Abstract
Protein cleavage inside the cell membrane triggers various patho-physiological signaling pathways, but the mechanism of catalysis is poorly understood. We solved ten structures of the Escherichia coli rhomboid protease in a bicelle membrane undergoing time-resolved steps that encompass the entire proteolytic reaction on a transmembrane substrate and an aldehyde inhibitor. Extensive gate opening accompanied substrate, but not inhibitor, binding, revealing that substrates and inhibitors take different paths to the active site. Catalysis unexpectedly commenced with, and was guided through subsequent catalytic steps by, motions of an extracellular loop, with local contributions from active site residues. We even captured the elusive tetrahedral intermediate that is uncleaved but covalently attached to the catalytic serine, around which the substrate was forced to bend dramatically. This unexpectedly stable intermediate indicates rhomboid catalysis uses an unprecedented reaction coordinate that may involve mechanically stressing the peptide bond, and could be selectively targeted by inhibitors.
INTRODUCTION
Four large and distinct superfamilies of intramembrane proteases evolved to regulate diverse signaling pathways from bacteria to mankind1,2. These unusual enzymes have been implicated in diseases ranging from Alzheimer’s disease to malaria, type II diabetes, cancer, Parkinson’s disease, cholera, and tuberculosis3–6.
The first described intramembrane protease, the site-2 protease, was discovered over twenty years ago as the enzyme that releases transcription factors from their transmembrane anchors in response to cholesterol depletion7. This unusual zinc-dependent metalloprotease defined an entire family of enzymes that were subsequently found to act as regulators of unrelated virulence circuits of diverse bacterial pathogens6. Most intensively studied are aspartyl intramembrane proteases including γ-secretase, which liberates the transcription factor domain of the Notch receptor as the ultimate consequence of ligand binding8. γ-secretase has long been pursued as a therapeutic target because it also delivers the final cut to generate the amyloid-β peptide that causes Alzheimer’s disease5. Last to come to light are glutamyl proteases defined by Ras-converting enzyme (Rce1), which removes the terminal tripeptide after prenylation of target proteins9.
Rhomboid proteases form the most pervasive enzyme superfamily that catalyze hydrolysis of peptide bonds inside the cell membrane10,11. These serine hydrolases probably originated in bacteria, and at least in one pathogenic species they facilitate communication between cells to organize population growth and development12. More is understood about rhomboid function in parasitic eukaryotes where they cleave transmembrane adhesins to dismantle junctions formed by the parasite with the host cell6. This dismantling plays a key role at the end of malaria invasion13,14, and in regulating cytoadherence by non-invasive parasites including trichomonads15. Animals encode multiple rhomboid proteases, some of which activate epidermal growth factor receptor ligands to initiate cell signaling16. All eukaryotic cells harbor a dedicated rhomboid protease in their mitochondria that maintains organelle health; failure of this function leads to Parkinsonian neurodegeneration17.
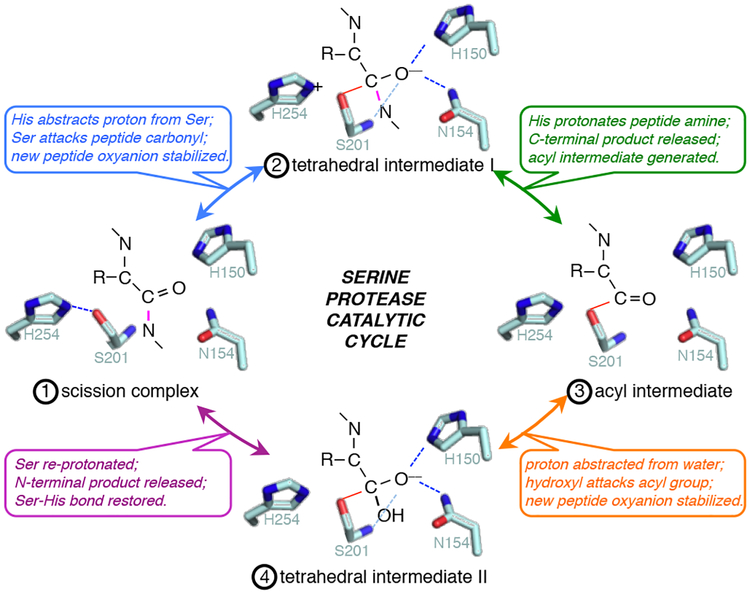
Despite their prominent patho-physiological roles, the steps underlying catalysis have never been observed directly for any intramembrane protease; current models only rationalize mechanisms by assuming parallels to their well-characterized soluble protease counterparts, and so it is for rhomboid enzymes. Serine proteases use a catalytic base, which is usually a histidine, to convert the benign serine into a powerful nucleophile by abstracting its proton (Fig. 1)18. Ensuing attack on the carbonyl carbon of the peptide bond results in a short-lived transition state in which the carbon becomes tetrahedral and forms an oxyanion that must be stabilized by the enzyme in order for this step to take place. Rapid protonation of the amide nitrogen by the catalytic base facilitates release of the carboxy-terminal peptide product, while the amino-terminal product simultaneously decomposes to an acyl intermediate covalently attached to the catalytic serine. The cycle repeats in the second half of the reaction, except this time with a water molecule, which is activated by the enzyme, attacking the carbonyl carbon, thereby releasing the amino-terminal product, and restoring a proton to the serine.
Fig. 1. Core chemical mechanism of serine protease catalysis.
Generalized major events of serine protease catalysis (numbered in order 1 through 4) are illustrated with E. coli rhomboid GlpG active-site residues: H254 and S201 catalyze nucleophilic peptide bond cleavage while H150 and N154 simultaneously conduct electrophilic catalysis by stabilizing the substrate oxyanion. The scissile peptide bond is colored in pink, while hydrogen bonds are depicted as dashed lines throughout.
While the general chemistry of peptide bond hydrolysis is likely to be similar between soluble serine proteases and rhomboid proteases, meaningful differences should be expected: rhomboid proteases evolved independently by convergent evolution and inside the membrane. Therefore it is important to delineate the mechanism of rhomboid catalysis directly. Time-resolved crystallography can visualize the discrete actions taken by enzymes during catalysis and thereby directly map out the reaction coordinate: reactions are made to occur in crystals and snapshots are taken in a time-dependent manner19. This approach has revealed some catalytic steps in atomic detail for a handful of soluble serine proteases20–22 and even such complicated enzymes as DNA polymerases23. However, this powerful approach has been difficult to implement with membrane enzymes. Transformative successes include witnessing the biosynthesis and extrusion of nature’s most abundant polymer, cellulose, from the membrane by a bacterial membrane-immersed enzyme24, light-driven steps of the bacteriorhodopsin proton-pumping cycle25, and water oxidation by massive plant photosystem complexes26,27. Time-resolved crystallography nevertheless remains rare with membrane enzymes, and has never been achieved with any membrane protease28. Given the remarkable power of this approach, we sought to examine whether rhomboid proteolysis inside the membrane might be amenable to time-resolved crystallography.
