SUMMARY
Structural lipids are mostly synthesized in the endoplasmic reticulum (ER), from which they are actively transported to the membranes of other organelles. Lipids can leave the ER through vesicular trafficking or non-vesicular lipid transfer and, curiously, both processes can be regulated either by the transported lipid cargos themselves or by different secondary lipid species. For most structural lipids, transport out of the ER membrane is a key regulatory component controlling their synthesis. Distribution of the lipids between the two leaflets of the ER bilayer or between the ER and other membranes is also critical for maintaining the unique membrane properties of each cellular organelle. How cells integrate these processes within the ER depends on fine spatial segregation of the molecular components and intricate metabolic channeling, both of which we are only beginning to understand. This review will summarize some of these complex processes and attempt to identify the organizing principles that start to emerge.
Keywords: Endoplasmic reticulum, phosphatidylinositol, phosphatidylserine, phosphatidylcholine, lipid transfer protein, membrane contact sites, non-vesicular lipid transfer
1. LOCALIZATION OF PHOSPHOLIPIDS VERSUS THEIR SYNTHESIZING ENZYMES
1.1. Localization of enzymes for de novo phosphatidic acid biosynthesis
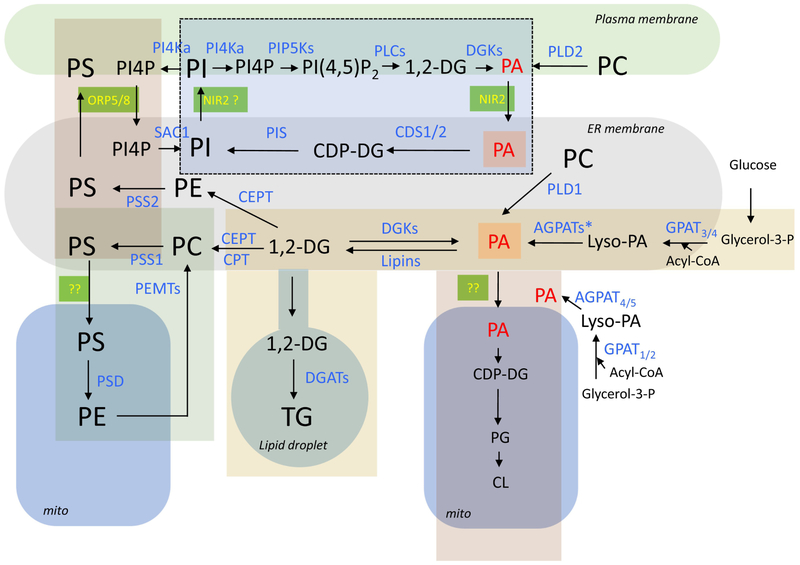
The biosynthetic pathways responsible for the synthesis of various phospholipids were already established between the late 1950s and early 1970s and have been thoroughly summarized in many excellent reviews (e.g. [1, 2] (Figure 1)). These intricate metabolic pathways became increasingly complicated in the genomic era after the identification of multiple genes encoding enzymes that seemingly carry out the same enzymatic reaction. While many of these enzymes are located in the ER, as expected, the localization and membrane topology of some of the lipid synthesizing enzymes remain puzzling. Moreover, as many of the emergent enzymes are identified and classified by sequence homology, it is not always clear which lipid substrates they use within the intact cell. Good examples include the growing family of GPATs (glycero-3-phosphate acyl transferases), which transfer fatty acyl chains to glycerol-3-phosphate (G3P) as the first step in de novo lipid synthesis. GPAT3 and GPAT4 are integral ER-resident membrane proteins, whereas GPAT1 and GPAT2 are localized to the outer mitochondrial membrane [3, 4]. Some of these enzymes, such as GPAT4 [5], are critical for triacylglycerol (TG) synthesis and lipid droplet (LD) biogenesis while others are involved in controlling phospholipid synthesis. However, the exact contribution of these enzymes to these respective pathways is still not all that clear. Similarly, some of the AGPAT (acyl-glycero-phosphate acyl transferase) enzymes that catalyze the acylation of lyso-phospholipids are found in the outer mitochondrial membrane (e.g. AGPAT4 and AGPAT5) [3], whereas others are found in the ER. AGPAT enzymes are mostly believed to use lyso-PA to generate PA in the de novo lipid synthetic pathway, but which isozymes are most important for phospholipid synthesis is not yet known. Interestingly, Agpat4-/- mice show significant decreases in phosphatidylinositol (PI), phosphatidylcholine (PC), and phosphatidylethanolamine (PE) but not in phosphatidic acid (PA), phosphatidylserine (PS), phosphatidylglycerol (PG) or cardiolipin levels in the brain [6]; suggesting that PA made on the outer surface of the mitochondria by this enzyme has to find its way to the ER, where the affected phospholipids are known to be made, and that not all phospholipid synthetic pathways are fed uniformly through the action of this enzyme.
Figure 1:
Enzymatic pathways of lipid synthesis and their membrane localization in a generic mammalian cell. The enzymes are labeled blue and the lipids are black except PA (red) which represents a major hub for several metabolic pathways. Major organelles are designated by the rounded shapes. Metabolic units that have been linked are designated by rectangles of different colors. The dotted lines denote inositol lipids that serve as signaling entities. Lipid transfer proteins working in contact sites are labeled yellow on green background. Note that lipids in the ER can be part of multiple metabolic units and how these are kept separated is one of the important questions yet to be understood. Abbreviations of enzymes: PI4K, PI 4-kinase; PIP5K, PI4P 5-kinase; PLC, phospholipase C; DGK, diacylglycerol kinase; PLD, phospholipase D; Sac1, Sac1 phosphatase; PIS, phosphatidylinositol synthase; CDS, CDP-DG synthase; PSS, phosphatidylserine synthase; CEPT, choline/ethanolamine phosphotransferase; CPT, CDP-choline:1,2-diacylglycerolphosphocholine transferase; PSD, phosphatidylserine decarboxylase; PEMT, Phosphatidylethanolamine N-methyltransferase; DGAT, diacylglycerol acyl transferase; AGPATs, 1-Acylglycerol-3-Phosphate O-Acyltransferase (* denotes that several forms of these enzymes exist in the ER. AGPAT4 and −5 are found in mitochondria); GPAT, Glycerol-3-Phosphate Acyltransferase. Abbreviation of lipids: PI, phosphatidylinositol; PI4P, phosphatidylinositol 4-phosphate; PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate; DG, diacylglycerol; PA, phosphatidic acid; PS, phosphatidylserine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; TG, triacylglycerol; PG, phosphatidylglycerol; CL, cardiolipin; CDP-DG, cytidine-diphosphate-diacylglycerol.
