Abstract
Graphene foam holds promise for tissue engineering applications. In this study, graphene foam was used as a three-dimension scaffold to evaluate cell attachment, cell morphology, and molecular markers of early differentiation. The aim of this study was to determine if cell attachment and elaboration of an extracellular matrix would be modulated by functionalization of graphene foam with fibronectin, an extracellular matrix protein that cells adhere well to, prior to the establishment of three-dimensional cell culture. The molecular dynamic simulation demonstrated that the fibronectin–graphene interaction was stabilized predominantly through interaction between the graphene and arginine side chains of the protein. Quasi-static and dynamic mechanical testing indicated that fibronectin functionalization of graphene altered the mechanical properties of graphene foam. The elastic strength of the scaffold increased due to fibronectin, but the viscoelastic mechanical behavior remained unchanged. An additive effect was observed in the mechanical stiffness when the graphene foam was both coated with fibronectin and cultured with cells for 28 days. Cytoskeletal organization assessed by fluorescence microscopy demonstrated a fibronectin-dependent reorganization of the actin cytoskeleton and an increase in actin stress fibers. Gene expression assessed by quantitative real-time polymerase chain reaction of 9 genes encoding cell attachment proteins (Cd44, Ctnna1, Ctnnb1, Itga3, Itga5, Itgav, Itgb1, Ncam1, Sgce), 16 genes encoding extracellular matrix proteins (Col1a1, Col2a1, Col3a1, Col5a1, Col6a1, Ecm1, Emilin1, Fn1, Hapln1, Lamb3, Postn, Sparc, Spp1, Thbs1, Thbs2, Tnc), and 9 genes encoding modulators of remodeling (Adamts1, Adamts2, Ctgf, Mmp14, Mmp2, Tgfbi, Timp1, Timp2, Timp3) indicated that graphene foam provided a microenvironment conducive to expression of genes that are important in early chondrogenesis. Functionalization of graphene foam with fibronectin modified the cellular response to graphene foam, demonstrated by decreases in relative gene expression levels. These findings illustrate the combinatorial factors of microscale materials properties and nanoscale molecular features to consider in the design of three-dimensional graphene scaffolds for tissue engineering applications.
Keywords: ATDC5, chondroprogenitor cells, graphene foam, fibronectin, extracellular matrix, differentiation, tissue engineering, three-dimensional cell culture, bioscaffold, molecular dynamic simulation, dynamic mechanical analysis
1. Introduction
Biophysical, biochemical, and biomechanical cues from the extracellular environment have a significant effect on cellular response. Synthetic materials can be tailored to mimic the extracellular matrix in a context-specific manner to allow an investigation into fundamental mechanisms that govern how cells sense and respond to their environment, which will aid in the design and development of biomaterials for tissue repair and regeneration. In this study, our goal was to investigate the cellular attachment and response to a graphene foam (GF) scaffold functionalized with the extracellular matrix molecule fibronectin. Our results highlight the suitability of GF as a scaffold for chondrogenesis and the influence of fibronectin in combination with a GF scaffold on such processes. Extracellular matrix functionalization can influence the measurable cellular response including cellular morphology, gene expression, and progress toward cellular differentiation outcomes.
Chondroprogenitor cells arise from several mesenchymal sources during vertebrate development, including the neural crest and the somites.1 Chondrogenesis is initiated as mesenchymal cells aggregate into condensations during skeletal development. During the process of condensation, the interactions among cells and between cells and matrix molecules are critical to the process.2,3 Cell–cell interactions are driven by cell surface receptors, and cell–matrix interactions are driven by extracellular matrix molecules, including collagens, proteoglycans, thrombospondins, laminins, and fibronectin.4,5 Prechondrogenic condensation is facilitated by extracellular matrix molecules, cell surface receptors and adhesion molecules.6 Fibronectin is essential in early embryogenesis and is upregulated in association with prechondrogenic condensations.7−9
Fibronectin, a ubiquitous extracellular matrix protein, is assembled into a fibrillar matrix through a cell-mediated process and links cells with other extracellular matrix proteins, including collagens.10 Fibronectin matrix assembly is essential for cell condensation during chondrogenesis. In the chondroprogenitor ATDC5 cell line, cell condensation, and induction of chondrogenesis are dependent on the assembly of the fibronectin matrix.8 During condensation, cells produce a unique transitional extracellular matrix that is rich in specific proteins including collagen type I and fibronectin.11 In contrast to the early matrix composition, the extracellular matrix molecules produced by mature differentiated chondrocytes is rich in collagen type II.12
Biomimetic scaffolds mimic the properties of a specific tissue environment. Three-dimensional scaffolds provide space in which cells can be trapped to foster cell–cell interaction, the establishment of a pericellular matrix and cell–matrix interactions, which together subsequently support cellular differentiation in response to local cues. In regenerative medicine, the biomimetic scaffold may be designed to recreate the native stem cell environment rather than that of the mature tissue. Novel scaffolds have been used to mimic the characteristics of the extracellular matrix of specific tissues, providing binding sites for ligands, timed-release of specific cytokines, and also the mechanical properties that are intrinsic to specific tissue types.13 GF as a biomimetic scaffold may provide a versatile platform upon which to design niche environments for stem cells and supply specific cues to drive differentiation of the cell and regeneration of the tissue.
The wide use of graphene scaffolds for stem cell investigation demonstrates the potential of graphene-based materials for the study of stem cell self-renewal, proliferation, and specific differentiation that will ultimately enable biomedical and regenerative medicine applications.14−19 To date, graphene-based materials have been used for the study of specific differentiation pathways including osteogenesis,20 neurogenesis,21 myogenesis,17,21,22 adipogenesis,23,24 chondrogenesis,15,25 and oligodendrogenesis.16 Although promising, what is not known currently are the conditions under which stem cell attachment can be fostered while also providing the specific molecular and biomechanical cues to promote differentiation along specific lineages to ultimately regenerate a functional tissue. This is particularly challenging in the field of cartilage tissue engineering due to cellular senescence, hypertrophy, and the dual potential for cells to convert to an osteoblast phenotype, resulting in mineralized tissue rather than cartilage.26,27
Previous studies have revealed the importance of surface roughness on cell–substrate interactions as well as the surface functionalization on cell attachment and behavior.28−30 Additionally, the effect of interfaces on cell attachment and differentiation have been extensively investigated, specifically the effect of surface rigidity and viscoelasticity.31−34 However, a need exists to increase our understanding of the interaction between stem cells and three-dimensional bioscaffolds and how the interaction influences cell morphology and gene expression patterns. Studies testing the combinatorial effects of graphene foam as a scaffold material plus a biological molecule such as fibronectin on both attachment and differentiation have not been performed for chondroprogenitor cells. A more in-depth and fundamental understanding will support future therapeutic applications in regenerative medicine.
The objective of this study was to identify differences between GF and fibronectin-derivatized GF with respect to cell attachment, cell morphology, and expression of genes encoding early indicators of differentiation. Here we show that chondroprogenitor ATDC5 cells adhere to fibronectin and GF. In response, cells adopt a distinct cellular morphology dependent on the presence or absence of fibronectin. Fibronectin on GF changed the elastic mechanical properties of the GF, yet no significant changes in the dynamic mechanical properties were detected. An additive effect was observed in the mechanical stiffness when the graphene foam was both coated with fibronectin and cultured with cells for 28 days. The fibronectin protein adhered to the GF surface via interactions involving arginine amino acid side groups. Cells responded to their environment by expressing specific genes in a differential manner that was dependent upon both the scaffold and the fibronectin. The results of this study indicate that GF in combination with ECM molecules to serve as a transitional matrix may provide the cellular niche to drive differentiation. An ECM molecule other than fibronectin will be required for the productive regeneration of challenging tissues such as cartilage.
2. Materials and Methods
2.1. Materials
Three dimensional GF was obtained from Graphene Laboratories (Graphene Laboratories Inc., Calverton, NY, U.S.A.). The scaffold used in these experiments comprised 7–10 atomic layers of graphene. The foam construct was two mm thick with a density of 4 mg/cm3, and a pore size of 580 μm. ATDC5 cells were obtained from Sigma-Aldrich (St. Louis, MO, U.S.A.). The ECM Select Array was obtained from Advanced BioMatrix (San Diego, CA). The prechondrogenic cell line ATDC5 was originally derived from the differentiating teratocarcinoma stem cell line AT805. ATDC5 cells undergo a sequential transition of phenotype in vitro, including stages from mesenchymal condensation to calcification.35 Bovine fibronectin protein solution was obtained from R & D Systems (Biotechne Corporation, Minneapolis, MN, U.S.A.) and diluted to a concentration of 100 μg/mL in Ca2+- and Mg2+-free phosphate-buffered saline (PBS). Paraformaldehyde and Triton X-100 were obtained from Sigma-Aldrich (St. Louis, MO). Block-Aid, Alexa Fluor 488 conjugated to phalloidin, and ProLong Gold Antifade with DAPI were obtained from Life Technologies (Carlsbad, CA). Glass bottom cell culture dishes were obtained from MatTek Corporation (Ashland, MA). Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM-F12) and fetal bovine serum (FBS) were obtained from Gibco by Life Technologies (Grand Island, NY). TRIzol reagent was obtained from Thermo Fisher Scientific (Hampton, NH).
2.2. Methods
2.2.1. ECM Protein Cell Attachment Assay
ECM Select Array was obtained from Advanced BioMatrix (San Diego, CA). The extracellular matrix screening array was composed of nine printed replicates of 400 μm diameter areas on glass functionalized with hydrogel printed with the extracellular matrix proteins at a concentration of 250 μg/mL. The following extracellular matrix proteins were screened for attachment: collagen I (COL I), collagen III (COL III), collagen IV (COL IV), collagen V (COL V), collagen VI (COL VI), fibronectin (FN), vitronectin (VTN), laminin (LMN), and tropoelastin (TE). Bovine serum albumin (BSA) was used as a negative control for attachment assays.
ATDC5 cells were seeded (5 × 104 cells/mL) to screen for cell adhesion to extracellular matrix proteins. After the ECM array was rinsed with PBS and conditioned for 5 min in the culture medium, five mL of cells suspended in culture medium was evenly distributed across the slide and incubated at 37 °C in 5% CO2. Attached cells were counted at 12 and 30 h for each extracellular matrix protein and each of nine replicates for each protein. Cell morphology and attachment were visualized using bright field microscopy. Cell counts were determined at 30 h and mean ± standard deviation was determined.
2.2.2. Molecular Dynamic Simulation of Fibronectin-Graphene Interaction
The PDB file for fibronectin type III domains 8–10 was obtained from the Protein Data Bank (PDB ID 1FNF).36 The fibronectin structure was placed atop of three 100 × 200 Å2 graphene sheets and neutralized in water using NaCl by assuming the height of the simulation box equal to 85 Å. The protocol was adopted from earlier molecular dynamics (MD) simulation on similar systems.37−40 Two different orientations of fibronectin on graphene were considered, resulting in two independent simulations. Each configuration was simulated for a total of 400 ns with an integrator time step of 2 fs under 1 bar pressure control, 310 K temperature control and using periodic boundaries using NAMD.41 Particle Mesh Ewald method was used to treat the long-range electrostatics42,43 with a cutoff distance of 1.2 Å. CHARMM 36 force field44−48 was used to model the interatomic interactions in both the protein and in the graphene sheet.
2.2.3. Culture Conditions for Seeding and Maintenance of ATDC5 Cells
GF coated with fibronectin was prepared by applying 700 μL of 100 μg/mL fibronectin solution to 1 cm2 × 2 mm GF, incubated at 37 °C for 1 h. Following the functionalization, GF scaffolds were conditioned for 24 h in cell culture medium.49
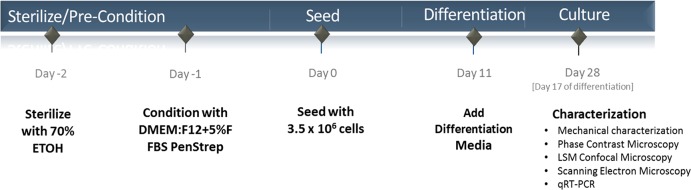
The GF scaffolds were seeded with 1.5 × 105 ATDC5 cells cultured for 24 h in DMEM/F-12 supplemented with 5% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in a humidified atmosphere, 5% CO2. During the seeding process, approximately 30% of the cells adhered to the GF or GF–fibronectin surface. Cells were maintained in parallel under 2-D culture conditions on glass-bottom tissue culture wells for comparison. At day 11 of the proliferation phase of the experiment, the growth medium was supplemented with 50 μg/mL ascorbate 2-phosphate, 10 mg/mL insulin, 5.5 mg/mL transferrin, and 6.7 μg/mL sodium selenite to induce chondrogenic differentiation. Samples for RNA extraction were collected 0, 3, and 7 days after initiation of differentiation from cells maintained in 2-D culture conditions and 17 days later for both 2-D and 3-D GF samples. Samples were collected on day 28 for the measurement of elastic and viscoelastic properties and for fluorescence imaging of the cytoskeleton. Final cell counts on GF scaffolds at day 28 was 8 × 105 per GF sample (n = 3). Cell proliferation was monitored and resulted in a 16-fold increase in cell number as cells underwent five cellular doublings during the proliferation phase prior to induction of differentiation. Representative bright-field images were collected using a Nikon TS-100 Microscope and SPOT R3 camera.
2.2.4. Confocal and Fluorescence Microscopy
Cells were fixed with a solution of 2% paraformaldehyde, permeabilized with 0.1% Triton X-100 (Sigma-Aldrich; St. Louis, MO), and treated to prevent nonspecific binding (BlockAid, Life Technologies; Carlsbad, CA). Cytoskeletal F-actin was detected with Alexa Fluor 488 conjugated to phalloidin, then mounted with ProLong Gold Antifade Mountant with DAPI (Life Technologies; Carlsbad, CA) to stain nuclei. Samples cured overnight before imaging. Slides were imaged with a Zeiss LSM 510 Meta system combined with the Zeiss Axiovert Observer Z2 inverted microscope and ZEN 2009 imaging software (Carl Zeiss, Inc., Thornwood, NY). Images were acquired in a single plane utilizing the Plan-Apochromat 20×/NA 0.8 and Fluar 40x/NA 1.30 Oil objectives. Transmitted light was collected on one channel during the z-stack acquisition to provide contrast to the GF structure. Confocal z-stack images were acquired utilizing the Plan-Apochromat 63X/NA 1.4 and alpha Plan-Fluar 100X/NA1.45 Oil objectives. All images were collected with a diode (405 nm) and Argon (488 nm) laser sources and the following band-pass emission filters: BP 420–480 BP 505–530. Images were processed with ZEN 2009 imaging software (Carl Zeiss, Inc., Thornwood, NY).
2.2.5. Scanning Electron Microscopy
Samples were fixed in 2.5% glutaraldehyde. After rinsing in deionized water, samples underwent dehydration using 50%, 70%, 90%, and 100% ethanol sequentially. After dehydration, the sample was taped to a silicon wafer for sputtering. The dehydrated GF with cells were sputter-coated with chromium using a CRC-150 (Torr Laboratories). A 12 nm coat was achieved after 75 s of exposure at 9.6 × 10–6 Torr and 50W. An FEI-Teneo scanning electron microscope set at 3.00 kV was used to collect images while utilizing the T2 detector by the Boise State Center for Materials Characterization.