RESULTS
Snapshots of catalysis on a peptide aldehyde inhibitor
Since rhomboid proteases target relatively short regions of their substrates29–32 and can take minutes to even hours to catalyze proteolysis of a single peptide bond33,34, we examined whether we might be able to visualize catalytic intermediates using a crystal soaking approach that we developed previously31. Our starting point was the Escherichia coli rhomboid GlpG that we recently crystallized in a bicelle membrane (Fig. 2). This was important for mechanistic studies, because the membrane modulates all known properties of rhomboid proteolysis35–37. In our crystals, GlpG is both active and assumes a partly open conformation that is able to accept peptide inhibitors. Specifically, the L5 loop that overlies that active site and must lift up to allow substrate access to the active site was entirely disordered in our starting structure. Having recently characterized the tetrahedral end product that resulted from catalysis by GlpG in crystals soaked with tetrapeptide aldehydes modeled on the classical substrate Gurken31, we sought to test whether we could visualize discrete steps in the inhibition pathway in atomic detail. We exposed crystals to the tetrapeptide aldehyde inhibitor of sequence VRMA (termed P4 to P1 in protease nomenclature) for varying lengths of time, and solved the resulting structures to 2.2–2.4 Å resolution (Table 1). We ultimately succeeded in witnessing five distinct enzymatic steps on this inhibitor, which we termed Snapshot-I1 through 4 (Fig. 3 and Supplementary Fig. 1).
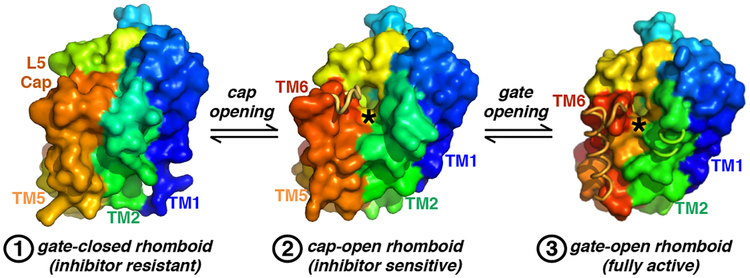
Fig. 2. Comparison of pre-catalytic GlpG conformers.
Lateral (membrane) views of structures in surface representation color-coded in rainbow from amino (blue) to carboxyl (red) termini. Disordered regions are depicted as ribbon-only, and the catalytic serine is marked with an asterisks. The closed form (leftmost) cannot accept either substrates or inhibitors. The cap-open structure (middle) that we visualized as a starting point in the bicelle can accept inhibitors but not substrates. Finally, the gate-open form (rightmost) can accept both substrates and inhibitors. These conformers correspond to the states numbered 1 (left), 2 (middle), and 3 (right) in the model (see Fig. 6).
Table 1 |.
Data collection and refinement statistics for inhibitor snapshots
| GlpG- VRMA-CHO Snapshot-I1 (PDB 6PJ4) |
GlpG- VRMA-CHO Snapshot-I2 (PDB 6PJ5) |
GlpG- VRMA-CHO Snapshot-I3 (PDB 6PJ7) |
GlpG- VRMA-CHO Snapshot-I4 (PDB 6PJ8) |
|
|---|---|---|---|---|
| Data collection | ||||
| Space group | C222(1) | C222(1) | C222(1) | C222(1) |
| Cell dimensions | ||||
| a, b, c (Å) | 70.91, 98.89, 62.69 | 70.77, 98.44, 62.64 | 70.84, 99.51, 62.70 | 70.57, 97.95, 62.55 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å)* | 62.69–2.30 (2.36–2.30) | 57.46–2.30 (2.38–2.30) | 57.72–2.30 (2.38–2.30) | 62.55–2.30 (2.39–2.30) |
| Rsym | 0.080 (0.285) | 0.137 (0.520) | 0.130 (0.549) | 0.190 (0.571) |
| I / σI | 14.0 (5.7) | 7.7 (2.9) | 7.6 (2.4) | 5.7 (2.3) |
| Completeness (%) | 99.9 (100.0) | 96.5 (96.4) | 95.5 (95.4) | 99.5 (99.4) |
| Redundancy | 6.1 (6.2) | 5.7 (5.9) | 4.9 (5.1) | 5.0 (5.1) |
| Refinement | ||||
| Resolution (Å) | 57.63–2.30 (2.36–2.30) | 50.01–2.40 (2.46–2.40) | 50.00–2.30 (2.36–2.40) | 50.01–2.40 (2.46–2.40) |
| No. reflections | 9,583 | 8,118 | 9,238 | 8,287 |
| Rwork / Rfree | 0.200/0.226 (0.194/0.307) | 0.234/0.275 (0.253/0.288) | 0.231/0.247 (0.344/0.377) | 0.234/0.263 (0.318/0.343) |
| No. atoms | 1503 | 1493 | 1491 | 1504 |
| Protein | 1461 | 1468 | 1456 | 1475 |
| Ligand/ion | 0 | 0 | 0 | 0 |
| Water | 42 | 25 | 35 | 29 |
| B-factors | 34.7 | 39.0 | 41.0 | 39.1 |
| Protein | 31.9 | 38.9 | 39.2 | 37.8 |
| Ligand/ion | 0 | 0 | 0 | 0 |
| Water | 37.3 | 43.2 | 44.4 | 41.0 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.019 | 0.013 | 0.016 | 0.012 |
| Bond angles (°) | 1.69 | 1.38 | 1.68 | 1.38 |
Values in parentheses are for highest-resolution shell.
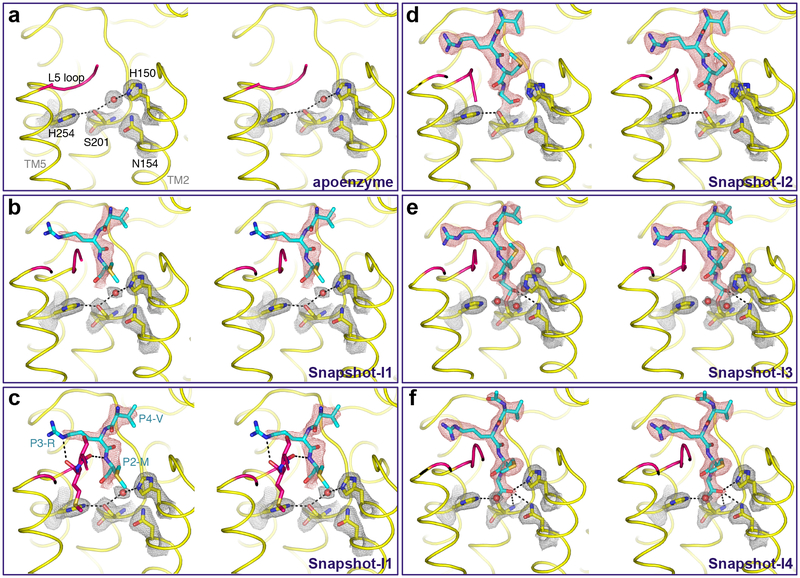
Fig. 3. Time-resolved X-ray crystallography of aldehyde inhibitor binding and catalysis.