1.2. Localization of enzymes required for phospholipid biosynthesis
The location of the enzymes directly involved in phospholipid synthesis is similarly puzzling. For example, PC, which is present in every membrane, is synthesized by either the Golgi-localized CDP-choline:1,2-diacylglycerolphosphocholine transferase (CPT) enzyme or by the ER-localized choline/ethanolamine phosphotransferase (CEPT) enzyme using diacylglycerol (DG) and CDP-choline as precursors [7, 8]. However, the enzymes that generate CDP-choline from phosphocholine and CTP (CTP:phosphocholine cytidylyltransferase, or CCTα and CCTβ) are soluble, peripheral membrane proteins, which are found in the nucleus in quiescent cells. In response to oleic acid exposure, CCTα associates with intranuclear canalicular membranes and subsequently moves out from the nucleus to associate with LDs and promote LD formation as well as lipid storage [7, 9, 10]. How PC gets to other membranes from its site of synthesis is far from being understood, although several PC transport proteins have been identified [11].
The PS synthesizing enzymes, phosphatidylserine synthase (PSS)-1 and −2, that are responsible for exchanging the headgroups of PC and PE, respectively, for serine are multi-membrane-spanning proteins localized to the ER [12], but their enzymatic activities have been found highest in mitochondria-associated membranes (MAMs) [13]. MAMs are special membrane compartments where the ER and mitochondrial outer membrane form close contacts and allow non-vesicular lipid transfer. Despite this localization and confined catalytic activity, the bulk of membrane PS is not found in these sites; rather, it is enriched within the inner leaflet of the PM.
The enzymes CDP-diacylglycerol synthase (CDS)-1 and −2 that generate CDP-diacylglycerol (CDP-DG) from PA are also localized to the ER [14], while a third CDS enzyme, Tam41, is a structurally-distinct peripheral membrane protein that associates with the mitochondrial inner membrane and generates CDP-DG, the precursor of mitochondrial PG and cardiolipin [15, 16]. The phosphatidylinositol synthase (PIS) enzyme (also called CDIPT for CDP-diacylglycerol-inositol 3-phosphotransferase) that synthesizes PI from CDP-DG and myo-inositol is unique in that it is not only found in the ER proper but is also located in a highly mobile ER-derived membrane compartment, which appears to make contacts with all other organelles, including the PM [14]. The unique localization of the PIS enzyme requires its enzymatic activity as several point mutations that render the enzyme catalytically inactive eliminate its localization to the rapidly moving compartment [14]. The significance of this localization feature of the enzyme for PI synthesis is not clear at this point nor is it understood what other proteins or lipids are found in this unique membrane structure. At the same time, the distribution of PI among the various subcellular membranes is still very poorly understood.
PE, like PC, is synthesized by the ER-localized enzyme CEPT, but it is also made in the mitochondria via decarboxylation of PS. This latter reaction requires PS transport from the ER to the mitochondria. On the other hand, PE made in the mitochondria can find its way to be converted to PC again within the ER by the PE-methyl transferase (PEMT) enzyme (Fig.1). A recent studies suggests that mitofusin2 (Mfn2), one of the proteins that plays a role in mitochondrial fusion, facilitates PS availability to the mitochondrial PSD enzyme [17]. Although Mfn2 was found to bind PS, the exact mechanism of how it works remains elusive.
Taken together, just by looking at these enzymatic pathways, it is obvious that lipid transfer between organelles is critical for the metabolic flow required for controlling lipid synthesis. Moreover, compartmentalization or metabolic channeling must exist in order to make sense of the otherwise seemingly futile or redundant enzymatic cascades.
2. MOST PHOSPHOLIPID SYNTHESIZING ENZYMES ARE UNDER FEED-BACK INHIBITION
While the amounts of the enzymes required for phospholipid synthesis are subject to transcriptional regulation, their activities are also directly controlled by the lipid composition of the very membranes to which they are localized. Selected examples of these regulatory processes will be discussed below.
2.1. Phosphatidylserine synthesis
It was already recognized 10 years ago that PS synthesis is under strong feedback inhibition [18]. Studies on the cloned and expressed PSS1 and PSS2 enzymes showed that it was PSS2, rather than PSS1, that showed the marked inhibition by the enzymatic product, PS [18, 19]. In light of these early studies, it was a surprising development that the PTDSS1 gene that encodes human PSS1 was found mutated in the rare disease of Lenz-Majewski Syndrome in a manner that removed its end-product inhibition [20]. Cells of these patients display massive PS synthesis and display PS on the outer leaflet of the PM and secrete large amounts of PS. This is most likely caused by the saturation of the PM-localized PS flippase that maintains the PS asymmetry between the inner and outer leaflets of the PM. Since extracellular PS is critical for the bone calcification process, excessive PS secretion is probably a major factor in the hyperostotic phenotype associated with affected patients. Importantly, cells expressing PSS1 harboring the patient mutations also displayed large amounts of PS on the cytoplasmic leaflet of the ER, a compartment where PS is not detected even in cells overexpressing the wild-type PSS1 enzyme [12]. The accumulation of PS in the cytoplasmic leaflet of the ER is caused by the overburdened non-vesicular PS transport machinery. Curiously, all of the identified PSS1 mutations are found in the luminal side of the multi-membrane spanning PSS1 enzyme based on topology predictions [20]. If the topology predictions are correct, it raises the question of which side of the ER membrane contains the enzyme active site and which part of the PSS1 enzyme is responsible for PS sensing. The corollary of this enzyme regulation is that the rate of PS synthesis is tightly coupled to PS removal from the ER and when this process is impaired, PS synthesis is rapidly inhibited [12].