2.2.6. Mechanical Testing of GF with Fibronectin and Cells
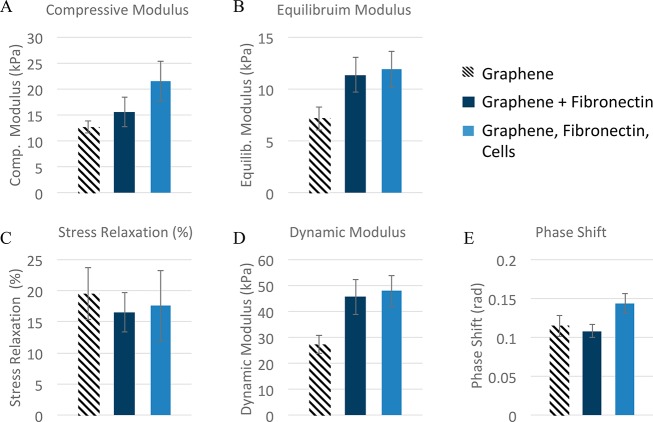
The dynamic mechanical analysis was carried out using the Instron ElectroPuls E-10000 mechanical test system (Instron, Norwood, MA) using previously described methods.49 In brief, at day 28, GF specimens (GF, GF + fibronectin, GF + fibronectin + cells) were subjected to cyclic preconditioning to 14% compression, quasi-static loading to 12% compression, 2 min of relaxation, and then 1 Hz cyclic compression at 1% amplitude, where compressive strain was calculated as the ratio of change in thickness to original thickness. The compressive elastic modulus, equilibrium modulus, stress relaxation, dynamic modulus, and phase shift were then calculated from the corresponding stress–strain waveform.
2.2.7. Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
RNA from each sample was extracted following the TRIzol protocol for RNA extraction (Thermo Fisher Scientific). Samples were flash-frozen with liquid nitrogen and then pulverized within the TRIzol reagent with an OMNI International TH homogenizer (Thomas Scientific). The RNA concentration was determined by measuring the absorbance at 260 and 280 nm. The RT2 First Strand synthesis method (Qiagen) was used to generate cDNA. Expression levels were measured by qRT-PCR using a Roche Lightcycler 96 (Roche). Genes analyzed included extracellular matrix proteins, matrix remodeling enzymes, and cell adhesion molecules. Relative gene expression levels, mean plus/minus standard deviation, were expressed with respect to housekeeping genes determined empirically for this study.
2.2.8. Selection of Housekeeping Genes
ActB and Hsp90ab1 were selected as the housekeeping gene for normalization in these experiments based on comparison to three other candidate housekeeping genes (Gapdh, B2m, and GusB) and were found to be stably expressed independent of experimental conditions based on minimal variance.50−52 Relative abundance values were calculated and reported here as mean plus/minus standard deviation.
2.2.9. Statistical Analysis
Cell attachment to extracellular matrix molecules was analyzed using the mean plus/minus standard deviation. The effect of culture time on the mechanical properties (compressive modulus, equilibrium modulus, stress relaxation, dynamic modulus, and phase shift) of the cellular graphene composites was analyzed using a one-way MANOVA in SPSS (p = 0.05) using the Least Significant Difference (LSD) correction for multiple comparisons. Selection of housekeeping genes for qRT-PCR was based on pairwise analysis of variance for differences between cycle threshold values for five candidate housekeeping genes from 15 samples within this study. Additionally, correlation analysis was carried out and data were fit to a trend line and R2 was determined. Relative expression of genes of interest was analyzed relative to average values for ActB and Hsp90ab1, and expressed as mean plus/minus standard deviation. Log transformed gene expression data was subject to a paired t test to determine if the differences in mean values for relative gene expression were statistically significant, setting significance at p < 0.05.
3. Results
3.1. ECM Protein–Cell Attachment Assay
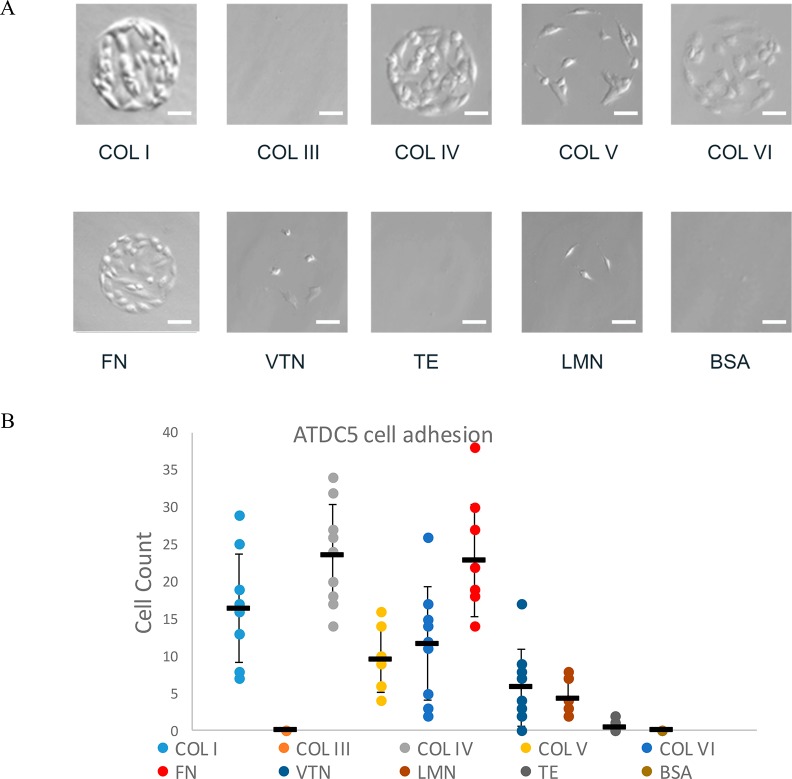
The chondroprogenitor cell line ATDC5 was derived from a mouse teratocarcinoma cell line. An extracellular matrix molecule array was utilized to screen specific extracellular matrix proteins for the ability of ATDC5 cells to adhere. Bright-field images were collected from each of the nine replicates of specific ECM proteins. Cell counts were determined at 12 and 30 h after initial cell plating. ATDC5 cells were found to adhere to collagen types I and IV, and fibronectin more extensively than other ECM molecules screened. The moderate affinity of ATDC5 to collagen types V and VI and little to no adherence of cells was observed for collagen III, vitronectin, tropoelastin, and laminin (Figure 1).
Figure 1.
ATDC5 cells adhere more extensively to fibronectin, collagen I, and collagen IV. ATDC5 cells were screened with extracellular matrix array printed with collagen I (COL I), collagen III (COL III), collagen IV (COL IV), collagen V (COL V), collagen VI (COL VI), fibronectin (FN), vitronectin (VTN), laminin (LMN), tropoelastin (TE), and BSA as a negative control. (A) Representative bright-field images of ATDC5 cells incubated for 30 h indicated differential binding of a number of extracellular proteins. Scale bar: 40 μm. (B) Attached cell counts determined for each of the nine replicates, as well as mean and standard deviation are shown (n = 9).
On the basis of this assay and information from published literature that indicates that fibronectin is essential for condensation during chondrogenesis,8 while in contrast, collagen type I is prevalent in dedifferentiated chondrocytes, bone, and other noncartilaginous tissues,53 and that collagen type IV, while present at low levels around chondrocytes, is a key marker for basement membranes,54,55 we chose fibronectin as a coating for GF to increase cellular adhesion of ATDC5 cells to the GF scaffold.
3.2. Fibronectin–graphene Interaction by Molecular Dynamic Simulation
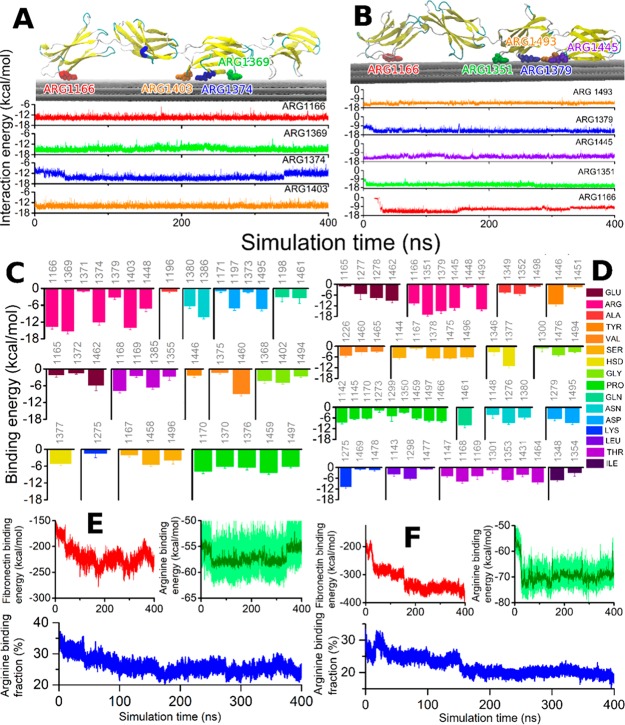
Prior to coating GF with fibronectin, we used molecular dynamics simulations to better understand the interaction of fibronectin with our GF scaffolds. The binding of fibronectin was investigated in silico in two different independent molecular dynamics simulations of 400 ns each. One simulation studied a random configuration, while the other simulation was set up to investigate the effects of the arginine-glycine-aspartic acid (RGD) tripeptide to the binding energy, as it has been theorized to be a major contributor to binding with integrin.56 The simulations revealed that in both simulated cases, fibronectin interacted with the graphene sheets; characteristic renderings of the binding motifs are shown in Figure 2A and B.
Figure 2.
Fibronectin interaction with graphene is stabilized by arginine residues. (A) Graphical rendering of the stabilized fibronectin atop the three graphene sheets with the four best arginine binders highlighted (Arg1166, Arg1369, Arg1374, Arg1403). The time evolution of the binding energy of these arginine residues with graphene is shown in the lower panel, color-coded for the amino acid residues. (B) Analogous to A but showing the data for the second studied configuration. This configuration features five arginine residue binders (Arg1166, Arg1351, Arg1379, Arg1445, Arg1493). (C) Binding energy with graphene computed for every amino acid with average binding energy above 1 kcal/mol, averaged over the 400 ns simulation. (D) Analogous to C, for the second studied configuration. The residue numbers are indicated, while the corresponding amino acid types are color-coded for both panels (C and D). (E and F) Time evolution of the fibronectin and arginine interaction energy with graphene for the two configurations. The lower plots in both panels show the fraction of arginine residue binding energy with respect to the total fibronectin-binding energy as a function of simulation time.
The computations demonstrated that in both cases, arginine significantly stabilized the binding as demonstrated in the plots directly beneath the graphical representations in Figure 2. Several arginine residues were identified to bind directly to the surface in both configurations with approximate binding energy of 15 kcal/mol. The binding of fibronectin to the graphene surface was due to a contribution from all amino acids, as summarized in Figure 2C and D. These plots stress that although arginine residues are only a small fraction of the total interacting amino acids, they provide the largest contribution to the fibronectin–graphene interaction, approximately 20–30% of the total binding energy as shown in Figure 2E and F.
The total fibronectin–graphene interaction energy was lower for the second configuration considered, approximately −400 kcal/mol, however, other binding configurations with even lower binding energies may be possible and should not be excluded based on the data presented here. Figure 2D also demonstrates that the contribution from the RGD tripeptide to the total binding energy of fibronectin to the graphene surface is −26.2 kcal/mol as the tripeptide consists of the residues Arg1493, Gly1494, and Asp1495. The RGD tripeptide therefore provides about 7% of the total binding energy. Note that in the performed analysis, the graphene sheet atoms were assumed neutral and no induced charge effects were considered. The binding energies are therefore, purely van der Waals in nature, and are expected to be even lower if polarization effects are accounted for.
3.3. Cellular Response to GF
Cells were seeded on GF according to the timeline shown in Figure 3. Cells seeded on GF were able to adhere to the surface of the GF as well as to other cells during an initial 24 h incubation period, forming small clusters of cells in and between the cavities of the foam scaffold during the subsequent growth and differentiation period (Figure 4).
Figure 3.
Cell seeding on GF overview.
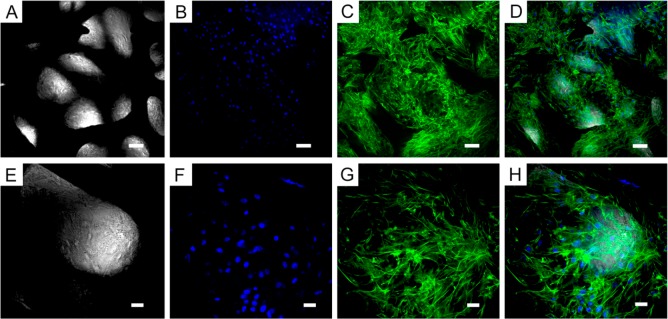
Figure 4.
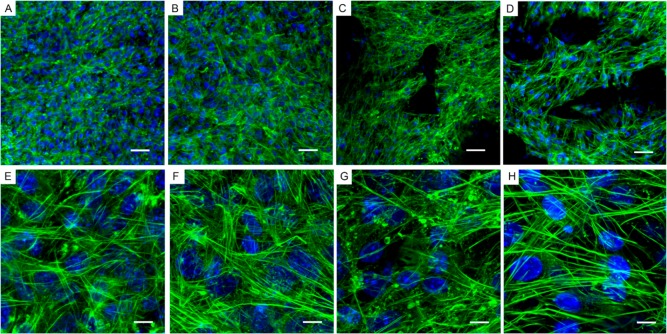
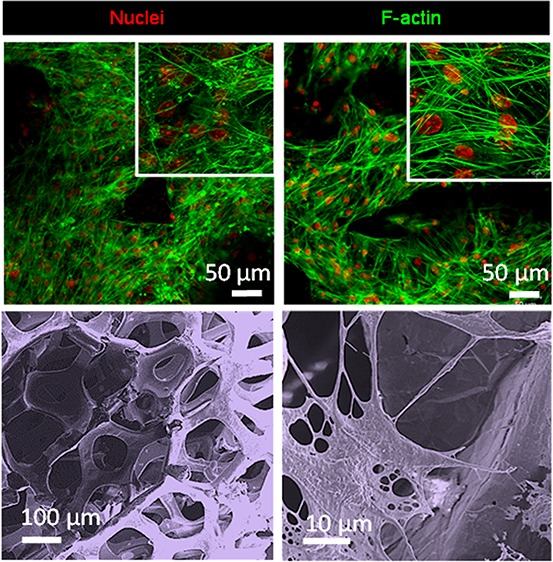
ATDC5 cell morphology on GF. Transmitted light and fluorescence microscopy of ATDC5 cells grown on bare three-dimensional GF for 28 days. (A and E) GF imaged by transmitted light microscopy, (B and F) Blue, nuclei (DAPI); (C and G) Green, F-actin (Alexa Fluor 488 phalloidin); (D and H) Overlay of transmitted light, DAPI, and phalloidin staining. (A–D) Scale-bar: 50 μm. (E–H) Scale-bar: 20 μm.