Stereo views of experimental electron density maps (2Fo-Fc at 1.2σ) for catalytic residues and waters (grey mesh), and substrate (red mesh). Waters appear as red spheres, while the L5 loop is colored in pink throughout for emphasis. To obtain these images, crystals of GlpG in a bicelle membrane were soaked with the Ac-VRMA-CHO peptide aldehyde for the indicated times prior to freezing and X-ray diffraction analysis. In addition to the catalytic steps, the interactions that formed with the L5 loop are detailed in the lower left image, which is the same data as the panel above it but includes L5 sidechain/mainchain interaction details. Also see Supplementary Figure 1 for an alternative, more simplified schematic.
The starting apoenzyme structure had a weak 3.29 Å hydrogen bond between the catalytic S201 and H254 residues, and a hydrogen bond between S201 and H150 (that stabilizes the oxyanion with N154) through a bridging water molecule with distances of 3.20 Å and 3.14 Å, respectively (Fig. 3a). The earliest difference we could ever see indicated some density beginning to appear for the inhibitor, but it was very weak and could not be modeled (not shown). Interestingly, the overlying L5 loop nevertheless already started to assume an ordered conformation. In our first snapshot we observed increased density and could model the amino terminal three residues of the peptide aldehyde, but the P1 alanine next to the catalytic residues remained too disordered to be observed (Fig. 3b). The catalytic residues maintained their positions similar to the apoenzyme structure (including presence of the bridging water between S201 and H150), but the L5 loop became ordered in all except four residues (L244-M247), and clearly established interactions with the P2, P3 and P4 residues of the inhibitor. Specifically, these included hydrogen bonds between M249 and P3-R at a distance of 3.29 Å, S248 and P2-M at a distance of 2.94 Å, and a hydrophobic interaction between P4-V and F146 (Fig. 3c).
Snapshot-I2 revealed strong and ordered electron density of the peptide aldehyde that allowed confident modeling of the entire peptide moiety (Fig. 3d). This was the first time that the P1 alanine residue became visible, making it the last element to bind stably. Despite this residue being essential for proteolysis, its late binding suggested that it does not make a strong interaction that contributes to affinity. This step was also accompanied by first changes in the catalytic residues. The water molecule that initially bridged S201 and H150 was pushed outwards by the aldehyde oxygen (the future oxyanion) of the inhibitor, and the sidechain of H150 assumed a new, flipped-out position by rotating 94.8° with its Nδ1 being displaced 3.93 Å from the future oxyanion. In fact, we could observe both the inward and flipped-out conformation of the H150 sidechain in approximately equal proportions at this timepoint. Notably, S201 and the peptide aldehyde remained too far apart for covalent linkage and without clear connecting electron density, indicating that catalysis had not yet taken place.
In Snapshot-I3 we witnessed first evidence of catalysis: clear electron density connected S201 and the peptide aldehyde thus mimicking the tetrahedral intermediate (Fig. 3e). No other changes in the substrate were evident along the length of the peptide. Of particular note was the change in position of the catalytic H254, which moved downward by rotating 38° and no longer interacted with S201, which itself rotated by 15°. Such a motion would be expected to accompany proton transfer from S201 during nucleophilic activation38. Surprisingly, we found the oxyanion interacting with only the sidechain of N154 at a distance of 3.19 Å, but became surrounded by three new water molecules, while H150 had assumed the flipped-out position fully.
The final snapshots all revealed a dramatic re-organization of the active site (Fig. 3f): H150 adopted the inward conformation, and all hydrogen bonds with the oxyanion were readjusted to form a tripartite interaction network consisting of the sidechains of H150 (at a distance of 3.03 Å) and N154 (at a distance of 3.17 Å), and the backbone of S201(at a distance of 3.30 Å). The catalytic H254 also shifted to restore its hydrogen bond with S201 (at a distance of 3.15 Å), which remained in covalent complex with the peptide aldehyde. This overall enzyme:inhibitor complex was stable in the bicelle crystal, because we could not see any further conformational changes even when we extended the incubation time.
Snapshots of catalysis on a transmembrane peptide substrate
Encouraged by the success of our approach with the short tetrapeptide inhibitor, we examined whether we could visualize any binding or catalytic steps on long peptide substrates (lacking any warheads that could help to stabilize them in the active site). We tested several peptide sequences, but ultimately obtained the best results with a 13-mer modeled on the sequence of the classical rhomboid substrate Gurken. The substrate sequence included 6 residues preceding the cleavage site (RKVRMA) and extended all the way to and including the deep transmembrane helix-breaking serine and proline residues that facilitates unwinding (AIVFSFP). In standard protease nomenclature, residues amino-terminal to the cleavage site are numbered from P1 onwards, while those carboxy-terminal to the cleavage site are similarly labeled but with prime designations39. Remarkably, we ultimately visualized the entire reaction, including intermediates, and in 6 snapshots of 2.3–2.6 Å resolution that we termed Snapshot-S1 through S6 (Table 2): gate-opening and substrate binding, catalysis forming the tetrahedral intermediate, formation of the acyl intermediate, and final product resolution (Fig. 4 and Supplementary Fig. 2).
Table 2 |.
Data collection and refinement statistics for substrate snapshots
| GlpG- RKVRMA AIVSFP Snapshot-S1 (PDB 6PJ9) |
GlpG- RKVRMA AIVSFP Snapshot-S2 (PDB 6PJA) |
GlpG- RKVRMA AIVSFP Snapshot-S3 (PDB 6PJP) |
GlpG- RKVRMA AIVSFP Snapshot-S4 (PDB 6PJQ) |
GlpG- RKVRMA AIVSFP Snapshot-S5 (PDB 6PJR) |
GlpG- RKVRMA AIVSFP Snapshot-S6 (PDB 6PJU) |
|
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space group | C222(1) | C222(1) | C222(1) | C222(1) | C222(1) | C222(1) |
| Cell dimensions | ||||||
| a, b, c (Å) | 71.26, 99.00, 63.21 | 72.14, 99.76, 63.14 | 73.03, 98.56, 62.97 | 71.74, 96.45, 62.57 | 71.28, 99.88, 63.18 | 71.50, 96.65, 62.57 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 57.84–2.50 (2.60–2.50) | 58.52–2.60 (2.72–2.60) | 58.75–2.40 (2.49–2.40) | 57.63–2.50 (2.60–2.50) | 58.02–2.30 (2.38–2.30) | 57.48–2.50 (2.60–2.50) |
| Rsym | 0.089 (0.687) | 0.065 (0.536) | 0.061 (0.275) | 0.041 (0.451) | 0.059 (0.277) | 0.042 (0.328) |
| I / σI | 4.5 (1.4) | 4.3 (1.8) | 6.4 (1.8) | 10.3 (2.1) | 5.2 (2.0) | 10.0 (1.2) |
| Completeness (%) | 99.2 (99.2) | 99.5 (99.4) | 95.3 (70.5) | 96.0 (77.0) | 99.7 (99.9) | 98.6 (98.2) |
| Redundancy | 3.9 (4.0) | 3.9 (3.9) | 5.2 (2.0) | 5.6 (3.7) | 4.1 (4.0) | 8.8 (8.6) |
| Refinement | ||||||
| Resolution (Å) | 57.84–2.50 (2.57–2.50) | 58.52–2.60 (2.67–2.60) | 58.75–2.45 (2.51–2.45) | 57.63–2.50 (2.57–2.50) | 58.02–2.40 (2.46–2.40) | 57.48–2.50 (2.57–2.50) |
| No. reflections | 7,588 | 6,804 | 8,007 | 7,304 | 8,667 | 7,282 |
| Rwork / Rfree | 0.269/0.278 (0.397/0.441) | 0.256/0.269 (0.367/0.428) | 0.250/0.261 (0.363/0.366) | 0.247/0.287 (0.341/0.275) | 0.236/0.248 (0.310/0.369) | 0.249/0.285 (0.340/0.291) |
| No. atoms | 1442 | 1448 | 1442 | 1419 | 1457 | 1395 |
| Protein | 1430 | 1438 | 1427 | 1403 | 1433 | 1386 |
| Ligand/ion | 0 | 0 | 0 | 0 | 0 | 0 |
| Water | 12 | 10 | 15 | 16 | 24 | 9 |
| B-factors | 60.2 | 56.3 | 47.0 | 69.5 | 49.7 | 70.6 |
| Protein | 60.3 | 56.4 | 47.4 | 69.7 | 49.6 | 70.8 |
| Ligand/ion | 0 | 0 | 0 | 0 | 0 | 0 |
| Water | 44.3 | 44.4 | 40.0 | 54.5 | 52.4 | 49.3 |
| R.m.s. deviations | ||||||
| Bond lengths (Å) | 0.006 | 0.006 | 0.007 | 0.006 | 0.006 | 0.006 |
| Bond angles (°) | 1.47 | 1.38 | 1.64 | 1.46 | 1.40 | 1.43 |
Values in parentheses are for highest-resolution shell.
