Abstract
Metastatic biopsy programmes combined with advances in genomic sequencing have provided new insights into the molecular landscape of castration-resistant prostate cancer (CRPC), identifying actionable targets, and emerging resistance mechanisms. The detection of DNA repair aberrations, such as mutation of BRCA2, could help select patients for poly(ADP-ribose) polymerase (PARP) inhibitor or platinum chemotherapy, and mismatch repair gene defects and microsatellite instability have been associated with responses to checkpoint inhibitor immunotherapy. Poor prognostic features, such as the presence of RB1 deletion, might help guide future therapeutic strategies. Our understanding of the molecular features of CRPC is now being translated into the clinic in the form of increased molecular testing for use of these agents and for clinical trial eligibility. Genomic testing offers opportunities for improving patient selection for systemic therapies and, ultimately, patient outcomes. However, challenges for precision oncology in advanced prostate cancer still remain, including the contribution of tumour heterogeneity, the timing and potential cooperation of multiple driver gene aberrations, and diverse resistant mechanisms. Defining the optimal use of molecular biomarkers in the clinic, including tissue-based and liquid biopsies, is a rapidly evolving field.
Prostate cancer is the most common non-cutaneous malignancy in men in the Western World1,2. Despite substantial advances in diagnosis and treatment, prostate cancer remains a leading cause of cancer mortality: >30,000 men die from prostate cancer per year in the USA2. Clinical challenges include distinguishing an indolent from an aggressive natural history in PSA-detected localized prostate cancer, determining the optimal sequencing of systemic therapies for metastatic castration-sensitive and treatment-resistant prostate cancer, and implementing biomarker-driven treatment approaches.
Prostate cancer initiation and disease progression are driven by androgen receptor (AR) signalling3, which has led to the use of androgen deprivation therapy (ADT) as the backbone of systemic therapy for patients with advanced disease for over 75 years4. In the past 5 years, data supporting the addition of potent AR pathway inhibitors (ARPIs) or docetaxel chemotherapy to ADT have improved clinical practice in patients with metastatic castration-sensitive disease5–8. Despite clinically significant responses to primary systemic therapy, castration resistance ensues, which occurs primarily through both ligand-dependent and ligand-independent AR signalling reactivation9. Potent ARPIs, such as abiraterone and enzalutamide, are also commonly used in patients with metastatic castration-resistant prostate cancer (mCRPC)10–13 and the next-generation ARPIs enzalutamide, apalutamide and darolutamide have demonstrated improved outcomes in men with non-metastatic CRPC (nmCRPC)14–16. In general, the sequential use of potent ARPIs in mCRPC is limited by cross-resistance between AR-targeted drugs17,18. Furthermore, with the early use and potentially long exposure to therapies that target the AR, downstream mechanisms of treatment resistance continue to evolve, potentially leading to an increase in diagnoses of non-AR-driven disease19,20. Identifying resistance mechanisms in individual patients has potential implications for personalization of systemic therapies, for determining the optimal sequence of drugs and for improving strategies to dynamically combat resistance mechanisms in the CRPC setting.
Resistance can be intrinsic and present before treatment, for example via TP53 mutations, or arise after therapeutic stress, for example via acquired AR amplification or mutations, or RB1 loss after ADT21. As only a few longitudinal studies have assessed different stages of disease progression, uncertainty remains regarding when specific alterations develop in an individual and how they continue to evolve over the course of subsequent therapies. In a biopsy study of metastatic lesions in 150 patients with mCRPC by the international Stand Up To Cancer-Prostate Cancer Foundation (SU2C-PCF) Dream Team22, the common recurrent somatic gene alterations in mCRPC included AR mutation or amplification (62.7%), TP53 mutation or deletion (53.3%), PTEN deletion (40.7%), RB1 loss (8.6%), BRCA1 or BRCA2 mutation or deletion (14.6%), and CDK12 mutation (4.7%); the most frequently altered pathways involved AR, PI3K, WNT, cell cycle regulation and DNA repair. These frequencies were similar in an updated analysis of nearly 500 tumours by the same team23. In addition to these recurrent aberrations, there exists a long tail of significantly mutated genes that occur in <5% of mCRPC patients, the biological and clinical significance of which remains uncertain24.
In addition to genomic aberrations, mCRPC tumours can evolve their phenotype during disease progression and treatment resistance manifests by changes in gene expression, epigenetics and/or tumour morphology. In a multi-institutional study evaluating 202 metastatic tumours from the West Coast SU2C-PCF Dream Team, 17% of patients with mCRPC developed small-cell neuroendocrine features at the time of resistance to enzalutamide or abiraterone20. Treatment-related small-cell neuroendocrine prostate cancer (tNEPC) is associated with distinct genomic, gene expression and epigenetic changes that might further inform therapy choices for patients25.
The molecular landscape of advanced disease
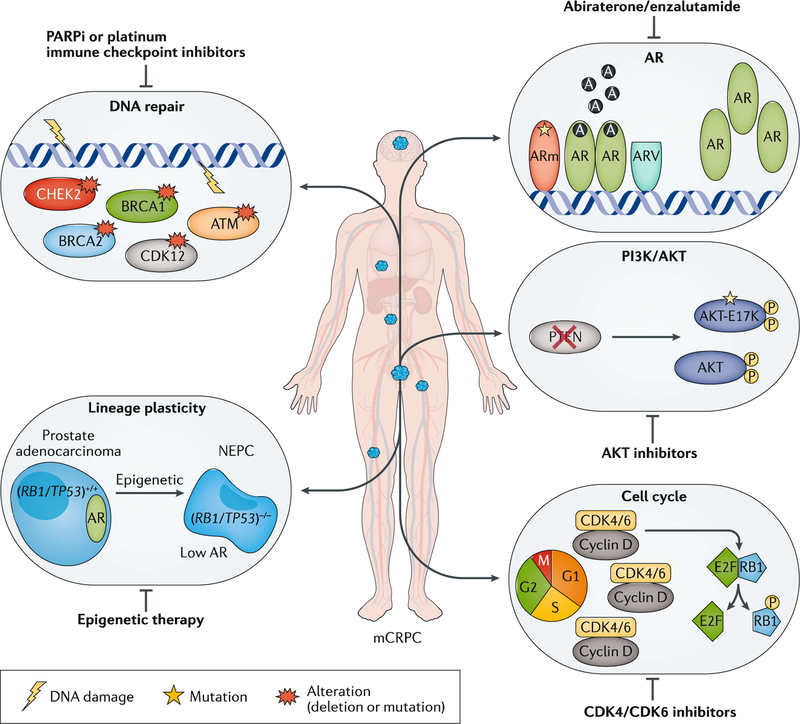
Data regarding the clinical significance of many of the molecular alterations observed in advanced prostate cancer are still emerging, and how best to test and act on these alterations in the clinic is an area of active research. Although a number of specific recurrent alterations have been documented (FIG. 1), these lesions do not always exist in isolation and much remains to be learned regarding the timing and potential cooperation of multiple driver gene aberrations and the role of less common alterations.
Fig. 1 |. Precision medicine in mCRPC.
Genomic alterations are often heterogeneous across patients with metastatic castration-resistant prostate cancer (mCRPC). Different alterations can have distinct biological roles in driving mCRPC progression and response, and resistance to therapies. By understanding each altered gene or pathway in an individual, precision medicine has the potential to guide unique therapeutic approaches for patients and improve clinical outcomes. A, androgen; AR, androgen receptor; ARm, mutant AR; ARV, AR splice variant; CDK, cyclin-dependent kinase; NEPC, neuroendocrine prostate cancer.