Using a combination of transmitted and fluorescence microscopy, DAPI was used to determine the location of cellular nuclei and phalloidin to label the cytoskeleton. Images shown in Figure 4 represent cells in culture on bare three-dimensional GF for 28 days. GF imaged by transmitted light microscopy images are shown in Figure 4A and E. Fluorescence microscopy of DAPI stained cells on GF to show nuclei are shown in Figure 4B and F. Fluorescence microscopy was also used to demonstrate the organization of the F-actin of the cytoskeleton using phalloidin labeled with Alexa Fluor 488 (Figure 4C and G). An overlay of transmitted light, DAPI, and phalloidin staining provides information about the relative location of the scaffold and the cells and is shown in Figure 4D and H.
3.4. Mechanical Properties of GF–FN+cells
The effect of fibronectin and cells on the mechanical properties (compressive modulus, equilibrium modulus, stress relaxation, dynamic modulus, and phase shift) of the cellular graphene composites was analyzed using a one-way MANOVA in SPSS (p = 0.05) using the LSD correction for multiple comparisons. The elastic properties of GF were enhanced by the addition of fibronectin (Figure 5A and B) and when fibronectin was used in cell culture, an additive effect was observed. The viscoelastic mechanical properties, phase shift and stress relaxation (Figure 5C and E), of GF were unaffected by the addition of fibronectin to the GF scaffold. The ratio of dynamic modulus (Figure 5D) to equilibrium modulus (Figure 5B) remained consistent between groups (∼4×). The effects of cells and fibronectin on the mechanical properties (compressive modulus, equilibrium modulus, stress relaxation, dynamic modulus, and phase shift) of the cellular graphene composites were analyzed using a one-way MANOVA in SPSS (p = 0.05) using the LSD correction for multiple comparisons. These results indicate a significant change due to the addition of fibronectin, even at this early stage in culture and may provide new insights on the structure–function relationships of GF.
Figure 5.
Mechanical properties. The measured quasi-static (A and B) and dynamic (C–E) properties of GF (hatched bars), GF coated in fibronectin (dark blue bars), and GF coated in fibronectin and cultured with ATDC5 cells (light blue bars) for 28 days. Fibronectin changed the elasticity of the composite (i.e., modulus values), but did not increase the viscoelastic properties (stress relaxation and phase shift).
Cytoskeletal organization within cells on GF was dependent on the presence or absence of fibronectin coating (Figure 6). Comparison of actin cytoskeletal arrangement on GF compared to control cultures grown on glass-bottom tissue culture wells confirmed that the cytoskeletal morphology was a function of the presence of fibronectin rather than the scaffold. Fluorescence micrographs demonstrate that cell growth on a surface in the presence of fibronectin resulted in an enhancement of stress fibers within the cytoskeleton accompanied by an absence of globular puncta of F-actin that were prevalent in control cultures without fibronectin (compare green Alexa Fluor 488 staining in Figure 6A, B and E, F). ATDC5 cells grown on GF in the absence and presence of fibronectin demonstrate similar findings to cells grown on the glass surface. Fibronectin coating resulted in alteration in the cytoskeletal organization in a manner that supported the formation of stress fibers on GF (compare Figure 6C, D and G, H). Globular puncta of F-actin is more prevalent in the absence of fibronectin on glass-bottomed tissue culture wells as well as on GF. The cytoskeletal arrangement is a key aspect of cellular phenotype during chondrocyte differentiation and has been shown to correlate to gene expression of chondrogenic markers.57−65
Figure 6.
Actin cytoskeleton of cells on GF and fibronectin-coated GF. Fluorescence of ATDC5 cells grown on glass-bottom tissue culture wells compared to GF, with or without fibronectin. Cell nuclei are stained blue (DAPI); Green, F-actin (Alexa Fluor 488 phalloidin); (A–D) ATDC5 cells were grown on glass-bottom tissue culture wells without (A and E) and with fibronectin (B and F); ATDC5 cells were grown on GF without (C and G) and with fibronectin (D and H). Note the prevalence of stress fibers and the absence of puncta in F and H compared to E and G, respectively. Additionally, note the relative abundance of puncta of actin which are more prevalent in the absence of fibronectin on glass-bottomed tissue culture wells as well as on GF. (A–D) Scale-bar: 50 μm. (E–H) Scale-bar: 10 μm.
3.5. Visualization of GF and Cell–GF Associations Using Scanning Electron Microscopy
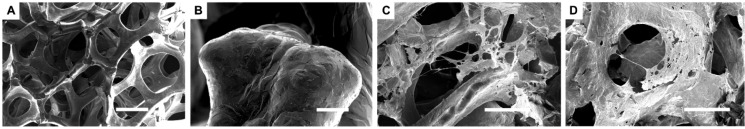
Cell–GF interactions were visualized using scanning electron microscopy. Figure 7 illustrates the 3D spaces available for cells to colonize (Figure 7A) and surface roughness characteristics of the GF (Figure 7B). The GF had a density of 4 mg/cm3, and pore size of 580 μm. Cells were able to adhere to bare GF (Figure 7C) as well as fibronectin-coated GF (Figure 7D). Cell adhesion may be supported by both surface roughness as well as the presence of fibronectin, consistent with studies from other laboratories that have investigated adhesion of nonchondrogenic cells as a function of surface roughness and fibronectin.28,66−68
Figure 7.
Cell–graphene interactions. Scanning electron microscope (SEM) images of bare graphene (A and B) and ATDC5 cells grown on graphene (C and D). SEM was operated at 2 kV with a beam current of 0.10 nA (A and B) or 13pA (C and D). (A) Scale-bar: 200 μm; (B) Scale-bar: 20 μm; (C, D) Scale-bar: 100 μm.
3.6. Gene Expression Analysis
3.6.1. Housekeeping Gene Selection
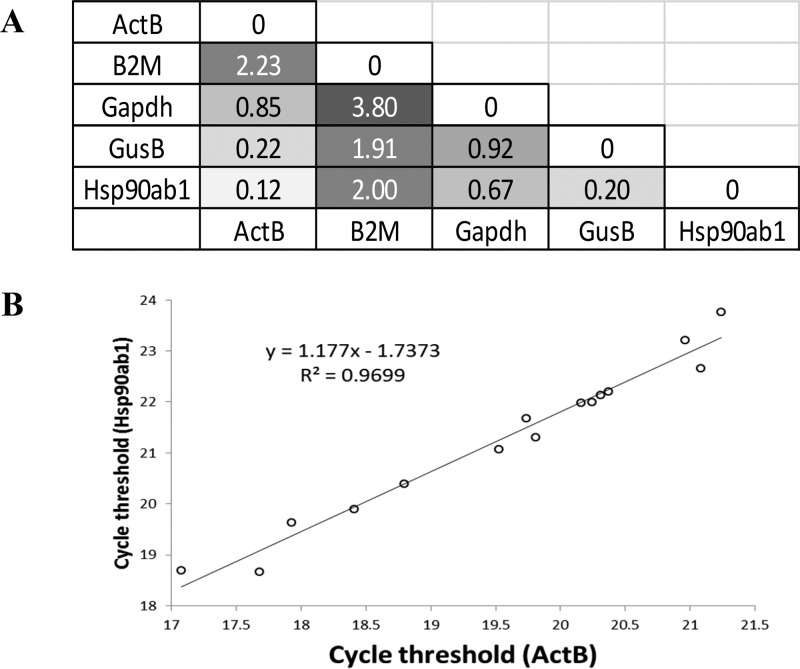
We used qRT-PCR gene expression analysis for the selection of housekeeping genes (HKGs). ActB, B2m, Gapdh, GusB, and Hsp90ab1 were analyzed for all samples in this study. ActB and Hsp90ab1 cycle threshold levels were most consistent among all samples analyzed for candidate HKGs considered, based on pairwise analysis of variance for differences between threshold values (Figure 8A) Correlation analysis resulted in a trend line with a slope close to 1 (1.177) and R2 close to 1 (0.9699) (n = 15) (Figure 8 B)
Figure 8.
ActB and Hsp90ab1 housekeeping genes. ActB and Hsp90ab1 are stably expressed by ATDC5 cells under all experimental conditions used in this study (i.e., on glass-bottom tissue culture wells, GF, and fibronectin-GF). (A) ActB and Hsp90ab1 cycle threshold levels were most consistent among all samples analyzed by qRT-PCR for candidate HKGs considered, based on pairwise analysis of variance for differences between threshold values, variance equal to 0.12. (B) Correlation analysis of cycle threshold values for Hsp90ab1 and ActB indicate a slope and an R2 value close to 1. (n = 15).
3.6.2. Gene Expression during Condensation and Prechondrocytic Differentiation of ATDC5 Cells
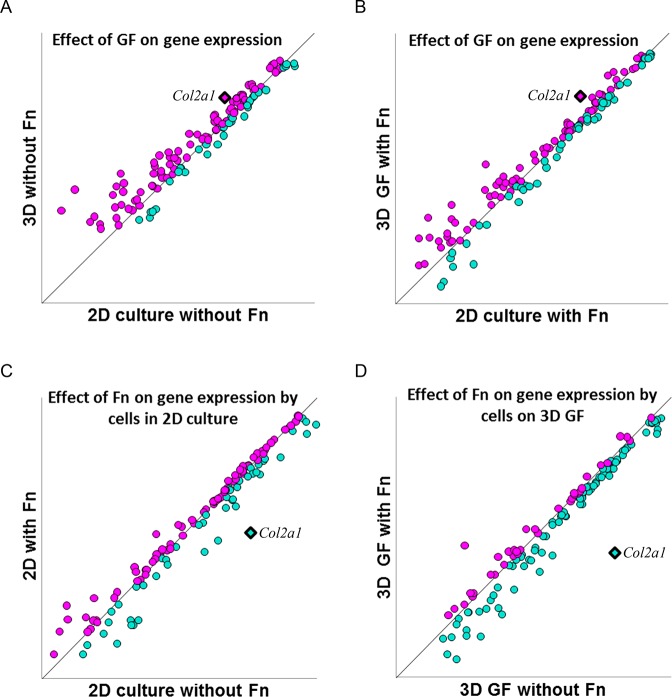
Sixty-four genes were analyzed for differential gene expression under experimental conditions used in this study, comparing 2D culture to cell culture on 3D GF in the presence and absence of fibronectin coating. Chondrogenic marker genes are presented in Tables 1, 2, and 3, and are grouped according to their functional roles. Figure 9 indicates a strong correlation between relative levels of expression for the majority of genes analyzed. Further analysis was carried out on those genes for which growth in 3D on GF supported a level of expression that met or exceeded expression levels during chondrogenic differentiation under control conditions as indicated by data points that fell above or on the diagonal lines in Figure 9. Results from the correlation analysis indicate that 3D GF without fibronectin supported the most robust relative gene expression compared to control or in the presence of fibronectin. Seventy percent of genes queried reached or exceeded a threshold level of expression on bare 3D GF compared to 2D culture controls (Figure 9A). Similarly, when cells were seeded on 3D GF pretreated with fibronectin, 65% of genes queried reached or exceeded the level of expression observed in control 2D cultures in the presence of fibronectin (Figure 9B). When investigating the effect of fibronectin in 2D cultures, 63% of genes reached an expression level similar to control 2D cultures (Figure 9C). However, in 3D GF cultures pretreated with fibronectin, only 29% of genes queried reached the level of expression observed in 3D GF without fibronectin (Figure 9D). The gold standard biomarker for chondrogenic differentiation, Col2a1, is upregulated in 3D GF cultures, indicated by bolded magenta diamonds in Figure 9A and B, and downregulated in the presence of fibronectin, indicated by bolded turquoise diamonds in Figure 9C and D. Tables 1–3 list representative genes for which the expression level was differentially upregulated or that met control chondrogenic levels by growing cells in 3D culture conditions on GF. Genes were assigned to the functional classifications of cell adhesion, extracellular matrix, and matrix remodeling based on established or suggested functions described previously in peer-reviewed publications. Taken together, these results suggest that 3D-GF without fibronectin supported chondrocyte differentiation to a greater extent than did GF pretreated with fibronectin.
Table 1. Cell Adhesion Genes Expressed during Chondroprogenitor Cell Differentiation Supported by Growth on GF.
| functional classification: cell adhesion |
||||
|---|---|---|---|---|
| gene symbol | protein name | function in chondrogenesis | reference | |
| 1 | Cd44 | Hyaluronate receptor | cell–matrix interactions during chondrogenesis and matrix assembly | Knudson 200369 |
| 2 | Ctnna1 | Catenin, alpha 1 | mediates functional mesenchymal cell condensation | Delise 20024 |
| 3 | Ctnnb1 | Catenin, beta 1 | mediates functional mesenchymal cell condensation | Delise 20024 |
| 4 | Itga3 | Integrin alpha 3 | mediates the connection between the cell and its external environment | Kim 200370 |
| 5 | Itga5 | Integrin alpha 5 | mediates chondrocyte adhesion to cartilage | Kurtis 200371 |
| 6 | Itgav | Integrin alpha V | mediates the connection between the cell and its external environment | Kurtis 200371 |
| 7 | Itgb1 | Integrin beta 1 | maintains the chondrocyte phenotype, prevents chondrocyte apoptosis, regulates chondrocyte-specific gene expression; mediates cell–matrix interactions; involved in chondrocyte mechanoreception | Kurtis 200371 Shakibaei 200872 |
| 8 | Ncam1 | Neural cell adhesion molecule | present in mesenchymal cell condensations; abundance increases during cell aggregation | Tavella 199473 |
| 9 | Sgce | Sarcoglycan epsilon | transmembrane protein linking cytoplasm to extracellular matrix | Rouillard 201674 |
Table 2. ECM Genes Expressed during Chondroprogenitor Cell Differentiation on GF.
| functional classification: ECM |
||||
|---|---|---|---|---|
| gene symbol | protein name | function in chondrogenesis | reference | |
| 1 | Col1a1 | Collagen α1(I) | major fibrillar collagen | Treilleux 199275 |
| 2 | Col2a1 | Collagen α1(II) | major fibrillar collagen | Liu 201376 Atsumi 199035 |
| 3 | Col3a1 | Collagen α1(III) | fibrillar collagen | Lodewyckx 201277 |
| 4 | Col5a1 | Collagen α1(V) | fibrillar collagen | Lodewyckx 201277 |
| 5 | Col6a1 | Collagen α1(VI) | pericellular collagen | Zelenski 201578 |
| 6 | Ecm1 | Extracellular matrix protein-1 | interacts with perlecan; regulates chondrogenesis | Kong 201679 Mongiat 200380 |
| 7 | Emilin1 | Elastin microfibril interface-located protein 1 | integrin binding activity; tissue remodeling in noncartilaginous tissues | This paper for chondrocyte differentiation* |
| 8 | Fn1 | Fibronectin | essential for early chondrocyte differentiation | White 200381 Singh 20148 |
| 9 | Hapln1 | Hyaluronan and proteoglycan link protein 1 | organizes extracellular matrix; links proteoglycan to hyaluronan | Xu 200882 |
| 10 | Lamb3 | Laminin subunit beta-3 | basement membrane protein; promotes chondrogenesis | Sun 201783 |
| 11 | Postn | Periostin | basement membrane protein; promotes chondrogenesis | Inaki 201884 |
| 12 | Sparc | Secreted protein acidic and rich in cysteine; Osteonectin | matricellular protein with calcium-binding properties; osteonectin, functions in growth and remodeling | Sage 198985 |
| 13 | Spp1 | Secreted phosphoprotein; Osteopontin Bone sialoprotein 1 | small integrin-binding ligand, N-linked glycoprotein | Shibata 200286 |
| 14 | Thbs1 | Thrombospondin 1 | matricellular protein; modulates cell–matrix interactions; Cartilage protection; | Miller 198887 DiCesare 199488 Pfander 200089 Maumus 201790 |
| 15 | Thbs2 | Thrombospondin 2 | matricellular protein; interacts with cell surface; regulates the bioavailability of proteases and growth factors in the pericellular environment | Jeong 201591 |
| 16 | Tnc | Tenascin | hexameric extracellular matrix glycoprotein prevalent in development; modulates cellular adhesion and interaction with fibronectin among other proteins | Gluhak 199692 Unno 201993 Mackie 198794 |
Table 3. Matrix Remodeling Genes Expressed during Chondroprogenitor Cell Differentiation Supported by Growth on GF.
