Abstract
RNA modifications are emerging as key determinants of development and of disease. However, compelling genetic demonstrations of their relevance to human disease are lacking. Here, we link rRNA 2’-O-methylation (2’-O-Me) to the etiology of dyskeratosis congenita (DC). We identify nucleophosmin (NPM1) as an essential regulator of 2’-O-Me on rRNA by directly binding C/D box small nucleolar RNAs (snoRNAs), thereby modulating translation. We demonstrate the importance of 2’-O-Me-regulated translation for cellular growth, differentiation and hematopoietic stem cell (HSC) maintenance, and show that Npm1 inactivation in adult HSCs results in bone marrow failure (BMF). We identify NPM1 germline mutations in DC patients presenting with BMF, and demonstrate that they are deficient in snoRNA binding. Mice harboring a DC germline NPM1 mutation recapitulate both hematological and non-hematological features of DC. Thus, our findings indicate that impaired 2’-O-Me can be pathogenic and etiological to human disease.
Introduction
Post-transcriptional RNA modifications, also known as the epitranscriptome, are crucial regulators of gene expression1-4. Ribosomal RNAs (rRNAs) are of the most heavily modified molecules in the cell, and require post-transcriptional modification to facilitate their processing and secondary structure formation. Therefore, rRNA modifications are essential for functional fidelity of the ribosome and proper gene expression5-7. 2’-O-methylation (2’-O-Me) is the most abundant rRNA modification as each ribosome harbors more than 100 2’-O-Me residues7-9. However, what role 2’-O-Me may play in human disease is unclear.
Nucleophosmin (NPM1) is an abundant and highly conserved phosphoprotein that mainly resides in nucleoli10. The NPM1 gene undergoes frequent genetic alterations in hematological diseases11. NPM1 bears the most frequent mutation in acute myeloid leukemia (AML) with ~30% of AML patients presenting with a frame-shift mutation that results in NPM1’s aberrant cytoplasmic localization (NPMc+)12. This suggests that nucleolar localization of NPM1 is essential for normal cellular function. While NPM1 is a bona fide RNA-binding protein13, little is known regarding its RNA-related function/s in the nucleolus and whether it contributes to pathogenesis.
By studying the RNA-binding function of NPM1 we demonstrate its key role in rRNA 2’-O-Me regulation through the direct binding of C/D box snoRNAs. We suggest that 2’-O-Me is crucial for hematopoietic stem cell maintenance and that deficiencies in 2’-O-Me contribute to pathogenesis.
Results
NPM1 regulates 2’-O-Me through snoRNA binding
We initiated our study by determining the repertoire of RNA species bound by NPM1. We performed a HITS-CLIP analysis using mouse embryonic fibroblasts (MEFs) from our previously established Npm1−/− mouse model14. Computational analysis revealed that NPM1 binds to several distinct sequence motifs (Fig. 1a), suggesting that it has a complex RNA binding capacity.
Figure 1. NPM1 regulates 2’-O-methylation through snoRNA binding.
a, RNA binding motifs of NPM1 identified by NPM1 HITS-CLIP. b, Functional annotation of the RNA species identified in NPM1 HITS-CLIP. c, Levels of specific 2’-O-Me residues (x-axis) mediated by C/D box snoRNAs in HITS-CLIP. Fold change was calculated Npm1−/− relative to Npm1+/+ MEFs. Data are presented as mean ± SD of n = 3 independent experiments and compared to levels of Ctrl modification. Ctrl is U1804 that is mediated by snoRD20, which was not identified in HITS-CLIP. d, FBL-IP was performed using Npm1+/+ and Npm1−/− MEFs. Snord enrichment was calculated relative to total input and fold enrichment in Npm1+/+ MEFs and compared to levels of enrichment of SnordCtrl, snoRD13 which was not identified to interact with NPM1 by HITS-CLIP. Bars present mean ± SD of n = 4 independent experiments. e, IP of either NPM1 (upper panel) or FBL (lower panel) was performed using nuclear extracts of Npm1+/+ and Npm1−/− MEFs. Shown is representative image out of 4 independent experiments. f, KEGG pathway analysis of Npm1+/− polysome-microarray analysis relative to Npm1+/+. Statistical significance was determined by gene set enrichment analysis. g, Cumulative distribution of minimum free energy (MFE) of 5’UTRs of polysome-depleted and -enriched transcripts, normalized to 5’UTR length. h, Diagram of the dual luciferase reporter testing for both cap-dependent and IRES-dependent translation. i, Npm1+/+, Npm1+/− and Npm1−/− MEFs were transfected with the Cap/IRES luciferase reporter (diagram in h). Bars represent SD of n = 6 independent experiments. j, Western blot of the IRES translated genes p27 and XIAP, in Npm1+/+ and Npm1−/− MEFs. RPL22 served as a loading control. Shown is representative data out of 6 independent experiments. k, 5’UTRs of Cdkn1b and Xiap lead to reduced translation of the luciferase reporter in Npm1−/− compared to Npm1+/+ MEFs. IRES versus Cap translation ratio was calculated relative to the activity in Npm1+/+. Data are presented as mean ± SD of 3 independent experiments, and compared to levels in Npm1+/+ MEFs. l, Transcript enrichment in Npm1−/− activated ribosomes (pS6-IP) was calculated relative to the enrichment in Npm1+/+ MEFs. Data are presented as mean ± SD of 4 independent experiments. m, IRES activity was calculated relative to the activity in Npm1+/+ MEFs. Data are presented as mean ± SD of 3 independent experiments. n, Analysis of specific 2’-O-Me in Clinical Remission (CR, n = 14) and NPMc+ AML patients (NPMc+, n = 16). Error bars represent SEM. Statistical significance was calculated by Mann-Whitney analysis. For all relevant panels, and unless otherwise stated, statistical significance was determined by one-tailed Student’s t test. Uncropped blot images are presented in Supplementary Figure 11.
SnoRNAs were the most abundant RNA species bound by NPM1 (Fig. 1b and Supplementary Table 1). The two main classes of snoRNAs are the C/D box and the H/ACA box snoRNAs, which associate with core proteins in ribonucleoprotein complexes (snoRNPs) to mediate rRNA modifications. Together with the methyltransferase fibrillarin (FBL), C/D box snoRNPs facilitate 2’-O-methylation9 (2’-O-Me), whereas H/ACA box snoRNPs associate with the pseudouridines synthase dyskerin (DKC1) to facilitate pseudouridylation7,8. Thus, our data suggested that NPM1 might have a role in facilitating post-transcriptional modifications of rRNA.
We first examined whether NPM1 deficiency would result in a deficit of pseudouridylation. However, no pseudouridine deficit was detected in Npm1−/− MEFs by Psi-seq15 (Supplementary Fig. 1a). Next, we measured by RTL-P16 the levels of individual rRNA 2’-O-Me residues targeted by the NPM1-interacting C/D box snoRNAs. In the absence of Npm1 we observed a significant reduction in the levels of five specific 2’-O-Me residues (Fig. 1c), and confirmed that NPM1 binds to the corresponding snoRNAs (Supplementary Fig.1b). Notably, we did not observe changes in FBL levels (Supplementary Fig.1c) or in the abundance of the specific snoRNAs (Supplementary Fig.1d).
Re-expression of NPM1 in Npm1−/− MEFs restored 2’-O-Me levels (Supplementary Fig. 1e), and led to an increase in other 2’-O-Me modifications (Supplementary Fig. 1e). This suggests that individual 2’-O-Me residues are differentially regulated and is in accordance with recent findings of 2’-O-Me regulation17-20.
In order to gain additional molecular insight, we focused on the five snoRNAs whose corresponding methylation was reduced in Npm1−/− MEFs (Fig. 1c) and tested whether their loading into snoRNPs was dependent on NPM1. In FBL-immunoprecipitation (IP) experiments we observed a significant reduction in FBL’s ability to interact with the NPM1-bound snoRNAs in Npm1−/− MEFs (Fig. 1d). Given the observed effect on snoRNP loading, and NPM1’s capacity to interact with a variety of proteins11, we hypothesized that NPM1 might interact directly with C/D box snoRNP to facilitate snoRNA loading. Reciprocal co-IP showed a robust interaction between NPM1 and FBL (Fig. 1e). This interaction is specific as neither NOP58 (a C/D box snoRNP component) nor DKC1 (a member of the H/ACA box snoRNP) were detected in the NPM1-IP (Supplementary Fig. 1f). In addition, we found that the NPM1-FBL interaction is RNA-independent as neither RNaseA nor micrococcal nuclease treatment released FBL to the supernatant (Supplementary Fig. 1g-h). These data demonstrate that NPM1 is a regulator of rRNA 2’-O-Me through interaction with C/D box snoRNAs and with FBL.
NPM1 controls IRES translation through 2’-O-Me regulation
The 2’-O-Me sites that are affected by Npm1 deletion are located in the 28S rRNA at nucleotide positions 1327, 3764, 3866, 3904, 4198 (numbers according to the human nomenclature, Fig. 1c). Notably, these residues are localized in highly structured regions20,21, with positions 3904 and 4198 located within the peptidyl transferase center and therefore have the potential to impact ribosome structure and function22. In view of this, we assessed whether global translation is compromised in the absence of Npm1. Both 35S-Met incorporation and L-Homopropargylglycine (HPG) metabolic labeling demonstrated that global translation in Npm1−/− MEFs was not defective (Supplementary Fig. 2a and 2b, respectively). In addition, Npm1+/+ and Npm1−/− MEFs were comparable in their cell size (Supplementary Fig. 2c), RNA content (Supplementary Fig. 2d) and rRNA processing (Supplementary Fig. 2e). Western blot analysis of the mTOR signaling pathway, a major pathway regulating translation23,24, did not find any significant differences between Npm1+/+ and Npm1−/− MEFs at steady state (Supplementary Fig. 2f). We have also previously shown that ribosome composition is not affected in the absence of NPM125. Taken together, these data support the conclusion that in spite of the compromised 2’-O-Me levels, global translation is unaffected.
However, ribosome alteration can cause gene-specific effects26,27. For example, low accessibility of otherwise functional ribosomes might affect translation of mRNAs that have low translation initiation and elongation rates, such as those that have a strong secondary structure of their 5’ untranslated region27,28 (5’ UTR). In order to test in an unbiased manner for gene-specific effects we performed microarray analysis of polysome-associated transcripts from Npm1+/+ and Npm1+/− MEFs (Npm1+/− provides a physiologically relevant model system to test translational skewing). Polysome microarray analysis revealed a skew in the distribution of polysome-associated transcripts upon loss of even just one NPM1 allele, with KEGG pathway analysis identifying the affected cellular pathways (Fig. 1f). As the 5’ UTR plays an important role in translational regulation, we carried out sequence analysis of enriched/depleted 5’ UTRs. While we did not detect any sequences or motifs that may lead to differential translation, minimum free energy (MFE) analysis found that Npm1+/− polysome-depleted transcripts display a higher MFE value (Fig. 1g), indicating that these transcripts have, on average, a more structured 5’ UTR.