2.2. Phosphatidylinositol synthesis
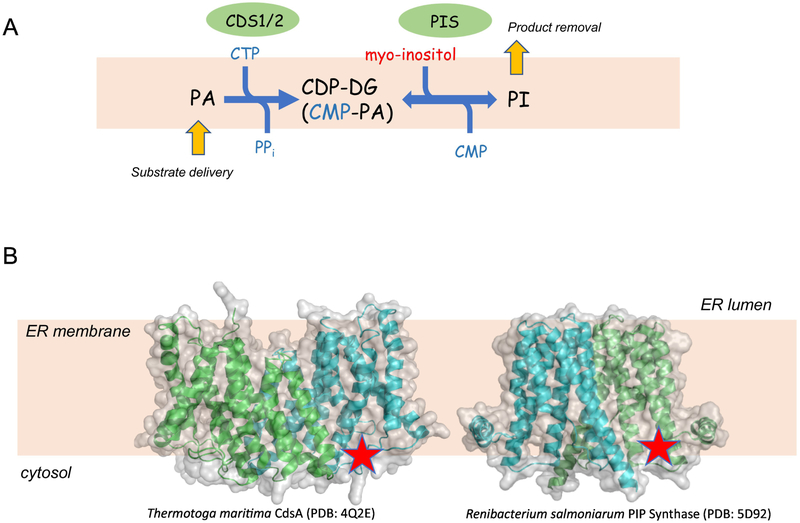
A different form of regulation is found with the PI synthesizing enzyme, PIS/CDIPT. This enzyme converts CDP-DG to PI using inositol in exchange for CMP. Curiously, the enzyme can also work in the reverse mode and, in the presence of PI and CMP, it will rapidly exchange the inositol headgroup; which explains the rapid [3H]-inositol labeling of PI in the presence of CMP [21, 22]. For a while, this inositol exchange activity, which is stimulated by Mn2+ in intact cells [23], was believed to be separate from the PI synthase, until the cloning of PIS proved that the two activities belong to the same protein [24, 25]. Although most of these studies have been done twenty or more years ago, they have important implications for understanding the control of PI synthesis, which is only now being realized. Namely, that the rate of PI synthesis is determined by the relative concentrations of the precursors CDP-DG and inositol as well as the enzymatic product, PI (Fig. 2A). This explains why the strongest stimulus of PI labeling with 3H-inositol is the phospholipase C (PLC)-mediated hydrolysis of PI(4,5)P2, which ultimate yields an increased supply of both CDP-DG and inositol [26]. This, however, also means that PI and PA transport between the ER and the PM is a critical determinant of the PI synthetic rate. This PI/PA exchange is mediated by the Nir2 protein [27-29] (Figure 1). PIS is an essential gene in yeast [30]; in vertebrates, PIS is required for survival of photoreceptor and lens epithelial cells [31]. There is no structural information available on mammalian PIS enzymes, but in yeast, the active site of the enzyme has been mapped facing the cytoplasm [32]. The X-ray structure of the Renibacterium salmoniarum PIP Synthase (the bacterial enzyme synthesizes PI3P from CDP-DG and inositol 3-phosphate) also suggests that the active site is facing the cytoplasm (Figure 2B).
Figure 2:
The enzymatic reactions of PI synthesis. (A) The two enzymes that work in tandem are CDS1/2 that convert PA to CDP-DG and, PIS, which converts CDP-DG to PI. Note that the PIS-catalyzed step is reversible and hence, the PI lipid product has to be removed in order to keep the reaction flowing to the right. Yellow arrows show that the direction of the reaction is determined by substrate delivery and product removal. (B) The X-ray structures of bacterial homologues (or ancestors) of the two enzymes are shown with the active site (red star) facing the cytoplasmic aspect of the ER membrane. The structures of the Thermotoga maritima CdsA (PDB: 4Q2E) (the closest bacterial homologue of the CDS enzymes) and the Renibacterium salmoniarum PIP Synthase (PDB: 5D92) (the closest bacterial homologue of the PIS enzyme) are shown using Pymol for rendering.
2.3. Phosphatidylcholine synthesis
The rate limiting step in PC biosynthesis is the generation of CDP-choline by the CCT enzymes. The two forms of CCT, CCTα and CCTβ, are encoded by separate genes, but CCTα is the more widely expressed form. CCTα is a soluble protein that is found primarily in the nucleoplasm, but it rapidly translocates to the nucleoplasmic reticulum, (a tubular membrane network connected to the nuclear envelope) when the relative PC content of membranes decreases (for original citations and more details on CCTs see [33]). CCTα also rapidly associates with LDs upon oleic acid loading, apparently through membrane-binding features that can detect the relatively PC-poor single layer phospholipid coat of LDs [7]. Specifically, membrane-association of CCTα is achieved through an amphipathic helix that somehow senses the ratio of PE/PC lipids present in membranes and membrane-association is important for increasing the enzyme’s catalytic activity [34]. The important question raised by these observations is how this localization of CCTα provides PC to the respective membranes given the fact that the final PC synthesizing step is mediated by the ER- and Golgi-localized CEPT and CPT enzymes, respectively.
2.4. Phosphatidylethanolamine synthesis
PE is generated by two independent pathways, one of which is analogous to that of PC synthesis, whereby CDP-ethanolamine is generated by the phosphoethanolamine cytidylyltransferase (ECT) enzyme, which is the rate-limiting step of this pathway. However, ECT it differs from CCTα in that its activity is not controlled by membrane association. The final step of this pathway is the conjugation of CDP-ethanolamine with DG by the ER-localized integral protein, CEPT [35]. The other major pathway of PE synthesis is PS decarboxylation by the phosphatidylserine decarboxylase (PSD) enzyme, which is located in the outer leaflet of the inner mitochondrial membrane. It is notable that yeast also contains another PSD, Psd2, within the ER [36]. Importantly, the PSD enzyme requires the transport of PS from the ER to the mitochondria and further from the outer into the inner mitochondrial membrane. This entire process was found to be ATP-dependent [37]; however, more recent studies have shown that PS transport from the ER to the mitochondria does not require ATP or GTP [38]. While the mechanism responsible for the transport of PS from the ER to the mitochondria is not known, the transport of PS between the outer and inner mitochondrial membranes is mediated by the Ups2-Mdm35 protein complex [39]. As mentioned before, Mfn2 was found to facilitate PS availability to the mitochondrial PSD enzyme [17]. Some of the mitochondrial PE has been shown to exit the mitochondria and is transported to the ER by a yet unknown mechanism where it can be converted to PC [40, 41]. These lipid transport steps between the ER and the mitochondria occur in the MAMs, the contact sites formed between the two organelles, as the efficiency of these transport-coupled enzymatic steps are greatly reduced in yeast cells lacking members of the organelle tethering ERMES (endoplasmic reticulum (ER)-mitochondria encounter structure) complex [39].
3. LIPID TRANSPORT IS CRITICAL FOR THE CONTROL OF LIPID SYNTHESIS
The picture that emerges from the above examples is that the synthesis of phospholipids must be tightly coupled to the transport processes that prevent accumulation of the lipid products at the site of synthesis. It is entirely possible, that the activity of lipid synthesizing enzymes were found highest in membrane preparations that contained contact sites, because the lipid products can be moved away from the ER in such preparations. Lipids can be transported out from the ER by lipid transfer proteins (LTPs) that often, although not always, function at membrane contact sites. Alternatively, lipids can also leave the ER by vesicular transport in the form of complete budded membranes. Non-vesicular lipid transport is much more selective than bulk membrane transport and has received an immense amount of attention in recent years.