| functional classification: remodeling |
||||
|---|---|---|---|---|
| gene symbol | protein name | function in chondrogenesis | reference | |
| 1 | Adamts1 | A disintegrin and metalloproteinase with thrombospondin motifs 1 | Aggrecanase and proteoglycanase; matrix rearrangement during chondrogenesis and cartilage regeneration | Boeuf 201295 Kelwick 201596 |
| 2 | Adamts 2 | A disintegrin and metalloproteinase with thrombospondin motifs 2 | Procollagen N-propeptidase; regulates structure and function of extracellular matrix collagen fibril assembly | Kelwick 201596 |
| 3 | Ctgf | Connective tissue growth factor (CCN2) | Cysteine-rich secreted protein with adhesive and chemotactic activities modulates matrix remodeling during skeletal development | Nakanishi 200097 Ivkovic 200398 |
| 4 | Mmp14 | Matrix metalloproteinase 14 | Matrix turnover during early chondrogenesis | Sekiya 200299 |
| 5 | Mmp2 | Matrix metalloproteinase 2 | Gelatinase; required for matrix remodeling during fracture repair and skeletal and craniofacial development | Arai 2016100 Lieu 2011101 Mosig 2007102 |
| 6 | Tgfbi | TGF-beta-induced 68 kDa protein | Binds to collagen type II fibrils, inhibits mineralization and maintains chondrocyte phenotype | Hashimoto 1997103 Huang 2010104 |
| 7 | Timp1 | Tissue inhibitor of matrix metalloproteinase 1 | Inhibitor of MMPs and ADAMTSs | Peterson 2006105 |
| 8 | Timp2 | Tissue inhibitor of matrix metalloproteinase 2 | Inhibitor of MMPs and ADAMTSs | Lin 2008106 |
| 9 | Timp3 | Tissue inhibitor of matrix metalloproteinase 3 | Inhibitor of MMPs and ADAMTSs | Lin 2008106 |
Figure 9.
GF supports or enhances gene expression levels. The effect of fibronectin, GF, and fibronectin in combination with GF on ATDC5 cell gene expression was investigated. Correlation analysis of relative expression levels was carried out to detect differential gene expression as a function of the cell culture substrate. The mRNA levels were compared for cells seeded on four distinct surfaces. Data points above the diagonal line indicate genes that are upregulated and data points below the diagonal line indicate genes that are downregulated. Data points falling on the diagonal line are not differentially expressed in experimental compared to control conditions. The effect of GF on gene expression is demonstrated in panels A and B. The effect of fibronectin on gene expression is demonstrated in panels C and D. (A) Relative gene expression levels in 2D cell culture conditions compared to cells grown in 3D on GF in the absence of fibronectin. (B) Relative gene expression levels in 2D cell culture conditions compared to cells grown in 3D on GF in the presence of fibronectin. (C) Relative gene expression levels in 2D cell culture conditions comparing the presence and absence of fibronectin. (D) Relative gene expression levels by cells grown in 3D on GF comparing the presence and absence of fibronectin. Genes for which expression levels met or exceeded the control are indicated in magenta, while those genes that were supported by substrate conditions are indicated by turquoise. Col2a1, a marker for chondrocyte differentiation, is shown as a diamond shape and bolded in each frame. Col2a1 is found above the diagonal line in A and B indicating upregulation as a function of 3D GF culture, and below the line in C and D, indicating downregulation as a function of fibronectin in either 2D or 3D culture. Genes included in this analysis are listed in Tables 1–3.
3.6.2.1. Cell Adhesion Molecules
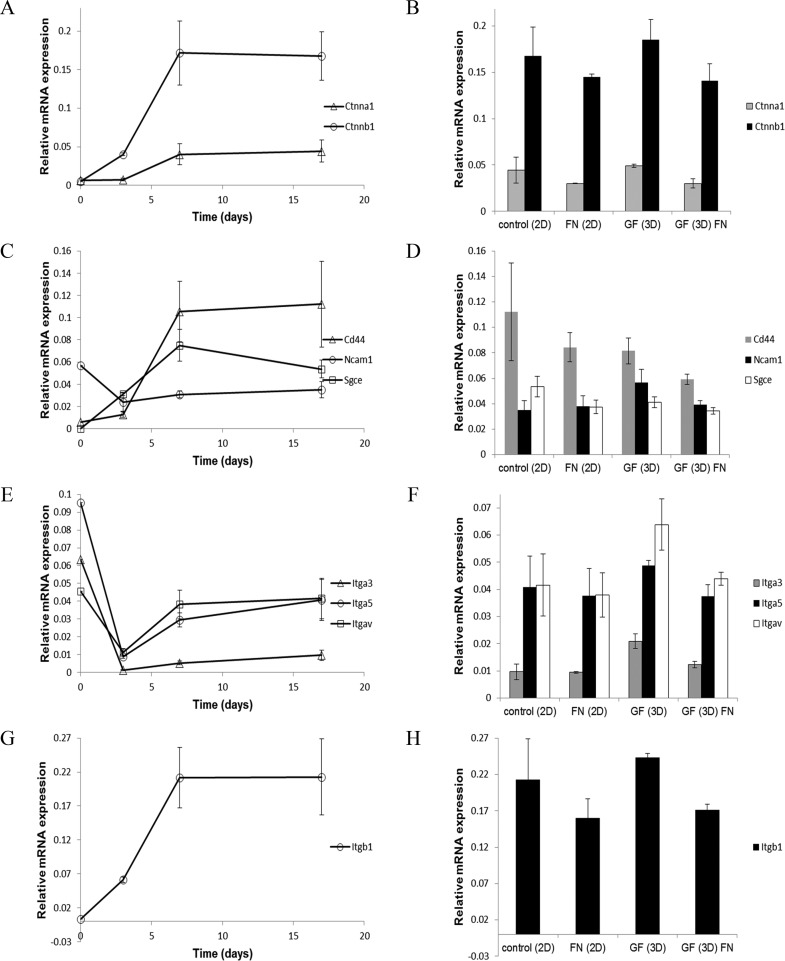
Nine genes encoding cell surface and cell adhesion molecules were analyzed for differential expression over time during chondrogenic differentiation (Table 1). Additionally, gene expression levels were assessed under 2-D and 3-D culture conditions on GF with and without fibronectin. Ctnna1 and Ctnnb1 increased over time during early chondrogenic differentiation and then plateaued under our experimental conditions (Figure 10A). Cells grown on GF expressed levels of Ctnna1 and Ctnnb1 at or above the threshold established in control cultures (Figure 10B), indicating that growth on GF supported gene expression patterns consistent with chondrogenic differentiation. The effect of fibronectin coating on GF was a slight reduction in the expression level of these markers. Cd44 and Sgce increased during early chondrogenesis and then plateaued under our experimental conditions (Figure 10C). Ncam1 expression levels decreased initially during early chondrogenic differentiation (Figure 10C). Cells grown on GF expressed levels of Cd44, Sgce, and Ncam1 meeting the threshold established in our control conditions (Figure 10D). Itga3, Itga5, and Itgav decreased during early chondrogenic differentiation followed by a gradual increase under our experimental conditions (Figure 10E). Growth on GF supported or enhanced gene expression levels for Itga3, Itga5, and Itgav, while the presence of fibronectin reduced this expression level (Figure 10F). Itgb1 expression increased initially during chondrogenic differentiation and plateaued between days 7 and 17 of our experiment (Figure 10G). Growth on GF supported the expression of Itgb1 at a level consistent with control conditions for chondrogenic differentiation (Figure 10H). These results suggest that 3D-GF supports chondrogenic differentiation and expression of adhesion molecules that serve as biomarkers for chondrocyte cells. Further, fibronectin alone or in combination with 3D-GF does not provide an advantage to 3D-GF alone.
Figure 10.
Expression of genes encoding mediators of cell attachment by ATDC5 cells on glass-bottom tissue culture wells, GF, and fibronectin-GF. (A) Time course of gene expression during chondrogenic differentiation for Ctnnal (triangle) and Ctnnb1 (circle). (B) Relative gene expression levels of Ctnnal (gray) and Ctnnb1 (black) at day 17 of chondrogenic differentiation in control 2D culture, 2D culture in the presence of fibronectin, 3D-GF, and 3D-GF coated with fibronectin. (C) Time course of gene expression during chondrogenic differentiation for Cd44 (triangle), Ncam1 (circle), and Sgce (square). (D) Relative gene expression levels of Cd44 (gray), Ncam1 (black), and Sgce (white) at day 17 in control 2D culture, 2D culture in the presence of fibronectin, 3D-GF, and 3D-GF coated with fibronectin. (E) Time course of gene expression during chondrogenic differentiation for Itga3 (triangle), Itga5 (circle), and Itgav (square). (F) Relative gene expression levels of Itga3 (gray), Itga5 (black), and Itgav (white) at day 17 in control 2D culture, 2D culture in the presence of fibronectin, 3D-GF, and 3D-GF coated with fibronectin. (G) Time course of gene expression during chondrogenic differentiation for Itgb1. (H) Relative gene expression levels of Itgb1 at day 17 in control 2D culture, 2D culture in the presence of fibronectin, 3D-GF, and 3D-GF coated with fibronectin. Error bars = Mean ± SD. These genes are listed in Table 1 with references from current literature indicating an association with chondrocyte differentiation.
3.6.2.2. Extracellular Matrix Molecules
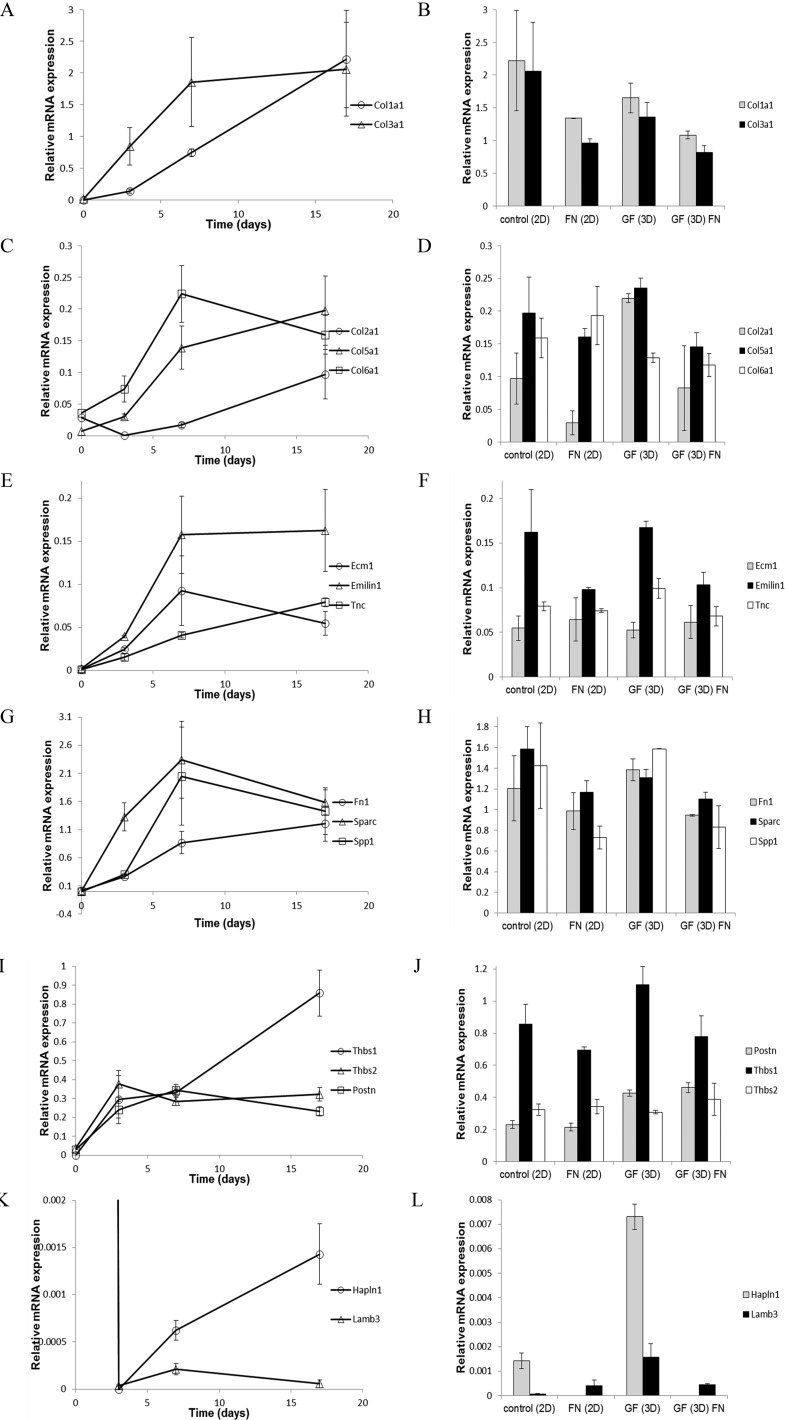
Sixteen genes encoding extracellular matrix molecules were analyzed for differential expression over time under 2-D and 3-D culture conditions on GF with and without fibronectin (see Table 2 for list of genes and description). Col1a1 and Col3a1 increased over time during early chondrogenic differentiation (Figure 11A). Cells grown on GF expressed levels of Col1a1 and Col3a1 similar to control cultures (Figure 11B), indicating that growth on GF supported gene expression patterns consistent with chondrogenic differentiation. The effect of fibronectin coating on GF was a reduction in the expression level of these markers to below the levels observed in our controls. Col2a1 and Col5a1 increased throughout chondrogenic differentiation, while Col6a1 increased during early chondrogenesis and then plateaued at later time points (Figure 11C). Cells grown on GF expressed levels of Col2a1, Col5a1, and Col6a1 similar to control conditions with an enhancement of Col2a1 expression by cells seeded onto bare GF (Figure 11D). Enhancement of Col2a1 expression was diminished in the presence of fibronectin coating in contrast to the enhancement observed on bare GF.
Figure 11.