One of the best characterized 5’ UTR structured elements is the internal ribosome entry site (IRES) element, which enables cap-independent translation initiation29. In order to assess whether Npm1 loss impairs IRES function, we first performed gene set enrichment analysis (GSEA) using a recently published dataset of IRES genes30 and observed a statistically significant negative enrichment of putative IRES genes (Supplementary Fig. 2g, top panel), a pattern that is in line with our findings. We also observed a negative trend with a more stringently filtered gene list, albeit not statistically significant (Supplementary Fig. 2g bottom panel). Taken together, this strongly suggests that IRES-dependent translation is affected in Npm1+/− polysomes.
In order to confirm this experimentally, we used a dual-luciferase reporter designed to evaluate cap- and IRES-mediated translation (Fig. 1h). We observed reduced IRES-dependent translation, in an NPM1 dose-dependent manner, whereas cap-dependent translation was unaffected (Fig. 1i and Supplementary Fig. 2h). As with Npm1−/− MEFs we did not observe any difference in snoRNA abundance in Npm1+/− (Supplementary Fig. 2i), however methylation of only 4 methylation residues was affected in Npm1+/− (Supplementary Fig. 2j). This difference might explain the gene-dosage-dependent reduction in IRES translation of the dual luciferase reporter (Fig. 1i).
We continued our investigation by looking at well characterized IRES-containing transcripts, such as the X-linked inhibitor of apoptosis protein (Xiap) and cyclin-dependent kinase inhibitor 1B (Cdkn1b/p27)31-35, and we use these as “endogenous IRES reporters” in subsequent analyses. Indeed we observed reduced protein levels of XIAP and p27 (Fig. 1j), with no changes in transcript abundance (Supplementary Fig. 2k), suggestive of a defective translational mechanism. This was confirmed by replacing the HCV IRES element in our reporter, with the 5’ UTRs of Cdkn1b or of Xiap (or of Actb as a non-IRES negative control) and demonstrating reduced activity with Cdkn1b and Xiap, but not with Actb (Fig. 1k). Polysome fractionation of Npm1+/+ and Npm1−/− MEFs demonstrated a shift to lighter polysome fraction of Cdkn1b and Xiap transcripts as well as of additional 5’ UTR structured transcripts (Supplementary Fig. 2l and 2m) in Npm1−/− MEFs. To support these findings even further, we performed IP of phosphorylated RPS6 (pS6-IP)36,37 and found that IRES containing transcripts but not the control transcript were depleted from Npm1−/− translating ribosomes (Fig. 1l).
We validated the involvement of snoRNAs in NPM1-mediated translational control, by over-expressing and by knocking-down (via GapmeRs) the NPM1-associated snoRNAs, Snord47 and Snord52. This led to increased and decreased 2’-O-Me levels respectively, with concomitant alterations to protein levels of XIAP and p27, with no changes in transcript levels (Supplementary Fig. 3a-g).
Intriguingly, expression of the cytoplasmic localized NPMc+ mutant (commonly found in AML), or of the NPM1-RARα fusion protein (found in APL and lacking a number of RNA binding domains) failed to rescue the IRES-dependent translation deficiency of the luciferase reporter (Fig. 1m and Supplementary Fig. 3h). Consistent with these findings, we observed impaired rRNA 2’-O-Me in human NPMc+ AML samples (Fig. 1n), as well as in the human leukemia cell line OCI-AML3 harboring the NPMc+ mutation (Supplementary Fig. 3i), while snoRNA levels were unaffected (Supplementary Fig. 3j). These data, together with previous reports of aberrant 2’-O-Me in leukemia38, strongly suggest that altered 2’-O-Me may contribute to leukemogenesis.
NPM1 regulates cellular growth and differentiation through 2’-O-Me
As NPM1 mutations are associated with hematological disease, we next investigated what effects perturbations of NPM1, snoRNAs and 2’-O-Me might have on growth and differentiation of hematopoietic cells using the human erythroleukemia cell line K562.
As expected, efficient NPM1 depletion (Supplementary Fig. 4a) resulted in decreased 2’-O-Me levels (Supplementary Fig. 4b), and reduced levels of IRES translated proteins, while FBL levels and snoRNAs’ abundance remained intact (Supplementary Fig. 4a and Supplementary Fig. 4c, respectively). NPM1 depletion also led to a substantial reduction in colony formation potential, demonstrating its role in clonogenic growth (Fig. 2a). Moreover, using hemin treatment to induce erythroid differentiation39 (Fig. 2b), we found that NPM1 depletion in K562 cells led to increased differentiation (Fig. 2c).
Figure 2. NPM1 regulates cellular growth and differentiation through 2’-O-Me.
a, NPM1-depleted K562 cells (shNPM1) and scrambled transduced cells (Ctrl), were plated in MethoCult. Data are presented as mean ± SD of n = 5 independent experiments. Shown are representative pictures. b, Illustration of Hemin treatment and time course. K562 cells that differentiated to erythrocytes were identified via positive Benzidine staining. c, NPM1 depletion (shNPM1) led to increased differentiation following Hemin treatment. Data are presented as mean ± SD of n = 4 independent experiments. d, Relative levels of snoRNA abundance and 2’-O-Me levels in snoRNA-inactivated cells were calculated relative to the levels in Ctrl cells (K562 transduced with an empty CRISPR/Cas9 vector, set as 1, not shown). Data are presented as mean ± SD of n = 3 independent experiments. e, Data are presented as mean ± SD of n = 4 independent experiments. f, SnoRNA-inactivated K562 cells were treated with Hemin and cellular differentiation was determined relative to Ctrl. Data are presented as mean ± SD of n = 4 independent experiments. g, Scheme of rescue experiments performed by overexpression of individual snoRNAs or of FBL in NPM1-depleted K562 cells. h, snoRNA levels and their corresponding 2’-O-Me levels were analyzed in the relevant RFP+ K562/shNPM1 cells. Data are relative to the levels in Ctrl cells (K562/shNPM1 transduced with an empty RFP vector). Data are presented as mean ± SD of n = 3 independent experiments. i, K562/shNPM1 cells, over-expressing specific snoRNAs (x-axis), were cultured in MethoCult and colony numbers were determined. Data are presented as mean ± SD of 4 independent experiments performed. Statistical significance was determined by two-tailed Student’s t test. j, K562/shNPM1 cells, over-expressing specific snoRNAs (x-axis), were treated with hemin and relative number of differentiated cells was determined by positive Benzidine staining. Data are presented as mean ± SD of n = 4 independent experiments. k, K562/shNPM1 cells, over-expressing FBL, were cultured in MethoCult. Data are presented as mean ± SD of n = 4 independent experiments performed. l, K562/shNPM1 cells, over-expressing FBL, were treated with Hemin and cellular differentiation was determined relative to Ctrl cells (K562/RFP). Data are presented as mean ± SD of n = 4 independent experiments performed. For all relevant panels, and unless otherwise stated, statistical significance was determined by one-tailed Student’s t test.
To understand whether C/D box snoRNAs have a role in these processes, we generated individual snoRNA-inactivated K562 cells by CRISPR/Cas9. Genome editing of snoRNAs was confirmed by Sanger sequencing in the bulk population (Supplementary Fig. 4d). SnoRNA inactivation led to a significant loss in specific snoRNA abundance (Fig. 2d) and to a consequent reduction in the specific rRNA methylation (Fig. 2d), while other unrelated methylations were preserved (Supplementary Fig. 4e). As was the case with NPM1 depletion, inactivation of SNORD15, SNORD47 and SNORD104 led to reduced colony formation (Fig. 2e). Inactivation of SNORD15, SNORD47, SNORD52 and SNORD58 led to increased erythroid differentiation (Fig. 2f). These data demonstrate that individual snoRNAs may have distinct roles in specific biological processes.
To investigate whether the NPM1 depletion phenotypes are mediated by 2’-O-Me, we performed rescue experiments by expressing each snoRNA in NPM1-depleted K562 cells (Fig. 2g). Cells were analyzed for individual snoRNA over-expression and specific 2’-O-Me levels (Fig. 2h). As SNORD47 failed to overexpress, with no change in the specific 2’-O-Me residue (Fig. 2h), we regarded these cells as an additional control.
In colony assays only SNORD52 was able to increase colony number (Fig. 2i). On the other hand, all snoRNAs except SNORD47 reduced the rate of differentiation of NPM1-depleted K562 upon hemin treatment (Fig. 2j). Whereas individual snoRNAs did partially correct the effects of NPM1 depletion, no individual snoRNA was able to fully rescue the phenotype of NPM1-depleted K562 cells. This suggests that the combined activity of several snoRNAs is required for a full rescue of NPM1 loss. As efficient co-expression of all five snoRNAs is technically challenging, we sought to simultaneously affect several methylation residues by overexpressing the snoRNA-dependent methyltransferase FBL (Fig. 2g). Overexpression of FBL led to increased methylation of most methylation sites (Supplementary Fig. 4f). Most importantly, functionally, overexpression of FBL in NPM1-depleted K562 cells fully rescued both colony formation (Fig. 2k) and differentiation (Fig. 2l). While it may be possible that rescue of such functional assays could be mediated by 2’-O-Me changes to other RNA species, such as mRNAs, 2’-O-Me modifications of mRNA is currently not ascribed to FBL, but to other snoRNA-independent methylatraferases40,41. Hence, these data support the notion that 2’-O-Me is a major mechanism through which NPM1 controls cell growth and differentiation.
Acute deletion of Npm1 in adult mHSCs leads to BMF
Previous studies have implicated aberrant ribosome biogenesis and function in the etiology of various hematological diseases and inherited syndromes, including myelodysplastic syndrome, Diamond Blackfan anemia and dyskeratosis congenita, hence classifying them as ribosomopathies42-47. These conditions share key features, particularly bone marrow failure (BMF) and cancer susceptibility45,48,49. In light of the in vitro phenotype observed in NPM1-depleted cells we aimed to assess the consequences of Npm1 inactivation on adult hematopoiesis in vivo. To this end we generated an Npm1-loxP mouse (Supplementary Fig. 5a) and crossed it with Mx1Cre transgenic mice to achieve a hematopoietic-specific conditional knockout of Npm1 (Supplementary Fig. 5b-d).