3.1. Non-vesicular lipid transport at membrane contact sites
Several excellent recent reviews have covered non-vesicular transport extensively [11, 42, 43]. Therefore, we will discuss this topic only from the standpoint of lipid dynamics at the ER. An early indication that phosphoinositide lipids are important for non-vesicular lipid transport came from studies that identified relatively specific binding of PI4P to several pleckstrin-homology (PH) domains found within LTPs [44, 45]. The connection to PI4P was further established, and the critical role of non-vesicular lipid transfer relative to vesicular trafficking was emphasized, by the discovery of the ceramide transfer protein, CERT, and its central importance in supplying ceramide from the ER for sphingomyelin biosynthesis in the Golgi [46]. However, the real breakthrough in understanding the importance of PI4P for non-vesicular lipid transfer came from the realization that OSBP and its yeast orthologue, Osh4/Kes4, use PI4P gradients between the Golgi and ER for driving cholesterol export from the ER to Golgi [47, 48]. This gradient is set up by phosphatidylinositol 4-kinase (PI4K) beta (PI4KB and Pik1 in yeast) on the Golgi side, and perhaps different PI4K, PI4K2A functioning in the TGN. In all cases, the Sac1 phosphatase that dephosphorylates PI4P in the ER ensures that PI4P is very low in the ER [48, 49]. This countertransport is based on the ability of the lipid transfer domain, which is named ORD in the case of OSBP and its homologues, to bind either PI4P or cholesterol, but not both at the same time, within the same lipid-binding pocket [47]. A recent study found that cholesterol transport from the ER directly to the PM is also mediated by another OSBP-related protein (ORP), ORP2, and suggested that the inositol lipid that is countertransported in the opposite direction is PI(4,5)P2, rather than PI4P [50].
The importance of PI4P gradients was subsequently shown in the context of PS transport from the ER to the PM. Here, the PI4KA enzyme generates PI4P in the PM and the Sac1 phosphatase facilitates the rapid removal PI4P within the ER. This process involves ORP5 and ORP8 in mammalian cells and Osh6 and perhaps some close relatives in yeast [51-53]. Due to the tight connection between PS transport and PI4KA function, inhibition of the PI4KA enzyme also halts PS synthesis [12]. In addition, PS has a strong stimulatory effect on the activity of the ER-localized Sac1 phosphatase [54]. The consequence of this regulation is that excess PS production, as in the case in the Lenz-Majewski PSS1 mutations, stimulates Sac1 activity and therefore affects the PI4P gradient across all ER contact sites causing secondary alterations in the transport of other lipids that rely upon the PI4P-driven countertransport mechanism [12].
The fact that PI4P can be removed from the PM by ORP5 and ORP8 has important implications to the production of the PI(4,5)P2 within the PM, for which PI4P also serves as a precursor. The fact that PM recruitment of the N-terminal PH domain and adjacent polybasic motifs of ORP5 and ORP8 relies on the levels of both PI4P and PI(4,5)P2 in the PM ensures that when PI4P or PI(4,5)P2 levels are low, these LTPs cease working. Alternatively, when PI(4,5)P2 levels are increasing, ORP8 joins ORP5 and by delivering more PI4P to the ER, they limit the amount of PI4P available for PI(4,5)P2 production [55]. In addition, ORP8 can directly remove PI(4,5)P2 from the PM [56]. These findings have highlighted the close connection between the regulation of PI4P and PI(4,5)P2 levels within the PM and the control of PS transport, which directly informs the rate of PS synthesis.
It has also been suggested that non-vesicular lipid transport at membrane contact sites was important for the control of PI synthesis and its integration into the cyclic model of inositol lipid-based signal transduction. As pointed out above in Section 2.2, PI synthesis in the ER is controlled by the relative abundance of both the substrate, PA (as a precursor of CDP-DG) and the product, PI. It has been known for a long time that the strongest stimulus of PI labeling by either 32P-phosphate or 3H-inositol was receptor-mediated activation of PLC [26]. Since PI is consumed in the PM as PLC hydrolyses PI(4,5)P2 that needs continuous replenishment from PI and PA is also generated in the PM during this process as DG is converted to PA, it has been postulated that a transport of PI from the ER to the PM and PA in the reverse direction was necessary for increased PI synthesis to occur in the ER [26]. As mentioned above, recent studies have found that the LTP Nir2 can fulfill both of these transport functions [27-29] as it is specifically recruited to ER-PM contact sites during PLC activation [57].
Additional mechanisms for PI transfer from the ER to the PM have been suggested by studies characterizing TMEM24, which tethers the ER to the PM and is important for Ca2+ signaling as well as insulin release from pancreatic beta cells [58, 59]. TMEM24 has an synaptotagmin-like mitochondrial-lipid-binding (SMP) domain, a characteristic feature of organelle-tethering proteins that are found in membrane contact sites [60, 61]. SMP domains are thought to act as hydrophobic tunnels capable of lipid transport along concentration gradients [62]. The SMP domain of TMEM24 was found to deliver PI from the ER to the PM during glucose-induced secretion [58]. In contrast, the SMP domain of the family of extended synaptotagmins (E-Syt1/2/3) appears to be important for DG transport from the PM to the ER during high Ca2+-induced PLC activation [63]. The unique localization of the PIS enzyme not only in the ER proper but in a highly mobile vesicular structure that contacts various membranes, and also spends extended periods (minutes) in repeated contact with the PM, raised the possibility that PI transfer may also take place in more dynamic contact sites between the PM and this unique organelle that function in addition to the more classical ER-PM contacts discussed above [14].
There is a common theme in these examples, namely the close connection between all these lipid transport processes and PI or its phosphorylated derivatives. The reliance of many lipid transport processes on PI4P gradients really calls for a better understanding of the transport pathway(s) responsible for the redistribution of PI between membranes. The long-known mammalian PI transfer proteins (PITPs) and the Sec14-related proteins in yeast have been the logical candidates to fulfill this function since they have been shown to be able to transport PI (and PC) between membranes in vitro. However, it has been more difficult to identify their PI transfer functions in vivo within intact cells despite large amount of work devoted to study these proteins. As already mentioned, the Class II PITPs, Nir2 and Nir3 (and their fly orthologue, RdgBα) has been identified as PA/PI lipid exchangers at ER-PM contacts during PLC activation, but it is not clear if that is all that these proteins do since Nir2 was also found to be critical for the maintenance of DG levels in the Golgi [64]. Whether Sec14 or the Class I PITPs work as bona fide PI transfer proteins, however, has been challenged. It has been suggested that these proteins work by presenting PI to specific PI kinases [65, 66] and even function in very specific biological contexts [67]. Most lipid transfer proteins work in membrane contact sites, but there is no reported enrichment of Class I PITPs in such sites, although the TGN localization of PITPβ [68] could reflect localization to ER-Golgi contact sites. It has also been speculated that the ER-derived mobile PIS organelle (see above) that makes ample contacts with the different membranes may be a way PI is delivered to those membranes. Since no membrane fusion has been detected between the PIS organelle with any membranes, the transfer of lipid would still require a PI transfer protein [14].