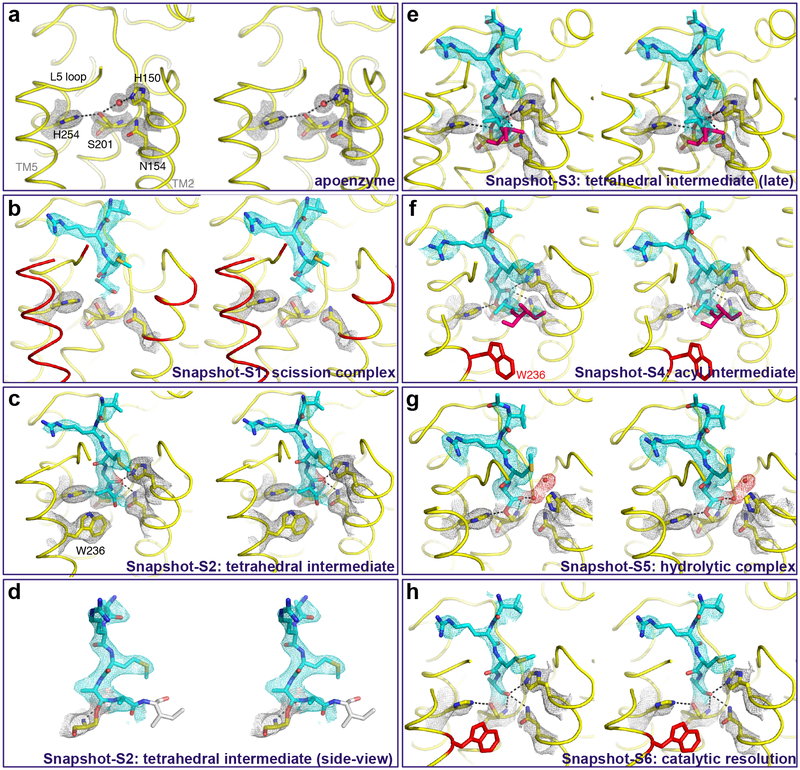
Fig. 4. Time-resolved X-ray crystallography of transmembrane substrate binding and cleavage.
Stereo views of experimental electron density maps (2Fo-Fc at 1.2σ) for catalytic residues (grey mesh), substrate (cyan mesh), and oxyanion and water (red spheres/mesh). To obtain these images, crystals of GlpG in a bicelle membrane were soaked with the Ac-RKVRMAAIVFSFP-amide peptide for differing lengths of time prior to freezing and X-ray diffraction analysis. The abrupt substrate bend is detailed in the lower left image, which is the same data as the panel above it but rotated ~90° to the right. Waters are rendered as red spheres, while disordered regions are shown in red (in b, e, f, h) or grey (in d) during the reaction steps. Also see Supplementary Figure 2 for an alternative, more simplified schematic.
In the earliest snapshot, soaking GlpG crystallized in a bicelle with the substrate resulted in dramatic changes in GlpG (Fig. 4b) that are indicative of gate-opening (Fig. 2). This was vastly more than we observed at any point with tetrapeptide aldehyde binding and catalysis. Remarkably, the entire TM5 helix and most of the overlying L5 loop became disordered (R227-M247), as did even sidechain residues on TM2 including L148-H150 as well as F153 (the key residue that interdigitates with TM5 in the gate-closed form40,41). Even residues within the active site itself became disorganized to the point that no hydrogen bond linked the catalytic serine and histidine (moved 3.55 Å apart).
In dramatic contrast, the first interactions with substrate were already forming and were quite similar to what we observed with the peptide aldehyde: the very distal part of the L5 loop (S248 and M249) became ordered and started interacting with the incoming peptide substrate at the P4, P3, and P2 positions (Fig. 4b). The rest of the substrate (P6-5, P1-P7’) remained entirely too disordered to model.
In the second snapshot we witnessed evidence of catalysis: the sidechain of S201 was in continuous electron density with the P1 carbonyl carbon, and the attack occurred from the uncommon Si-face (Fig. 4c). TM5 and the L5 loop assumed an ordered conformation in all except four residues (L244-M247), and the gating sidechain W236 on TM5 adopted an ordered and inward-facing conformation identical to its position in the apoenzyme structure. Such an extensive interaction network with substrate is often key precisely at the moment of catalysis. Importantly, we could also for the first time observe substrate electron density for the mainchain on the prime side, which has never been observed. Also notable was how weak electron density was for more distal prime-side residues, and no electron density whatsoever was evident for any sidechain or for the mainchain of residues P4’-P7’. The lack of strong electron density and no sidechain interactions at residues following the cleavage site suggests that no specific binding pockets exist for substrates at these distal sites.
However, to our astonishment, at this timepoint electron density was actually continuous between the catalytic serine and both the P1 and P1’ residues, which indicates that we captured the tetrahedral intermediate that has proven so elusive to image directly with natural substrates18,21,22. Accordingly, the oxyanion itself was stabilized by all three hydrogen bonds with GlpG (at distances of 3.19 Å with H150, 3.20 Å with N154, and weakly with the backbone of S201 at a distance of 3.41 Å). S201 and H254 also restored their hydrogen bond at a distance of 3.14 Å, which is radically different from prior structures of tetrahedral-mimicking phosphonates that induced a non-catalytic complex where H254 swung away completely and made no bond with S20142,43. The substrate in the active site formed a remarkably abrupt angle precisely at the catalytic serine, adopting a dramatic >90° bend between the carboxy-terminal and amino-terminal halves (Fig. 4d). This structure indicates that mechanically stressing the peptide bond, and/or stabilizing the tetrahedral geometry once formed, is a key strategy for rhomboid enzymes.