Androgen receptor.
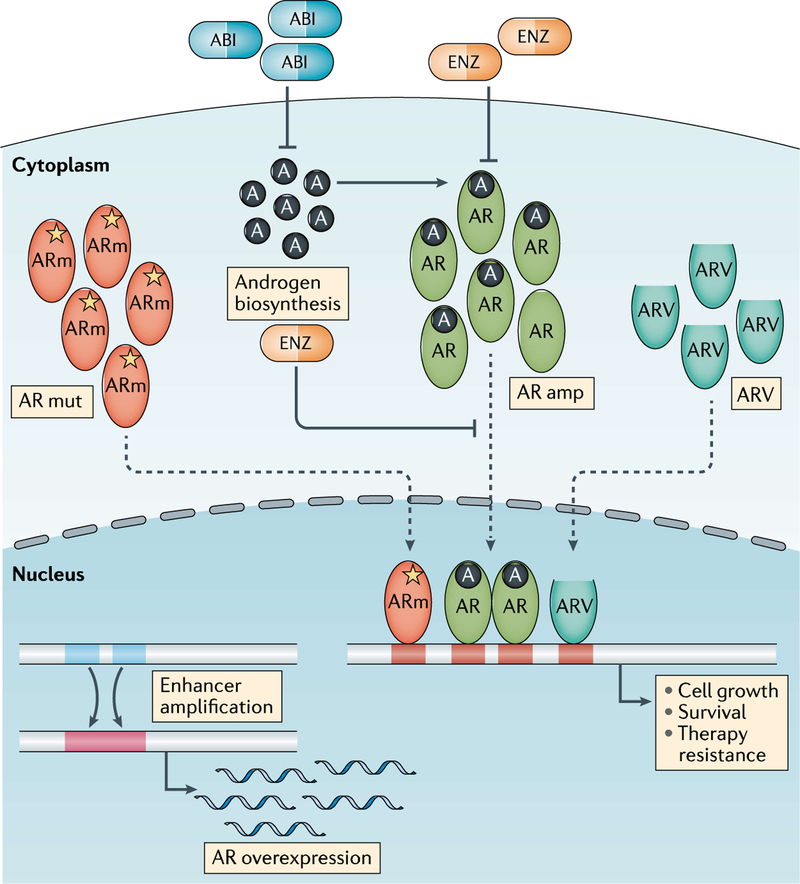
The most common genomic alterations in mCRPC that occur in the majority (50–70%) of patients involve AR and include AR amplification, AR activating mutations (for example, L702H, W742C, F877L and T878A) and AR structure rearrangements26 including deletion, duplication, inversion and translocation, all of which lead to the reactivation of AR signalling9 (FIG. 2). Whole-genome studies have also identified frequent amplification of upstream enhancers of AR in mCRPC that results in AR overexpression26–28. Specific mutations involving the AR ligand-binding domain lead to AR promiscuity and activation by adrenal androgens or steroids such as cortisol or progesterone9. AR mutations such as W742C, W742L and F877L are associated with resistance to bicalutamide29 and enzalutamide30 by causing the AR antagonists to instead act as agonists31. In addition, patients receiving abiraterone plus prednisone can acquire L702H mutations, causing promiscuous activation of the AR by prednisone21,32. AR mutations or amplification in circulating tumour DNA (ctDNA) of patients with mCRPC have been associated with reduced rates of decline in PSA levels and shorter time to progression in patients treated with potent ARPIs compared with those without detectable AR alterations33–35. In addition to AR genomic alterations, other molecular mechanisms that also drive AR signalling activation include AR splice variants (ARVs), cofactors that lead to increased AR stability and intratumoural androgen biosynthesis. ARVs lack the ligand-binding domain on the C terminus and continuously activate downstream AR signalling without androgen stimuli9. Detection of the most frequent ARV, AR-V7, in circulating tumour cells (CTCs) and tissue biopsy samples has been associated with inferior outcomes in patients with mCRPC treated with potent ARPIs compared with those without detectable AR-V7, potentially owing to persistent AR activation36–38. How best to use AR ctDNA or AR-V7 testing in CTCs in the clinic, and when, have not been fully established.
Fig. 2 |. Altered AR signalling in mCRPC.
Alterations in androgen receptor (AR) signalling are the most prevalent biological events in metastatic castration-resistant prostate cancer (mCRPC) resulting in persistent AR activation. These alterations include AR amplification (amp), mutations, AR splice variants (ARVs), intratumoural androgen biosynthesis and AR enhancer amplification. Enzalutamide and abiraterone acetate are two FDA-approved drugs that target AR signalling in mCRPC. Enzalutamide is an AR antagonist that also blocks AR translocation and function, whereas abiraterone inhibits androgen biosynthesis. A, androgen; ABI, abiraterone; ARm, mutant AR; ENZ, enzalutamide; mut, mutation.
Androgen biosynthesis also occurs in CRPC tumours and is enhanced by the presence of a gain of function mutation of the 3β-hydroxysteroid dehydrogenase type 1 (3βHSD1) gene HSD3B1, which encodes a key enzyme for the conversion of adrenal-derived steroids to dihydrotestosterone39. In a multi-cohort study with genotype data from 443 patients, homozygous mutation of 1245 A>C HSD3B1 was predictive of decreased metastasis-free survival and overall survival after prostatectomy, and decreased progression-free survival (PFS) in patients on ADT40, suggesting that HSD3B1 might be a useful biomarker to stratify patients for therapy intensification by identifying those more resistant to ADT41.
Overall these data supporting the reactivation of AR signalling in mCRPC has led to the development of potent therapeutic strategies to target the AR. Sequential therapy with ARPIs, such as abiraterone followed by enzalutamide or vice versa, is associated with limited responses in the mCRPC setting with PSA response rates typically <30%17,18,42, indicating cross-resistance between the currently available androgen biosynthesis inhibitors and AR antagonists. Furthermore, adding abiraterone at the time of enzalutamide resistance in the phase II PLATO trial43 was also not effective, with no differences in PFS observed between the combination therapy group and those switching to abiraterone alone. Combining abiraterone and enzalutamide upfront in patients with mCRPC is also not beneficial, as observed in the phase III Alliance A031201 trial44, in which no differences in overall survival were observed in patients receiving abiraterone plus enzalutamide in the first-line mCRPC setting versus enzalutamide alone. These data suggest that either there is an overlap in AR-driven resistance mechanisms such that resistance is not sufficiently overcome by combining our current therapies, or other downstream effects of the AR or bypass pathways are driving tumour progression. To offset the possibility of persistent AR-driven disease, alternative ARPIs are in development to specifically block the amino-terminal domain45 or the DNA-binding domain46 as a means to more effectively target AR signalling, disrupt co-activator or chaperone recruitment47, or directly suppress other androgen biosynthesis and/or steroidogenesis enzymes. The ability of high-dose testosterone or alternating and/or rapid cycling of ADT and androgen therapy (‘bipolar androgen therapy’, or BAT) to induce supraphysiological levels of testosterone thereby potentially restoring responses to enzalutamide has been investigated in the mCRPC setting48. In a phase II trial, 9 of 30 men with mCRPC achieved a PSA response of ≥50% on BAT after progression on enzalutamide and 15 of 21 patients responded to subsequent rechallenge with enzalutamide48. As testosterone is also a driver of prostate cancer growth, careful patient selection is important. Some tumours might be more sensitive to this approach, such as those with underlying DNA repair aberrations owing to AR-mediated induction of DNA double-strand breaks, cell cycle arrest and cellular senescence49.