PITPs have been covered over the years in many excellent reviews (for two recent ones see [69, 70]) and will not be discussed in further details.
3.2. Lipid regulation of vesicular transport at the ER
The assembly of COPII coat components and their roles in ER membrane budding in the process of Golgi-destined vesicular carrier generation has been elegantly worked out [71]. What is less understood, however, is how lipids contribute to the control of this process. That said, the lipid-modifying enzyme, phospholipase D (PLD) was found to be critical for COPII vesicle formation and cargo exit from the ER [72]. It has also been shown that Sar1, the small GTPase that is first recruited to the ER and marks the site of COPII coat formation, stimulates PLD activity thereby enriching PA at these sites [73]. Sar1 activation, which includes insertion of its N-terminal helix into the ER membrane, recruits the Sec23-Sec24 heterodimer as well as additional coat proteins that function to generate the curvature of the growing bud (for details see [71]). Importantly, a Sec23-interacting protein (Sec23ip, also called p125A), which is also enriched in ER exit sites, is a member of a family of three proteins that possess DDHD domains. (DDHD domains are 180 amino acid protein modules that selectively binds PA and can also be found in the lipid transfer proteins, Nir2/3 [57]). The other two proteins of this family, DDHD1 and DDHD2 are phospholipase A1 (PLA) enzymes capable of generating lyso-PI and lyso-PA, respectively (PLA activity has not been demonstrated for Sec23ip/p125 [74]. That said, Sec23ip/p125 was found to be important in the functional assembly of ER exit sites [75, 76] and was shown to depend on both PA and PI4P [77]. Sar1 was also shown to promote the local generation of PI4P, a lipid that promoted Sar1-induced membrane constriction and COPII assembly in an in vitro ER-budding assay, presumably through the activity of one class of the four PI 4-kinases (PI4Ks), namely the type II PI4Ks [73]. Another report found that the number of ER exit sites was decreased in cells depleted in another PI4K, the type III PI4K, PI4KA [78], although this effect could be secondary as many changes to cellular lipid levels have been shown to follow PI4KA depletion [79]. These findings suggest that lipid remodeling by lyso-lipid generating enzymes likely plays a role in helping to shape the membrane at ER exit sites. Indeed, a recent study showed that lyso-lipids, and Lyso-PI (16:0) in particular, could rescue Sec-14 temperature-sensitive mutant yeast strains and that this lipid also increased binding of COPII proteins to membranes as well as lowered the rigidity of reconstituted in giant unilamellar vesicles (GUVs) [80].
Another interesting piece of information related to the ER exit process is the finding that expression of PIS-GFP, but not mutant forms that eliminate enzymatic activity, generate rapidly moving ER-derived vesicles that do not contain any of the classical ER-markers but are enriched in the PIS enzyme itself [14]. This process also requires Sar1 and Arf1 [14]. This dynamic membrane compartment was found to contain other lipid synthesizing enzymes, such as CEPT1 and is regulated by Rab10 [81]. Another report showed that the LYCAT enzyme, which remodels acyl-chains in PI and specifically contributes to the enrichment of PI(4,5)P2 in arachidonic acid at the sn-2 position, also co-localizes with PIS in this unique membrane structure [82]. The importance of these observations is not clear at present, but they also point to the possible role of lyso-lipids and lipid remodeling in membrane budding processes.
CONCLUDING REMARKS
The specific lipid composition of membranes is critical for defining organelle identity and the proper assembly and function of protein complexes that work on those organelles. Lipid synthesis and distribution are fine-tuned to maintain the unique lipid composition of membranes. The product inhibition characterized in most lipid synthesizing enzymes makes lipid transport processes not only important for lipid distribution, but also for the dynamic regulation of lipid synthesis. Non-vesicular lipid transfer, often confined to membrane contact sites, provides both the speed and specificity requisite for the intricate control of lipid transport and synthesis. It is also important that non-vesicular lipid transport only samples the cytoplasmic leaflet of the membrane bilayer, which clearly contrasts vesicular transport, which carries both membrane layers. However, it is still not clear whether tethering proteins only form the molecular architecture of membrane contact sites or if they also play direct role(s) in the lipid-transfer process. Tethering proteins that have SMP domains possess the structural features that could serve as hydrophobic lipid tunnels bridging the aqueous phase between two membranes but do not explain how specificity of lipid transport can be achieved. The recent developments pointing to the importance of PI4P gradients in driving many non-vesicular lipid transport processes raise the question of how general this principle is among the various LTPs. Additionally, even for LTPs with defined lipid-binding pockets, there are still outstanding questions concerning cargo specificity and many LTPs may be more promiscuous in their selection of lipid cargoes than currently believed. In fact, it is becoming apparent that the local composition of the membranes between which LTPs operate may ultimately determine what lipid transport processes they support. This possibility places a special emphasis on methods that analyze lipid transfer processes in situ using intact cells, which will complement the in vitro lipid transfer assays already employed by many studies. Another important point to consider is the speed of these processes, which makes it extremely difficult to analyze them inside living cells as a new steady state can be established extremely quickly when lipid changes are induced. These challenges will require improvement of methods as well as a new way of thinking about old problems in order to make significant progress in this exciting research field.
Highlights.
Lipid synthesizing enzymes are under product inhibition
Lipid transport from the ER helps maintain lipid synthesis
Inositol lipids control lipid transport between membranes
ACKNOWLEDGEMENT:
We would like to thank Dr. Joshua Pemberton for many fruitful discussions and for his suggestions during the writing of this manuscript. The work of the authors is supported by the intramural research program of the Eunice Kennedy Shriver NICHD, at the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The Authors declare no conflict of interest related to this work.