The next two snapshots progressively led to acyl-intermediate formation. First, in Snapshot-S3, density for the P1’ residue elongated by shifting slightly further from P1 but they remained continuous (Fig. 4e). In Snapshot-S4 we observed first evidence of enzyme acylation: while we could still visualize the P1 residue in continuous electron density with S201, the electron density of the P1’ residue had weakened and clearly moved too far (2.58 Å) to be covalently attached to P1 (Fig. 4f). This was accompanied by W236 on TM5 assuming a disordered conformation.
The second half of the catalytic cycle that resolves the acyl intermediate is driven by hydrolysis18. In Snapshot-S5 (Fig. 4g), substrate interactions with the enzyme again became pronounced such that most of the L5 loop became ordered again (only residues D243-G246 were disordered) and W236 on TM5 also assumed an ordered position. In contrast, this was the first time that we observed H150 flipping outwards: this motion was blocked by steric clash with the prime side of the substrate. In fact, until this point H150 was bracketed by the sidechains of P2 and P1’ and F146 immediately above it. Dissociation of the carboxy-terminal product now liberated H150, which, relative to the aldehyde structures, established a longer distance from the oxyanion (6.65 Å) but through a smaller rotational angle (76.6°). Two new water molecules occupied the space left by H150, perhaps in preparation for hydrolysis. N154 also moved and was too far away (3.70 Å) to make a stabilizing interaction with the substrate carbonyl of the acyl intermediate.
In our sixth and final snapshot (Fig. 4h), we observed evidence of catalytic resolution: substrate was no longer covalently attached to S201 and substrate electron density weakened indicative of dissociation, but nevertheless the cleaved product remained bound via hydrogen bonds with all three of the oxyanion stabilizing groups (at distances of 2.88 Å with H150, 2.96 Å with N154, and 3.17 Å with the backbone of S201), and maintained prominent interactions with the overlying L5 loop. The catalytic serine and histidine restored the original hydrogen bond (at a distance of 3.09 Å) that readies them for the next round of catalysis.
A critical role for the L5 loop in catalysis
An entirely unexpected finding that emerged from our ability to visualize rhomboid catalysis from start to finish was the continuous role played by the extracellular L5 loop. This loop ‘caps’ the active site in the gate-closed state, and, as such, it is assumed that its only role is to lift away for substrate to access the active site. However, witnessing L5 loop actions at nearly every step of catalysis raised the exciting possibility that it plays a direct role in catalysis by progressively positioning substrate through the various reaction steps.
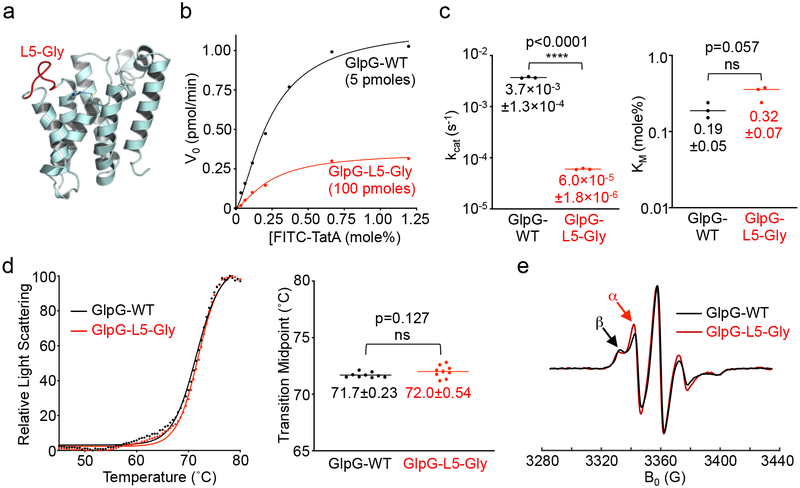
To distinguish between a purely gating role versus catalytic roles for the L5 loop, we installed glycines into the L5 loop (Fig. 5a) to enhance its mobility, and assessed the effect on proteolysis. If its role was purely in gating, increasing loop dynamics should facilitate L5 cap opening and thus increase the overall catalytic rate. Conversely, if the L5 loop does indeed repeatedly position substrate for catalysis, then introducing glycines should enhance its initial opening but compromise its subsequent interactions with substrate and lead to an overall decrease in catalytic rate.
Fig. 5. Enzyme analysis validates a catalytic role for the L5 loop in intramembrane proteolysis.
a, Lateral view (cytosol down) of E coli GlpG (PDB 2NRF) illustrating the L5 loop residues that were mutated to glycine (red). The catalytic serine and histidine are shown in ball-and-stick representation with the hydrogen bond linking them depicted with a dashed green line. b, Real-time steady-state kinetic analysis of wildtype and the L5 loop glycine mutant (L5-Gly) GlpG catalysis in reconstituted liposomes. Note that due to its low protease activity 20× more of the mutant enzyme was used in the analysis. c, Kinetic constants kcat (turnover rate) and KM (Michaelis constant, in mole percent relative to phospholipid) derived for WT and L5-Gly mutant GlpG were graphed on scatter plots with a log scale (Y-axis). Mean ± s.d. are indicated for three separate experiments and an unpaired t-test was used to calculate significance. d, Thermostability analysis of wildtype and L5-Gly mutant GlpG (left graph), and statistical analysis of the resulting transition midpoints (right scatter plot). Indicated are the mean ± s.d. of nine total replicates analyzed in two separate experiments, and an unpaired t-test was used to calculate significance. The same protein preparations were used for kinetic and thermostability analyses. e, Electron paramagnetic resonance spectra of wildtype and L5-Gly mutant GlpG. The central M247 position of the L5 loop was labeled with a nitroxide spin probe, and the spectra were recorded at 310 K (37°C). The red arrow denotes an increase in the characteristic fast mobility α component peak (immobile β component denoted by black arrow).
Consistent with a key role in catalysis, installing glycines into the L5 loop reduced proteolytic activity in reconstituted proteoliposomes by two orders of magnitude (Fig. 5b). This dramatic reduction was entirely reflected in the catalytic rate constant kcat, without any significant change in initial substrate binding (KM) (Fig. 5c). Importantly, the glycine mutant exhibited no change in its structural stability as revealed in a very sensitive and quantitative thermostability assay40 (Fig. 5d). This is consistent with the folding nucleus being centered near the cytosolic apex of TM244, and subsequent L5 interactions in the closed state not contributing to GlpG’s structural stability40. Finally, by installing a spin probe at the central M247 position of the L5 loop, we were able to use electron paramagnetic resonance spectroscopy to confirm that the glycines did indeed noticeably enhance loop dynamics (Fig. 5e). Therefore, as predicted by time-resolved crystallography, the L5 loop plays a critical, yet entirely unanticipated, role directly in progressively ushering substrate through the steps of catalysis.
