreum (150), Arabidopsis (114) and Therobroma cacao (159) were chosen for phylogenetic analysis. First, the genes of these species are all homologous; second, use these genes to better cluster similar genes in Gossypium hirsutum L.; third, to explore the results of existing studies in other species such as Arabidopsis and Therobroma cacao for comparative analysis. 274 UGT genes in Gossypium hirsutum L. were clustered into 10 groups based on phylogenetic analysis. In contrast, the UGT gene family in other plants was divided into 14 groups in Arabidopsis or even 16 groups in peach. This was probably caused by the differences of the methods used to identify UGT genes.
Based on the 274 upland cotton UGTs members’ sequence, this study analyzed introns, positions, motif, structure and expression to understand the evolution of gene family within the phylogenetic groups, among which 47.08% only had one intron, and 39.42% had two introns. Furthermore, 17 genes had one intron in the H subgroup.
To predict and understand the roles of these UGT genes in various tissue types, gene expression pattern analysis assisted in identifying which gene family members were expected to perform distinct or similar roles. To this end, the study analyzed the expression of the upland cotton UGTs in subgroup H using transcriptome sequencing data and RT-qPCR. The experimental materials for the transcriptome in the early stage of the experiment were as follows: CCRI45 and CCSLs 7747. To match transcriptome data from different periods of fiber samples of CCRI45 and CCSL 7747 varieties were chosen for qRT-PCR analysis. CCSLs 7747 had better fiber quality than CCRI45. Therefore, the results could use transcriptome and qRT-PCR results to determine whether these genes were related to the fiber quality of cotton.
The study found that many genes in the H subfamily might be related to cotton fiber yield and quality traits. For example, the GhUGT203 gene was located in the qFL-chr16-1 and qFS-chr16-3 regions. What is more, the QTLs were stable and could be detected in two environments (Liu et al. 2018). GhUGT6, GhUGT50 and GhUGT150 were separately located in the regions of qFL01.1, qFL05.2 and qFS17.1, and the three QTLs could also be detected in two environments (Tan et al. 2018). GhUGT6, GhUGT105 and GhUGT207 were related to qBW-chr01-1, qBW-chr12-7 and qBW-chr24-1, respectively, and qBW-chr01-1 and qBW-chr12-7 were stable QTLs(Zhang et al. 2016). GhUGT6 and GhUGT177 were, respectively, related to the stable QTLs of qFS-c1-1 and qFS-c19-1(Zhang et al. 2017). GhUGT16 could also be detected related to qGhFS-c2-2 by the research of Huang (Huang et al. 2017). In our laboratory research, it was found that the SSR marker HAU0734 had a significant effect on qFL-12-5 (Lu 2017), and in the physical position of Gossypium hirsutum L., GhUGT103 was closely linked to this marker, and the gene expression indicated that it was related to fiber development. The information of mapping subgroup H genes in QTL intervals is shown in Supplementary Table S3.
With gene expression and QTL results combined, GhUGT105 might be related to fiber yield, and GhUGT16 and GhUGT103 might be related to cotton fiber development, especially GhUGT6 was associated with the three traits. This indicated that some of the GhUGT genes could be important for the study of cotton fiber development and improvement.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Pengyun Chen for assistance in phylogenetic analysis, and synthetic analysis.
Author contributions
YY and HZ conceived and designed the experiments. XX and QL performed the experiments. YS, JG, AL, HS, WG, QG and JL contributed reagents/materials/analysis tools. Resources were provided by PL, XD, SL, QC, QZ, and DN. XX, QL and RL wrote and revised the paper.
Funding
This study was funded by the National Natural Science Foundation of China (U1804103, 31101188), the National Key R & D Program for Crop Breeding (2016YFD0100306) and the Agricultural Science and Technology Innovation Program for CAAS (CAAS-ASTIP-ICRCAAS).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Xianghui Xiao and Quanwei Lu contributed equally to this work.
Contributor Information
Xianghui Xiao, Email: xiaoxianghui4953@163.com.
Quanwei Lu, Email: 13707667581@163.com.
Ruixian Liu, Email: ruixianliu6@126.com.
Juwu Gong, Email: gongjuwu@caas.cn.
Wankui Gong, Email: gongwankui@cass.cn.
Aiying Liu, Email: liuaiying@caas.cn.
Qun Ge, Email: gequn@caas.cn.
Junwen Li, Email: lijunwen@caas.cn.
Haihong Shang, Email: shanghaihong@caas.cn.
Pengtao Li, Email: lipengtao1056@126.com.
Xiaoying Deng, Email: dengxiaoying@caas.cn.
Shaoqi Li, Email: li_shaoqi@foxmail.com.
Qi Zhang, Email: qizhangyx@126.com.
Doudou Niu, Email: 82101175018@cass.cn.
Quanjia Chen, Email: chqjia@126.com.
Yuzhen Shi, Email: shiyuzhen@caas.cn.
Hua Zhang, Email: hazelzhang@163.com.
Youlu Yuan, Email: yylcri@126.com.