In addition to understanding resistance mechanisms, the identification of additional biomarkers of ARPI response might also guide future therapy choice. Point mutations involving SPOP are present in up to 10% of localized disease but only ~5% of mCRPC and might be associated with improved prognosis and sensitivity to ARPIs50,51. In patients with localized prostate tumours treated with radical prostatectomy, SPOP mutations have been associated with fewer adverse pathological features, and improved biochemical progression-free survival, metastasis-free survival and prostate cancer-specific mortality compared with wild-type SPOP tumours51. In patients with mCRPC treated with abiraterone, SPOP mutations have been associated with improved overall survival compared with those with wild-type SPOP50. A preclinical study using genetically engineered mouse models suggested that SPOP mutations maintain AR signalling by blocking reciprocal negative feedback mediated by the PI3K-mTOR pathway, providing a biological rationale for SPOP mutation status as a potential biomarker of response to AR-targeted drugs52.
The PTEN-PI3K-AKT pathway.
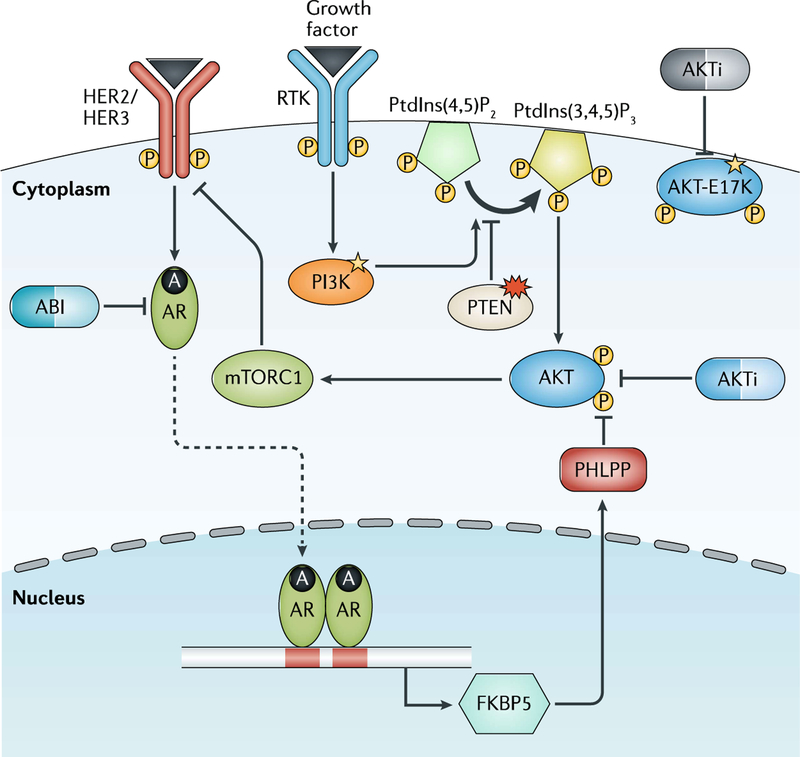
Alterations involving genes within the PTEN-PI3K-AKT pathway are commonly observed in prostate cancer23 (FIG. 3). The tumour suppressor PTEN, for example, is deleted in ~50% of mCRPC tumours. Amplification or activating mutations of PIK3CA, PIK3CB, PIK3R1 and AKT1 are less common, being observed in <15% of patients22. Alterations in these genes are predicted to activate the PI3K-AKT pathway and, therefore, drugs targeting this pathway might be effective in some patients. The E17K hotspot mutation in the AKT1 gene, which is also observed in breast, colorectal, and ovarian cancer, increases recruitment of AKT1 to the cell membrane leading to AKT activation53. A basket trial, in which 58 patients with different refractory tumour types that harboured the same E17K mutation received the same treatment with the AKT inhibitor AZD5363 (capivasertib), demonstrated encouraging antitumour activity after a median five lines of prior therapy with a median PFS of 6.6 months54. Further investigation of capivasertib in AKT1 (E17K) mutated prostate cancer is warranted.
Fig. 3 |. Dysregulated PI3K-AKT signalling in mCRPC.
Genomic alterations involving the PTEN-PI3K-AKT pathway occur in ~50% of metastatic castration-resistant prostate cancers (mCRPCs) resulting in PI3K-AKT pathway activation. Loss of PTEN has been associated with shorter time on androgen receptor (AR) pathway inhibitor (ARPI) treatment potentially due to reciprocal negative feedback of the PI3K and AR signalling pathways. Drugs that target AKT or PI3K inhibitors are currently being tested in clinical trials as monotherapy (for AKT-mutated tumours) or in combination with ARPIs. A, androgen; ABI, abiraterone; AKTi, AKT inhibitor; FKBP5, FK506 binding protein 5; PHLPP, PH domain leucine-rich repeat protein phosphatase; Ptdlns(4,5)P2, phosphatidylinositol 4,5-bisphosphate; Ptdlns(3,4,5)P3, phosphatidylinositol 3,4,5-trisphosphate; RTK, receptor tyrosine kinase.
PTEN loss has also been associated with relative resistance to ARPIs55. In an analysis of 144 patients treated with abiraterone after receiving docetaxel for mCRPC, 40% of whom had loss of PTEN expression in their tumour, a correlation was observed between PTEN loss and shorter overall survival (14 versus 21 months) as well as time on abiraterone. This outcome might be due to crosstalk between PI3K signalling and AR signalling, whereby AR transcriptional output is inhibited in tumours with PTEN loss through a reciprocal negative feedback loop56 (FIG. 3). This crosstalk observation led to the development of combination therapy strategies, including the combination of AKT inhibitor ipatasertib and abiraterone. The randomized phase III IPATential150 trial ()57 is currently recruiting men with previously untreated mCRPC to receive ipatasertib plus abiraterone and prednisone versus abiraterone and prednisone, with radiographic PFS being specifically analysed in patients with PTEN loss by immunohistochemistry against the intent-to-treat population.
DNA repair.
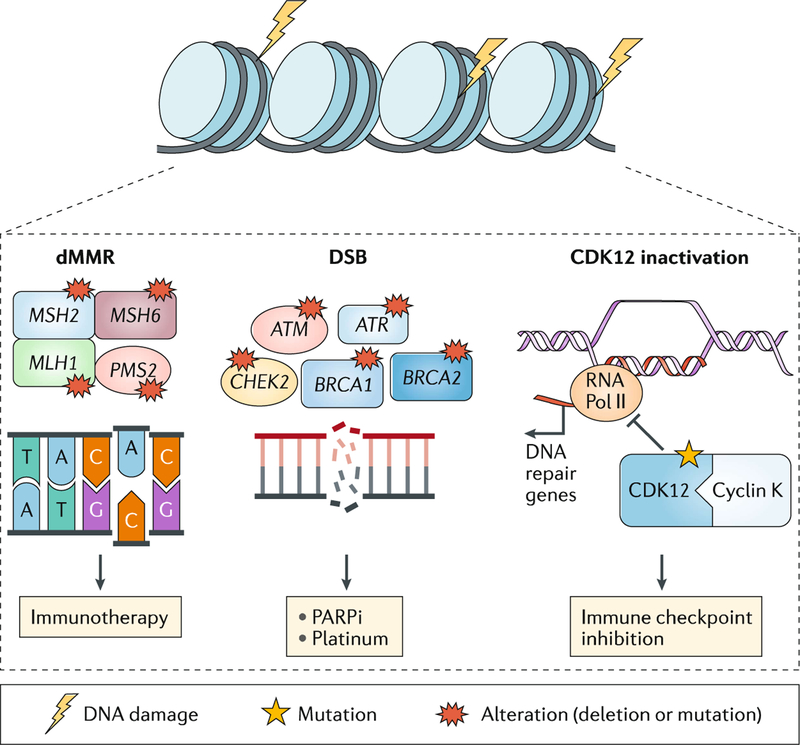
DNA repair alterations are observed in ~20% of mCRPC, most commonly mutations in homologous recombination (HR) genes such as BRCA2, BRCA1 and ATM23 (FIG. 4). Importantly these alterations can occur at either the somatic (tumour) or the germline level23,58. Owing to a high frequency of germline alterations (in up to 12% of men with advanced prostate cancer, even patients unselected for age or family history)58, germline testing is now recommended by the National Comprehensive Cancer Network for all patients with metastatic prostate cancer59. This high frequency of mutations has important implications not only for men with prostate cancer, but also for their family members, as they are at increased risk of developing certain other cancers60.