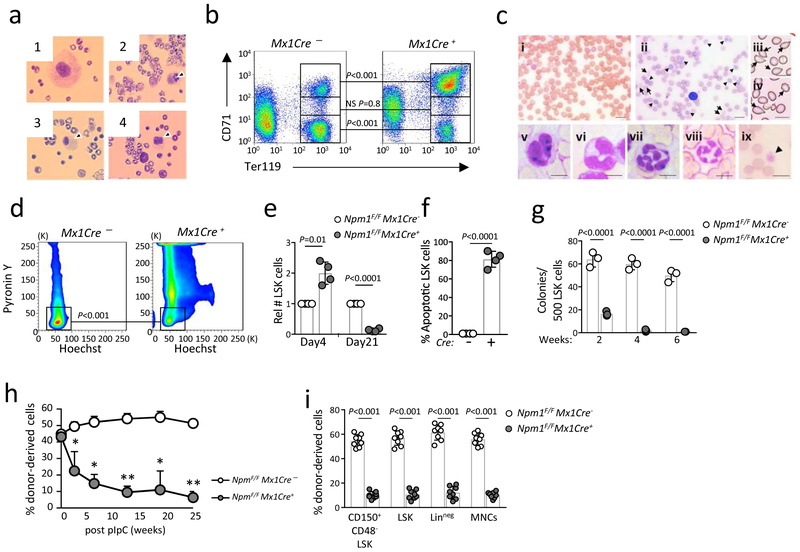
Seven to 10 days after Npm1 deletion, 20 out of 20 Npm1F/F;Mx1Cre+ mice exhibited dysmegakaryopoiesis (Fig. 3a), defective erythroid maturation (Fig. 3b), dysplastic and low platelet counts (Fig. 3c and Supplementary Fig. 6a), and dysplastic neutrophils (Fig. 3c), which are all features observed in human ribosomopathies50. Flow cytometry analysis of bone marrow (BM) cells revealed that acute deletion of Npm1 resulted in exit from quiescence of hematopoietic stem/progenitor cells (HSPCs: Lineage-;Sca1+;cKit+, referred to as LSK cells, Supplementary Fig. 6b), as well as of the HSC compartment (CD150+CD48- cells, Fig. 3d). Exit from quiescence was found to be cell autonomous, as it was also observed ex vivo (Supplementary Fig. 6c).
Figure 3. Acute deletion of Npm1 in adult mHSCs leads to BMF.
a-c, Acute loss of Npm1 in the hematopoietic system leads to human ribosomopathy phenotype. 7-10 days after pIpC injection, dysmegakaryopoiesis (a) and erythroid developmental defects (b) were observed, while 28 days after pIpC injection peripheral blood shows dysplastic features (c). a, Representative morphology (n = 20 biologically independent samples) of Npm1-deficient megakaryocytes (panel 2-4) 10 days after pIpC injection. b, Staining for TER119/CD71 positivity (n = 4 biologically independent samples) 10 days after Npm1 deletion is shown. c, Smears of peripheral blood of Npm1F/F;Mx1Cre− (c, panel i) or Npm1F/F;Mx1Cre+ mice (c, panels ii-ix) 4 weeks after pIpC administration show images of dysplastic erythroid cells (polychromasia, ii; poikilocytosis, iii,iv, arrowhead), dysplastic neutrophils (v,vi; hypersegmented neutrophils, vii,viii) and dysplastic platelets (giant platelet, ix). Scale bars, 10 μm. Shown are representative blood smear out of n = 20 independent biological samples. d, Npm1 deletion in adult HSCs leads to impairment of maintenance of quiescence. Percent of cells in G0 phase in Npm1-deleted HSCs (gated on LSK;CD48-;CD150+) 4 days after pIpC injection (n = 4 biologically independent samples). e, Relative number (± SD) of LSK cells 4 days and 21 days after Npm1 deletion, compared with pIpC-treated Npm1F/FMx1Cre− mice (n = 4 biologically independent samples per group). f, Npm1 deletion leads to increased apoptosis of LSK cells 21 days after pIpC injection. Data are presented as mean ± SD (n = 4 biologically independent samples per group). g, Results depict mean colony numbers ± SD/500 KSL cells (n = 3 biologically independent samples for each group). h-i, Reconstitution of donor cells in peripheral blood and bone marrow was monitored by staining blood cells with antibodies against CD45.2 (donor) and CD45.1. Data are presented as mean ± SD, (n = 8 biologically independent samples for each group h, and n = 8 biologically independent samples for each group, i). h, * P < 0.01, ** P < 0.0001. For all relevant panels, and unless otherwise stated, statistical significance was determined by one-tailed Student’s t test.
The impact of acute Npm1 loss on proliferation of LSK cells was then determined in vivo by using a competitive bone marrow transplantation (BMT) assay. When compared to control LSK cells, Npm1-ablated LSK cells showed increased cell cycle entry (Supplementary Fig. 6d). Consistent with this, the number of LSK cells was increased 4 days after Npm1 ablation (Fig. 3e), followed by a significant loss of LSK cells at day 21 (Fig. 3e) due to extensive apoptosis (Fig. 3f).
In vitro Npm1-ablated LSK cells had a decreased capacity to form short-term (Supplementary Fig. 6e), and long-term colonies (Fig. 3g and supplementary Fig. 6f). To test the repopulating capacity of Npm1-deficient HSCs in vivo we carried out competitive BMT experiments. Within 5 weeks following Npm1 ablation, Npm1-deficient HSCs were significantly reduced in recipient mice and were eventually out-competed by wild-type HSCs, confirming BMF and exhaustion (Fig. 3h and 3i). Thus, we show that Npm1 deletion in adult HSCs leads to BMF. Furthermore, in line with the fact that one of the hallmarks of BMF syndromes is increased susceptibility to cancer51, Npm1 deletion in adult HSCs in conjunction with Trp53 deletion led to the development of an aggressive and fatal form of myelocytic leukemia (Supplementary Fig. 6g-i).
NPM1 germline mutations identified in dyskeratosis congenita patients
Given the importance of NPM1 for adult murine hematopoiesis, we investigated whether mutations in NPM1 may be associated with other BMF disorders.
DC is an inherited disease characterized by mucocutaneous abnormalities, lung fibrosis, BMF and predisposition to cancer49. DC exhibits considerable clinical and genetic heterogeneity. The most common genetic lesion discovered in DC is in the X-linked dyskerin gene (DKC1), the pseudouridine synthase that associates with H/ACA box snoRNAs. However, in more than 40% of DC cases no mutation has been identified49.
Mutant DKC1 was shown by others and by us to underlie reduced levels of rRNA pseudouridylation47, as well as aberrant translation52. Given the shared molecular and cellular features of NPM1 loss-of-function described above, with those of mutations of DKC1, we wondered whether germline mutations of NPM1 might also cause DC. A review of whole-exome sequence datasets of DC and DC-related patients (after excluding patients with known DC-causing mutations), led us to uncover NPM1 variants in two patients with classic DC.
One patient, who presented with severe growth defects at birth, thumb abnormalities and thrombocytopenia, harbored an NPM1 missense mutation, NG_016018.1(NM_001355006.1):c.532G>C, leading to D178H substitution. A second patient presented with skin pigmentation abnormalities, nail dystrophy, microcephaly, developmental delay, short stature, skeletal abnormalities in the radius, and BMF by the age of 6, harbored an in-frame deletion, NG_016018.1(NM_001355006.1):c.538_540del (D180del). Interestingly, both mutations are located in an acidic D/E repeat region that regulates the specificity of NPM1 binding to RNA by its C-terminal basic domain53 (Fig. 4a).
Figure 4. NPM1 germline mutations identified in dyskeratosis congenita patients.
a, Schematic illustration of NPM1. Presented are the amino acid sequences of the acidic domain (pink) in wild type NPM1 (upper sequence), and in the two NPM1 mutants, NPM1D178H and NPM1D180del identified in DC patients. b, RNA affinity purification of biotinylated Snord15 in NPM-FLAG-expressing cells. Blot is a representative of n = 3 independent experiments. c, FBL-IP to evaluate snoRNAs enrichment in snoRNPs in CM108 and BV311 cells. Enrichment in CM108 cells was calculated relative to total input and fold enrichment in control BV311 cells. Data are presented as mean ± SD of n = 3 independent experiments. Significance was calculated relative to the enrichment of SNORDCtrl (SNORD13, which was not identified in NPM1 HITS-CLIP). d, Analysis of specific 2’-O-Me (x-axis) in CM108 compared to BV311 cells. Data are presented as mean ± SD of n = 3 independent experiments. 2’-O-Me at 1804 18S rRNA served as a control (Ctrl). e, Data are presented as mean ± SD of n = 4 independent experiments. f, Reduced IRES activity in CM108 patient cells compared to BV311 control cells. Data are presented as the raw luminescence of each individual luciferase of n = 6 independent experiments. Black squares represent the mean ± SD of all six experiments. g, WB analysis of IRES translated genes in BV311, CM108 and CM108 over-expressing FBL. αTUBULIN served as loading control. Shown is a representative blot of n = 3 independent experiments performed. h, Ribosome enrichment (by pS6-IP) was calculated relative to BV311 cells. ACTB served as a non-IRES control. Data are presented as mean ± SD of n = 3 independent experiments. i-j, K562/shNPM1 cells were transduced with either, empty vector (EV), NPMwt, NPMD178H, or NPMD180del. Cells were assayed for clonogenic growth (i) and hemin-induced differentiation (j). i, Data are presented as mean ± SD, of n = 3 independent experiments. j, Data are presented as mean ± SD, of n = 3 independent experiments. For all relevant panels, and unless otherwise stated, statistical significance was determined by one-tailed Student’s t test. Uncropped blot images are presented in Supplementary Figure 11.