REFRENCES
- [1].Kent C, Eukaryotic phospholipid biosynthesis, Annu Rev Biochem, 64 (1995) 315–343. [DOI] [PubMed] [Google Scholar]
- [2].Yang Y, Lee M, Fairn GD, Phospholipid subcellular localization and dynamics, J Biol Chem, 293 (2018) 6230–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gonzalez-Baro MR, Coleman RA, Mitochondrial acyltransferases and glycerophospholipid metabolism, Biochim Biophys Acta, 1862 (2017) 49–55. [DOI] [PubMed] [Google Scholar]
- [4].Wang H, Airola MV, Reue K, How lipid droplets "TAG" along: Glycerolipid synthetic enzymes and lipid storage, Biochim Biophys Acta, 1862 (2017) 1131–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Frohlich F, Liu X, Buhman KK, Coleman RA, Bewersdorf J, Farese RV Jr., Walther TC, Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets, Dev Cell, 24 (2013) 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bradley RM, Marvyn PM, Aristizabal Henao JJ, Mardian EB, George S, Aucoin MG, Stark KD, Duncan RE, Acylglycerophosphate acyltransferase 4 (AGPAT4) is a mitochondrial lysophosphatidic acid acyltransferase that regulates brain phosphatidylcholine, phosphatidylethanolamine, and phosphatidylinositol levels, Biochim Biophys Acta, 1851 (2015) 1566–1576. [DOI] [PubMed] [Google Scholar]
- [7].Krahmer N, Guo Y, Wilfling F, Hilger M, Lingrell S, Heger K, Newman HW, Schmidt-Supprian M, Vance DE, Mann M, Farese RV Jr., Walther TC, Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase, Cell Metab, 14 (2011) 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Henneberry AL, Wright MM, McMaster CR, The major sites of cellular phospholipid synthesis and molecular determinants of Fatty Acid and lipid head group specificity, Mol Biol Cell, 13 (2002) 3148–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Aitchison AJ, Arsenault DJ, Ridgway ND, Nuclear-localized CTP:phosphocholine cytidylyltransferase alpha regulates phosphatidylcholine synthesis required for lipid droplet biogenesis, Mol Biol Cell, 26 (2015) 2927–2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Haider A, Wei YC, Lim K, Barbosa AD, Liu CH, Weber U, Mlodzik M, Oras K, Collier S, Hussain MM, Dong L, Patel S, Alvarez-Guaita A, Saudek V, Jenkins BJ, Koulman A, Dymond MK, Hardie RC, Siniossoglou S, Savage DB, PCYT1A Regulates Phosphatidylcholine Homeostasis from the Inner Nuclear Membrane in Response to Membrane Stored Curvature Elastic Stress, Dev Cell, 45 (2018) 481–495 e488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wong LH, Gatta AT, Levine TP, Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes, Nat Rev Mol Cell Biol, (2018). [DOI] [PubMed] [Google Scholar]
- [12].Sohn M, Ivanova P, Brown HA, Toth DJ, Varnai P, Kim YJ, Balla T, Lenz-Majewski mutations in PTDSS1 affect phosphatidylinositol 4-phosphate metabolism at ER-PM and ER-Golgi junctions, Proc Natl Acad Sci U S A, 113 (2016) 4314–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Stone SJ, Vance JE, Phosphatidylserine synthase-1 and −2 are localized to mitochondria-associated membranes, J Biol Chem, 275 (2000) 34534–34540. [DOI] [PubMed] [Google Scholar]
- [14].Kim YJ, Guzman-Hernandez ML, Balla T, A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes, Dev Cell, 21 (2011) 813–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tamura Y, Harada Y, Nishikawa S, Yamano K, Kamiya M, Shiota T, Kuroda T, Kuge O, Sesaki H, Imai K, Tomii K, Endo T, Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria, Cell Metab, 17 (2013) 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Blunsom NJ, Gomez-Espinosa E, Ashlin TG, Cockcroft S, Mitochondrial CDP-diacylglycerol synthase activity is due to the peripheral protein, TAMM41 and not due to the integral membrane protein, CDP-diacylglycerol synthase 1, Biochim Biophys Acta Mol Cell Biol Lipids, 1863 (2018) 284–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hernandez-Alvarez MI, Sebastian D, Vives S, Ivanova S, Bartoccioni P, Kakimoto P, Plana N, Veiga SR, Hernandez V, Vasconcelos N, Peddinti G, Adrover A, Jove M, Pamplona R, Gordaliza-Alaguero I, Calvo E, Cabre N, Castro R, Kuzmanic A, Boutant M, Sala D, Hyotylainen T, Oresic M, Fort J, Errasti-Murugarren E, Rodrigues CMP, Orozco M, Joven J, Canto C, Palacin M, Fernandez-Veledo S, Vendrell J, Zorzano A, Deficient Endoplasmic Reticulum-Mitochondrial Phosphatidylserine Transfer Causes Liver Disease, Cell, 177 (2019) 881–895 e817. [DOI] [PubMed] [Google Scholar]
- [18].Tomohiro S, Kawaguti A, Kawabe Y, Kitada S, Kuge O, Purification and characterization of human phosphatidylserine synthases 1 and 2, Biochem J, 418 (2009) 421–429. [DOI] [PubMed] [Google Scholar]
- [19].Stone SJ, Vance JE, Cloning and expression of murine liver phosphatidylserine synthase (PSS)-2: differential regulation of phospholipid metabolism by PSS1 and PSS2, Biochem J, 342 (Pt 1) (1999) 57–64. [PMC free article] [PubMed] [Google Scholar]
- [20].Sousa SB, Jenkins D, Chanudet E, Tasseva G, Ishida M, Anderson G, Docker J, Ryten M, Sa J, Saraiva JM, Barnicoat A, Scott R, Calder A, Wattanasirichaigoon D, Chrzanowska K, Simandlova M, Van Maldergem L, Stanier P, Beales PL, Vance JE, Moore GE, Gain-of-function mutations in the phosphatidylserine synthase 1 (PTDSS1) gene cause Lenz-Majewski syndrome, Nat Genet, 46 (2014) 70–76. [DOI] [PubMed] [Google Scholar]
- [21].Agranoff BW, Bradley RM, Brady RO, The enzymatic synthesis of inositol phosphatide, J.Biol.Chem, 233 (1958) 1077–1083. [PubMed] [Google Scholar]
- [22].Cubitt AB, Gershengorn MC, CMP activates reversal of phosphatidylinositol synthase and base exchange by distinct mechanisms in rat pituitary GH3 cells, Biochem J, 272 (1990) 813–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schoepp DD, Manganese stimulates the incorporation of [3H]inositol into a pool of phosphatidylinositol in brain that is not coupled to agonist-induced hydrolysis, J Neurochem, 45 (1985) 1481–1486. [DOI] [PubMed] [Google Scholar]
- [24].Tanaka S, Nikawa J, Imai H, Yamashita S, Hosaka K, Molecular cloning of rat phosphatidylinositol synthase cDNA by functional complementation of the yeast Saccharomyces cerevisiae pis mutation, FEBS Lett, 393 (1996) 89–92. [DOI] [PubMed] [Google Scholar]
- [25].Lykidis A, Jackson PD, Rock CO, Jackowski S, The role of CDP-diacylglycerol synthetase and phosphatidylinositol synthase activity levels in the regulation of cellular phosphatidylinositol content, J Biol Chem, 272 (1997) 33402–33409. [DOI] [PubMed] [Google Scholar]