Expression of genes encoding extracellular matrix proteins by ATDC5 cells on glass-bottom tissue culture wells, GF, and fibronectin-GF. (A) Time course of gene expression during chondrogenic differentiation for Col1a1 (circle) and Col3a1 (triangle). (B) Relative gene expression levels of Col1a1 (gray) and Col3a1 (black) at day 17 of chondrogenic differentiation in control 2D culture, 2D culture in the presence of fibronectin, 3D-GF, and 3D-GF coated with fibronectin. (C) Time course of gene expression during chondrogenic differentiation for Col2a1 (circle), Col5a1 (triangle), and Col6a1 (square). (D) Relative gene expression levels of Col2a1 (gray), Col5a1 (black), and Col6a1 (white) at day 17 in control 2D culture, 2D culture in the presence of fibronectin, 3D-GF, and 3D-GF coated with fibronectin. (E) Time course of gene expression during chondrogenic differentiation for Ecm1 (circle), Emilin1 (triangle), and Tnc (square). (F) Relative gene expression levels of Ecm1 (gray), Emilin1 (black), and Tnc (white) at day 17 in control 2D culture, 2D culture in the presence of fibronectin, 3D-GF, and 3D-GF coated with fibronectin. (G) Time course of gene expression during chondrogenic differentiation for Fn (circle), Sparc (triangle), and Spp1 (square). (H) Relative gene expression levels of Fn (gray), Sparc (black), and Spp1 (white) at day 17 in control 2D culture, 2D culture in the presence of fibronectin, 3D-GF, and 3D-GF coated with fibronectin. (I) Time course of gene expression during chondrogenic differentiation for Thbs1 (circle), Thbs2 (triangle), and Postn (square). (J) Relative gene expression levels of Thbs1 (black), Thbs2 (white), and Postn (gray) at day 17 in control 2D culture, 2D culture in the presence of fibronectin, 3D-GF, and 3D-GF coated with fibronectin. (K) Time course of gene expression during chondrogenic differentiation for Hapln1 (circle) and Lamb3 (triangle). (L) Relative gene expression levels of Hapln1 (gray) and Lamb3 (black) at day 17 in control 2D culture, 2D culture in the presence of fibronectin, 3D-GF, and 3D-GF coated with fibronectin. Error bars = Mean ± SD Table 2 lists extracellular matrix genes with description, function, and literature citations that corroborate an upregulation during early chondrogenic differentiation.
Ecm1, Emilin1, Sparc, Spp1, Thbs2, and Postn increased at early chondrogenic time points and then plateaued, while Tnc, Fn, and Thbs1 increased throughout the time course of the experiment (Figure 11E, G, and I). Growth on GF supported gene expression levels for Ecm1, Emilin1, Tnc, Fn, Sparc, Spp1, Thbs1, Thbs2, and Postn consistent with or enhanced compared to levels observed under control conditions for chondrogenesis, and these levels were slightly reduced in the presence of fibronectin (Figure 11F, H, and J). Thbs1 and Postn expression were enhanced when cells were grown on bare GF. Hapln1 and Lamb3 mRNA levels initially dropped significantly during chondrogenic differentiation, followed by an increase in the case of Hapln1 and relative plateau for Lamb3 (Figure 11K). Cells seeded on GF expressed Hapln1 and Lamb3 at enhanced levels compared to control conditions, while the presence of fibronectin diminished the observed enhancement (Figure 11L). Taken together, these results suggest that 3D-GF supports chondrogenic differentiation and the expression of genes encoding extracellular matrix molecules that serve as biomarkers for chondrocyte cells. Further, fibronectin alone or in combination with 3D-GF does not provide an advantage to 3D-GF alone.
3.6.2.3. Matrix Remodeling Genes
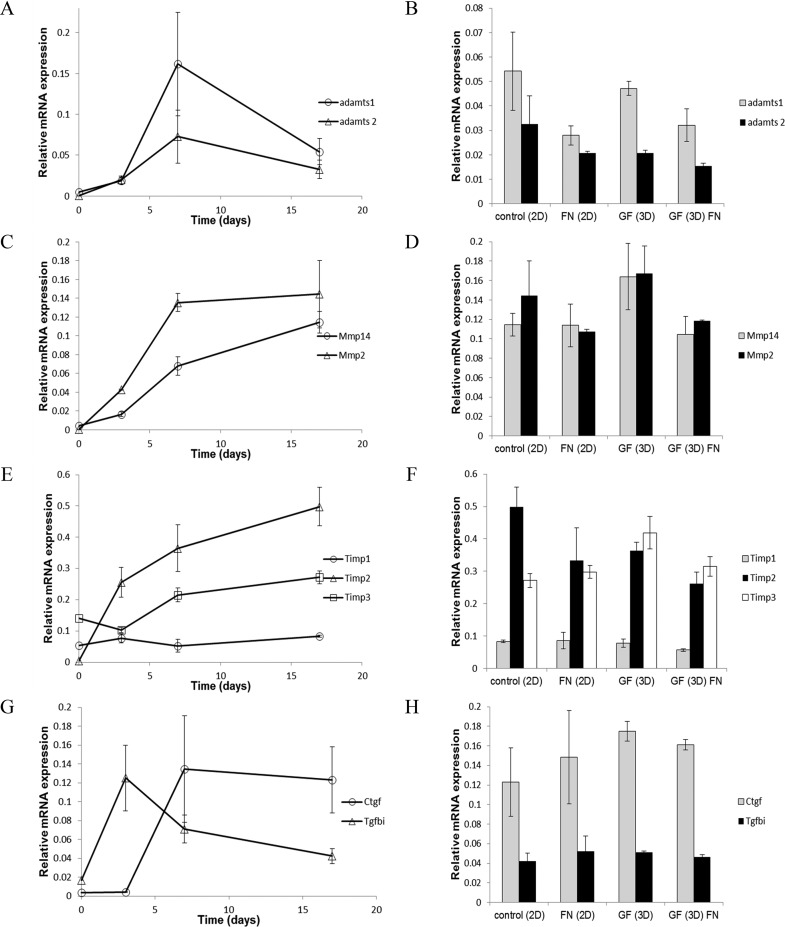
Nine genes encoding remodeling enzymes their endogenous inhibitors, and mediators of remodeling were analyzed for differential expression over time under 2-D and 3-D culture conditions, and on GF with and without fibronectin (see Table 3 for a description of genes encoding matrix remodeling molecules). Adamts1 and Adamts2 increased over time during early chondrogenic differentiation and then plateaued or decreased later in our time course of chondrogenic differentiation (Figure 12A). Cells grown on GF expressed levels of Adamts1 and Adamts2 similar to control cultures (Figure 12B), indicating that growth on GF supported gene expression patterns consistent with chondrogenic differentiation. The effect of fibronectin coating on GF was a reduction in the expression level of these markers to below the levels observed in our controls. Mmp2 increased early in chondrogenesis and then plateaued while Mmp14 increased throughout chondrogenic differentiation (Figure 12C). Cells grown on GF expressed levels of Mmp2 and Mmp14 similar to control conditions (Figure 12D). Expression levels were decreased in the presence of fibronectin.
Figure 12.
Expression of genes encoding matrix remodeling proteins and their endogenous inhibitors by ATDC5 cells on glass-bottom tissue culture wells, GF, and fibronectin-GF. (A) Time course of gene expression during chondrogenic differentiation for Adamts1 (circle) and Adamts2 (triangle). (B) Relative gene expression levels of Adamts1 (gray) and Adamts2 (black) at day 17 of chondrogenic differentiation in control 2D culture, 2D culture in the presence of fibronectin, 3D-GF, and 3D-GF coated with fibronectin. (C) Time course of gene expression during chondrogenic differentiation for Mmp2 (triangle) and Mmp14 (circle). (D) Relative gene expression levels of Mmp2 (black) and Mmp14 (gray) at day 17 in control 2D culture, 2D culture in the presence of fibronectin, 3D-GF, and 3D-GF coated with fibronectin. (E) Time course of gene expression during chondrogenic differentiation for Timp1 (circle), Timp2 (triangle), and Timp3 (square). (F) Relative gene expression levels of Timp1 (gray), Timp2 (black), and Timp3 (white) at day 17 in control 2D culture, 2D culture in the presence of fibronectin, 3D-GF, and 3D-GF coated with fibronectin. (G) Time course of gene expression during chondrogenic differentiation for Ctgf (circle) and Tgfbi (triangle). (H) Relative gene expression levels of Ctgf (gray) and Tgfbi (black) at day 17 in control 2D culture, 2D culture in the presence of fibronectin, 3D-GF, and 3D-GF coated with fibronectin. Error bars = Mean ± SD Table 3 lists matrix remodeling genes analyzed in this study with descriptions and literature citations that have demonstrated a link between increases in gene expression and chondrogenic differentiation.
Timp1 expression levels remained constant over time during chondrogenesis while Timp2 and Timp3 expression levels increased throughout the time course (Figure 12E). Cells grown on GF expressed mRNA for Timp2 and reduced levels, Timp3 at enhanced levels, and Timp1 and a level consistent with expression levels observed under control conditions (Figure 12F). Reduced expression levels were observed in the presence of fibronectin coating. Expression levels of Ctgf and Tgfbi increased during chondrogenic differentiation followed by a decrease or plateau level (Figure 12G). While cells seeded on GF expressed Ctgf and Tgfbi at levels consistent with chondrogenesis, they were not influenced by the presence of fibronectin (Figure 12H), unlike other genes investigated in this study. These results suggest that 3D-GF supports chondrogenic differentiation and the expression of genes encoding matrix remodeling molecules that can serve as biomarkers for chondrocyte cells. Further, fibronectin alone or in combination with 3D-GF does not provide an advantage to 3D-GF alone.
4. Discussion
In this study, we used GF as a three-dimension scaffold to support chondroprogenitor cell attachment and differentiation. Our results indicate that cell morphology can be modified by the functionalization of GF with fibronectin. The molecular dynamic simulation demonstrated that arginine residue side chains play a stabilizing role in the graphene–fibronectin interaction. Cells adhered to GF and GF functionalized with fibronectin. GF provided a microenvironment compatible with chondroprogenitor gene expression as indicated in Tables 1–3 and in Figures 9–12, and in some cases, enhanced the expression of key chondrogenic markers. However, fibronectin influenced the cellular morphology as well as the gene expression patterns, resulting in decreased gene expression levels for the majority of genes analyzed.
We note that previous studies have revealed the importance of surface roughness on cell–substrate interactions.28−32 Our graphene foams exhibit wrinkles on the order of several nanometers to 10s of nanometers in good agreement with previous studies.17,49,107 Furthermore, previous studies have shown the importance of surface functionalization on cell culture.108−110 While we have focused on the impact of protein functionalization of graphene–cell interfaces, further investigations are needed to better understand the time-dependent biochemical nature of such interfaces.
We analyzed only the early cellular responses in the differentiation pathway rather than later events and the formation of mature cartilage tissue. The GF used in this study was made by chemical vapor deposition processing on a nickel foam template, which was removed prior to use. Although our results were consistent among all GF used, it is possible that lot-to-lot variability may exist and therefore precaution should be taken to confirm lot or batch effects. The manufacturing process used to prepare the scaffold may influence the outcome of cell-based investigations and may alter both cytoskeletal organization as well as gene expression profiles.
The interactions between fibronectin and the graphene surface may be stabilized by the π electron cloud in graphene, which is capable of interacting with the hydrophobic protein core. Alternatively, as we investigated here, the interaction may be stabilized by arginine side chains. Note that in the performed analysis, the graphene sheet atoms were assumed neutral and no induced charge effects were considered. The binding energies are therefore, purely van der Waals in nature, and are expected to be even lower if polarization effects are accounted for.
Prior testing by Yocham and colleagues49 demonstrated an increase in the GF’s elastic modulus after 28 days of cell culture without the use of fibronectin. When compared to this study, the compressive elastic modulus measured previously and the compressive elastic modulus reported here is not significantly different.49 This suggests that the elastic strength contributed to the GF scaffold by the fibronectin coating is similar to the strength contributed to the scaffold after 28 days of cell growth. The compressive modulus of GF coated with fibronectin and then cultured with ATDC5 cells for 28 days was significantly higher than either one individually, suggesting that fibronectin coating and cell growth contribute to the resulting elastic strength of the GF additively. Neither cell growth nor fibronectin coating affected the viscoelastic properties phase shift and stress relaxation. This may be due to insufficient chondrogenic differentiation to maintain an increase in interstitial fluid pressure. As in the prior study of GF without fibronectin, the ratio of dynamic modulus to equilibrium modulus of this study remains consistent between groups, reinforcing the conclusion that the time-dependent mechanisms are unchanged by cell culture and fibronectin coating, and these factors primarily affect the elastic strength of the scaffold. One potential time-dependent mechanism is protein adsorption, as a study by Lee and colleagues observed that GF absorbed 8% of serum proteins after 24 h in tissue culture media24 which may contribute to the greater load dissipation by ripple effect as described by Nautiyal and colleagues.111
Cells exist in unique microenvironments in vivo that influence their survival and differentiation and gene expression patterns. Here, we took measures to provide extracellular matrix cues to support prechondrogenic cells. While the optimal in vitro matrix environment is not known for ATDC5 cells, it is known that both materials properties as well as biochemical signals play critical roles. Here we used fibronectin to promote cell attachment and early condensation. Col I and Col IV were not used for this study because Col I is associated with nonchondrogenic tissues as well as dedifferentiated chondrocytes53 and Col IV is a marker for basement membranes.54,55
The clonal mouse embryonic cell line ATDC5 was used in this study as a chondroprogenitor cell line. Originally isolated from an embryonal carcinoma, ATDC5 cells demonstrate all phases of chondrocyte differentiation from early cell attachment and condensation, through a proliferative phase, a chondrogenic differentiation phase marked by increased levels of cartilage matrix constituent production, and finally, differentiation into hypertrophic chondrocytes that produce an extracellular matrix suitable for mineralization.35 Differentiation of chondroprogenitor cells depends upon fibronectin for the early stages of condensation and differentiation.7,8 In addition, epithelial to mesenchymal transitions depend on fibronectin.112 Enhancement of chondrogenesis of ATDC5 cells has been demonstrated by using an RGD-functionalized scaffold.113
Because of the importance of cell adhesion in the condensation and differentiation process, cell adhesion molecules involved in chondrogenic differentiation including Cd44,69Ctnna1,4Ctnnb1,4Itga3,70Itga5,71Itgav,71Itgb1,71Ncam1,73 and Sgce(74) were analyzed in our study. Cell adhesion molecules play critical roles at various stages during chondrogenic differentiation. For example, an increase in the cell–cell adhesion molecule Ncam1 is a hallmark of prechondrogenic condensation, and subsequently decreases during differentiation.6,73 We found that 3D-GF supported the gene expression levels of these cell adhesion markers for chondrogenic differentiation and that fibronectin did not provide an advantage over 3D-GF alone.
Fibronectin is an extracellular matrix protein that plays an important role in bringing cells together at the earliest stage of mesenchymal cell differentiation in chondrocytes. Fibronectin matrix also acts as a platform for type I collagen deposition, and may also serve this role for type II collagen. ECM molecules that play a role in chondrogenic differentiation analyzed in this study included Col1a1,75Col2a1,35,76Col3a1,77Col5a1,77Col6a1,78Ecm1,79,80Emilin1, Fn1,8,81Hapln1,82Lamb3,83Postn,84Sparc,85Spp1,86Thbs1,87−90Thbs2,91 and Tnc.92−94 The upregulation of these genes supports our conclusion that 3D-GF provides an environment supporting of chondrogenic differentiation and that while fibronectin facilitates cell adhesion, it does not improve the cellular response.
Matrix remodeling molecules play essential roles in the formation of cartilage and the remodeling of the ECM during the differentiation of cells that give rise to tissues such as cartilage. Our analysis included Adamts1,95,96Adamts2,96Ctgf,97,98Mmp14,99Mmp2,100−102Tgfbi,103,104Timp1,105Timp2,106 and Timp3.106 In each case, we demonstrated the increase in gene expression levels that occur during chondrogenic differentiation of ATDC5 cells can be supported by 3D-GF compared to control conditions. We found that pretreatment of 3D-GF with fibronectin did not improve the gene expression of matrix remodeling molecules during chondrogenic differentiation.