In view of this, we tested NPM1D178H and NPM1D180del for their capacity to bind snoRNAs, and found that it was reduced in both (Fig. 4b and Supplementary Fig. 7a); whereas their ability to interact with FBL is unaltered (Supplementary Fig. 7b). By using skin fibroblasts from the NPM1D178H patient (referred to as CM108), and from an aged-matched healthy control (referred to as BV311), we found that although snoRNAs levels were similar (Supplementary Fig.7c), binding of FBL to the NPM1-bound snoRNAs was reduced in patient cells (Fig. 4c). Concomitantly, 2’-O-Me levels at the previously identified sites were reduced in DC patient cells (Fig. 4d). As post-transcriptional rRNA modifications affect rRNA secondary structure, they can affect the binding site of ribosome-targeting antibiotics (e.g. puromycin and anisomycin)54,55, and hence affect sensitivity to antibiotic treatment. Indeed, DC patient cells were hyposensitive to antibiotic treatment (Fig. 4e), further indicating that NPM1D178H affects ribosome structure and function. Using the cap/IRES luciferase reporter we demonstrated a reduction in IRES-dependent activity in patient cells (Fig. 4f), validated by reduced protein levels of XIAP and p27, while NPM1 and FBL levels were unaltered (Supplementary Fig. 7d). Importantly, overexpression of FBL in DC patient cells rescued the reduced levels of XIAP and p27 (Fig. 4g). pS6-IP experiments demonstrated that XIAP and CDKN1B transcripts were indeed depleted from actively translating ribosomes in patient cells (Fig. 4h), while transcript abundance did not change (Supplementary Fig. 7e), indicative of translational dysregulation. Global protein synthesis analysis (Supplementary Fig. 7f), mTOR signaling (Supplementary Fig. 7g), rRNA processing analysis () and polysome fractionation curves (Supplementary Fig. 7i) did not show any significant differences between patient and control cells, indicating that global translation is not affect. Interestingly, NPM1D178H function was preserved in other respects. NPM1D178H retained nucleolar localization (Supplementary Fig. 7j), as well as regulation of p53 (Supplementary Fig. 7k) and of centrosome numbers (Supplementary Fig. 7l). Thus, this particular NPM1 mutation specifically compromises its snoRNA binding capacity, leading to altered 2’-O-Me levels and aberrant IRES translation in DC patient cells.
Next, we functionally analyze NPM1D180del. To this end, since patient-derived cells were unavailable, we generated NPM1D180del MEFs by CRISPR/Cas9. NPM1D180del MEFs were similar to NPM1D178H patient cells in all respects: ribosomal profiles (Supplementary Fig. 8a), reduced IRES translation (Supplementary Fig. 8b), reduced XIAP and p27 protein levels, while NPM1 levels were not affected (Supplementary Fig. 8c). Similar to the NPM1D178H mutant, the NPM1D180del mutant also showed reduced loading of snoRNAs onto C/D box-snoRNPs (Supplementary Fig. 8d), and defective regulation of 2’-O-Me (Supplementary Fig. 8e).
As a further demonstration that these NPM1 mutations entail loss-of-function, we found that while wild type NPM1 fully rescued the NPM1 depletion phenotype in K562 cells (Fig. 2), NPM1D178H or NPM1D180del were unable to do so (Fig. 4i-j and Supplementary Fig. 8f). Thus, we demonstrate that NPM1D178H and NPM1D180del, two NPM1 mutants found in the germline of DC patients, have low snoRNA binding capacity and are dysfunctional in regulating cellular functions such as growth and differentiation.
NPM1D180del mice show multi-organ features of DC
Next we sought to establish a causal link between NPM1 germline mutations and DC. To this end we generated a knock-in NPM1D180del allele using CRISPR/Cas956. NPM1D180del heterozygous and homozygous mice were born in Mendelian ratios, had no overt developmental or behavioral abnormalities and appeared healthy. However, at 2 months of age both heterozygous and homozygous mice were found to show features of aberrant hematopoiesis, which developed into BMF by 6 months of age (Supplementary Fig. 9 and Fig. 5, respectively). As we found no significant difference in the phenotype of NPM1D180del heterozygous and homozygous, both genotypes of mice were analyzed together as NPM1D180del mice.
Figure 5. NPM1D180del mice show multi-organ features of DC.
a, Bone H&E staining demonstrating reduced cellularity in NPM1D180del mice (Heterozygous n = 2, Homozygous n = 2 and control mice n = 4 independent animals). Data are shown as mean ± SD. Scale bar, 100 μm. b, HSC compartment analysis of NPM1D180del mice and control littermates (Ctrl n = 5, Heterozygous n = 3, Homozygous n = 3). Data are shown as mean ± SD. c, Progenitors analysis of NPM1D180del mice and control littermates (Ctrl n = 5, Heterozygous n = 3, Homozygous n = 3). Data are shown as mean ± SD. d, Analysis of % Gr1+CD11b+ myeloid population in the bone marrow of NPM1D180del compared to littermates mice (Ctrl n = 5, Heterozygous n = 3, Homozygous n = 3). Data are shown as mean ± SD. e, HSPC cells were sorted and plated in semisolid media. Data are presented as mean ± SD of n = 4 independent experiments. f-g, HSPC cells from NPM1D180del mice and littermates (Ctrl) were sorted and transduced to overexpress FBL. Presented is data from second plating demonstrating full rescue of colony growth (f) and differentiation (g) by FBL expression. Data are presented as mean ± SD of n = 5 independent experiments. h, Spleen weight of 6 months old NPM1D180del and littermates control (Ctrl n = 5 (white circles), Heterozygous n = 3 (purple circles), Homozygous n = 3 (blue circles)). Data are presented as mean ± SD. i, H&E staining of 6 months old NPM1D180del mice spleen show aberrant architecture with no clear red or white pulp zones. Picture shown is a representative of n = 6 biological samples. Scale bar, 50 μm. j, Spleen IHC demonstrating increase TER119+ cells in NPM1D180del spleen. Picture shown is a representative of n = 4 biological samples. Scale bar, 50 μm. k, Erythroblast analysis in the spleen. ProE were gated on CD71highTER119dim cells, EryA were gated as TER119highCD71highFSChigh cells. Data are presented as mean ± SD of Ctrl n = 5, and of Heterozygous n = 3, Homozygous n = 3. l, pS6-IP was performed using total BM cells from heterozygous NPM1D180del mice (n = 2) and control littermates (n = 3). Enrichment was calculated relative to the enrichment in littermates. m, Analysis of specific 2’-O-Me (x-axis) in BM cells of heterozygous NPM1D180del compared to littermates (n = 3, each). 2’-O-Me at 1804 18S rRNA served as a control (Ctrl). n, Representative skin H&E staining (out of n = 4 mice) of NPM1D180del and littermates control, demonstrating thickening of the epidermis. o, Analysis of NPM1D180del and littermate testicular size. NPM1D180del mice present with reduced testicular size, Data are presented as mean ± SD, n = 11 of each group. p, Representative testicle H&E staining (out of n = 4 mice) showing atrophic seminiferous tubules in NPM1D180del testes (yellow arrows). For all relevant panels, and unless otherwise stated, statistical significance was determined by one-tailed Student’s t test.
At 2 months of age NPM1D180del mice demonstrated low numbers of long-term (LT)-HSCs (Supplementary Fig. 9a), as well as increased megakaryocyte-erythroid progenitors (MEPs, Supplementary Fig. 9b) and increased erythroblasts (ProE and EryA, Supplementary Fig. 9c). Accordingly, red blood counts (RBC) and platelets (PLT) were elevated in the peripheral blood (Supplementary Fig. 9d).
At 6 months of age NPM1D180del mice displayed extramedullary hematopoiesis, probably as a consequence of BMF. Specifically, we observed reduced BM cellularity (Fig. 5a), as well as reduced LT-HSC, short-term (ST)-HSC and multipotent progenitors (MPPs, Fig. 5b). Progenitor cells analysis revealed a decrease in common myeloid progenitors (CMPs) and MEPs, together with a concomitant increase in granulocyte-monocyte progenitors (GMPs, Fig. 5c). Flow cytometry analysis of BM cells validated an increase in the myeloid Gr1+CD11b+ population in the BM of NPM1D180del mice (Fig. 5d).
The above data indicate that the HSC compartment of NPM1D180del mice undergoes exhaustion. To further evaluate this, we carried out in vitro colony assays and serial plating of 6-month old mice HSPCs. Initially, we observed no difference in total colony number, but a higher percentage of myeloid (CFU-GM) colonies was found in NPM1D180del plates (Fig. 5e). However, second replating identified fewer total colonies in NPM1D180del plates, and a lower percentage of erythroid (BFU-E) colonies (Fig. 5e). Upon a third replating, despite normal growth in control plates, no colonies were observed in the NPM1D180del plates (Fig. 5e). Once again, expression of FBL in NPM1D180del HSPCs was able to rescue colony growth (Fig. 5f) and differentiation (Fig. 5g). Taken together these data further substantiate the exhaustion of NPM1D180del HSPCs, and suggest that defective 2’-O-Me as the underlying mechanism.
Interestingly, NPM1D180del mice were also found to have enlarged spleens (Fig. 5h). Spleen architecture was abnormal with increased red pulp (Fig. 5i and 5j), as well as increased erythroid blasts populations (Fig. 5k), reflecting extramedullary hematopoiesis probably arising in response to HSPC exhaustion.
Peripheral blood analysis identified abnormal red blood cells and dysplastic neutrophils in a number of NPM1D180del mice (Supplementary Fig. 9e); with one these mice progressing to an overt myeloid proliferation disorder (Supplementary Fig. 9f-i).
In line with our previous data (Supplementary Fig. 8), BM cells from NPM1D180del mice exhibited aberrant translation, whereby XIAP and p27 levels were reduced (Supplementary Fig. 9j), and their transcripts were depleted from actively translating ribosomes (Fig. 5l). Finally, while snoRNAs levels were comparable between NPM1D180del and in control mice (Supplementary Fig. 9k), 2’-O-Me levels were low in NPM1D180del HSPCs (Fig. 5m).
Consistent with DC being a multi-organ syndrome, NPM1D180del mice presented with several non-hematopoietic abnormalities. A third (4/12) displayed acanthosis, a thickening of the skin that is frequently observed in DC (Fig. 5n). Additionally, NPM1D180del males demonstrate reduced testicular size (Fig. 5o) with atrophic seminiferous tubules (Fig. 5p), also features associated with DC. Thus, NPM1D180del mutant mice recapitulate the phenotype of DC beyond BMF. These data further substantiate the causal role of this mutation in human DC.
Discussion
By identifying NPM1 mutations in the germline of DC patients and modeling these mutations in the mouse, we suggest that aberrant 2’-O-Me is a pathogenic mechanism. Our findings lay the ground to speculate that additional RNA or protein components of the 2’-O-Me snoRNP complex may also be targeted for dysregulation in DC, and possibly in other human disorders.
Ribosomopathies are a heterogeneous group of human disorders, sharing characteristics of BMF and cancer susceptibility. With the ribosome being a ubiquitous and essential cellular engine it is difficult to understand the mechanism driving the tissue-specific features of these diseases. Two main and non-contradictory views try to explain the pathological specificity of ribosomopathies27. The first argues that ribosome concentration and hence availability is crucial for translation and that mRNAs are differentially translated depending on ribosome availability/concentration57-60. The second view, referred to as the “specialized” ribosome hypothesis, argues that ribosome heterogeneity and diversity in either protein and/or RNA composition can be critical in regulating protein expression of specific mRNAs within a specific cell type20,61-65. Regardless of the underlying mechanism, it is commonly thought that ribosome dysfunction would be most detrimental in certain specific cells or tissues that either require the expression of specific rate-limiting gene/s or high protein synthesis rates27.