Fig. 4 |. DNA repair pathway in mCRPC.
Germline or somatic mutations involving DNA repair genes, such as BRCA1, BRCA2, ATM and MSH2 are present in 20% of metastatic castration-resistant prostate cancers (mCRPCs). Loss of homologous recombination genes (such as BRCA2) has been associated with response to poly(ADP-ribose) polymerase (PARP) inhibitor (PARPi) treatment and platinum chemotherapy. Mutations in DNA mismatch repair (MMR) genes (for example, MSH2) results in hypermutation and microsatellite instability. Loss of CDK12 or deficiency in mismatch repair genes (dMMR) has been associated with response to checkpoint inhibitor therapy. CDK, cyclin-dependent kinase; DSB, double-strand breaks; RNA Pol II, RNA polymerase II.
Tumours that lose the HR pathway are preferentially sensitive to inhibition of poly (ADP-ribose) polymerase (PARP) or administration of a DNA-damaging agent such as platinum chemotherapy through a mechanism of synthetic lethality61. PARP inhibitors and platinum chemotherapy are effective in other cancer types with HR gene aberrations62. In mCRPC, significant responses have been observed to treatment with the PARP inhibitor olaparib63 or platinum chemotherapy64–66 in patients with HR deficiency (somatic or germline). For instance, 88% of patients (14/16) who harboured HR defects in a phase II trial of olaparib responded to therapy63. A phase III clinical trial of olaparib versus ARPI in men with mCRPC and a HR gene mutation who have progressed on prior ARPI (the PROfound trial) was reported in a press release (August 2019) to have met its primary end point of radiographic PFS in mCRPC patients with BRCA1, BRCA2 or ATM genomic alterations. The full data have not yet been released, but testing for these gene aberrations is likely to become more frequent. Although no prospective clinical trial data have been reported for platinum-based chemotherapies in patients with HR deficiency, exceptional responses have been reported65–67. Data regarding which genes to test and which alterations respond best to PARP inhibitors and platinum chemotherapy are still needed. Furthermore, a deeper understanding of treatment resistance is required, as well as an understanding of how to identify from among patients who develop resistance those who would benefit from sequential therapy with platinum after a PARP inhibitor or vice versa68. Resistance to PARP inhibitors and platinum chemotherapy are not completely overlapping but might both involve reversion mutations that cause the DNA repair genes to restore their normal open reading frame and reverse sensitivity to DNA-damaging agents69,70.
In addition to HR genes, approximately 3–5% of patients with prostate cancer harbour a deficiency in mismatch repair genes (dMMR) such as MSH2, MSH6, PMS2 and MLH1, which typically leads to hypermutation and microsatellite instability (MSI)71. Mutations in MMR genes can also occur at the somatic or germline level, and loss of mismatch repair protein expression is often detectable in tumours by immunohistochemistry. The presence of dMMR in prostate tumours has been associated with increased expression of neoantigens and PD-L1, and immune infiltration including upregulation of genes associated with recruitment of dendritic cells, macrophages and other myeloid cells, and T cells72. Identifying patients with dMMR is important because this subset of patients is potentially amenable to checkpoint inhibitor immunotherapy — responses to checkpoint inhibition have been reported in other tumour types71, and pembrolizumab, an anti-PD-1 antibody, is approved by the FDA for all dMMR and MSI cancer types including prostate cancer73. However, responses in dMMR and MSI prostate cancer are not universal; in one study, only 6 of 11 CRPC patients with MSI-high/dMMR tumours who were treated with anti-PD-1 therapy achieved a PSA decrease of >50%71. More data are needed to understand why some patients do not respond and to determine the optimal assay to assess MMR loss and MSI in prostate cancer. Nonetheless, the approval of pembrolizumab for this biomarker-selected population has led to increased clinical testing to identify patients for this treatment.
Inactivating mutations of the cyclin-dependent kinase CDK12, which are present in up to 7% of mCRPC tumours, have also been associated with response to immune checkpoint inhibitors. Cyclin-dependent kinase 12 (CDK12) is a transcription factor that forms a complex with cyclin K to regulate gene expression in the DNA repair pathway; inactivation results in focal tandem duplications, increased gene fusions and neoantigen production74. Exceptional PSA responses have been observed (two of four patients) in men with mCRPC with CDK12 mutations treated with an anti-PD-1 immune checkpoint inhibitor74, suggesting that CDK12 mutations might be a potential biomarker of response to immune checkpoint inhibition. A phase II clinical trial of ipilimumab and nivolumab for CDK12-mutated mCRPC is underway ()75.
Cell cycle.
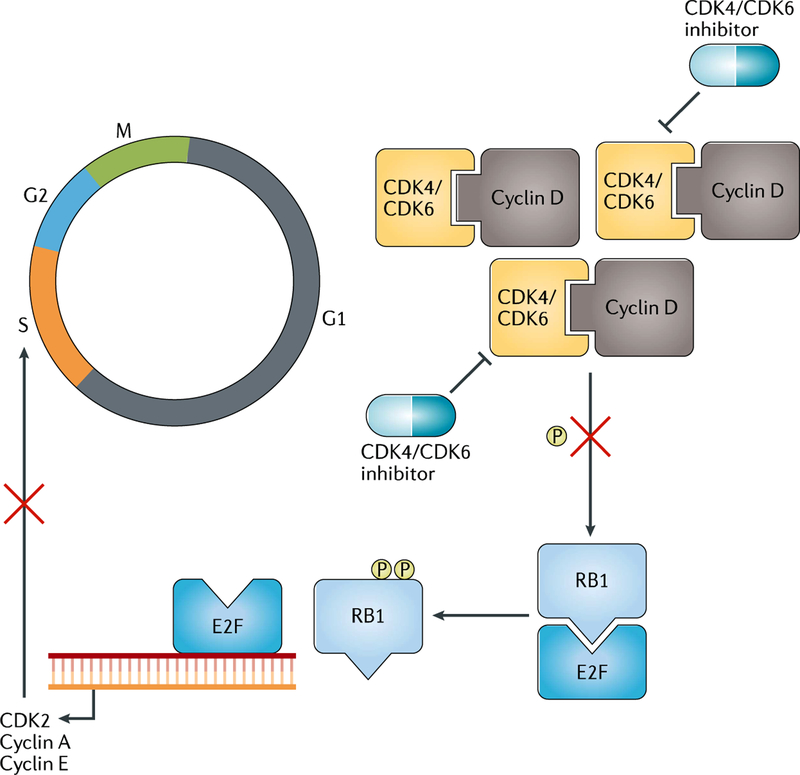
Cell cycle machinery governs cell division, and disruptions of this pathway can lead to uncontrolled cell proliferation, as is the case in cancer76. In mCRPC, genomic alterations leading to disruption of the cell cycle regulators RB1 and/or CDKs are present in up to 25% of patients22 (FIG. 5). The retinoblastoma gene RB1 is a cell cycle gatekeeper that restrains E2F from driving cyclins and CDKs to advance to S phase; loss of RB1 or gain of CDKs therefore results in uncontrolled cellular proliferation. Palbociclib, a selective CDK4 and CDK6 inhibitor, induces cell cycle arrest in RB1 wild-type preclinical models77. Phase II clinical trials of the use of palbociclib in the treatment of RB1-expressing mCRPC are ongoing, including its use as a single agent ()78 and in combination with ARPI ()79, as well as in the metastatic, hormone-naive setting ()80.