Graphene-based scaffolds have been widely investigated for numerous applications including their effect on stem cell commitment. Graphene coated with laminin was shown to support neural stem cell attachment and differentiation, as well as accelerate myogenesis of C2C12 cells on GF.17,114−116 Chondrogenic differentiation of placenta-derived and tonsil-derived mesenchymal stem cells on graphene-based scaffold/hydrogel was reported by Park and colleagues117 Differentiation and long-term survival of neural and mesenchymal stem cells in an undifferentiated state has been accomplished using graphene foam.118,119 These examples from current literature demonstrate the use of GF to enhance osteogenesis and facilitate neurogenesis and astrocytogenesis of neuronal stem cells.
GF in conjunction with extracellular matrix proteins may provide tissue functionality during the transient regeneration phase of cartilage healing and repair. Additionally, the electrical conductivity may provide the advantage of stimulating cells to produce more matrix. GF in combination with a hydrogel scaffold may be ideally suited for bone/cartilage repair in the case of osteochondral defects. With an improved understanding of the influences of scaffold and biochemical factors, an ideal microenvironment can be designed.
5. Conclusions
Future studies are warranted to investigate the role of other extracellular matrix molecules and three-dimensional scaffolds to determine cell fate in tissue engineering and regenerative medicine applications. Damaged articular cartilage repair is a challenging issue in regenerative medicine, due in part to the limited ability for cartilage to heal. According to the World Health Organization, the United Nations has categorized OA as a priority disease in need of research on potential therapies. Given that between 2015 and 2050, the proportion of the world’s population over 60 years will nearly double from 12% to 22%, an estimated 130 million people will suffer from OA worldwide (WHO, 2018). Existing methodologies to treat OA are palliative, nonreparative, nonrestorative, reparative, restorative, and transplantation strategies. Autologous chondrocyte transplantation shows promise in clinical treatment, however the process involves the harvest, culture, and transplant of cells grown in a monolayer (2-D culture). Under these culture conditions, the risk of dedifferentiation of the chondrocyte phenotype before use is a major concern of tissue engineering.120 Unlike chondrocyte cells, mesenchymal stem cells may maintain their chondrogenic potential if provided the proper biochemical, biophysical, and mechanical cues during proliferation and subsequent differentiation to regenerate cartilage tissue.
Cartilage engineering approaches need to consider the cell source, biomaterial scaffold, and a conducive environment to promote the formation of functional tissues, promoting the very early stages of chondrogenic commitment to the later differentiation stages of chondrocytes during which they produce high levels of cartilage biomarkers, while providing a scaffold that can provide functionality during the various stages of the regeneration process. GF in combination with a transitional extracellular matrix may provide the necessary niche environment in which to support all phases of mesenchymal stem cell differentiation, chondrocyte differentiation, and cartilage production by mature chondrocytes. On the basis of the findings of this investigation, we conclude that because cell differentiation is regulated by a combination of molecular and materials properties of the underlying scaffold, both the characteristics of the scaffold and the nature of the ECM protein used for functionalization must be considered carefully to align with the tissue-specific goals of the application.
Acknowledgments
The authors wish to thank Jason Stonick for technical assistance in cell culture. Scanning electron microscopy was carried by the Boise State Center for Materials Characterization. The Biomolecular Research Center provided access to cell culture and confocal microscopy.
Glossary
Abbreviations
- Cd44
Hyaluronate receptor
- Ctnna1
catenin, alpha 1
- Ctnnb1
catenin, beta 1
- Itga3
integrin alpha 3
- Itga5
Integrin alpha 5
- Itgav
integrin alpha V
- Itgb1
Integrin beta 1
- Ncam1
neural cell adhesion molecule
- Sgce
Sarcoglycan epsilon
- Col1a1
Collagen α1(I)
- Col2a1
Collagen α1(II)
- Col3a1
Collagen α1(III)
- Col5a1
Collagen α1(V)
- Col6a1
Collagen α1(VI)
- DAPI
4′,6-Diamidino-2-phenylindole
- Ecm1
Extracellular matrix protein-1
- Emilin1
Elastin microfibril interface-located protein 1
- Fn
Fibronectin
- GF
Graphene foam
- Hapln1
Hyaluronan and proteoglycan link protein 1
- Lamb3
Laminin subunit beta-3
- Postn
Periostin
- Sparc
Secreted protein acidic and rich in cysteine; Osteonectin
- Spp1
Secreted phophoprotein; Osteopontin; Bone sialoprotein 1
- Thbs1
Thrombospondin 1
- Thbs2
Thrombospondin 2
- Tnc
Tenascin
- Adamts1
A distintegrin and metalloproteinase with thrombospondin motifs 1
- Adamts2
A distintegrin and metalloproteinase with thrombospondin motifs 2
- Ctgf
Connective tissue growth factor (CCN2)
- Mmp14
Matrix metalloproteinase 14
- Mmp2
Matrix metalloproteinase 2
- Tgfbi
TGF-beta-induced 68 kDa protein
- Timp1
Tissue inhibitor of matrix metalloproteinase 1
- Timp2
Tissue inhibitor of matrix metalloproteinase 2
- Timp3
Tissue inhibitor of matrix metalloproteinase 3
- COL I
Collagen type I protein
- COL III
Collagen type III protein
- COL IV
Collagen type IV protein
- COLV
Collagen type V protein
- COL VI
Collagen type VI protein
- FN
Fibronectin protein
- VTN
Vitronectin protein
- LMN
Laminin protein
- TE
Tropoelastin protein
- MANOVA
Multivariate analysis of variance
- LSD
Least significant difference
Author Contributions
Substantial contributions to the conception and design of the work (JTO, DE, TJL, IAS, CMS, RJB); acquisition, analysis, and interpretation of data (JCR, SMF, AF, KF, RSB, Jr, CMS); drafting the work and revising it critically for intellectual content (JTO, DE, TJL, SMF, RJB, IAS). All authors have approved the final approval of the version to be published and agree to be accountable for all aspects of the work.
Funding is acknowledged from the Institutional Development Awards (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under Grants P20GM103408 (DE, TJL, JTO, RSB) and P20GM109095 (DE, TJL, JTO, RSB), and R01AR47985 and K02AR48672 from the National Institute of Arthritis, Musculoskeletal, and Skin Diseases to JTO, National Science Foundation, Grants 0619793 and 0923535; MJ Murdock Charitable Trust, Duane and Lori Stueckle endowment, Idaho State Board of Education, Biomolecular Sciences Graduate Program, and the Micron School of Materials Science Graduate Program. D.E. acknowledges support under NSF CAREER Award 1848516. I.A.S. acknowledges support from the Volkswagen Stiftung (Lichtenberg professorship), the DFG (GRK1885), the Lundbeck Foundation, and the Danish Council for Independent Research. The authors are also grateful to DeiC National HPC Center (SDU) for providing computational resources necessary for the calculations.
The authors declare no competing financial interest.
References
- Nitzan E.; Kalcheim C. Neural Crest and Somitic Mesoderm as Paradigms to Investigate Cell Fate Decisions during Development. Dev. Growth Differ. 2013, 55 (1), 60–78. 10.1111/dgd.12004. [DOI] [PubMed] [Google Scholar]
- Hall B. K.; Miyake T. The Membranous Skeleton: The Role of Cell Condensations in Vertebrate Skeletogenesis. Anat. Embryol. 1992, 186 (2), 107–124. 10.1007/BF00174948. [DOI] [PubMed] [Google Scholar]
- Hall B. K.; Miyake T. Divide, Accumulate, Differentiate: Cell Condensation in Skeletal Development Revisited. Int. J. Dev. Biol. 1995, 39 (6), 881–893. [PubMed] [Google Scholar]
- Delise A. M.; Tuan R. S. Analysis of N-Cadherin Function in Limb Mesenchymal Chondrogenesis in Vitro. Dev. Dyn. 2002, 225 (2), 195–204. 10.1002/dvdy.10151. [DOI] [PubMed] [Google Scholar]
- Gao Y.; Liu S.; Huang J.; Guo W.; Chen J.; Zhang L.; Zhao B.; Peng J.; Wang A.; Wang Y.; Xu W.; Lu S.; Yuan M.; Guo Q. The ECM-Cell Interaction of Cartilage Extracellular Matrix on Chondrocytes. BioMed Res. Int. 2014, 2014, 1–8. 10.1155/2014/648459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. K.; Miyake T. All for One and One for All: Condensations and the Initiation of Skeletal Development. BioEssays 2000, 22 (2), 138–147. . [DOI] [PubMed] [Google Scholar]
- Kulyk W. M.; Upholt W. B.; Kosher R. A. Fibronectin Gene Expression during Limb Cartilage Differentiation. Development 1989, 106 (3), 449–455. [DOI] [PubMed] [Google Scholar]
- Singh P.; Schwarzbauer J. E. Fibronectin Matrix Assembly Is Essential for Cell Condensation during Chondrogenesis. J. Cell Sci. 2014, 127 (20), 4420–4428. 10.1242/jcs.150276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard C. M.; Fuld H. M.; Frenz D. A.; Downie S. A.; Massagué J.; Newman S. A. Role of Transforming Growth Factor-Beta in Chondrogenic Pattern Formation in the Embryonic Limb: Stimulation of Mesenchymal Condensation and Fibronectin Gene Expression by Exogenenous TGF-Beta and Evidence for Endogenous TGF-Beta-like Activity. Dev. Biol. 1991, 145 (1), 99–109. 10.1016/0012-1606(91)90216-P. [DOI] [PubMed] [Google Scholar]
- Moursi A. M.; Globus R. K.; Damsky C. H. Interactions between Integrin Receptors and Fibronectin Are Required for Calvarial Osteoblast Differentiation in Vitro. J. Cell Sci. 1997, 110 (18), 2187–2196. [DOI] [PubMed] [Google Scholar]
- Dessau W.; von der Mark H.; von der Mark K.; Fischer S. Changes in the Patterns of Collagens and Fibronectin during Limb-Bud Chondrogenesis. J. Embryol. Exp. Morphol. 1980, 57, 51–60. [PubMed] [Google Scholar]
- Von Der Mark K.; von der Mark H. Immunological and Biochemical Studies of Collagen Type Transition during in Vitro Chondrogenesis of Chick Limb Mesodermal Cells. J. Cell Biol. 1977, 73 (3), 736–747. 10.1083/jcb.73.3.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palchesko R. N.; Zhang L.; Sun Y.; Feinberg A. W. Development of Polydimethylsiloxane Substrates with Tunable Elastic Modulus to Study Cell Mechanobiology in Muscle and Nerve. PLoS One 2012, 7 (12), e51499 10.1371/journal.pone.0051499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Nayak T. R.; Hong H.; Cai W. Graphene: A Versatile Nanoplatform for Biomedical Applications. Nanoscale 2012, 4 (13), 3833–3842. 10.1039/c2nr31040f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. C.; Lim C. H.; Su K. C.; Loh K. P.; Lim C. T.. Cell-Assembled Graphene Biocomposite for Enhanced Chondrogenic Differentiation. Small 2015, 11, 963–969 10.1002/smll.201401635. [DOI] [PubMed] [Google Scholar]
- Duran M.; Luzo A. C. M.; de Souza J. G.; Favaro W. J.; Garcia P.; Duran N.. Graphene Oxide as Scaffolds for Stem Cells: An Overview. Curr. Mol. Med. 2018, 17, 619. 10.2174/1566524018666180308111915. [DOI] [PubMed] [Google Scholar]
- Krueger E.; Chang A. N.; Brown D.; Eixenberger J.; Brown R.; Rastegar S.; Yocham K. M.; Cantley K. D.; Estrada D. Graphene Foam as a Three-Dimensional Platform for Myotube Growth. ACS Biomater. Sci. Eng. 2016, 2 (8), 1234–1241. 10.1021/acsbiomaterials.6b00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M.; Moon H. J.; Ko D. Y.; Jeong B. Composite System of Graphene Oxide and Polypeptide Thermogel As an Injectable 3D Scaffold for Adipogenic Differentiation of Tonsil-Derived Mesenchymal Stem Cells. ACS Appl. Mater. Interfaces 2016, 8 (8), 5160–5169. 10.1021/acsami.5b12324. [DOI] [PubMed] [Google Scholar]
- Lim J.; Stoll H.; Kwon I. Material and Mechanical Factors: New Strategy in Cellular Neurogenesis. Neural Regener. Res. 2014, 9 (20), 1810. 10.4103/1673-5374.143426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak T. R.; Andersen H.; Makam V. S.; Khaw C.; Bae S.; Xu X.; Ee P.-L. R.; Ahn J.-H.; Hong B. H.; Pastorin G.; Özyilmaz B. Graphene for Controlled and Accelerated Osteogenic Differentiation of Human Mesenchymal Stem Cells. ACS Nano 2011, 5 (6), 4670–4678. 10.1021/nn200500h. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Klausen L. H.; Chen M.; Dong M. Electroactive Scaffolds for Neurogenesis and Myogenesis: Graphene-Based Nanomaterials. Small 2018, 14, 1801983.November 10.1002/smll.201801983. [DOI] [PubMed] [Google Scholar]
- Patel A.; Xue Y.; Mukundan S.; Rohan L. C.; Sant V.; Stolz D. B.; Sant S. Cell-Instructive Graphene-Containing Nanocomposites Induce Multinucleated Myotube Formation. Ann. Biomed. Eng. 2016, 44 (6), 2036–2048. 10.1007/s10439-016-1586-6. [DOI] [PubMed] [Google Scholar]
- Kim J.; Choi K. S.; Kim Y.; Lim K. T.; Seonwoo H.; Park Y.; Kim D. H.; Choung P. H.; Cho C. S.; Kim S. Y.; Choung Y. H.; Chung J. H.. Bioactive Effects of Graphene Oxide Cell Culture Substratum on Structure and Function of Human Adipose-Derived Stem Cells. J. Biomed. Mater. Res., Part A 2013, 101, 3520. 10.1002/jbm.a.34659. [DOI] [PubMed] [Google Scholar]
- Lee W. C.; Lim C. H. Y. X.; Shi H.; Tang L. A. L.; Wang Y.; Lim C. T.; Loh K. P. Origin of Enhanced Stem Cell Growth and Differentiation on Graphene and Graphene Oxide. ACS Nano 2011, 5 (9), 7334–7341. 10.1021/nn202190c. [DOI] [PubMed] [Google Scholar]