Here we find that ribosome function is regulated by a distinct pattern of 2’-O-Me on discrete rRNA residues, and that alteration to snoRNP complex-associated proteins can result in ribosome dysfunction. We propose a model in which 2’-O-Me levels are differentially regulated by NPM1 to maintain an optimal translational program. We hypothesize that the presence of mutant NPM1 in the FBL methylation complex reduces its functionality (but does not abolish it) by limiting snoRNA loading to snoRNPs, leading to aberrant translation and resulting in dysfunctional HSCs and BMF (Supplementary Fig. 10a and 10b). Nevertheless, it remains possible that mutant NPM1 also affects other proteins in the snoRNP complexes. Interestingly, heterozygosity for specific NPM1 mutations is sufficient to cause DC. Since NPM1 is known to pentamerize13, it is likely that one or more abnormal monomers make the entire pentamer dysfunctional, in line with a dominant-negative mechanism of action (Supplementary Fig. 10a and 10b).
In addition, we have established that NPM1 function as a 2’-O-Me modulator is impaired in several NPM1-mutated malignancies. It is interesting to note that NPMc+ mutation and Npm1 deletion KO share some similarities, most importantly both lead to increased HSC proliferation (Fig. 3d-e and 66). While in the Npm1 conditional KO model this brisk HSC expansion is followed by their rapid exhaustion, in the NPMc+ model this increased proliferation ultimately leads to leukemia development. Interestingly, however, combining Npm1 conditional KO with additional genetic lesion/s (i.e. Trp53 deletion) also triggers to the development of aggressive leukemia (Supplementary Fig. 6g-i). These findings further support a role for aberrant 2’-O-Me in the development of hematological malignancies at large. In agreement with this notion, it was recently demonstrated that induction of C/D box snoRNA/RNP function by AML1-ETO translocation constitutes an important mechanism towards leukemogenesis38. Intriguingly, TCGA analysis highlights mutations in the acidic repeats of NPM1 in a variety of human cancers (Supplementary Fig. 10c), suggesting that aberrant 2’-O-Me may underlie the development of cancers of various histological origins.
Our findings therefore provide genetic support to the notion that an aberrant epitranscriptome can be pathological, and suggest a role for 2’-O-Me in human disease.
Methods
NPM1 HITS-CLIP
UV-Crosslinking and cell lysis
Npm1+/+Trp53−/− MEFs or Npm1−/−Trp53−/− MEFs were grown to 80% confluency in DMEM 10% FBS, 5% PS. Before UV-Crosslinking the medium was aspirated and dishes containing cells were placed in ice. UV-Crosslinking was performed in a hybridization oven at setting 400 mJ/cm, followed by an additional 200 mJ/cm after one minute of recovery. Cross-linked cells were washed once with ice-cold PBS, collected and pelleted for 5 min at 4 °C. PBS was aspirated and cells were snap-frozen on dry ice and stored in −80 °C until further processing. Cross-linked cells were lysed in 1× PXL buffer (1× PBS, 0.1% SDS, 0.5% DOC, 0.5% NP-40) for 5 min on ice, followed by the addition of RQ1 DNase, which was incubated at 37 °C for 10 minutes. To partially digest the crosslinked RNAs, RNaseA was added to the lysates at a 1:10,000 dilution and incubated at 37 °C for 5 min (a titration experiment was performed beforehand to determine optimal concentration). Next, the lysates were spun at 14,000 rpm in a table top centrifuge for 20 min at 4 °C.
Immunoprecipitation and CIP treatment
The NPM antibody (clone FC82291) was coupled to Dynabeads Protein G according to the manufacturer’s protocol. After coupling the NPM antibody to the magnetic beads, the supernatant of the spun lysate was added to the beads and incubated at 4 °C for 2 h. After immunoprecipitation, the magnetic beads were washed twice for 5 minutes each in wash buffers (5× PXL: 5× PBS, 0.1% SDS, 0.5% DOC, 0.5% NP-40, PNK-buffer: 50 mM Tris-Cl, pH 7.4, 10 mM MgCl2, 0.5% NP-40). Next, the RNA was treated with 1.5 μl CIP FAST-AP (alkaline phosphatase) according to the manufacturer’s instructions for 20 min at 37 °C, followed by washing steps: once in PNK-buffer, once in PNK-buffer + 20 mM EGTA, and twice in PNK-buffer.
Linker Ligation:
The RNA linker (SRA3) was obtained from Dharmacon, and ligated using T4 RNA ligase I (Fermentas) according to the manufacturers protocol at 16 °C overnight, followed by 3 washed in PNK buffer.
Phosphorylation of RNA tags
Next, RNA tags were radioactively labeled using 32P-γ-ATP (3,000 Ci mmol and T4 PNK (NEB) according to the manufacturer’s protocol for 20 min at 37 °C, followed by 3 washes in PNK buffer.
SDS-PAGE and transfer to nitrocellulose membrane
Beads with radioactively labeled RNAs were eluted with 60 μl Elution buffer (30 μl 1× PNK + 30 μl LDS loading buffer) and eluates were incubated at 70 °C for 10 minutes, followed by SDS-PAGE on a 10% Bis-Tris Novex gel and transfer to a nitrocellulose membrane at 30V for 2 h. Next the membrane was washed in 1× PBS and exposed on film for 15 min.
RNA isolation from nitrocellulose membrane
RNA bands were excised from the nitrocellulose membrane ~20 kDa above the expected molecular weight of the protein. The slice was cut into very small pieces and put into Eppendorf tube. Then, 200 μl of of Proteinase K solution (4 mg/ml PK stock was diluted 1:5 in 1× Proteinase K buffer: 100 mM Tris-Cl pH 7.5, 50 mM NaCl, 10 mM EDTA) was added to the membrane pieces and incubated for 20 min at 37 °C. Next, 200 μl of Proteinase K buffer with 7 M Urea was added and incubated for 20 min at 37 °C, followed by the addition of 400 μl RNA Phenol + 130 μl Chloroform+isoamyl alcohol, which was incubated in the thermomixer at 37 °C at 1,000 rpm for 20 min, followed by RNA extraction by ethanol/isopropanol precipitation in the presence of glycogen.
RT-Reaction
Beads for Brdu-IP were prepared as follows: ProtG beads: three washes with Ab binding buffer (composition as in manufacturers protocol), followed by the addition of 20 μl of l× Ab binding buffer, 5 μl of 50× Denhardt’s Solution and 25 μl (5 μg) of αBrdU antibody (Santa Cruz, sc-32323). Rotation at RT for at least 45 min, followed by 3 washes in 1× IP Buffer (0.3× SSPE, 1 mM EDTA, 0.05% Tween-20). In the meantime, the RNA pellet was resuspended in water, denatured at 65 °C for 5 min (in microfuge tube), and put on ice. The reverse transcription reaction was performed using Superscript II according to the manufacturer’s protocol, in the presence of 1 μl Br-dUTP/reaction (8.2 mM; Sigma, B0631) and 1 μl of a RNA linker reverse-complement containing RT Primer (25 μM). Cycling conditions: 45 min at 50 °C, 15 min at 55 °C, 5 min at 85 °C, 4 °C hold. Primers used in this experiment also contained an index for identifying a particular experiment and enabling the multiplexing of experiment during sequencing and a barcode to identify PCR artifacts. Please see Supplementary Table 2 for RT primers used in this experiment. Following the RT reaction, 1 μl RNase H (2U/μl, Invitrogen 18021-071) was added and incubated for 20 minutes at 37 °C, and spun through a G-25 column according to the manufacturer’s instructions. Next, the volume of the flow through was measured and water up to 40 μl was added, followed by 10 μl 50× Denhardt’s Solution (Sigma, D2532); and 50μl 2× IP Buffer (0.6× SSPE, 2 mM EDTA, 0.1% Tween-20). The mix was incubated at for 5 min at 70 °C, and 2 min at 25 °C, and afterwards added to the prepared tube of BrdU-coated beads. The IP was performed for 30 min at RT, followed by the following washes: 1 time with 1× IP Buffer with 5× Denhardt’s, twice with Nelson Low Salt Buffer (15 mM Tris pH 7.5, 5 mM EDTA, 1× Denhardt’s), twice with Nelson Stringent Buffer (15 mM Tris-HCl pH 7.5, 5 mM EDTA, 2.5 mM EGTA, 1% Triton X-100, 1% NaDOC, 0.1% SDS, 120 mM NaCl, 25 mM KCl 1× Denhardt’s) and twice with 1× IP Buffer. To elute the IP’d cDNA, 50 μl of 100 μM BrdU (Sigma, B5002) was added in 1× IP Buffer and incubated at RT for 30 min. The eluate was collected using a magnet and spun through a G25 column according to the manufacturer’s instructions. Afterwards, the volume was measured, and water was added up to 97.5 μl, in addition to 37.5 μl 4× IP buffer (1.2× SSPE, 4 mM EDTA, 0.2% Tween-20) and 15 μl 50× Denhardt’s Solution.
Circularization, Re-linearization, PCR Amplification and HTS
Next, the cDNA was added to BrdU-ProtG beads and circularized using 50U CircLigase (Epicentre, CL9021K) according to the manufacturer’s protocol at 60 °C for 1 h. The following washes were performed: 2 times with Nelson Low Salt Buffer, 2 times with Nelson Stringent Buffer, and 2 times with APE1 wash buffer (50 mM Potassium Acetate, 20 mM Tris-Acetate, 10 mM Magnesium Acetate (pH 7.9)). For re-linearization the enzyme Ape1 was added (10U/μl, NEB M0282S) according to the manufacturer’s instructions for 1 h at 37 °C, followed by the following washes: 2 times with Nelson Low Salt Buffer, 2 times with Nelson Stringent Buffer, 2 times with Phusion wash buffer (50 mM Tris (pH 8.0)). Lastly, there-linearized protocols were amplified using Phusion DNA Polymerase (NEB, M0530) in the presence of 0.5 μl of 50× SYBR Green using the following cycling conditions: 98 °C for 30 s, then cycling 98 °C for 10 s, 60 °C for 15 s, 72 °C for 20 s until the RFU reaches ~ 1,000. Primers used are listed in Supplementary Table 2. PCR products were purified using Agencourt AMPure XP beads (Beckman Coulter) according to manufacturer’s instructions.