Fig. 5 |. Dysregulated cell cycle in mCRPC.
Cell cycle machinery is governed by cyclins and cyclin-dependent kinases (CDKs) at different phases. For instance, at the G1 phase, CDK4/6 binds to cyclin D to phosphorylate RB1 to release E2F. This enables E2F, a transcription factor, to relocate onto DNA to drive gene expression, such as those encoding CDK2, cyclin A and cyclin E, to advance cells to S phase. Loss of RB1 and/or amplification of CDKs are more common in metastatic castration-resistant prostate cancer (mCRPC) than in localized prostate cancer. CDK4/6 inhibitors are being tested in clinical trials in RB1 wild-type mCRPC as a monotherapy and in combination with androgen receptor pathway inhibitors.
RB1 loss can also lead to changes in the E2F1 cistrome that are distinct from its canonical role in the cell cycle to regulate differentiation, DNA repair and other cellular programmes81. Functional impairment of RB1 can occur genomically or via phosphorylation or methylation81. Notably, RB1 loss is also enriched in small-cell NEPC and, along with TP53 mutation or deletion82, drives loss of AR dependence and lineage plasticity83,84. Thus, RB1 deficiency might have additional context-dependent functions in the setting of low AR signalling or co-occurring TP53 alterations.
Lineage plasticity.
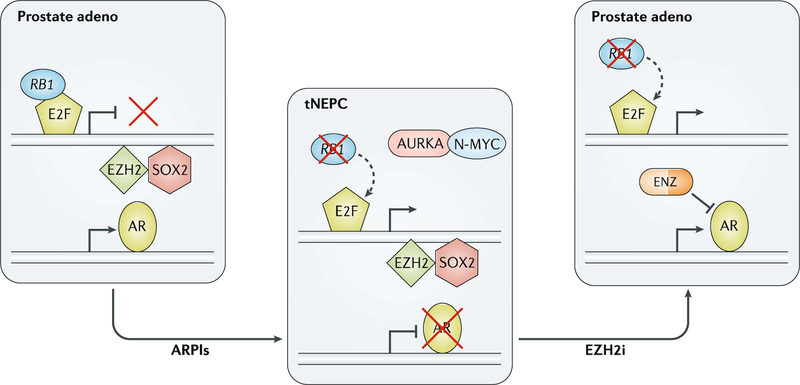
A subset of mCRPC tumours lose AR dependence during the course of tumour progression and therapy resistance19,25,85. One mechanism involves lineage plasticity associated with loss of AR signalling and activation of alternative lineage programmes including neuronal and neuroendocrine pathways25. In extreme cases, tumours can transition to a small-cell carcinoma or neuroendocrine histology86 (FIG. 6). After evaluating metastatic tumour biopsies of 148 patients progressing on ARPIs, one study found that 17% of tumours harboured pathological features of small-cell NEPC, which was associated with inferior overall survival compared with those with typical adenocarcinoma histology20. NEPC tumours are associated with low AR expression and AR signalling, combined loss of the tumour suppressors TP53 and RB1, upregulation of plasticity, developmental and pluripotency genes such as SOX2, and significant epigenomic alterations including changes in DNA methylation and upregulation of EZH2 (REF.25). Men who develop treatment-related small-cell NEPC confirmed by metastatic biopsy are often managed in the same way as those with small-cell lung cancer and treated with platinum-based combination chemotherapy59. Repeated tumour biopsies are, therefore, sometimes performed in patients with mCRPC who develop aggressive disease and/or atypical metastatic patterns in the setting of low PSA levels to look for small-cell neuroendocrine transformation. NEPC is typically diagnosed by tumour morphology, and immunohistochemistry for classic neuroendocrine markers (for example, synaptophysin and chromogranin) can be used to support the diagnosis87. Molecular biomarkers to identify patients developing lineage plasticity and small-cell transformation — such as combined loss of RB1 and TP53 or epigenetic changes — are under investigation85. Emerging therapies that target lineage plasticity and NEPC include drugs targeting the cell cycle kinase Aurora kinase A (which indirectly targets upregulation of N-MYC88 and loss of RB1 (REF.89), which both commonly occur in NEPC) and EZH2 (REFS25,83) (to target epigenetic changes), and drugs investigated in small-cell lung cancer such as those targeting DLL3 (REF.90) and immunotherapy85.
Fig. 6 |. Lineage plasticity in mCRPC.
Lineage plasticity has been increasingly observed in patients with metastatic castration-resistant prostate cancer (mCRPC) treated with androgen receptor pathway inhibitors (ARPIs) to drive prostate cancer from an adenocarcinoma histology towards a neuroendocrine prostate cancer (NEPC) phenotype. This change is associated with loss of androgen receptor (AR) expression, combined loss of RB1 and TP53, and distinct epigenetic changes. In preclinical studies, inhibition of the epigenetic regulator EZH2 has a potential role in reversing the phenotype back towards an AR+ adenocarcinoma to regain responses to enzalutamide (ENZ). adeno, adenocarcinoma; EZH2i, EZH2 inhibition; tNEPC, treatment-related NEPC.
Future considerations.
Targeted and whole-exome sequencing studies have identified recurrent alterations in mCRPC involving coding genes, but the full genomic landscape of structural variations including deletions, insertions, inversions and translocations involving non-coding regions are not captured by this approach. Whole-genome sequencing (WGS) has shown that primary prostate cancer is characterized by complex genomic rearrangements often involving both coding and non-coding regions91,92, and these arise through a series of events that dysregulate genes coordinately or simultaneously91. In mCRPC, WGS studies have also uncovered structural variations in the non-coding mCRPC genome that are biologically informative26,28,93. For instance, WGS has shown that ≤70–87% of mCRPC tumours harbour amplification of an upstream enhancer of the AR gene resulting in AR overexpression and likely contributing to ARPI treatment resistance26–28. Although WGS is not currently used clinically, it could be feasible and informative in the future, given the additional information about prostate tumours uncovered by this approach.