- Shen H.; Lin H.; Sun A. X.; Song S.; Zhang Z.; Dai J.; Tuan R. S.. Chondroinductive Factor-Free Chondrogenic Differentiation of Human Mesenchymal Stem Cells in Graphene Oxide-Incorporated Hydrogels. J. Mater. Chem. B 2018, 6, 908. 10.1039/C7TB02172K. [DOI] [PubMed] [Google Scholar]
- Chen S.; Fu P.; Cong R.; Wu H.; Pei M. Strategies to Minimize Hypertrophy in Cartilage Engineering and Regeneration. Genes Dis 2015, 2 (1), 76–95. 10.1016/j.gendis.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Pei M.. Cell Senescence: A Challenge in Cartilage Engineering and Regeneration. Tissue Eng., Part B 2012, 18, 270. 10.1089/ten.teb.2011.0583. [DOI] [PubMed] [Google Scholar]
- Zareidoost A.; Yousefpour M.; Ghaseme B.; Amanzadeh A. The Relationship of Surface Roughness and Cell Response of Chemical Surface Modification of Titanium. J. Mater. Sci.: Mater. Med. 2012, 23 (6), 1479–1488. 10.1007/s10856-012-4611-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P.; Wang C.; Zhou J.; Jiang L.; Xue J.; Li W. Influence of Surface Properties on Adhesion Forces and Attachment of Streptococcus Mutans to Zirconia In Vitro. BioMed Res. Int. 2016, 2016, 1–10. 10.1155/2016/8901253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodolov V. I.; Kuznetsov A. P.; Nicolaeva O. A.; Shayakhmetova E. Sh.; Makarova L. G.; Shabanova I. N.; Khokhriakov N. V.; Volkova E. G. Bioengineered Material Surfaces for Medical Applications. Surf. Interface Anal. 2001, 32, 10–14. 10.1002/sia.996. [DOI] [Google Scholar]
- Jia X.; Minami K.; Uto K.; Chang A. C.; Hill J. P.; Ueki T.; Nakanishi J.; Ariga K.. Modulation of Mesenchymal Stem Cells Mechanosensing at Fluid Interfaces by Tailored Self-Assembled Protein Monolayers. Small 2019, 15 ( (5), ), 1804640. 10.1002/smll.201804640. [DOI] [PubMed] [Google Scholar]
- Minami K.; Mori T.; Nakanishi W.; Shigi N.; Nakanishi J.; Hill J. P.; Komiyama M.; Ariga K.. Suppression of Myogenic Differentiation of Mammalian Cells Caused by Fluidity of a Liquid–Liquid Interface. ACS Appl. Mater. Interfaces 2017, 930553. 10.1021/acsami.7b11445. [DOI] [PubMed] [Google Scholar]
- Hosseini M.-S.; Katbab A. A. Effects of Surface Viscoelasticity on Cellular Responses of Endothelial Cells. Reports Biochem. Mol. Biol. 2014, 3 (1), 20–28. [PMC free article] [PubMed] [Google Scholar]
- Wells R. G. The Role of Matrix Stiffness in Regulating Cell Behavior. Hepatology 2008, 47, 1394–1400. April 10.1002/hep.22193. [DOI] [PubMed] [Google Scholar]
- Atsumi T.; Ikawa Y.; Miwa Y.; Kimata K. A Chondrogenic Cell Line Derived from a Differentiating Culture of AT805 Teratocarcinoma Cells. Cell Differ. Dev. 1990, 30 (2), 109–116. 10.1016/0922-3371(90)90079-C. [DOI] [PubMed] [Google Scholar]
- Leahy D. J.; Aukhil I.; Erickson H. P. 2.0 A Crystal Structure of a Four-Domain Segment of Human Fibronectin Encompassing the RGD Loop and Synergy Region. Cell 1996, 84 (1), 155–164. 10.1016/S0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- Samuelsen S. V.; Solov’yov I. A.; Balboni I. M.; Mellins E.; Nielsen C. T.; Heegaard N. H. H.; Astakhova K. Synthetic Oligonucleotide Antigens Modified with Locked Nucleic Acids Detect Disease Specific Antibodies. Sci. Rep. 2016, 6 (1), 35827. 10.1038/srep35827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis I.; Sjulstok E.; Solov’yov I. A. Computational Reconstruction Reveals a Candidate Magnetic Biocompass to Be Likely Irrelevant for Magnetoreception. Sci. Rep. 2017, 7 (1), 13908. 10.1038/s41598-017-13258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber D. E. Cellular Mechanotransduction: Putting All the Pieces Together Again. FASEB J. 2006, 20 (7), 811–827. 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- Solov’yov I. A.; Korol A. V.; Solov’yov A. V.. Multiscale Modeling of Complex Molecular Structure and Dynamics with MBN Explorer; Springer International Publishing: Cham, 2017 10.1007/978-3-319-56087-8. [DOI] [Google Scholar]
- Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kalé L.; Schulten K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26 (16), 1781–1802. 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T. A.; Pedersen L. G. Molecular Modeling: An Experimental Tool. Environ. Health Perspect. 1993, 101 (5), 410–412. 10.1289/ehp.93101410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solov’yov I. A.; Yakubovich A. V.; Nikolaev P. V.; Volkovets I.; Solov’yov A. V. MesoBioNano Explorer--a Universal Program for Multiscale Computer Simulations of Complex Molecular Structure and Dynamics. J. Comput. Chem. 2012, 33 (30), 2412–2439. 10.1002/jcc.23086. [DOI] [PubMed] [Google Scholar]
- MacKerell A. D.; Bashford D.; Bellott M.; Dunbrack R. L.; Evanseck J. D.; Field M. J.; Fischer S.; Gao J.; Guo H.; Ha S.; Joseph-McCarthy D.; Kuchnir L.; Kuczera K.; Lau F. T.; Mattos C.; Michnick S.; Ngo T.; Nguyen D. T.; Prodhom B.; Reiher W. E.; Roux B.; Schlenkrich M.; Smith J. C.; Stote R.; Straub J.; Watanabe M.; Wiórkiewicz-Kuczera J.; Yin D.; Karplus M. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J. Phys. Chem. B 1998, 102 (18), 3586–3616. 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Mackerell A. D.; Feig M.; Brooks C. L. Extending the Treatment of Backbone Energetics in Protein Force Fields: Limitations of Gas-Phase Quantum Mechanics in Reproducing Protein Conformational Distributions in Molecular Dynamics Simulations. J. Comput. Chem. 2004, 25 (11), 1400–1415. 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- Best R. B.; Zhu X.; Shim J.; Lopes P. E. M.; Mittal J.; Feig M.; Mackerell A. D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone φ, ψ and Side-Chain χ(1) and χ(2) Dihedral Angles. J. Chem. Theory Comput. 2012, 8 (9), 3257–3273. 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.; MacKerell A. D. CHARMM36 All-Atom Additive Protein Force Field: Validation Based on Comparison to NMR Data. J. Comput. Chem. 2013, 34 (25), 2135–2145. 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.; Rauscher S.; Nawrocki G.; Ran T.; Feig M.; de Groot B. L.; Grubmüller H.; MacKerell A. D. CHARMM36m: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods 2017, 14 (1), 71–73. 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yocham K. M.; Scott C.; Fujimoto K.; Brown R.; Tanasse E.; Oxford J. T.; Lujan T. J.; Estrada D. Mechanical Properties of Graphene Foam and Graphene Foam-Tissue Composites. Adv. Eng. Mater. 2018, 20 (9), 1800166. 10.1002/adem.201800166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mane V. P.; Heuer M. A.; Hillyer P.; Navarro M. B.; Rabin R. L. Systematic Method for Determining an Ideal Housekeeping Gene for Real-Time PCR Analysis. J. Biomol. Technol. 2008, 19 (5), 342–347. [PMC free article] [PubMed] [Google Scholar]
- Andersen C. L.; Jensen J. L.; Ørntoft T. F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64 (15), 5245–5250. 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W.; Tichopad A.; Prgomet C.; Neuvians T. P. Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper--Excel-Based Tool Using Pair-Wise Correlations. Biotechnol. Lett. 2004, 26 (6), 509–515. 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Caron M. M. J.; Emans P. J.; Coolsen M. M. E.; Voss L.; Surtel D. A. M.; Cremers A.; van Rhijn L. W.; Welting T. J. M. Redifferentiation of Dedifferentiated Human Articular Chondrocytes: Comparison of 2D and 3D Cultures. Osteoarthr. Cartil. 2012, 20 (10), 1170–1178. 10.1016/j.joca.2012.06.016. [DOI] [PubMed] [Google Scholar]
- Kvist A. J.; Nyström A.; Hultenby K.; Sasaki T.; Talts J. F.; Aspberg A. The Major Basement Membrane Components Localize to the Chondrocyte Pericellular Matrix — A Cartilage Basement Membrane Equivalent?. Matrix Biol. 2008, 27 (1), 22–33. 10.1016/j.matbio.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Foldager C. B.; Toh W. S.; Gomoll A. H.; Olsen B. R.; Spector M. Distribution of Basement Membrane Molecules, Laminin and Collagen Type IV, in Normal and Degenerated Cartilage Tissues. Cartilage 2014, 5 (2), 123–132. 10.1177/1947603513518217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer A.; Craig D.; Thomas W. E.; Schulten K.; Vogel V. A Structural Model for Force Regulated Integrin Binding to Fibronectin’s RGD-Synergy Site. Matrix Biol. 2002, 21 (2), 139–147. 10.1016/S0945-053X(01)00197-4. [DOI] [PubMed] [Google Scholar]
- Sasazaki Y.; Seedhom B. B.; Shore R. Morphology of the Bovine Chondrocyte and of Its Cytoskeleton in Isolation and in Situ: Are Chondrocytes Ubiquitously Paired through the Entire Layer of Articular Cartilage?. Rheumatology 2008, 47 (11), 1641–1646. 10.1093/rheumatology/ken341. [DOI] [PubMed] [Google Scholar]
- Durrant L. A.; Archer C. W.; Benjamin M.; Ralphs J. R. Organisation of the Chondrocyte Cytoskeleton and Its Response to Changing Mechanical Conditions in Organ Culture. J. Anat. 1999, 194 (3), 343–353. 10.1046/j.1469-7580.1999.19430343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. A; Knight M. M; Bolton J. F.; Idowu B. D; Kayser M. V; Bader D. L Chondrocyte Deformation within Compressed Agarose Constructs at the Cellular and Sub-Cellular Levels. J. Biomech. 2000, 33 (1), 81–95. 10.1016/S0021-9290(99)00160-8. [DOI] [PubMed] [Google Scholar]
- Langelier E.; Suetterlin R.; Hoemann C. D.; Aebi U.; Buschmann M. D. The Chondrocyte Cytoskeleton in Mature Articular Cartilage: Structure and Distribution of Actin, Tubulin, and Vimentin Filaments. J. Histochem. Cytochem. 2000, 48 (10), 1307–1320. 10.1177/002215540004801002. [DOI] [PubMed] [Google Scholar]
- Blain E. J.; Gilbert S. J.; Hayes A. J.; Duance V. C. Disassembly of the Vimentin Cytoskeleton Disrupts Articular Cartilage Chondrocyte Homeostasis. Matrix Biol. 2006, 25 (7), 398–408. 10.1016/j.matbio.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Blain E. J. Involvement of the Cytoskeletal Elements in Articular Cartilage Homeostasis and Pathology. Int. J. Exp. Pathol. 2009, 90, 1–15. 10.1111/j.1365-2613.2008.00625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; February
- Brown P. D.; Benya P. D. Alterations in Chondrocyte Cytoskeletal Architecture during Phenotypic Modulation by Retinoic Acid and Dihydrocytochalasin B-Induced Reexpression. J. Cell Biol. 1988, 106 (1), 171–179. 10.1083/jcb.106.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman P.; Watt F. M. Influence of Cytochalasin D-Induced Changes in Cell Shape on Proteoglycan Synthesis by Cultured Articular Chondrocytes. Exp. Cell Res. 1988, 178 (2), 199–210. 10.1016/0014-4827(88)90391-6. [DOI] [PubMed] [Google Scholar]
- Archer C. W.; Rooney P.; Wolpert L. Cell Shape and Cartilage Differentiation of Early Chick Limb Bud Cells in Culture. Cell Differ. 1982, 11 (4), 245–251. 10.1016/0045-6039(82)90072-0. [DOI] [PubMed] [Google Scholar]
- Degasne I.; Baslé M. F.; Demais V.; Huré G.; Lesourd M.; Grolleau B.; Mercier L.; Chappard D. Effects of Roughness, Fibronectin and Vitronectin on Attachment, Spreading, and Proliferation of Human Osteoblast-like Cells (Saos-2) on Titanium Surfaces. Calcif. Tissue Int. 1999, 64 (6), 499–507. 10.1007/s002239900640. [DOI] [PubMed] [Google Scholar]
- Dolatshahi-Pirouz A.; Jensen T.; Kraft D. C.; Foss M.; Kingshott P.; Hansen J. L.; Larsen A. N.; Chevallier J.; Besenbacher F. Fibronectin Adsorption, Cell Adhesion, and Proliferation on Nanostructured Tantalum Surfaces. ACS Nano 2010, 4 (5), 2874–2882. 10.1021/nn9017872. [DOI] [PubMed] [Google Scholar]
- Lee M. H.; Ducheyne P.; Lynch L.; Boettiger D.; Composto R. J. Effect of Biomaterial Surface Properties on Fibronectin-Alpha5beta1 Integrin Interaction and Cellular Attachment. Biomaterials 2006, 27 (9), 1907–1916. 10.1016/j.biomaterials.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Knudson C. B.Hyaluronan and CD44: Strategic Players for Cell–Matrix Interactions during Chondrogenesis and Matrix Assembly. Birth Defects Res., Part C 2003, 69, 174. 10.1002/bdrc.10013. [DOI] [PubMed] [Google Scholar]
- Kim S. J.; Kim E. J.; Kim Y. H.; Hahn S. B.; Lee J. W.. The Modulation of Integrin Expression by the Extracellular Matrix in Articular Chondrocytes. Yonsei Med. J. 2003, 44, 493. 10.3349/ymj.2003.44.3.493. [DOI] [PubMed] [Google Scholar]
- Kurtis M. S.; Schmidt T. A.; Bugbee W. D.; Loeser R. F.; Sah R. L.. Integrin-Mediated Adhesion of Human Articular Chondrocytes to Cartilage. Arthritis Rheum. 2003, 48, 110. 10.1002/art.10704. [DOI] [PubMed] [Google Scholar]
- Shakibaei M.; Csaki C.; Mobasheri A. Diverse Roles of Integrin Receptors in Articular Cartilage. Adv. Anat., Embryol. Cell Biol. 2008, 197, 1–60. 10.1007/978-3-540-78771-6_1. [DOI] [PubMed] [Google Scholar]
- Tavella S.; Raffo P.; Tacchetti C.; Cancedda R.; Castagnola P.. N-CAM and N-Cadherin Expression during in Vitro Chondrogenesis. Exp. Cell Res. 1994, 215, 354. 10.1006/excr.1994.1352. [DOI] [PubMed] [Google Scholar]
- Rouillard A. D.; Gundersen G. W.; Fernandez N. F.; Wang Z.; Monteiro C. D.; McDermott M. G.; Ma’ayan A.. The Harmonizome: A Collection of Processed Datasets Gathered to Serve and Mine Knowledge about Genes and Proteins. Database (Oxford); 2016 10.1093/database/baw100. [DOI] [PMC free article] [PubMed]