DISCUSSION
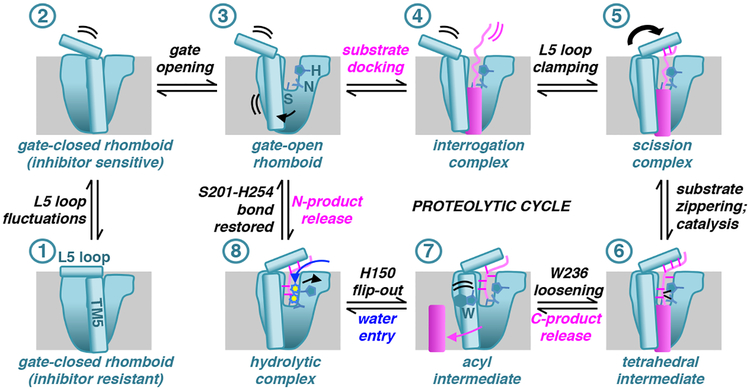
In summary, we collected ten time-resolved snapshots of rhomboid proteolysis inside the membrane that encompassed the entire reaction from substrate binding to hydrolytic enzyme deacylation (Fig. 6). In addition to broadly mapping out the reaction coordinate from start to finish, these snapshots revealed three counter-intuitive features as defining hallmarks of rhomboid catalysis.
Fig. 6. Model of enzymatic actions underlying rhomboid intramembrane proteolysis.
Illustrated are landmark steps of rhomboid catalysis on a substrate viewed laterally from the membrane. All of the steps have been visualized crystallographically except the interrogation complex, which is predicted to be highly dynamic. Rhomboid/substrate/water actions proceeding from one step to the next (in numerical order) are described above/below arrows (all steps are formally reversible). Double curved lines indicate dynamics, while arrows denote conformational changes that occurred to get to that step. Individual enzyme elements or residues are labeled the first time they appear in the schematic.
Our first surprise was the extent of gate opening that facilitates binding of even a relatively short (13 residue) transmembrane substrate peptide: in addition to the L5 loop, the entire TM5 segment, as well as its interacting residues on top of TM2, became disordered. This structural characterization puts to rest the idea that gating deep inside the membrane cannot occur during substrate proteolysis. On the contrary, it reveals that gating involves considerably greater lateral opening than is currently accepted45. Importantly, no such changes occurred with short peptidic inhibitors of identical sequence that can enter the active site directly, revealing that natural substrates and inhibitors/short peptides take different paths to the active site31,46.
Another unanticipated feature was the return of the L5 loop after substrate had already entered the active site. The current view is that the L5 loop only has to move away for substrates to access the active site; all subsequent catalytic steps are thought to be caused by local active site residue motions45,47. While this opening is an incontrovertible first step, L5 returning was actually the key step that “seeded” substrate binding in the active site itself through stabilizing interactions. This new insight supports a controversial view of intramembrane proteolysis centered on substrate dynamics: rhomboid enzymes initially identify their substrates by interrogating transmembrane helix dynamics where unstable helices that spontaneously unwind will be cut, while those that maintain their secondary structure will not30,35. Our time-resolved crystallography now reveals that L5 loop “clamping down” restrains substrates into a catalytically competent conformation for proteolysis, thereby converting the “interrogation” complex to a “scission” complex, as predicted by inhibition kinetics31.
Perhaps even more revealing were L5 motions that progressively formed and loosened distal interactions with substrate throughout the reaction. These unexpected features suggest that it is primarily the actions of the remote, extramembraneous L5 loop that progressively re-positions substrate as a means to ‘usher’ it through the entire reaction coordinate. This is strongly supported by the dramatic ~100-fold decrease specifically in catalytic rate kcat displayed by the glycine variant of GlpG, which renders the L5 loop more mobile and thus compromised in its ability to control the position of the substrate during catalysis. Local active site residues also changed position during catalysis, some of which prior mutagenesis experiments support as key for catalysis, while others less so. For example, we and others observed various interactions of W236 with both substrates and inhibitors that imply a role in catalysis42,43,45. Yet mutagenesis of W236 results in a 5–10-fold increase in proteolysis40,41,48. Thus, in contrast to the L5 loop, the gating role of W236 dominates its role in the catalytic cycle, with subsequent substrate interactions having minor, if any, meaningful functions during catalysis. More enigmatic is H150, which is both key in stabilizing the oxyanion, but also undergoes a dramatic outward flipping motion. While this may facilitate water entry during the second stage of catalysis, the precise and/or full function(s) of this flipping motion remains unclear, because H150 mutants cannot be studied using an enzymatic approach as they render GlpG catalytically inert40,41,48. Similar limitations hinder dissecting the multifaceted roles of H254 and N15440,41,48.
The third and most surprising discovery of our study was visualizing the elusive tetrahedral intermediate in two separate snapshots as a stable entity. This is extraordinarily unexpected, because in other serine proteases the tetrahedral intermediate is a short-lived transition state18,21,22. Visualizing it during rhomboid catalysis suggests that rhomboid is the only serine protease known to have evolved the tetrahedral intermediate to be a stable stalling point, and thus not the transition state. Achieving this unprecedented stability may result from serine attack from the rare Si face, tripartite oxyanion stabilization that only rhomboid is known to employ, and/or the effect of accommodating a dramatic >90° bend of substrate at the point of catalysis. These features have never, to our knowledge, been observed in any other protease18,20–22. Since serine protease mechanisms historically provided the foundation on which our modern understanding of enzyme theory is based, characterizing the unusual reaction coordinate that our structures mapped out could add a new dimension to our understanding of general enzyme mechanisms. Although measuring energetics is challenging with membrane enzymes, these studies could expose new vulnerabilities for designing novel inhibitors to target rhomboid enzymes selectively49.
Finally, while this is the first structural glimpse of proteolysis inside the membrane, prototypic members of other intramembrane protease families have also been crystallized, are able to act on short peptides, and have comparably slow catalytic rates28,50–52. Implementing our time-resolved soaking strategy should therefore also enable delineating their gating and catalytic cycles in atomic detail.
METHODS
Crystallization, Data Collection, and Refinement.