HITS-CLIP Computational Analysis
The CLIP reads had a 4-nt sample multiplexing index and a 8-nt random barcode at the 5′ end. The libraries generated with BrdU-CLIP adaptors were first filtered according to their quality scores. We required a minimum average quality score of 20 for the read. As NPM is known to bind to rRNAs and rRNA would confound the alignment due to its repetitive nature in the genome, we computationally subtracted all sequences that aligned to the 5S, 5.8S, 18S and 28S rRNAs. The remaining read sequences (actual RNA tags without index and barcodes) were mapped to the reference genomes (mm9) using Novoalign program (http://www.novocraft.com) with no more than two mismatches (substitutions, insertions, or deletions) allowed per read. We only kept reads that mapped uniquely to the genome. To remove PCR duplicates, we analyzed barcode sequences of all reads that mapped to the same genomic position. Initially, we collapsed all reads with identical barcodes into one read. Next we compared all different barcodes at the same position and only kept reads for which at least two out of eight of the barcode nucleotides were different from all other barcode sequences of reads at that position. These steps remove PCR duplicates while retaining genuinely unique reads representing independent events. Next, we calculated peaks, which were defined as regions that contain overlapping reads. The peak intensity refers to the number of independent reads that falls within the boundary of the peak. Statistical significance was calculated using the program Piranha as previously described67.
Psi-Seq
For analysis of pseudouridine levels within the rRNA, psi-seq was performed, essentially as described in15, on total RNA extracted, in triplicates, from either WT or Npm1 knockout MEF cells. Ψ-ratios, capturing the ratio between the number of reads beginning at a position (and indicative of reverse-transcription termination) divided by the number of reads overlapping that position were calculated for each of the annotated sites in rRNA, obtained from MODOMICS68. Plotted are the mean values, per each position, obtained in Npm1+/+ versus Npm1−/− samples.
RNA Motif scanning
We identified all unique peaks amongst the bona fide peak sets. As a part of this process we merged peaks that overlapped by at least 25 bp using an in-house Perl script. The genomic regions were initially scanned for the novo motif discovery using Homer suite69 using the hypergeometric distribution, searching for motifs of length from six to twelve nucleotides. The resulting significant motifs were further collapsed together by comparing the Euclidean distances of the PWMs.
CRISPR-mediated generation of NPMD180del mice and MEFs
Mice were generated as described in ref. 56. Injections to C57BL/6 zygotes were performed at the Beth Israel Deaconess Transgenic Core Facility. Single-stranded oligo-deoxynucleotide (ssODN) for homologous recombination was purchased from IDT. For genotyping, a DraI restriction site was inserted into the ssODN by mutating the sequence TTCAAA to TTTAAA. Cas9 for injection as well as the sgRNAs were purchased from PNAbio. For the generation of NPM1D180del CRISPR MEFs, all of the above reagents were transfected to Trp53−/− MEFs and single colonies were screened with DraI restriction enzyme for positive clones.
Additional Mice
Npm1F/F mice were crossed with Mx1-Cre transgenic mice (purchased from the Jackson Laboratory) and deletion of exon 1-6 of the floxed Npm1 allele was detected by PCR. C57BL/6 mice (B6-CD45.2) and C57BL/6 mice congenic for the CD45 locus (B6-CD45.1) were purchased from The Jackson Laboratory. For Mx1 promoter induction polyinosine-polycytidine (pIpC; Sigma) was resuspended in Dulbecco's PBS at 2 mg ml− and passed through a 0.22-μm filter. Mice received 25 μg of pIpC per gram of body mass every other day for two weeks.
BM isolation
Long bones from 8 weeks old mice were collected and crashed. The isolated cells were treated with ACK for 2 minutes, washed, and then subjected to further analysis. Animal experiments were performed in accordance with the guidelines of the Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee.
Flow cytometry and antibodies
For flow cytometric analyses, we used monoclonal antibodies specific for the following: CD41 (eBioMWRag30), Flt-3 (Avas12a1), CD34 (RAM34), c-Kit (2B8), Sca-1 (E13-161.7), CD3e (145-2C11), CD4 (L3T4), CD8 (53-6.72), B220 (RA3-6B2), TER-119 (TER-119), Gr-1 (RB6-8C5), CD11b (M1/70), IgM (II/41), CD19 (eBio1D3) and NK-1.1 (PK136); all were from eBioscience. Anti-CD150 (TC15-12F12.2) and CD48 (HM48-1) antibodies were from BioLegend. We used a mixture of monoclonal antibodies against CD4, CD8, CD3e, B220, TER-119, CD11b, Gr-1, IgM, CD19 and NK-1.1 as a lineage marker (Lineage). Pyronin Y staining was performed as follows: cells were stained with 10 μg/ml of Hoechst 33342 (Sigma-Aldrich) in Hoechst staining buffer (Hank's Balanced Salt Solution + 3% fetal bovine serum [FBS] + 10 mM HEPES) at 37 °C for 45 min. Then Pyronin Y (1 μg/ml, Sigma-Aldrich) was added to the buffer, and the cells were further incubated for 45 min. For additional immunolabeling, the antibodies were added to the Hoechst staining buffer supplemented with Hoechst/PryoninY. Apoptosis was measured by flow cytometry using APC Annexin V Apoptosis detection kit (BioLegend). Cells were gated and only double-positive cells were regarded as apoptotic cells. The anti-NPM1 antibody used in the HITS-CLIP assay and WB was purchased from Sigma Aldrich clone FC82291. Also, anti-FBL (H-140, for WB), anti-p53 (clone DO-1) and anti-p21 were purchased from Santa-Cruz. Anti-gamma tubulin (GTU-88), anti-FLAG M2 (F1804), anti-GAPDH (G9545) and anti-RPL22 (SAB2107796) were purchased from Sigma-Aldrich. anti-pS6, anti-XIAP (3B6), were purchased from CST. Also used, anti-FBL (Abcam, ab5821, in IP and IF experiments) and anti-p27 (BD transduction laboratories 610242).
Long-term cultures and colony-forming assays
For long-term cultures, we co-cultured LSK cells with irradiated OP9 cells in Minimum Essential Media, alpha modification (αMEM Sigma) containing 12.5% FCS (JRH Bioscience), 12.5% horse serum (Gibco BRL). After 2, 4, or 6 weeks of culture with OP9 cells, we harvested cells and used them for hematopoietic colony forming assays as described70.
Competitive reconstitution assay
We sorted 1.5 × 103 LSK cells from NpmF/F Mx1Cre+ or Mx1Cre− mice (CD45.2), and transplanted cells into lethally irradiated CD45.1 congenic mice in competition with BMMNCs from CD45.1 mice. Reconstitution of donor (CD45.2) cells was monitored by staining blood cells with antibodies against CD45.2 and CD45.1. For inducible Npm1 inactivation in the bone marrow-reconstituted recipient mice, after confirmation of engraftment of the donor-derived cells in recipient mice 8 weeks after BMT, we subsequently treated recipient mice with pIpC (25 μg of pIpC per gram of body mass every other day for two weeks).
Lentivirus
Human NPM, NPMc+, NPM-RARa, NPMD178H and NPMD180del (all FLAG-tagged) were cloned into the 3rd generation lentiviral vector pLJMI-eGFP using AgeI and EcoRI restriction site and the excision of the eGFP cassette.
For rescue experiments FBL was cloned into EF.CMV.RFP which was a gift from Linzhao Cheng (Addgene plasmid # 17619). A week after transductions, RFP+ cells were sorted.
SnoRNAs overexpression
For the expression of mouse snoRNAs we used the PiggyBac Transposons system (SBI). Introns of mouse snoRNAs were cloned into the PB513B-1 vector expressing a GFP cassette and puromycin resistance. MEFs were transfected together with the transposase vector and cultured with selection for two weeks. For rescue experiments (Fig. 2) the introns encoding the relevant human snoRNAs were cloned into EF.CMV.RFP which was a gift from Linzhao Cheng (Addgene plasmid # 17619). A week after transductions, RFP+ cells were sorted.
CRISPR inactivation of snoRNAs in K562 cell line
Guide RNAs targeting the human snoRNAs were design by the CRISPR design tool at http://crispr.mit.edu/. The gRNAs were cloned into lentiCRISPRv2-puro, which was a gift from Brett Stringer (Addgene plasmid # 98290).
Nuclear extract for Co-immunoprecipitation
Cells were washed with PBS, dislodged and pelleted by centrifugation and resuspended in the cell lysis buffer: 10 mM HEPES; pH 7.5, 10 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol (DTT), 0.5% Nonidet-P40 with the protease inhibitor cocktail (Sigma) and allowed to swell on ice for 15-20 min with intermittent mixing. Tubes were vortexed to disrupt cell membranes and then centrifuged at 12,000 g at 4 °C for 10 min. The pelleted nuclei were washed three with the cell lysis buffer and resuspended in the nuclear extraction buffer containing 20 mM HEPES (pH 7.5), 300 mM NaCl, 1 mM EDTA, 1 mM DTT and protease inhibitor cocktail, incubated in ice for 30 min. Nuclear extract is collected by centrifugation at 12,000 g for 15 min at 4 °C. Protein concentration of the nuclear extract is estimated using Bradford’s reagent (BioRad, USA).
Protein extraction
Cells were lysed with lysis buffer (50 mM Tris pH 7.5, 10% Glycerol, 5 mM MgCl2, 150mM NaCl, 0.2% NP-40) supplemented with Complete protease inhibitor cocktail tablets (Roche) and Phosphatase inhibitor cocktail tablets (Roche). When continuing to RNA extraction the lysis buffer was also supplemented with Recombinant RNasin Ribonuclease inhibitor (Promega). IP was performed using either protein A or protein G Dynabeads (Invitrogen).
RNA extraction, DNase treatment and RT
RNA extraction was performed using the Direct-zol RNA MiniPrep Plus kit (ZymoResearch). On-column DNase treatment was performed for all samples. For the isolation of RNA after IP standard TRizol (Ambion) isolation was perform using the manufacturer’s protocols.
Collection of human samples
A written informed consent for analysis of bone marrow/peripheral blood samples was obtained from each patient. Approval from the local ethical committees was obtained at Perugia Hospital, Institute of Hematology-Centro di Ricerche Emato-Oncologiche (CREO), University of Perugia, Italy.
Polysome fractionation
For polysome preparation, cells were then incubated with cycloheximide at a final concentration of 100 mg/ml for a period of 15 min. Plates were then washed with ice-cold PBS containing 100 mg/ml cycloheximide (PBS/CHX), scraped, and collected in ice-cold PBS/CHX. Cells were pelleted by centrifugation and subsequently lysed in polysome lysis buffer (20 mM Tris-HCl pH 7.4; 5 mM MgCl2; 150 mM NaCl; 1% Triton X-100; 1% deoxycholate; 2.5 mM DTT; 200 U/ml RNasin; 100 mg/ml cycloheximide; complete, EDTA-free protease inhibitor cocktail [Roche]; protease inhibitor set [without EDTA] [G-Biosciences]; α1-antitrypsin [EMD Biosciences]) and incubated on ice for 10 min with occasional mixing. Lysates were centrifuged at 7,000 rpm for 5 min at 4 °C, and the supernatant carefully removed. Protein concentrations for lysates were measured by Bradford assay, and equal amounts of protein loaded on a 15%–50% sucrose gradient containing 100 mg/ml cycloheximide, 0.2 mg/ml heparin, and 1 mM DTT. Gradients were centrifuged at 36,000 rpm for 3 h at 4 °C in a Beckman SW40 rotor and subsequently fractionated using an ISCO-Foxy Jr. fraction collector. Polysome profiles were reordered using a UA-6 absorbance detector connected to the fraction collector and measuring absorbance at 254 nm. RNA was subsequently extracted from each fraction using Trizol and the RNeasy Mini Kit (QIAGEN).