Epigenetic mechanisms that edit chromatin structure, such as DNA methylation and histone modifications, can control gene expression and have a critical role in cancer development and treatment resistance94. These are also not yet in clinical use, but epigenetic biomarkers might also provide future insights for therapy selection in patients with advanced prostate cancer. Drugs targeting epigenetic pathways have been tested preclinically and are in clinical trials, albeit mostly without molecular biomarker selection. For instance, the epigenetic regulator EZH2, a histone methyltransferase that adds three methyl groups onto the histone 3 lysine 27 tail, is upregulated in CRPC, resulting in transcriptional repression95. Moreover, EZH2 has been indicated as a key factor for driving lineage plasticity in prostate cancer by rewiring the transcriptome96,97, and repressing its function with small molecules has shown antitumour activity and reversal of lineage plasticity programmes83, suggesting that EZH2 inhibitor therapy could be used to treat a subset of advanced prostate cancers, potentially including those developing lineage plasticity. Two clinical trials ( and )98,99 are ongoing to test EZH2 inhibitors alone or in combination with enzalutamide or abiraterone and prednisone in patients with mCRPC.
Epigenetic drugs targeting bromodomain and extra-terminal (BET) family of chromatin readers, such as BRD2, BRD3 and BRD4 are also in clinical development100. BRD4 has been shown to be a co-regulator of AR that facilitates downstream transcription and maintains AR signalling in mCRPC101,102. Preclinical studies have shown that drugs targeting BRD4 disrupt the recruitment of BRD4 to AR binding loci, suppress signalling, down-regulate ARV expression and, alone and in combination with enzalutamide, demonstrate antitumour activity101–103. A phase I/II trial is ongoing to examine the effects of the BET inhibitor ZEN003694 in combination with enzalutamide in patients with mCRPC104. Lysine-specific demethylase 1 (LSD1) is a histone demethylase that removes methyl groups from lysine 4 and lysine 9 tails of histone H3 leading to gene silence or activation105,106. LSD 1-targeted drugs are also in development for mCRPC, as LSD1 is highly expressed in castration-resistant disease and functions as a co-activator of AR to control downstream gene expression107,108. These studies suggest that epigenetic therapies, such as targeting histone methyltransferases or demethylases or readers, might be an effective approach in a subset of advanced prostate cancers, and predictive biomarkers for these drugs require further study.
Tumour heterogeneity and biomarker strategies
In addition to ‘wide’ genomic studies examining the landscape of mutations in a broad group of patients, ‘deep’ studies have focused on fewer patients, analysing multiple cancer metastases and primary tumour samples from the same individuals to elucidate tumour evolution during cancer progression64. Rapid autopsy studies have provided valuable insights into tumour heterogeneity across different anatomical sites of metastases at the time of lethal progression64,109–111. These studies have suggested a monoclonal origin of lethal prostate cancer with early driver genomic alterations, such as TMPRSS2-ERG fusion, commonly shared between metastases in an individual. However, metastasis-to-metastasis spread might also occur in later stages leading to intrapatient tumoural heterogeneity64. Identifying early versus late events has clinical implications for selecting samples to test to look for targetable alterations in patients (for example, whether to test for DNA repair gene defects in the primary prostate tumour or in a metastatic lesion). To date, metastatic tumour biopsy has been the preferred approach for collecting information regarding mCRPC tumour features, including genomics, protein expression and histology. However, biopsies of metastatic lesions are not always feasible, as they might be in locations that are not safe or amenable to biopsy. Single-site biopsies also do not capture intraindividual heterogeneity that might occur across metastases in an individual or changes with time or disease progression. The development of a liquid biopsy approach might overcome these challenges by capturing the relative contribution of different anatomical sites of metastases in the bloodstream, providing a non-invasive and safer means for serial tumour sampling112. As described above, the detection of AR gene aberrations in cell-free ctDNA of plasma or AR-V7 expression in CTCs has been associated with inferior responses to ARPIs32,34,112–114. Combining AR liquid biopsy analysis with other features such as serum neuroendocrine markers or TP53 or RB1 aberrations in ctDNA might also help identify non-AR-driven resistance35,115. Concordance between ctDNA and biopsies is high and captures clinically relevant prostate cancer alterations116. Phenotypic tumour heterogeneity can also be captured by diverse CTC size, cell density, AR localization and/or various morphological features117,118. Although these approaches are promising, the sensitivity and specificity of liquid biopsy approaches in detecting clinically significant prostate cancer alterations across various disease states and their optimal use in the clinic have not been fully established. Nonetheless, several commercial and research assays are available and these are sometimes used in situations in which tumour biopsy is not feasible.
Conclusions
Data have begun to accumulate regarding the molecular background of mCRPC. A broader application of metastatic biopsies and liquid biopsy approaches has brought new insights into the clinical effects of molecular alterations on prognosis and response to systemic therapies. Today, molecular testing is increasingly being performed in specific clinical scenarios: testing for HR defects (somatic or germline) to identify patients who could benefit from PARPi treatment or platinum chemotherapy and to assess cancer risk in family members (germline); testing for mismatch repair deficiency and MSI to identify patients who could benefit from pembrolizumab; and metastatic biopsy to look for small-cell NEPC transformation to select patients for platinum combination chemotherapy using small-cell lung cancer regimens. Emerging data support the use of other promising biomarkers, such as AR, SPOP, PTEN, AKT, RB1 and CDK12 for treatment selection in the clinic, and additional studies are ongoing. Tumour evolution means that metastatic biopsy is still the preferred method for testing. However, analysis of primary tumours or liquid biopsies is reasonable when biopsies are not feasible or safe. Multiple biomarker-driven studies are underway, which will continue to inform the effective translation of these findings into routine practice.
Precision oncology in advanced prostate cancer presents a number of opportunities, but is also faced with challenges, including detailed investigation of the less common ‘tail’ alterations, the contribution and effect of co-occurring lesions, and the development of non-genomic biomarkers that might help refine the development of more precise, molecularly driven treatment strategies and combination approaches for patients with advanced prostate cancer.
Key points.
Studies investigating the genomic landscape of metastatic prostate cancer have identified targetable molecular alterations and emerging resistance mechanisms.
Alterations in the androgen receptor (AR) gene are a key driver of castration resistance in prostate cancer; AR mutation, amplification and the V7 splice variant can be detected non-invasively in patients, and have been associated with resistance to AR pathway inhibitors.
A subset of advanced prostate cancers harbour germline or somatic alterations involving DNA repair genes; homologous repair gene DNA repair defects have been associated with platinum chemotherapy and poly(ADP-ribose) polymerase (PARP) inhibitor sensitivity. Mismatch repair gene and CDK12 loss have been associated with responses to immunotherapy.
Combined loss of tumour suppressors RB1 and TP53 has been associated with lineage plasticity and the development of non-AR driven therapy resistance, which is enriched in tumours with small-cell and/or neuroendocrine pathological features on metastatic biopsy and aggressive clinical features.
Several biomarker-driven clinical trials are underway in patients with advanced prostate cancer that might ultimately lead to increasingly precise therapeutic strategies in patients.
Acknowledgements
The authors acknowledge research support from the Prostate Cancer Foundation (S.-Y.K., M.E.G., H.B.), the Terry Fox Research Institute (M.E.G.), Prostate Cancer Canada (M.E.G.), the National Cancer Institute SPORE (H.B) and the Department of Defense Prostate Cancer Research Program (H.B).