Crystals of E. coli rhomboid GlpG (the Y205F variant with the N-terminal 87 residues removed) were obtained in a bicelle composed of DMPC/CHAPSO (2.8:1) exactly as described previously31. Reservoir buffer contained 0.1 M NaOAc pH 5.5, 3 M NaCl, and 5 % ethylene glycol. For time-resolved analysis with the peptide aldehyde inhibitor (Ac-VRMA-CHO), apoenzyme crystals were incubated in 2.5 mM peptide aldehyde dissolved in the same reservoir buffer at room temperature for a series of time points, and flash-frozen in a nitrogen stream for data collection. Specifically, the time-points used to generate the inhibitor snapshots in this study were: 30 min (Snapshot-I1), 3 hours (Snapshot-I2), 5 hours (Snapshot-I3), and 7 hours (Snapshot-I4). For the time-resolved analysis with the peptide substrate (Ac-RKVRMAAIVFSFP-NH2), the peptide was dissolved at 5 mM in crystallization buffer (a 1:4 mixture of bicelle and reservoir buffer) overnight and undissolved peptide was removed by ultracentrifugation. The apoenzyme crystals were soaked in the peptide solution at room temperature for a series of time points and then flash-frozen in a nitrogen stream for data collection. Specifically, the time-points used to generate the substrate snapshots in this study were: 5 min (Snapshot-S1), 2 hours (Snapshot-S2), 5 hours (Snapshot-S3), 6 hours (Snapshot-S4), 7 hours (Snapshot-S5), and 3 days (Snapshot-S6). All X-ray diffraction data sets were collected at the F1 station of the Cornell High Energy Synchrotron Source, and were processed with iMosflm 7.2.2 in the CCP4 program suite53. Structures were solved by molecular replacement using Molrep and further refined by refmac5 and PHENIX with iterative manual model building using COOT54. Final Ramachandran statistics were: 92.2% favored, 0% outliers (Snapshot-I1); 93.5% favored, 0% outliers (Snapshot-I2); 92.0% favored, 0% outliers (Snapshot-I3); 93.1% favored, 0.52% outliers (Snapshot-I4); 100% favored, 0% outliers (Snapshot-S1); 98.3% favored, 0.6% outliers (Snapshot-S2); 97.5% favored, 0.5% outliers (Snapshot-S3); 97.7% favored, 0% outliers (Snapshot-S4); 98.8% favored, 0% outliers (Snapshot-S5); and 97.6% favored, 0% outliers (Snapshot-S6).
Enzymatic Analysis of Intramembrane Proteolysis in Proteoliposomes.
Full-length GlpG proteins were analyzed for protease activity under steady-state conditions, thermostability, and protein dynamics as established previously. Briefly, site-directed mutagenesis was used to engineer a GlpG variant with L5 loop residues D243, L244, F245, M247, S248, M249, and A250 substituted with glycine (L5-Gly), and verified by sequencing the entire GlpG open reading frame. Expression of this variant and the wild-type GlpG control (amino-terminally tagged with GST and HA tags) was induced in parallel in E. coli C43(DE3) cultures with 50 μM IPTG at 16°C overnight40. Membranes were isolated from cell lysates by ultracentrifugation at ~385,000g for 30 minutes at 10°C and then solubilized for 1 h at room temperature with 2 % n-Dodecyl-β-D-maltoside (DDM; Anatrace). GlpG proteins were affinity purified using Glutathione Sepharose 4B (GE Healthcare) and eluted by on-column cleavage of the GST-tag with PreScission protease in 50 mM Tris pH 7.5, 90 mM NaCl, 1 mM EDTA, 1 mM DTT, and 0.1 % DDM.
Intramembrane proteolytic activity was measured in a real-time fluorogenic assay with a 34-residue FITC-TatA transmembrane peptide substrate con-reconstituted with GlpG into liposomes formed from E. coli polar lipids55. Briefly, increasing amounts of FITC-TatA substrate, ranging from 37.6 pmoles to 1.28 nmoles, were co-reconstituted at pH 4 in 1 mg/mL liposomes with 5 pmoles of wild-type GlpG or 100 pmoles of the L5-Gly variant. Substrate cleavage was monitored for 30 minutes at 37°C using a BioTek Synergy H4 plate reader to detect FITC fluorescence. Slopes from linear reaction time courses plotted against substrate concentration in mole percent (relative to liposome phospholipids) were fit using the Michaelis-Menten equation to derive KM and Vmax for each enzyme. Average KM and kcat values were calculated from three separate biological replicate experiments.
Spectroscopic Analyses.
Thermostability analysis of wildtype and L5-Gly GlpG proteins was performed in a StarGazer™ instrument (Harbinger Biotechnology) as described40. Proteins were diluted to a concentration of ~2.5 μM in elution buffer, placed in wells of an optical-bottom black 384-well plate (Nunc, 242764), and exposed to increasing temperature from 25°C to 85°C at a heating rate of 1°/min. Differential static light scattering was detected from a 620 nm LED light source and imaged by CCD photography every 30 seconds. Scattering intensity data was extracted from each image and plotted against temperature to generate curves that were fit by a Boltzmann sigmoidal equation (GraphPad Prism). The transition midpoints derived from curve fits were averaged from nine replicates for each protein. Nitroxide spin labeling and continuous-wave electron paramagnetic resonance spectroscopy was performed on GlpG proteins (lacking their amino-terminal 87 residues and harboring a C104A mutation) reconstituted into liposomes formed from E. coli polar lipids exactly as has been described in great detail very recently56.
Reporting summary statement.
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data and materials availability.
Coordinates of all structures have been deposited into the Protein Data Bank under the accession codes 6PJ4 (Snapshot-I1), 6PJ5 (Snapshot-I2), 6PJ7 (Snapshot-I3), 6PJ8 (Snapshot-I4), 6PJ9 (Snapshot-S1), 6PJA (Snapshot-S2), 6PJP (Snapshot-S3), 6PJQ (Snapshot-S4), 6PJR (Snapshot-S5), and 6PJU (Snapshot-S6). Source data for Figure 5b–d are available with the paper online. Any other data are available upon reasonable request from the authors.
Supplementary Material
Acknowledgements
We are grateful to current and past members of the Urban Lab for discussions and support, as well as the expert, friendly, and helpful staff at the Cornell High Energy Synchrotron Source (CHESS) for access to beamtime. This work was supported by National Institutes of Health (NIH) grant R01AI066025 (to SU). All X-ray diffraction data were collected using instruments at CHESS (beamline supported by National Science Foundation grant DMR-0936384 and NIH grant GM-103485).