- [26].Michell RH, Inositol phospholipids and cell surface receptor function, Biochim.Biophys.Acta, 415 (1975) 81–147. [DOI] [PubMed] [Google Scholar]
- [27].Kim YJ, Guzman-Hernandez ML, Wisniewski E, Balla T, Phosphatidylinositol-Phosphatidic Acid Exchange by Nir2 at ER-PM Contact Sites Maintains Phosphoinositide Signaling Competence, Dev Cell, 33 (2015) 549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chang CL, Hsieh TS, Yang TT, Rothberg KG, Azizoglu DB, Volk E, Liao JC, Liou J, Feedback regulation of receptor-induced ca(2+) signaling mediated by e-syt1 and nir2 at endoplasmic reticulum-plasma membrane junctions, Cell Rep, 5 (2013) 813–825. [DOI] [PubMed] [Google Scholar]
- [29].Yadav S, Garner K, Georgiev P, Li M, Gomez-Espinosa E, Panda A, Mathre S, Okkenhaug H, Cockcroft S, Raghu P, RDGBalpha, a PtdIns-PtdOH transfer protein, regulates G-protein-coupled PtdIns(4,5)P2 signalling during Drosophila phototransduction, J Cell Sci, 128 (2015)3330–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nikawa J, Kodaki T, Yamashita S, Primary structure and disruption of the phosphatidylinositol synthase gene of Saccharomyces cerevisiae, J Biol Chem, 262 (1987) 4876–4881. [PubMed] [Google Scholar]
- [31].Murphy TR, Vihtelic TS, Ile KE, Watson CT, Willer GB, Gregg RG, Bankaitis VA, Hyde DR, Phosphatidylinositol synthase is required for lens structural integrity and photoreceptor cell survival in the zebrafish eye, Exp Eye Res, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bochud A, Conzelmann A, The active site of yeast phosphatidylinositol synthase Pis1 is facing the cytosol, Biochim Biophys Acta, 1851 (2015) 629–640. [DOI] [PubMed] [Google Scholar]
- [33].Cornell RB, Ridgway ND, CTP:phosphocholine cytidylyltransferase: Function, regulation, and structure of an amphitropic enzyme required for membrane biogenesis, Prog Lipid Res, 59 (2015) 147–171. [DOI] [PubMed] [Google Scholar]
- [34].Lee J, Taneva SG, Holland BW, Tieleman DP, Cornell RB, Structural basis for autoinhibition of CTP:phosphocholine cytidylyltransferase (CCT), the regulatory enzyme in phosphatidylcholine synthesis, by its membrane-binding amphipathic helix, J Biol Chem, 289 (2014) 1742–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].van der Veen JN, Kennelly JP, Wan S, Vance JE, Vance DE, Jacobs RL, The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease, Biochim Biophys Acta Biomembr, 1859 (2017) 1558–1572. [DOI] [PubMed] [Google Scholar]
- [36].Trotter PJ, Wu WI, Pedretti J, Yates R, Voelker DR, A genetic screen for aminophospholipid transport mutants identifies the phosphatidylinositol 4-kinase, Stt4p, as an essential component in phosphatidylserine metabolism, J.Biol.Chem, 273 (1998) 13189–13196. [DOI] [PubMed] [Google Scholar]
- [37].Voelker DR, Phosphatidylserine translocation to the mitochondrion is an ATP-dependent process in permeabilized animal cells, Proc Natl Acad Sci U S A, 86 (1989) 9921–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kojima R, Endo T, Tamura Y, A phospholipid transfer function of ER-mitochondria encounter structure revealed in vitro, Sci Rep, 6 (2016) 30777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Miyata N, Watanabe Y, Tamura Y, Endo T, Kuge O, Phosphatidylserine transport by Ups2-Mdm35 in respiration-active mitochondria, J Cell Biol, 214 (2016) 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kainu V, Hermansson M, Hanninen S, Hokynar K, Somerharju P, Import of phosphatidylserine to and export of phosphatidylethanolamine molecular species from mitochondria, Biochim Biophys Acta, 1831 (2013) 429–437. [DOI] [PubMed] [Google Scholar]
- [41].Friedman JR, Kannan M, Toulmay A, Jan CH, Weissman JS, Prinz WA, Nunnari J, Lipid Homeostasis Is Maintained by Dual Targeting of the Mitochondrial PE Biosynthesis Enzyme to the ER, Dev Cell, 44 (2018) 261–270 e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lahiri S, Toulmay A, Prinz WA, Membrane contact sites, gateways for lipid homeostasis, Curr Opin Cell Biol, 33 (2015) 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mesmin B, Antonny B, The counterflow transport of sterols and PI4P, Biochim Biophys Acta, 1861 (2016) 940–951. [DOI] [PubMed] [Google Scholar]
- [44].Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR, Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities, Biochem.J, 351 (2000) 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Levine TP, Munro S, Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components, Curr.Biol, 12 (2002) 695–704. [DOI] [PubMed] [Google Scholar]
- [46].Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M, Molecular machinery for non-vesicular trafficking of ceramide, Nature, 426 (2003) 803–809. [DOI] [PubMed] [Google Scholar]
- [47].de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G, Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers, J Cell Biol, 195 (2011) 965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, Antonny B, A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP, Cell, 155 (2013) 830–843. [DOI] [PubMed] [Google Scholar]
- [49].Mesmin B, Bigay J, Polidori J, Jamecna D, Lacas-Gervais S, Antonny B, Sterol transfer, PI4P consumption, and control of membrane lipid order by endogenous OSBP, EMBO J, 36 (2017) 3156–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang H, Ma Q, Qi Y, Dong J, Du X, Rae J, Wang J, Wu WF, Brown AJ, Parton RG, Wu JW, Yang H, ORP2 Delivers Cholesterol to the Plasma Membrane in Exchange for Phosphatidylinositol 4, 5-Bisphosphate (PI(4,5)P2), Mol Cell, (2018). [DOI] [PubMed] [Google Scholar]
- [51].Moser von Filseck J, Copic A, Delfosse V, Vanni S, Jackson CL, Bourguet W, Drin G, INTRACELLULAR TRANSPORT. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate, Science, 349 (2015) 432–436. [DOI] [PubMed] [Google Scholar]
- [52].Maeda K, Anand K, Chiapparino A, Kumar A, Poletto M, Kaksonen M, Gavin AC, Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins, Nature, 501 (2013) 257–261. [DOI] [PubMed] [Google Scholar]