Competing interests
H.B. has received research funding from Janssen, Abbvie Stemcentryx, Astellas, Eli Lilly and Millennium, and has served as advisor/consultant for Janssen, Astellas, Amgen, Astra Zeneca and Sanofi Genzyme. M.E.G is listed as inventor on patents granted to the University of British Columbia on antisense and small-molecule inhibitors of HSP27 for the treatment of cancer. S.-Y. K. declares no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ferlay J et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD & Jemal A Cancer statistics, 2019. CA Cancer J. Clin 69, 7–34 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Lonergan PE & Tindall DJ Androgen receptor signaling in prostate cancer development and progression. J. Carcinog 10, 20 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huggins C & Hodges CV Studies on prostatic cancer. I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J. Clin 22,232–240(1972). [DOI] [PubMed] [Google Scholar]
- 5.Sweeney CJ et al. Chemohormonal therapy in metastatic hormone-sensitive prostate cancer. N. Engl. J. Med 373, 737–746 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.James ND et al. Abiraterone for prostate cancer not previously treated with hormone therapy. N. Engl. J. Med 377,338–351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fizazi K et al. Abiraterone plus prednisone in metastatic, castration-sensitive prostate cancer.N. Engl. J. Med 377, 352–360 (2017). [DOI] [PubMed] [Google Scholar]
- 8.James ND et al. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet 387, 1163–1177 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watson PA, Arora VK & Sawyers CL Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer 15, 701–711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan CJ et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N. Engl. J. Med 368, 138–148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Bono JS et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med 364, 1995–2005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beer TM et al. Enzalutamide in metastatic prostate cancer before chemotherapy N. Engl. J. Med 371, 424–433 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scher HI et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med 367, 1187–1197 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Smith MR et al. Apalutamide treatment and metastasis-free survival in prostate cancer. N. Engl. J. Med 378, 1408–1418 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Hussain M et al. Enzalutamide in men with nonmetastatic, castration-resistant prostate cancer.N. Engl. J. Med 378, 2465–2474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fizazi K et al. Darolutamide in nonmetastatic, castration-resistant prostate cancer. N. Engl. J. Med 380, 1235–1246 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Loriot Y et al. Antitumour activity of abiraterone acetate against metastatic castration-resistant prostate cancer progressing after docetaxel and enzalutamide (MDV3100). Ann. Oncol 24, 1807–1812 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Noonan KL et al. Clinical activity of abiraterone acetate in patients with metastatic castration-resistant prostate cancer progressing after enzalutamide.Ann. Oncol 24, 1802–1807 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Bluemn EG et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 32, 474–489.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aggarwal R et al. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: a multi-institutional prospective study. J. Clin. Oncol 36, 2492–2503 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carreira S et al. Tumor clone dynamics in lethal prostate cancer. Sci. TransI Med 6, 254ral 25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson D et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abida W et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl Acad. Sci. USA 116, 11428–11436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armenia J et al. The long tail of oncogenic drivers in prostate cancer. Nat. Genet 50, 645–651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beltran H et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med 22, 298–305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viswanathan SR et al. Structural alterations driving castration-resistant prostate cancer revealed by linked-read genome sequencing. Cell 174, 433–447.el 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeda DY et al. A somatically acquired enhancer of the androgen receptor is a noncoding driver in advanced prostate cancer. Cell 174,422–432.el 3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quigley DA et al. Genomic hallmarks and structural variation in metastatic prostate Cancer. Cell 174, 758–769.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bohl CE, Gao W, Miller DD, Bell CE& Dalton JT Structural basis for antagonismand resistance of bicalutamide in prostate cancer. Proc. Natl Acad. Sci. USA 102, 6201–6206 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korpal M et al. An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide). Cancer Discov 3, 1030–1043 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Lallous N et al. Functional analysis of androgen receptor mutations that confer anti-androgen resistance identified in circulating cell-free DNA from prostate cancer patients. Genome Biol 17,10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romanel A et al. Plasma AR and abiraterone-resistant prostate cancer. Sci. Transl Med 7, 312re310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azad AA et al. Androgen receptor gene aberrations in circulating cell-free DNA: biomarkers of therapeutic resistance in castration-resistant prostate cancer.Clin. Cancer Res 21, 2315–2324 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Conteduca V et al. Androgen receptor gene status in plasma DNA associates with worse outcome on enzalutamide or abiraterone for castration-resistant prostate cancer: a multi-institution correlative biomarker study. Ann. Oncol 28, 1508–1516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Annala M et al. Circulating Tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 8, 444–457 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Antonarakis ES et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer.N. Engl. J. Med 371, 1028–1038 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armstrong AJ et al. Prospective multicenter validation of androgen receptor splice variant 7 and hormone therapy resistance in high-risk castration-resistant prostate cancer: the PROPHECY study.J. Clin. Oncol 37, 1120–1129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharp A et al. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J. Clin. Invest 129, 192–208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang KH et al. A gain-of-function mutation in DHT synthesis in castration-resistant prostate cancer.Cell 154, 1074–1084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hearn JWD et al. HSD3B1 and resistance to androgen-deprivation therapy in prostate cancer: a retrospective, multicohort study. Lancet Oncol 17, 1435–1444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hettel D & Sharifi N HSD3B1 status as a biomarker of androgen deprivation resistance and implications for prostate cancer. Nat. Rev. Urol 15, 191–196 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Azad AA, Eigl BJ, Murray RN, Kollmannsberger C & Chi KN Efficacy of enzalutamide following abiraterone acetate in chemotherapy-naive metastatic castration-resistant prostate cancer patients. Eur. Urol 67,23–29 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Attard G et al. Abiraterone alone or in combination with enzalutamide in metastatic castration-resistant prostate cancer with rising prostate-specific antigen during enzalutamide treatment. J. Clin. Oncol 36, 2639–2646 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris MJ et al. Alliance A031201: a phase III trial of enzalutamide (ENZ) versus enzalutamide, abiraterone, and prednisone (ENZ/AAP) for metastatic castration resistant prostate cancer (mCRPC) [abstract]. J. Clin. Oncol 37 (Suppl. 15), 5008 (2019). [Google Scholar]
- 45.Andersen RJ et al. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell 17, 535–546 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Dalal K et al. Selectively targeting the DNA-binding domain of the androgen receptor as a prospective therapy for prostate cancer. J. Biol. Chem 289, 26417–26429 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zoubeidi A et al. Cooperative interactions between androgen receptor (AR) and heat-shock protein 27 facilitate AR transcriptional activity. Cancer Res 67, 10455–10465 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Teply BA et al. Bipolar androgen therapy in men with metastatic castration-resistant prostate cancer after progression on enzalutamide: an open-label, phase 2, multicohort study. Lancet Oncol 19, 76–86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chatterjee P et al. Supraphysiological androgens suppress prostate cancer growth through androgen receptor-mediated DNA damage. J. Clin. Invest https://doi.org/10.!172/JCI127613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boysen G et al. SPOP-mutated/CHD 1 -deleted lethal prostate cancer and abiraterone sensitivity. Clin. Cancer Res 24,5585–5593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu D et al. Impact of the SPOP mutant subtype on the interpretation of clinical parameters in prostate cancer. JCO Precis. Oncol 2, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blattner M et al. SPOP mutation drives prostate tumorigenesis in vivo through coordinate regulation of PI3K/mTOR and AR signaling. Cancer Cell 31,436–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carpten JD et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 448, 439–444 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Hyman DM et al. AKT inhibition in solid tumors with AKT1 mutations. J. Clin. Oncol 35,2251–2259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferraldeschi R et al. PTEN protein loss and clinical outcome from castration-resistant prostate cancer treated with abiraterone acetate. Eur. Urol 67, 795–802 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carver BS et al. Reciprocal feedback regulationof PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19, 575–586 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.US National Library of Medicine. CIinicalTrials.gov https://clinicaltrials.gov/show/NCT03072238 (2019).