- Treilleux I.; Mallein-Gerin F.; Guellec D. Le; Herbage D.. Localization of the Expression of Type I, II, III Collagen, and Aggrecan Core Protein Genes in Developing Human Articular Cartilage. Matrix 1992, 12, 221. 10.1016/S0934-8832(11)80065-X. [DOI] [PubMed] [Google Scholar]
- Liu H.; Zhao Z.; Clarke R. B.; Gao J.; Garrett I. R.; Margerrison E. E. C.. Enhanced Tissue Regeneration Potential of Juvenile Articular Cartilage. Am. J. Sports Med. 2013, 41, 2658. 10.1177/0363546513502945. [DOI] [PubMed] [Google Scholar]
- Lodewyckx L.; Cailotto F.; Thysen S.; Luyten F. P.; Lories R. J.. Tight Regulation of Wingless-Type Signaling in the Articular Cartilage - Subchondral Bone Biomechanical Unit: Transcriptomics in Frzb-Knockout Mice. Arthritis Res. Ther. 2012, 14, R16. 10.1186/ar3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenski N. A.; Leddy H. A.; Sanchez-Adams J.; Zhang J.; Bonaldo P.; Liedtke W.; Guilak F.. Type VI Collagen Regulates Pericellular Matrix Properties, Chondrocyte Swelling, and Mechanotransduction in Mouse Articular Cartilage. Arthritis Rheumatol.. 2015, 67, 1286. 10.1002/art.39034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L.; Zhao Y. P.; Tian Q. Y.; Feng J. Q.; Kobayashi T.; Merregaert J.; Liu C. J.. Extracellular Matrix Protein 1, a Direct Targeting Molecule of Parathyroid Hormone-Related Peptide, Negatively Regulates Chondrogenesis and Endochondral Ossification via Associating with Progranulin Growth Factor. FASEB J.. 2016, 30, 2741. 10.1096/fj.201600261R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongiat M.; Fu J.; Oldershaw R.; Greenhalgh R.; Gown A. M.; Iozzo R. V.. Perlecan Protein Core Interacts with Extracellular Matrix Protein 1 (ECM1), a Glycoprotein Involved in Bone Formation and Angiogenesis. J. Biol. Chem. 2003, 278, 17491. 10.1074/jbc.M210529200. [DOI] [PubMed] [Google Scholar]
- White D. G.; Hershey H. P.; Moss J. J.; Daniels H.; Tuan R. S.; Bennett V. D.. Functional Analysis of Fibronectin Isoforms in Chondrogenesis: Full-Length Recombinant Mesenchymal Fibronectin Reduces Spreading and Promotes Condensation and Chondrogenesis of Limb Mesenchymal Cells. Differentiation 2003, 71, 251. 10.1046/j.1432-0436.2003.7104502.x. [DOI] [PubMed] [Google Scholar]
- Xu J.; Wang W.; Ludeman M.; Cheng K.; Hayami T.; Lotz J. C.; Kapila S.. Chondrogenic Differentiation of Human Mesenchymal Stem Cells in Three-Dimensional Alginate Gels. Tissue Eng., Part A 2008, 14, 667. 10.1089/tea.2007.0272. [DOI] [PubMed] [Google Scholar]
- Sun Y.; Wang T. L.; Toh W. S.; Pei M.. The Role of Laminins in Cartilaginous Tissues: From Development to Regeneration. Eur. Cells Mater. 2017, 34, 40. 10.22203/eCM.v034a03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaki R.; Fujihara Y.; Kudo A.; Misawa M.; Hikita A.; Takato T.; Hoshi K.. Periostin Contributes to the Maturation and Shape Retention of Tissue-Engineered Cartilage. Sci. Rep. 2018 10.1038/s41598-018-29228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage H.; Vernon R. B.; Decker J.; Funk S.; Iruela-Arispe M. L.. Distribution of the Calcium-Binding Protein SPARC in Tissues of Embryonic and Adult Mice. J. Histochem. Cytochem. 1989, 37, 819. 10.1177/37.6.2723400. [DOI] [PubMed] [Google Scholar]
- Shibata S.; Fukada K.; Suzuki S.; Ogawa T.; Yamashita Y.. In Situ Hybridization and Immunohistochemistry of Bone Sialoprotein and Secreted Phosphoprotein 1 (Osteopontin) in the Developing Mouse Mandibular Condylar Cartilage Compared with Limb Bud Cartilage. J. Anat. 2002, 200, 309. 10.1046/j.1469-7580.2002.00033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. R.; McDevitt C. A.. Thrombospondin Is Present in Articular Cartilage and Is Synthesized by Articular Chondrocytes. Biochem. Biophys. Res. Commun. 1988, 153, 708. 10.1016/S0006-291X(88)81152-5. [DOI] [PubMed] [Google Scholar]
- DiCesare P. E.; Morgelin M.; Mann K.; Paulsson M.. Cartilage Oligomeric Matrix Protein and Thrombospondin 1: Purification from Articular Cartilage, Electron Microscopic Structure, and Chondrocyte Binding. Eur. J. Biochem. 1994, 223, 927. 10.1111/j.1432-1033.1994.tb19070.x. [DOI] [PubMed] [Google Scholar]
- Pfander D.; Cramer T.; Deuerling D.; Weseloh G.; Swoboda B.. Expression of Thrombospondin-1 and Its Receptor CD36 in Human Osteoarthritic Cartilage. Ann. Rheum. Dis. 2000, 59, 448. 10.1136/ard.59.6.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maumus M.; Manferdini C.; Toupet K.; Chuchana P.; Casteilla L.; Gachet M.; Jorgensen C.; Lisignoli G.; Noël D.. Thrombospondin-1 Partly Mediates the Cartilage Protective Effect of Adipose-Derived Mesenchymal Stem Cells in Osteoarthritis. Front. Immunol. 2017 10.3389/fimmu.2017.01638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S. Y.; Ha J.; Lee M.; Jin H. J.; Kim D. H.; Choi S. J.; Oh W.; Yang Y. S.; Kim J. S.; Kim B. G.; Chang J. H.; Cho D. H.; Jeon H. B.. Autocrine Action of Thrombospondin-2 Determines the Chondrogenic Differentiation Potential and Suppresses Hypertrophic Maturation of Human Umbilical Cord Blood-Derived Mesenchymal Stem Cells. Stem Cells 2015, 33, 3291. 10.1002/stem.2120. [DOI] [PubMed] [Google Scholar]
- Gluhak J.; Mais A.; Mina M.. Tenascin-C Is Associated with Early Stages of Chondrogenesis by Chick Mandibular Ectomesenchymal Cells in Vivo and in Vitro. Dev. Dyn. 1996, 205, 24.. [DOI] [PubMed] [Google Scholar]
- Unno H.; Hasegawa M.; Suzuki Y.; Iino T.; Imanaka-Yoshida K.; Yoshida T.; Sudo A.. Tenascin-C Promotes the Repair of Cartilage Defects in Mice. J. Orthop. Sci. 2019 10.1016/j.jos.2019.03.013. [DOI] [PubMed] [Google Scholar]
- Mackie E. J.; Thesleff I.; Chiquet-Ehrismann R.. Tenascin Is Associated with Chondrogenic and Osteogenic Differentiation in Vivo and Promotes Chondrogenesis in Vitro. J. Cell Biol. 1987, 105, 2569. 10.1083/jcb.105.6.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeuf S.; Graf F.; Fischer J.; Moradi B.; Little C. B.; Richter W.. Regulation of Aggrecanases from the ADAMTS Family and Aggrecan Neoepitope Formation during in Vitro Chondrogenesis of Human Mesenchymal Stem Cells. Eur. Cells Mater. 2012, 23, 320. 10.22203/eCM.v023a25. [DOI] [PubMed] [Google Scholar]
- Kelwick R.; Desanlis I.; Wheeler G. N.; Edwards D. R.. The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin Motifs) Family. Genome Biol.. 2015 10.1186/s13059-015-0676-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T.; Nishida T.; Shimo T.; Kobayashi K.; Kubo T.; Tamatani T.; Tezuka K.; Takigawa M.. Effects of CTGF/Hcs24, a Product of a Hypertrophic Chondrocyte-Specific Gene, on the Proliferation and Differentiation of Chondrocytes in Culture. Endocrinology 2000, 141, 264. 10.1210/endo.141.1.7267. [DOI] [PubMed] [Google Scholar]
- Ivkovic S.Connective Tissue Growth Factor Coordinates Chondrogenesis and Angiogenesis during Skeletal Development. Development 2003, 130, 2779. 10.1242/dev.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya I.; Vuoristo J. T.; Larson B. L.; Prockop D. J.. In Vitro Cartilage Formation by Human Adult Stem Cells from Bone Marrow Stroma Defines the Sequence of Cellular and Molecular Events during Chondrogenesis. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 4397. 10.1073/pnas.052716199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai Y.; Park S.; Choi B.; Ko K. W.; Choi W. C.; Lee J. M.; Han D. W.; Park H. K.; Han I.; Lee J. H.; Lee S. H.. Enhancement of Matrix Metalloproteinase-2 (MMP-2) as a Potential Chondrogenic Marker during Chondrogenic Differentiation of Human Adipose-Derived Stem Cells. Int. J. Mol. Sci. 2016, 17, 963. 10.3390/ijms17060963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieu S.; Hansen E.; Dedini R.; Behonick D.; Werb Z.; Miclau T.; Marcucio R.; Colnot C.. Impaired Remodeling Phase of Fracture Repair in the Absence of Matrix Metalloproteinase-2. Dis. Models & Mech. 2011, 4, 203. 10.1242/dmm.006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosig R. A.; Dowling O.; DiFeo A.; Ramirez M. C. M.; Parker I. C.; Abe E.; Diouri J.; Aqeel A. A.; Wylie J. D.; Oblander S. A.; et al. Loss of MMP-2 Disrupts Skeletal and Craniofacial Development and Results in Decreased Bone Mineralization, Joint Erosion and Defects in Osteoblast and Osteoclast Growth. Hum. Mol. Genet. 2007, 16 (9), 1113–1123. 10.1093/hmg/ddm060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K.; Noshiro M.; Ohno S.; Kawamoto T.; Satakeda H.; Akagawa Y.; Nakashima K.; Okimura A.; Ishida H.; Okamoto T.; Pan H.; Shen M.; Yan W.; Kato Y. Characterization of a Cartilage-Derived 66-KDa Protein (RGD-CAP/Big-H3) That Binds to Collagen. Biochim. Biophys. Acta, Mol. Cell Res. 1997, 1355 (3), 303–314. 10.1016/S0167-4889(96)00147-4. [DOI] [PubMed] [Google Scholar]
- Huang A. H.; Stein A.; Mauck R. L. Evaluation of the Complex Transcriptional Topography of Mesenchymal Stem Cell Chondrogenesis for Cartilage Tissue Engineering. Tissue Eng., Part A 2010, 16 (9), 2699–2708. 10.1089/ten.tea.2010.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J. T.The Importance of Estimating the Therapeutic Index in the Development of Matrix Metalloproteinase Inhibitors. Cardiovasc. Res.. 2006, 69, 677. 10.1016/j.cardiores.2005.11.032. [DOI] [PubMed] [Google Scholar]
- Lin Z.; Fitzgerald J. B.; Xu J.; Willers C.; Wood D.; Grodzinsky A. J.; Zheng M. H.. Gene Expression Profiles of Human Chondrocytes during Passaged Monolayer Cultivation. J. Orthop. Res. 2008, 26, 1230. 10.1002/jor.20523. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Ren W.; Gao L.; Liu B.; Pei S.; Cheng H. M. Three-Dimensional Flexible and Conductive Interconnected Graphene Networks Grown by Chemical Vapour Deposition. Nat. Mater. 2011, 10 (6), 424–428. 10.1038/nmat3001. [DOI] [PubMed] [Google Scholar]
- Grigoriou E.; Cantini M.; Dalby M. J.; Petersen A.; Salmeron-Sanchez M. Cell Migration on Material-Driven Fibronectin Microenvironments. Biomater. Sci. 2017, 5 (7), 1326–1333. 10.1039/C7BM00333A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A.; Saxena V.; Pandey L. M. Surface Functionalization of Ti6Al4V via Self-Assembled Monolayers for Improved Protein Adsorption and Fibroblast Adhesion. Langmuir 2018, 34 (11), 3494–3506. 10.1021/acs.langmuir.7b03152. [DOI] [PubMed] [Google Scholar]
- Kuddannaya S.; Chuah Y. J.; Lee M. H. A.; Menon N. V.; Kang Y.; Zhang Y. Surface Chemical Modification of Poly(Dimethylsiloxane) for the Enhanced Adhesion and Proliferation of Mesenchymal Stem Cells. ACS Appl. Mater. Interfaces 2013, 5 (19), 9777–9784. 10.1021/am402903e. [DOI] [PubMed] [Google Scholar]
- Nautiyal P.; Boesl B.; Agarwal A. The Mechanics of Energy Dissipation in a Three-Dimensional Graphene Foam with Macroporous Architecture. Carbon 2018, 132, 59–64. 10.1016/j.carbon.2018.02.028. [DOI] [Google Scholar]
- Park J.; Schwarzbauer J. E. Mammary Epithelial Cell Interactions with Fibronectin Stimulate Epithelial-Mesenchymal Transition. Oncogene 2014, 33 (13), 1649–1657. 10.1038/onc.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y.; Zeng L.; Huang Y. The Enhancement of Chondrogenesis of ATDC5 Cells in RGD-Immobilized Microcavitary Alginate Hydrogels. J. Biomater. Appl. 2016, 31 (1), 92–101. 10.1177/0885328216640397. [DOI] [PubMed] [Google Scholar]
- Park J. S.; Chu J. S.; Tsou A. D.; Diop R.; Tang Z.; Wang A.; Li S. The Effect of Matrix Stiffness on the Differentiation of Mesenchymal Stem Cells in Response to TGF-Beta. Biomaterials 2011, 32 (16), 3921–3930. 10.1016/j.biomaterials.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhavan O.; Ghaderi E. Flash Photo Stimulation of Human Neural Stem Cells on Graphene/TiO2 Heterojunction for Differentiation into Neurons. Nanoscale 2013, 5 (21), 10316. 10.1039/c3nr02161k. [DOI] [PubMed] [Google Scholar]
- Yang J.-W.; Hsieh K. Y.; Kumar P. V.; Cheng S.-J.; Lin Y.-R.; Shen Y.-C.; Chen G.-Y. Enhanced Osteogenic Differentiation of Stem Cells on Phase-Engineered Graphene Oxide. ACS Appl. Mater. Interfaces 2018, 10 (15), 12497–12503. 10.1021/acsami.8b02225. [DOI] [PubMed] [Google Scholar]
- Park J.; Kim I. Y.; Patel M.; Moon H. J.; Hwang S. J.; Jeong B.. 2D and 3D Hybrid Systems for Enhancement of Chondrogenic Differentiation of Tonsil-Derived Mesenchymal Stem Cells. Adv. Funct. Mater. 2015, 25, 2573. 10.1002/adfm.201500299. [DOI] [Google Scholar]
- Crowder S. W.; Prasai D.; Rath R.; Balikov D. A.; Bae H.; Bolotin K. I.; Sung H. J.. Three-Dimensional Graphene Foams Promote Osteogenic Differentiation of Human Mesenchymal Stem Cells. Nanoscale 2013, 5, 4171. 10.1039/c3nr00803g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N.; Zhang Q.; Gao S.; Song Q.; Huang R.; Wang L.; Liu L.; Dai J.; Tang M.; Cheng G. Three-Dimensional Graphene Foam as a Biocompatible and Conductive Scaffold for Neural Stem Cells. Sci. Rep. 2013, 3 (1), 1604. 10.1038/srep01604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka K. M.; Dahlin R. L.; Meretoja V. V.; Kasper F. K.; Mikos A. G. Enhancing Chondrogenic Phenotype for Cartilage Tissue Engineering: Monoculture and Coculture of Articular Chondrocytes and Mesenchymal Stem Cells. Tissue Eng., Part B 2014, 20 (6), 641–654. 10.1089/ten.teb.2014.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]