References
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barvkar VT. Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC genomics. 2012;13(1):175. doi: 10.1186/1471-2164-13-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bönisch F, Frotscher J, Stanitzek S, Rühl E, Wüst M, Bitz O, Schwab W. A UDP-Glucose: monoterpenol glucosyltransferase adds to the chemical diversity of the grapevine metabolome. Plant Physiol. 2014;165(2):561. doi: 10.1104/pp.113.232470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles D, Lim EK, Poppenberger B, Vaistij FE. Glycosyltransferases of lipophilic small molecules. Annurevplant Biol. 2006;57(57):567–597. doi: 10.1146/annurev.arplant.57.032905.105429. [DOI] [PubMed] [Google Scholar]
- Caputi L, Malnoy M, Goremykin V, Nikiforova S, Martens S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012;69(6):1030–1042. doi: 10.1111/j.1365-313X.2011.04853.x. [DOI] [PubMed] [Google Scholar]
- Cheng J, Wei G, Zhou H, Gu C, Vimolmangkang S, Liao L, Han Y. Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach. Plant Physiol. 2014;166(2):1044–1058. doi: 10.1104/pp.114.246876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MK, Bland JM, Shockey JM, Cao H, Hinchliffe DJ, Fang DD, Naoumkina M. A transcript profiling approach reveals an abscisic acid-specific glycosyltransferase (UGT73C14) induced in developing fiber of ligon lintless-2 mutant of cotton (Gossypium hirsutum L.) PloS one. 2013;8(9):e75268. doi: 10.1371/journal.pone.0075268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Hereditas. 2007;29(8):1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- Huang C, Nie X, Shen C, You C, Li W, Zhao W, Zhang X, Lin Z. Population structure and genetic basis of the agronomic traits of upland cotton in China revealed by a genome-wide association study using high-density SNPs. Plant Biotechnol J. 2017;15(11):1374–1386. doi: 10.1111/pbi.12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Vogt T. Glycosyltransferases in secondary plant metabolism: tranquilizers and stimulant controllers. Planta. 2001;213(2):164–174. doi: 10.1007/s004250000492. [DOI] [PubMed] [Google Scholar]
- Jr SH. Plant metabolism of xenobiotics. Trends Biochem Sci. 1992;17(2):82. doi: 10.1016/0968-0004(92)90507-6. [DOI] [PubMed] [Google Scholar]
- Jung KH, An G, Ronald PC. Towards a better bowl of rice: assigning function to tens of thousands of rice genes. Nat Rev Genet. 2008;9:91–101. doi: 10.1038/nrg2286. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Baldauf S, Lim EK, Bowles DJ. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem. 2001;276(6):4338. doi: 10.1074/jbc.M007447200. [DOI] [PubMed] [Google Scholar]
- Lim EK, Bowles DJ. A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 2004;23(15):2915. doi: 10.1038/sj.emboj.7600295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JS, Huang XX, Li Q, Cao Y, Bao Y, Meng XF, Li YJ, Fu C, Hou BK. UDP-glycosyltransferase 72B1 catalyzes the glucose conjugation of monolignols and is essential for the normal cell wall lignification in Arabidopsis thaliana. Plant J. 2016;88(1):26–42. doi: 10.1111/tpj.13229. [DOI] [PubMed] [Google Scholar]
- Liu R, Gong J, Xiao X, Zhang Z, Li J, Liu A, Lu Q, Shang H, Shi Y, Ge Q, Iqbal MS, Deng X, Li S, Pan J, Duan L, Zhang Q, Jiang X, Zou X, Hafeez A, Chen Q, Geng H, Gong W, Yuan Y. GWAS analysis and QTL identification of fiber quality traits and yield components in upland cotton using enriched high-density SNP markers. Front Plant Sci. 2018;9:1067. doi: 10.3389/fpls.2018.01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif) 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu Q (2017) Fine mapping and candidate gene identification of qFL-12-2 in chromosome introgression line carrying Gossypium barbadense chromosomal segments in Gossypium hirsutum background. PhD. Shanxi Agriculture University
- Lu Q, Shi Y, Xiao X, Li P, Gong J, Gong W, Liu A, Shang H, Li J, Ge Q. Transcriptome analysis suggests that chromosome introgression fragments from sea Island cotton (Gossypium barbadense) increase fiber strength in upland cotton (Gossypium hirsutum) G3. 2017;7(10):3469–3479. doi: 10.1534/g3.117.300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie PI, Owens IS, Burchell B, Bock KW, Bairoch A, Bélanger A, Fournelgigleux S, Green M, Hum DW, Iyanagi T. The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics. 1997;7(4):255. doi: 10.1097/00008571-199708000-00001. [DOI] [PubMed] [Google Scholar]