- 58.Pritchard CC et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer.N. Engl. J. Med 375, 443–453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology (NCCN guidelines®). Prostate cancer version 4.2019. NCCN https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf (2019).
- 60.Cheng HH, Sokolova AO, Schaeffer EM, Small EJ & Higano CS Germline and somatic mutations in prostate cancer for the clinician.J. Natl. Compr. Cane. Netw 17, 515–521 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Lord CJ & Ashworth A PARP inhibitors: synthetic lethality in the clinic. Science 355, 1152–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Konstantinopoulos PA, Ceccaldi R, Shapiro GI& D’Andrea AD Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov 5, 1137–1154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mateo J et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med 373, 1697–1708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar A et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat. Med 22, 369–378(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng HH, Pritchard CC, Boyd T,Nelson PS & Montgomery B Biallelic inactivation of BRCA2 in platinum-sensitive metastatic castration-resistant prostate cancer. Eur. Urol 69, 992–995 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pomerantz MM et al. The association between germline BRCA2 variants and sensitivity to platinum-based chemotherapy among men with metastatic prostate cancer. Cancer 123, 3532–3539 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zafeiriou Z et al. Genomic analysis of three metastatic prostate cancer patients with exceptional responses to carboplatin indicating different types of DNA repair deficiency. Eur. Urol 75, 184–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.D’Andrea AD Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair 71, 172–176 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Goodall J et al. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov 7, 1006–1017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quigley D et al. Analysis of circulating cell-free DNA identifies multiclonal heterogeneity of BRCA2 reversion mutations associated with resistance to PARP inhibitors. Cancer Discov 7, 999–1005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abida W et al. Analysis of the prevalence of microsatellite instability in prostate cancer and response to immune checkpoint blockade. JAMA Oncol. S, 471–478 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nava Rodrigues D et al. Immunogenomic analyses associate immunological alterations with mismatch repair defects in prostate cancer. J. Clin. Invest 128, 4441–4453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boyiadzis MM et al. Significance and implications of FDA approval of pembrolizumab for biomarker-defined disease. J. Immunother Cancer 6,35 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu YM et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell 173, 1770–1782.e 14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/show/NCT03570619 (2019).
- 76.Malumbres M & Barbacid M Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9, 153–166 (2009). [DOI] [PubMed] [Google Scholar]
- 77.Comstock CE et al. Targeting cell cycle and hormone receptor pathways in cancer. Oncogene 32,5481–5491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/show/NCT02905318 (2019).
- 79.US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/show/NCT02555189 (2019).
- 80.US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/show/NCT02059213 (2019).
- 81.Chinnam M & Goodrich DW RB 1, development, and cancer. Curr. Top. Dev. Biol 94, 129–169 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aparicio AM et al. Combined tumor suppressor defects characterize clinically defined aggressive variant prostate cancers. Clin. Cancer Res 22,1520–1530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ku SY et al. Rb 1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355, 78–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mu P et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53-and RB1 -deficient prostate cancer. Science 355, 84–88 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beltran H et al. The role of lineage plasticity in prostate cancer therapy resistance. Clin. Cancer Res 10.1158/1078-0432.CCR-19-1423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Palmgren JS, Karavadia SS & Wakefield MR Unusual and underappreciated: small cell carcinoma of the prostate. Semin. Oncol 34, 22–29 (2007). [DOI] [PubMed] [Google Scholar]
- 87.Epstein JI et al. Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am. J. Surg. Pathol 38, 756–767 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Beltran H et al. A phase 2 study of the aurora kinase A inhibitor alisertib for patients with neuroendocrine prostate cancer (NEPC) [abstract]. Ann. Oncol 27 (Suppl. 6), LBA29 (2016). [Google Scholar]
- 89.Gong X et al. Aurora A kinase inhibition is synthetic lethal with loss of the RB 1 tumor suppressor gene. Cancer Discov 9, 248–263 (2019). [DOI] [PubMed] [Google Scholar]
- 90.Puca L et al. Delta-like protein 3 expression and therapeutic targeting in neuroendocrine prostate cancer. Sci. Transl Med 11,eaav0891 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baca SC et al. Punctuated evolution of prostate cancer genomes. Cell 153, 666–677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Taylor RA et al. Germline BRCA2 mutations drive prostate cancers with distinct evolutionary trajectories. Nat. Commun 8, 13671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nava Rodrigues D et al. RB 1 heterogeneity in advanced metastatic castration-resistant prostate cancer. Clin. Cancer Res 25,687–697 (2019). [DOI] [PubMed] [Google Scholar]
- 94.Easwaran H, Tsai HC & Baylin SB Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol. Cell 54, 716–727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Varambally S et al. The polycomb group protein EZH2 is involved in progression of prostate cancer.Nature 419, 624–629 (2002). [DOI] [PubMed] [Google Scholar]
- 96.Beltran H et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov 1,487–495 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dardenne E et al. N-myc Induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell 30, 563–577 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.US National Library of Medicine. ClinicalTrials.gov https://cIinicaitrials.gov/show/NCT03480646 (2019).
- 99.US National Library of Medicine. ClinicalTriais.gov https://clinicaltrials.gov/show/NCT03460977 (2019).
- 100.Stathis A & Bertoni F BET proteins as targets for anticancer treatment. Cancer Discov 8, 24–36 (2018). [DOI] [PubMed] [Google Scholar]
- 101.Asangani IA et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 510, 278–282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Welti J et al. Targeting bromodomain and extra-terminal (BET) family proteins in castration-resistant prostate cancer (CRPC). Clin. Cancer. Res 24, 3149–3162(2018). [DOI] [PubMed] [Google Scholar]
- 103.Asangani IA et al. BET bromodomain inhibitors enhance efficacy and disrupt resistance to AR antagonists in the treatment of prostate cancer.Mol. Cancer. Res 14, 324–331 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/show/NCT02711956 (2019).
- 105.Shi Y et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD 1. Cell 119, 941–953 (2004). [DOI] [PubMed] [Google Scholar]
- 106.Metzger E et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437,436–439 (2005). [DOI] [PubMed] [Google Scholar]
- 107.Sehrawat A et al. LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc. N atl Acad. Sci. USA 115, E4179–E4188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cai C et al. Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep 9, 1618–1627(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Haffner MC et al. Tracking the clonal origin of lethal prostate cancer. J. Clin. Invest 123, 4918–4922 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gundem G et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Aryee MJ et al. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci. Transl Med 5, 169ra 110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wyatt AW et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol 2, 1598–1606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Salvi S et al. Circulating AR copy number and outcome to enzalutamide in docetaxel-treated metastatic castration-resistant prostate cancer. Oncotarget 7, 37839–37845 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Conteduca V et al. Plasma androgen receptor (pAR) status and activity of taxanes in metastatic castration resistant prostate cancer (mCRPC) [abstract]. J. Clin. Oncol 36 (15 Suppl.), 5074–5074 (2018). [Google Scholar]
- 115.Conteduca V et al. Plasma androgen receptor and serum chromogranin 8, A in advanced prostate cancer.Sci. Rep 15442 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wyatt AW et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J. Natl Cancer Inst 109, djx 118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Beltran H et al. The initial detection and partial characterization of circulating tumor cells in neuroendocrine prostate cancer. Clin. Cancer Res 22, 1510–1519(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lambros MB et al. Single-cell analyses of prostate cancer liquid biopsies acquired by apheresis. Clin. Cancer Res 24, 5635–5644 (2018). [DOI] [PubMed] [Google Scholar]