Affymetrix GeneChip and analysis
Polysome and total RNA of Npm1+/+ and Npm1+/− MEFs was extracted as described in Polysome fractionation section. The extracted RNA was then subjected to analysis via the Affymetrix Mouse Genome 430 2.0 Array. Double-stranded cDNA was synthesized from total or polysomal RNA (1 to 10 μg). Linear amplification with T7-RNA polymerase (Ambion) and biotin labeling (ENZO) were performed by in vitro transcription by according to manufacturer’s instructions. The resulting biotin-labeled cRNA was fragmented and hybridized to the Affymetrix Murine Genome 430 2.0 Array oligonucleotide 12,488-gene microarray chip for 16 h at 45 °C. Following hybridization, the probe array was washed and stained on a fluidics station and immediately scanned on a Hewlett-Packard GeneArray Scanner. Raw microarray data were imported into R and subjected to robust multi-array analysis (RMA) using the Affy package. Using the probe annotation information downloaded from the product website, RMA-transformed probe intensities corresponding to each RefSeq transcript ID were aggregated by arithmetic mean. The log2 fold changes in polysome-associated RNA intensity were calculated between the heterozygous and wild-type samples by taking the difference of the RMA-transformed probe intensities and then normalized by subtracting these values with the corresponding log2 fold change of total RNA intensities. Pathway analysis was performed using the fgsea package and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations 71 .
Dual Luciferase assay
Cells were seeded in 80% confluence, after 24 h 250 ng per well of the Cap/IRES reporter were transfected using Lipofectamine2000 (Thermo Fisher Scientific). The ratio of IRES-dependent translation (Firefly) was calculated relative to cap-dependent translation (Renilla) normalized to control cells. The reporter used contains a Renilla luciferase under the CMV promoter, followed by a Hepatitis C virus IRES element that drives the expression of the Firefly reporter. For cloning of 5’UTRs of ACTB, CDKN1B and XIAP, HCV IRES was excised using BamHI and EcoRI restriction enzymes and inserts were cloned via Gibson assembly master mix, according to manufacturer’s instructions.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde for 10 min, washed with PBS, and permeabilized with PBS, Triton-X100 0.2% for 10 min. Blocking before antibodies was performed in PBS, Triton-X100 0.2% and 10% Goat serum for 30 min. Primary antibodies were incubated overnight in blocking buffer, and secondary antibodies were diluted 1:500 (Jackson ImmuneResearch) and incubated for 2 h at room temperature. DAPI was used for nucleus staining.
2’-O-methylation detection
Measurement of 2’-O-methylation levels were measured via the RTL-P as described in16 with the following modifications. For the detection of 2′-O-methylated sites in the target rRNAs, RT-PCR was performed in a 20-μl reaction mixture containing 25-50 ng of total RNA, 1 μl (10 mM) specific RT primers and a low (0.5 μM) or high (1 mM) concentration of dNTPs. The primer/RNA mixture was denatured at 70 °C for 5 min and then chilled on ice. Following an initial annealing step at 42 °C for 10 min, 200 U of M-MLV reverse transcriptase (Invitrogen) and 0.5 U RNaseOUT (Invitrogen) were added. The reaction was incubated at 37 °C for 1 h and then heated at 75 °C for 15 min to deactivate the reverse transcriptase.
Hemin treatment
Cells were cultured with 30 μM Hemin (Sigma-Aldrich 51280) for 72 h. Cell were collected, washes once with PBS and resuspended in a ratio of 1:1 with Benzidine solution (14.6 ml acetic acid, 485.4 ml H2O, 1 g benzidine dihydrochloride) and a final concentration of 0.1% of hydrogen peroxide.
Colony Formation assay
1,500 cells were plated in MethoCult™ H4034 optimum medium in 35 mm Petri dishes (Falcon, Cockeysville, MD, USA) in duplicate. Plates were incubated at 37 °C with 5% CO2 and 95% humidity for 7 days. Colonies were scored using an inverted microscope. To visualize the colonies MTT reagent (Thermo Fisher Scientific) was added for 1 h at 37 °C with 5% CO2.
35S-methionine incorporation
Cells were plated at 70% confluence. The next day, the cells were washed three times with pre-warmed PBS to remove any residual methionine and cultured for 30 min in Dulbecco’s Modified Eagle’s Medium (DMEM) without L-methionine (Sigma Aldrich, D0422). Then 100 μCi 35S labeled methionine was added to the media for a pulse labeling of 15 min. Cells were harvested after 30 min incubation with unlabeled media.
Northern Blotting
Northern blotting was carried out according to the manufacturer’s protocol (DIG Northern Starter Kit, Roche). 2 μg total RNA was loaded on native Agarose gels. Digoxigenin (Dig) labeled antisense probes were generated using T7 RNA polymerase by in vitro transcription with the RiboMAX™ Large Scale RNA Production System (Promega). Please see Supplementary Table 2 for probe primers.
SnoRNA biotinylation and RNA affinity purification
SnoRNA15a was cloned into the MCS of pBSII-KS. In vitro transcription (Megascript-T7 in-vitro transcription kit, Ambion) was performed using linearized plasmid and in the presence of Biotin-16-UTPs (BU6105H, lucigen) overnight at 37 °C. 260 pmol of purified biotin-snoRNA15a were incubated overnight at 4 °C with total lysate. Next day, 50 μl of GE-Healthcare streptavidin agarose beads (cat: 17-5113-01, pre-blocked with BSA, glycogen and yeast tRNA) were added for at least 4 h at 4 °C. Beads were washed three times in high-salt buffer, resuspended in protein loading sample buffer and loaded on an acrylamide gel.
Click-IT™ L-Homopropargylglycine (HPG) metabolic labeling and detection
Npm1+/+ and Npm1−/− MEFs were plated at 70% confluence. The next day cell were washed with warm PBS and incubated in Methionine-free DMEM media (Sigma-Aldrich, D0422) for at least 30 min. Then HPG was added to the media according to manufacturer’s instructions and incubated for 1 h. Cells were harvested, processed and labeled with AlexaFluor-647(azide) according to manufacturer’s instructions.
SnoRNA qPCR detection
Total RNA was isolated by TRI reagent (Invitrogen). Poly(A) tailing reaction (Thermo Fisher Scientific) was performed using 250 ng of total RNA. Reverse transcription (Invitrogen, MMLV) was preformed using adapter primer following manufacturer’s instructions. SnoRNA detection was performed using a mix of a snoRNA specific primer and an abridged RT primer.
IRES GSEA
We determined the set of genes tested by Wintergarten-Gabbay et al. 2016, for enrichment in our polysome enrichment dataset. Genes from this set were selected first based on eGFP levels above background (NaN), resulting in 3,694 putative IRES genes. This gene set included XIAP and CDKN1B, well-annotated IRES genes; more stringent filtering resulted in the loss of these positive control genes. We used this set of genes for gene set enrichment analysis with the fgsea package in R.
WES analysis
WES by J.S. (NPM1D178H mutation) was performed on DNA from cultured skin fibroblast cells as described in 72 (including an extensive description of analysis pipeline). Briefly, libraries were prepared using SureSelect XT Target Enrichment (Agilent Technologies) and sequenced on NextSeq500 system (Illumina Inc.). Data analysis was performed using standard tools to identify point mutations, insertions or deletions, as well as copy number alterations. The pathogenic effect of the variants was assessed using guidelines from the American College of Medical Genetics and Genomics (ACMG). A list of suspected and known IBMF genes was selected as an assisting tool and the broad WES data were also manually investigated for potential novel IBMF-causing genes, including the NPM1 gene. WES data of I.D. (NPM1D180del mutation) were processed according to the pipeline described in 73.
Statistical Analysis
Data are presented as mean ± standard deviation (SD) unless otherwise specified. At least three independent repetitions were performed for all experiments, and Student’s t test was utilized to determine statistical significance unless otherwise specified.
Data Availability Statement
Raw and preprocessed sequencing and microarray data may be accessed from the Gene Expression Omnibus with accession number GSE135726. As the informed consent obtained from DC patients does not allow for public deposition of the data, WES data from patient CM108 (NPMD178H) and healthy controls can be communicated upon reasonable request to J.S. and WES data from patient harboring NPM1D180del mutation can be communicated upon reasonable request to I.D. and T.J.V..
Code Availability Statement
Code for microarray analysis is available in Supplementary information 1 file. Code for WES analysis is available at https://github.com/UCLGeneticsInstitute/DNASeq_pipeline.
Supplementary Material
Acknowledgments
D.N. was supported by an EMBO long-term fellowship EMBO-LTF498–2014. K.I. was supported by NIH grants. A.H.B. was supported by The Jake Wetchler Foundation and the Damon Runyon Cancer Research Foundation. This work was supported in part by the European Research Council Consolidator grant no. 311660 and the Cancéropole Ile-de-France, Paris (IDF) to J.S.. MRC,UK (MR/PO18440/1) and Bloodwise, UK (14032) to I.D.. The Associazione Italiana Ricerca Cancro (AIRC) IG 2016 n.18568 and ERC Advanced Grant 2016 no 740230 to B.F., and by an Outstanding Investigator Award R35 CA197529 grant and the SHINE 5R01DK115536 grant awarded by NIH to P.P.P..