- [53].Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB, Narayanaswamy P, Wenk MR, Nakatsu F, De Camilli P, INTRACELLULAR TRANSPORT. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts, Science, 349 (2015) 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhong S, Hsu F, Stefan CJ, Wu X, Patel A, Cosgrove MS, Mao Y, Allosteric activation of the phosphoinositide phosphatase Sac1 by anionic phospholipids, Biochemistry, 51 (2012) 3170–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sohn M, Korzeniowski M, Zewe JP, Wills RC, Hammond GRV, Humpolickova J, Vrzal L, Chalupska D, Veverka V, Fairn GD, Boura E, Balla T, PI(4,5)P2 controls plasma membrane PI4P and PS levels via ORP5/8 recruitment to ER-PM contact sites, J Cell Biol, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ghai R, Du X, Wang H, Dong J, Ferguson C, Brown AJ, Parton RG, Wu JW, Yang H, ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane, Nat Commun, 8 (2017) 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kim S, Kedan A, Marom M, Gavert N, Keinan O, Selitrennik M, Laufman O, Lev S, The phosphatidylinositol-transfer protein Nir2 binds phosphatidic acid and positively regulates phosphoinositide signalling, EMBO Rep, 14 (2013) 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lees JA, Messa M, Sun EW, Wheeler H, Torta F, Wenk MR, De Camilli P, Reinisch KM, Lipid transport by TMEM24 at ER-plasma membrane contacts regulates pulsatile insulin secretion, Science, 355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pottekat A, Becker S, Spencer KR, Yates JR 3rd, Manning G, Itkin-Ansari P, Balch WE, Insulin biosynthetic interaction network component, TMEM24, facilitates insulin reserve pool release, Cell Rep, 4 (2013) 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Toulmay A, Prinz WA, A conserved membrane-binding domain targets proteins to organelle contact sites, J Cell Sci, 125 (2012) 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kopec KO, Alva V, Lupas AN, Homology of SMP domains to the TULIP superfamily of lipid-binding proteins provides a structural basis for lipid exchange between ER and mitochondria, Bioinformatics, 26 (2010) 1927–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Reinisch KM, De Camilli P, SMP-domain proteins at membrane contact sites: Structure and function, Biochim Biophys Acta, 1861 (2016) 924–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Saheki Y, Bian X, Schauder CM, Sawaki Y, Surma MA, Klose C, Pincet F, Reinisch KM, De Camilli P, Control of plasma membrane lipid homeostasis by the extended synaptotagmins, Nat Cell Biol, 18 (2016) 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Litvak V, Dahan N, Ramachandran S, Sabanay H, Lev S, Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function, Nat.Cell Biol, 7 (2005) 225–234. [DOI] [PubMed] [Google Scholar]
- [65].Schaaf G, Ortlund EA, Tyeryar KR, Mousley CJ, Ile KE, Garrett TA, Ren J, Woolls MJ, Raetz CR, Redinbo MR, Bankaitis VA, Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily, Mol Cell, 29 (2008) 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kular GS, Chaudhary A, Prestwich G, Swigart P, Wetzker R, Cockcroft S, Co-operation of phosphatidylinositol transfer protein with phosphoinositide 3-kinase gamma in vitro, Adv Enzyme Regul, 42 (2002) 53–61. [DOI] [PubMed] [Google Scholar]
- [67].Grabon A, Khan D, Bankaitis VA, Phosphatidylinositol transfer proteins and instructive regulation of lipid kinase biology, Biochim Biophys Acta, 1851 (2015) 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Phillips SE, Ile KE, Boukhelifa M, Huijbregts RP, Bankaitis VA, Specific and nonspecific membrane-binding determinants cooperate in targeting phosphatidylinositol transfer protein beta-isoform to the mammalian trans-Golgi network, Mol Biol Cell, 17 (2006) 2498–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Grabon A, Bankaitis VA, McDermott MI, The Interface Between Phosphatidylinositol Transfer Protein Function and Phosphoinositide Signaling in Higher Eukaryotes, J Lipid Res, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Cockcroft S, Raghu P, Phospholipid transport protein function at organelle contact sites, Curr Opin Cell Biol, 53 (2018) 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R, COPII and the regulation of protein sorting in mammals, Nat Cell Biol, 14 (2011) 20–28. [DOI] [PubMed] [Google Scholar]
- [72].Pathre P, Shome K, Blumental-Perry A, Bielli A, Haney CJ, Alber S, Watkins SC, Romero G, Aridor M, Activation of phospholipase D by the small GTPase Sar1p is required to support COPII assembly and ER export, EMBO J, 22 (2003) 4059–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Blumental-Perry A, Haney CJ, Weixel KM, Watkins SC, Weisz OA, Aridor M, Phosphatidylinositol 4-phosphate formation at ER exit sites regulates ER export, Dev Cell, 11 (2006) 671–682. [DOI] [PubMed] [Google Scholar]
- [74].Inoue H, Baba T, Sato S, Ohtsuki R, Takemori A, Watanabe T, Tagaya M, Tani K, Roles of SAM and DDHD domains in mammalian intracellular phospholipase A1 KIAA0725p, Biochim Biophys Acta, 1823 (2012) 930–939. [DOI] [PubMed] [Google Scholar]
- [75].Ong YS, Tang BL, Loo LS, Hong W, p125A exists as part of the mammalian Sec13/Sec31 COPII subcomplex to facilitate ER-Golgi transport, J Cell Biol, 190 (2010) 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Shimoi W, Ezawa I, Nakamoto K, Uesaki S, Gabreski G, Aridor M, Yamamoto A, Nagahama M, Tagaya M, Tani K, p125 is localized in endoplasmic reticulum exit sites and involved in their organization, J Biol Chem, 280 (2005) 10141–10148. [DOI] [PubMed] [Google Scholar]
- [77].Klinkenberg D, Long KR, Shome K, Watkins SC, Aridor M, A cascade of ER exit site assembly that is regulated by p125A and lipid signals, J Cell Sci, 127 (2014) 1765–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Farhan H, Weiss M, Tani K, Kaufman RJ, Hauri HP, Adaptation of endoplasmic reticulum exit sites to acute and chronic increases in cargo load, EMBO J, 27 (2008) 2043–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Nakatsu F, Baskin JM, Chung J, Tanner LB, Shui G, Lee SY, Pirruccello M, Haio M, Ingolia NT, Wenk MR, De Camilli P, PtdIns4P synthesis by PI4KIIIa at the plasma membrane and its impact on plasma membrane identity, J Cell Biol, 199 (2012) 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Melero A, Chiaruttini N, Karashima T, Riezman I, Funato K, Barlowe C, Riezman H, Roux A, Lysophospholipids Facilitate COPII Vesicle Formation, Curr Biol, 28 (2018) 1950–1958 e1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].English AR, Voeltz GK, Rab10 GTPase regulates ER dynamics and morphology, Nat Cell Biol, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Bone LN, Dayam RM, Lee M, Kono N, Fairn GD, Arai H, Botelho RJ, Antonescu CN, The acyltransferase LYCAT controls specific phosphoinositides and related membrane traffic, Mol Biol Cell, 28 (2017) 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]