Footnotes
Competing interests
The authors declare no competing interests.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
REFERENCES
- 1.Brown MS, Ye J, Rawson RB & Goldstein JL Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100, 391–8. (2000). [DOI] [PubMed] [Google Scholar]
- 2.Urban S SnapShot: Cartography of Intramembrane Proteolysis. Cell 167, 1898–1898 e1 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Verhelst SHL Intramembrane proteases as drug targets. FEBS J 284, 1489–1502 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Dusterhoft S, Kunzel U & Freeman M Rhomboid proteases in human disease: Mechanisms and future prospects. Biochim Biophys Acta 1864, 2200–2209 (2017). [DOI] [PubMed] [Google Scholar]
- 5.De Strooper B & Chavez Gutierrez L Learning by failing: ideas and concepts to tackle gamma-secretases in Alzheimer’s disease and beyond. Annu Rev Pharmacol Toxicol 55, 419–37 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Urban S Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms. Nature Reviews Microbiology 7, 411–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rawson RB et al. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell 1, 47–57. (1997). [DOI] [PubMed] [Google Scholar]
- 8.De Strooper B et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–22 (1999). [DOI] [PubMed] [Google Scholar]
- 9.Manolaridis I et al. Mechanism of farnesylated CAAX protein processing by the intramembrane protease Rce1. Nature 504, 301–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urban S, Lee JR & Freeman M Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell 107, 173–82 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Kinch LN & Grishin NV Bioinformatics perspective on rhomboid intramembrane protease evolution and function. Biochim Biophys Acta 1828, 2937–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stevenson LG et al. Rhomboid protease AarA mediates quorum-sensing in Providencia stuartii by activating TatA of the twin-arginine translocase. Proc Natl Acad Sci U S A 104, 1003–8 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker RP, Wijetilaka R & Urban S Two Plasmodium Rhomboid Proteases Preferentially Cleave Different Adhesins Implicated in All Invasive Stages of Malaria. PLoS Pathog 2, e113: 922–932 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Donnell RA et al. Intramembrane proteolysis mediates shedding of a key adhesin during erythrocyte invasion by the malaria parasite. J Cell Biol 174, 1023–33 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riestra AM et al. A Trichomonas vaginalis Rhomboid Protease and Its Substrate Modulate Parasite Attachment and Cytolysis of Host Cells. PLoS Pathog 11, e1005294 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lastun VL, Grieve AG & Freeman M Substrates and physiological functions of secretase rhomboid proteases. Semin Cell Dev Biol 60, 10–18 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Spinazzi M & De Strooper B PARL: The mitochondrial rhomboid protease. Semin Cell Dev Biol 60, 19–28 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Hedstrom L Serine protease mechanism and specificity. Chem Rev 102, 4501–24 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Samara NL, Gao Y, Wu J & Yang W Detection of Reaction Intermediates in Mg(2+)-Dependent DNA Synthesis and RNA Degradation by Time-Resolved X-Ray Crystallography. Methods Enzymol 592, 283–327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radisky ES, Lee JM, Lu CJ & Koshland DE Jr. Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates. Proc Natl Acad Sci U S A 103, 6835–40 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B, Schofield CJ & Wilmouth RC Structural analyses on intermediates in serine protease catalysis. J Biol Chem 281, 24024–35 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Wilmouth RC et al. X-ray snapshots of serine protease catalysis reveal a tetrahedral intermediate. Nat Struct Biol 8, 689–94 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Raper AT, Reed AJ & Suo Z Kinetic Mechanism of DNA Polymerases: Contributions of Conformational Dynamics and a Third Divalent Metal Ion. Chem Rev 118, 6000–6025 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Morgan JL et al. Observing cellulose biosynthesis and membrane translocation in crystallo. Nature 531, 329–34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nango E et al. A three-dimensional movie of structural changes in bacteriorhodopsin. Science 354, 1552–1557 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Kupitz C et al. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature 513, 261–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suga M et al. Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL. Nature 543, 131–135 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Sun L, Li X & Shi Y Structural biology of intramembrane proteases: mechanistic insights from rhomboid and S2P to gamma-secretase. Curr Opin Struct Biol 37, 97–107 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Strisovsky K, Sharpe HJ & Freeman M Sequence-specific intramembrane proteolysis: identification of a recognition motif in rhomboid substrates. Mol Cell 36, 1048–59 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urban S & Freeman M Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol Cell 11, 1425–34 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Cho S, Dickey SW & Urban S Crystal Structures and Inhibition Kinetics Reveal a Two-Stage Catalytic Mechanism with Drug Design Implications for Rhomboid Proteolysis. Mol Cell 61, 329–40 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zoll S et al. Substrate binding and specificity of rhomboid intramembrane protease revealed by substrate-peptide complex structures. EMBO J 33, 2408–21 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dickey SW, Baker RP, Cho S & Urban S Proteolysis inside the Membrane Is a Rate-Governed Reaction Not Driven by Substrate Affinity. Cell 155, 1270–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ticha A et al. Sensitive Versatile Fluorogenic Transmembrane Peptide Substrates for Rhomboid Intramembrane Proteases. J Biol Chem 292, 2703–2713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moin SM & Urban S Membrane immersion allows rhomboid proteases to achieve specificity by reading transmembrane segment dynamics. eLife 1, e00173 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urban S & Moin SM A Subset of Membrane-Altering Agents and gamma-Secretase Modulators Provoke Nonsubstrate Cleavage by Rhomboid Proteases. Cell Rep 8, 1241–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urban S & Wolfe MS Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proceedings of the National Academy of Sciences of the United States of America 102, 1883–8 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y & Zhang Y Serine protease acylation proceeds with a subtle re-orientation of the histidine ring at the tetrahedral intermediate. Chem Commun (Camb) 47, 1577–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schechter I Mapping of the active site of proteases in the 1960s and rational design of inhibitors/drugs in the 1990s. Curr Protein Pept Sci 6, 501–12 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Baker RP & Urban S Architectural and thermodynamic principles underlying intramembrane protease function. Nature Chemical Biology 8, 759–68 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker RP, Young K, Feng L, Shi Y & Urban S Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc Natl Acad Sci U S A 104, 8257–62 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue Y & Ha Y Catalytic mechanism of rhomboid protease GlpG probed by 3,4-dichloroisocoumarin and diisopropyl fluorophosphonate. Journal of Biological Chemistry 287, 3099–107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue Y et al. Conformational change in rhomboid protease GlpG induced by inhibitor binding to its S’ subsites. Biochemistry 51, 3723–31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paslawski W et al. Cooperative folding of a polytopic alpha-helical membrane protein involves a compact N-terminal nucleus and nonnative loops. Proc Natl Acad Sci U S A 112, 7978–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue Y & Ha Y Large lateral movement of transmembrane helix S5 is not required for substrate access to the active site of rhomboid intramembrane protease. J Biol Chem 288, 16645–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baker RP & Urban S Cytosolic extensions directly regulate a rhomboid protease by modulating substrate gating. Nature 523, 101–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y & Ha Y Open-cap conformation of intramembrane protease GlpG. Proc Natl Acad Sci U S A 104, 2098–102 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urban S & Baker RP In vivo analysis reveals substrate-gating mutants of a rhomboid intramembrane protease display increased activity in living cells. Biological Chemistry 389, 1107–15 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drag M & Salvesen GS Emerging principles in protease-based drug discovery. Nat Rev Drug Discov 9, 690–701 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamp F et al. Intramembrane Proteolysis of beta-Amyloid Precursor Protein by gamma-Secretase Is an Unusually Slow Process. Biophys J 108, 1229–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szaruga M et al. Alzheimer’s-Causing Mutations Shift Abeta Length by Destabilizing gamma-Secretase-Abetan Interactions. Cell 170, 443–456 e14 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Bolduc DM, Montagna DR, Gu Y, Selkoe DJ & Wolfe MS Nicastrin functions to sterically hinder gamma-secretase-substrate interactions driven by substrate transmembrane domain. Proc Natl Acad Sci U S A 113, E509–18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collaborative Computational Project N The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50, 760–3 (1994). [DOI] [PubMed] [Google Scholar]
- 54.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallographica. Section D: Biological Crystallography 60, 2126–32 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Baker RP & Urban S An Inducible Reconstitution System for the Real-Time Kinetic Analysis of Protease Activity and Inhibition Inside the Membrane. Methods Enzymol 584, 229–253 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kreutzberger AJB, Ji M, Aaron J, Mihaljevic L & Urban S Rhomboid distorts lipids to break the viscosity-imposed speed limit of membrane diffusion. Science 363, 497 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.