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Davalos V, Blanco S & Esteller M SnapShot: Messenger RNA Modifications. Cell 174, 498–498 e491, doi: 10.1016/j.cell.2018.06.046 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Esteller M & Pandolfi PP The Epitranscriptome of Noncoding RNAs in Cancer. Cancer discovery 7, 359–368, doi: 10.1158/2159-8290.CD-16-1292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frye M, Harada BT, Behm M & He C RNA modifications modulate gene expression during development. Science 361, 1346–1349, doi: 10.1126/science.aau1646 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Safra M et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551, 251–255, doi: 10.1038/nature24456 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Chawla M, Oliva R, Bujnicki JM & Cavallo L An atlas of RNA base pairs involving modified nucleobases with optimal geometries and accurate energies. Nucleic acids research 43, 9573, doi: 10.1093/nar/gkv925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polikanov YS, Melnikov SV, Soll D & Steitz TA Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nature structural & molecular biology 22, 342–344, doi: 10.1038/nsmb.2992 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma S & Lafontaine DL 'View From A Bridge': A New Perspective on Eukaryotic rRNA Base Modification. Trends in biochemical sciences 40, 560–575, doi: 10.1016/j.tibs.2015.07.008 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Kiss T Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109, 145–148 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Watkins NJ & Bohnsack MT The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley interdisciplinary reviews. RNA 3, 397–414, doi: 10.1002/wrna.117 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Borer RA, Lehner CF, Eppenberger HM & Nigg EA Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 56, 379–390 (1989). [DOI] [PubMed] [Google Scholar]
- 11.Grisendi S, Mecucci C, Falini B & Pandolfi PP Nucleophosmin and cancer. Nature reviews. Cancer 6, 493–505, doi: 10.1038/nrc1885 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Falini B et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. The New England journal of medicine 352, 254–266, doi: 10.1056/NEJMoa041974 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Box JK et al. Nucleophosmin: from structure and function to disease development. BMC molecular biology 17, 19, doi: 10.1186/s12867-016-0073-9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grisendi S et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature 437, 147–153, doi: 10.1038/nature03915 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Schwartz S et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162, doi: 10.1016/j.cell.2014.08.028 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong ZW et al. RTL-P: a sensitive approach for detecting sites of 2'-O-methylation in RNA molecules. Nucleic acids research 40, e157, doi: 10.1093/nar/gks698 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krogh N et al. Profiling of 2'-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic acids research 44, 7884–7895, doi: 10.1093/nar/gkw482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Natchiar SK, Myasnikov AG, Hazemann I & Klaholz BP Visualizing the Role of 2'-OH rRNA Methylations in the Human Ribosome Structure. Biomolecules 8, doi: 10.3390/biom8040125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natchiar SK, Myasnikov AG, Kratzat H, Hazemann I & Klaholz BP Visualization of chemical modifications in the human 80S ribosome structure. Nature 551, 472–477, doi: 10.1038/nature24482 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Sharma S, Marchand V, Motorin Y & Lafontaine DLJ Identification of sites of 2'-O-methylation vulnerability in human ribosomal RNAs by systematic mapping. Scientific reports 7, 11490, doi: 10.1038/s41598-017-09734-9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steitz TA A structural understanding of the dynamic ribosome machine. Nature reviews. Molecular cell biology 9, 242–253, doi: 10.1038/nrm2352 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Liang XH, Liu Q & Fournier MJ Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. Rna 15, 1716–1728, doi: 10.1261/rna.1724409 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Proud CG mTORC1 signalling and mRNA translation. Biochem Soc Trans 37, 227–231, doi: 10.1042/BST0370227 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Wullschleger S, Loewith R & Hall MN TOR signaling in growth and metabolism. Cell 124, 471–484, doi: 10.1016/j.cell.2006.01.016 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Reschke M et al. Characterization and analysis of the composition and dynamics of the mammalian riboproteome. Cell reports 4, 1276–1287, doi: 10.1016/j.celrep.2013.08.014 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig LS et al. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat Med 20, 748–753, doi: 10.1038/nm.3557 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mills EW & Green R Ribosomopathies: There's strength in numbers. Science 358, doi: 10.1126/science.aan2755 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Bugaud O et al. Kinetics of CrPV and HCV IRES-mediated eukaryotic translation using single-molecule fluorescence microscopy. Rna 23, 1626–1635, doi: 10.1261/rna.061523.117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonenberg N & Hinnebusch AG Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745, doi: 10.1016/j.cell.2009.01.042 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weingarten-Gabbay S et al. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351, doi: 10.1126/science.aad4939 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Holcik M & Korneluk RG Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Molecular and cellular biology 20, 4648–4657 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holcik M, Lefebvre C, Yeh C, Chow T & Korneluk RG A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nature cell biology 1, 190–192, doi: 10.1038/11109 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Jiang H, Coleman J, Miskimins R, Srinivasan R & Miskimins WK Cap-independent translation through the p27 5'-UTR. Nucleic acids research 35, 4767–4778, doi: 10.1093/nar/gkm512 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kullmann M, Gopfert U, Siewe B & Hengst L ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5'UTR. Genes & development 16, 3087–3099, doi: 10.1101/gad.248902 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miskimins WK, Wang G, Hawkinson M & Miskimins R Control of cyclin-dependent kinase inhibitor p27 expression by cap-independent translation. Molecular and cellular biology 21, 4960–4967, doi: 10.1128/MCB.21.15.4960-4967.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knight ZA et al. Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell 151, 1126–1137, doi: 10.1016/j.cell.2012.10.039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O E et al. Integrin alphavbeta3 is not significantly implicated in the anti-migratory effect of anti-angiogenic urokinase kringle domain. Oncology reports 20, 631–636 (2008). [PubMed] [Google Scholar]
- 38.Zhou F et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nature cell biology 19, 844–855, doi: 10.1038/ncb3563 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Baliga BS, Mankad M, Shah AK & Mankad VN Mechanism of differentiation of human erythroleukaemic cell line K562 by hemin. Cell Prolif 26, 519–529 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Ayadi L, Galvanin A, Pichot F, Marchand V & Motorin Y RNA ribose methylation (2'-O-methylation): Occurrence, biosynthesis and biological functions. Biochim Biophys Acta Gene Regul Mech 1862, 253–269, doi: 10.1016/j.bbagrm.2018.11.009 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Dai Q et al. Nm-seq maps 2'-O-methylation sites in human mRNA with base precision. Nat Methods 14, 695–698, doi: 10.1038/nmeth.4294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dauwerse JG et al. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nature genetics 43, 20–22, doi: 10.1038/ng.724 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Draptchinskaia N et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nature genetics 21, 169–175, doi: 10.1038/5951 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Luzzatto L & Karadimitris A Dyskeratosis and ribosomal rebellion. Nature genetics 19, 6–7, doi: 10.1038/ng0598-6 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Narla A & Ebert BL Ribosomopathies: human disorders of ribosome dysfunction. Blood 115, 3196–3205, doi: 10.1182/blood-2009-10-178129 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ridanpaa M et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell 104, 195–203 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Ruggero D et al. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 299, 259–262, doi: 10.1126/science.1079447 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Armistead J & Triggs-Raine B Diverse diseases from a ubiquitous process: the ribosomopathy paradox. FEBS letters 588, 1491–1500, doi: 10.1016/j.febslet.2014.03.024 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Dokal I Dyskeratosis congenita. Hematology / the Education Program of the American Society of Hematology. American Society of Hematology. Education Program 2011, 480–486, doi: 10.1182/asheducation-2011.1.480 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Danilova N & Gazda HT Ribosomopathies: how a common root can cause a tree of pathologies. Disease models & mechanisms 8, 1013–1026, doi: 10.1242/dmm.020529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fumagalli S & Thomas G The role of p53 in ribosomopathies. Seminars in hematology 48, 97–105, doi: 10.1053/j.seminhematol.2011.02.004 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Yoon A et al. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science 312, 902–906, doi: 10.1126/science.1123835 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Hisaoka M, Nagata K & Okuwaki M Intrinsically disordered regions of nucleophosmin/B23 regulate its RNA binding activity through their inter- and intra-molecular association. Nucleic acids research 42, 1180–1195, doi: 10.1093/nar/gkt897 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunkle JA et al. Molecular recognition and modification of the 30S ribosome by the aminoglycoside-resistance methyltransferase NpmA. Proceedings of the National Academy of Sciences of the United States of America 111, 6275–6280, doi: 10.1073/pnas.1402789111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gutierrez B, Douthwaite S & Gonzalez-Zorn B Indigenous and acquired modifications in the aminoglycoside binding sites of Pseudomonas aeruginosa rRNAs. RNA biology 10, 1324–1332, doi: 10.4161/rna.25984 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quadros RM et al. Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome biology 18, 92, doi: 10.1186/s13059-017-1220-4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Vos D, Bruggeman FJ, Westerhoff HV & Bakker BM How molecular competition influences fluxes in gene expression networks. PloS one 6, e28494, doi: 10.1371/journal.pone.0028494 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khajuria RK et al. Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell 173, 90–103 e119, doi: 10.1016/j.cell.2018.02.036 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lodish HF Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature 251, 385–388 (1974). [DOI] [PubMed] [Google Scholar]
- 60.Schwanhausser B et al. Global quantification of mammalian gene expression control. Nature 473, 337–342, doi: 10.1038/nature10098 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Buszczak M, Signer RA & Morrison SJ Cellular differences in protein synthesis regulate tissue homeostasis. Cell 159, 242–251, doi: 10.1016/j.cell.2014.09.016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dinman JD Pathways to Specialized Ribosomes: The Brussels Lecture. Journal of molecular biology 428, 2186–2194, doi: 10.1016/j.jmb.2015.12.021 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi Z & Barna M Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. Annual review of cell and developmental biology 31, 31–54, doi: 10.1146/annurev-cellbio-100814-125346 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Simsek D et al. The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell 169, 1051–1065 e1018, doi: 10.1016/j.cell.2017.05.022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sloan KE et al. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA biology, 1–16, doi: 10.1080/15476286.2016.1259781 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vassiliou GS et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nature genetics 43, 470–475, doi: 10.1038/ng.796 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Method-only references
- 67.Uren PJ et al. Site identification in high-throughput RNA-protein interaction data. Bioinformatics 28, 3013–3020, doi: 10.1093/bioinformatics/bts569 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Machnicka MA et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic acids research 41, D262–267, doi: 10.1093/nar/gks1007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 38, 576–589, doi: 10.1016/j.molcel.2010.05.004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arai F et al. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 118, 149–161, doi: 10.1016/j.cell.2004.07.004 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Kanehisa M, Furumichi M, Tanabe M, Sato Y & Morishima K KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic acids research 45, D353–D361, doi: 10.1093/nar/gkw1092 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bluteau O et al. A landscape of germ line mutations in a cohort of inherited bone marrow failure patients. Blood 131, 717–732, doi: 10.1182/blood-2017-09-806489 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Pontikos N et al. Phenopolis: an open platform for harmonization and analysis of genetic and phenotypic data. Bioinformatics 33, 2421–2423, doi: 10.1093/bioinformatics/btx147 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and preprocessed sequencing and microarray data may be accessed from the Gene Expression Omnibus with accession number GSE135726. As the informed consent obtained from DC patients does not allow for public deposition of the data, WES data from patient CM108 (NPMD178H) and healthy controls can be communicated upon reasonable request to J.S. and WES data from patient harboring NPM1D180del mutation can be communicated upon reasonable request to I.D. and T.J.V..