- Montefiori M, Espley RV, Stevenson D, Cooney J, Datson PM, Saiz A, Atkinson RG, Hellens RP, Allan AC. Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis) Plant J. 2011;65(1):106–118. doi: 10.1111/j.1365-313X.2010.04409.x. [DOI] [PubMed] [Google Scholar]
- Moon S, Kim SR, Zhao G, Yi J, Yoo Y, Jin P, Lee SW, Jung KH, Zhang D, An G. Rice glycosyltransferase1 encodes a glycosyltransferase essential for pollen wall formation. Plant Physiol. 2013;161(2):663–675. doi: 10.1104/pp.112.210948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono E, Homma Y, Horikawa M, Kunikane-Doi S, Imai H, Takahashi S, Kawai Y, Ishiguro M, Fukui Y, Nakayama T. Functional differentiation of the glycosyltransferases that contribute to the chemical diversity of bioactive flavonol glycosides in grapevines (Vitis vinifera) Plant Cell. 2010;22(8):2856–2871. doi: 10.1105/tpc.110.074625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette S, Møller BL, Bak S. On the origin of family 1 plant glycosyltransferases. Phytochemistry. 2003;62(3):399–413. doi: 10.1016/S0031-9422(02)00558-7. [DOI] [PubMed] [Google Scholar]
- Paquette SM, Jensen K, Bak S. A web-based resource for the Arabidopsis P450, cytochromes b5, NADPH-cytochrome P450 reductases, and family 1 glycosyltransferases. Phytochemistry. 2009;70(17–18):1940–1947. doi: 10.1016/j.phytochem.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Research. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren ZY, Yu DQ, Yang ZE, Li CF, Qanmber G, Li Y, Li J, Liu Z, Lu LL, Wang LL. Genome-wide identification of the MIKC-Type MADS-Box gene family in Gossypium hirsutum L. Unravels their roles in flowering. Front. Plant Sci. 2017;8:384. doi: 10.3389/fpls.2017.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Hong X, Zhao S, Liu J, Schulenburg K, Huang FC, Franz-Oberdorf K, Schwab W. Glucosylation of 4-Hydroxy-2,5-Dimethyl-3(2H)-Furanone, the key strawberry flavor compound in strawberry fruit. Plant Physiol. 2016;171(1):139–151. doi: 10.1104/pp.16.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Zhao S, Hong X, Liu J, Schulenburg K, Schwab W. A UDP-glucosyltransferase functions in both acylphloroglucinol glucoside and anthocyanin biosynthesis in strawberry (Fragaria x ananassa) Plant J Cell Mol Biol. 2016;85(6):730–742. doi: 10.1111/tpj.13140. [DOI] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18(1):207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- Tan Z, Zhang Z, Sun X, Li Q, Sun Y, Yang P, Wang W, Liu X, Chen C, Liu D. Genetic map construction and fiber quality QTL mapping using the CottonSNP80K array in upland cotton. Front Plant Sci. 2018;9:225. doi: 10.3389/fpls.2018.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Gao L, Gao J, Xu Y, Liu H, Cao X, Zhang B, Chen K. Genome-Wide identification, expression patterns, and functional analysis of UDP glycosyltransferase family in peach (Prunus persica L. Batsch). Front. Plant Sci. 2017;8:389. doi: 10.3389/fpls.2017.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauk YK, Ged C, Wang MY, Matich AJ, Tessarotto L, Cooney JM, Chervin C, Atkinson RG. Manipulation of flavour and aroma compound sequestration and release using a glycosyltransferase with specificity for terpene alcohols. Plant J. 2015;80(2):317–330. doi: 10.1111/tpj.12634. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Hanada K. An evolutionary view of functional diversity in family1 glycosyltransferases. Plant J Cell Mol Biol. 2011;66(1):182–193. doi: 10.1111/j.1365-313X.2011.04493.x. [DOI] [PubMed] [Google Scholar]
- Yonekurasakakibara K, Fukushima A, Nakabayashi R, Hanada K, Matsuda F, Sugawara S, Inoue E, Kuromori T, Ito T, Shinozaki K. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 2012;69(1):154–167. doi: 10.1111/j.1365-313X.2011.04779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Shang H, Shi Y, Huang L, Li J, Ge Q, Gong J, Liu A, Chen T, Wang D, Wang Y, Palanga KK, Muhammad J, Li W, Lu Q, Deng X, Tan Y, Song W, Cai J, Li P, Rashid H, Gong W, Yuan Y. Construction of a high-density genetic map by specific locus amplified fragment sequencing (SLAF-seq) and its application to Quantitative Trait Loci (QTL) analysis for boll weight in upland cotton (Gossypium hirsutum) BMC Plant Biol. 2016;16:79. doi: 10.1186/s12870-016-0741-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ge Q, Liu A, Li J, Gong J, Shang H, Shi Y, Chen T, Wang Y, Palanga KK, Muhammad J, Lu Q, Deng X, Tan Y, Liu R, Zou X, Rashid H, Iqbal MS, Gong W, Yuan Y. Construction of a high-density genetic map and its application to qtl identification for fiber strength in upland cotton. Crop Sci. 2017;57(2):774. doi: 10.2135/cropsci2016.06.0544. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.