Abstract
We generated parietal cortex RNA-seq data from individuals with and without Alzheimer disease (AD; ncontrol = 13; nAD = 83) from the Knight-ADRC. Using this and an independent (MSBB) AD RNA-seq dataset, we quantified cortical circular RNA (circRNA) expression in the context of AD. We identified significant associations between circRNA expression and AD diagnosis, clinical dementia severity, and neuropathological severity. We demonstrated that a majority of circRNA AD-associations are independent from changes in cognate linear mRNA expression or brain cell-type proportions. We provided evidence for circRNA expression changes occurring early in pre-symptomatic AD, and in autosomal dominant AD. We also observed AD-associated circRNAs co-expressing with known AD genes. Finally, we identified potential microRNA binding sites in AD-associated circRNAs for microRNAs predicted to target AD genes. Together, these results highlight the importance of analyzing non-linear RNAs and support future studies exploring the potential roles of circRNAs in AD pathogenesis.
Circular RNAs (circRNAs) are a class of RNAs that result from backsplicing events, in which the 3’ ends of transcripts are covalently spliced with the 5’ ends thereby forming continuous loops1,2. As RNA sequencing has become widespread, thousands of circRNAs have been identified across eukaryotes3–8. These studies have found circRNAs to be are highly expressed in the nervous system and enriched in synapses3,4,8–10. In the brain, circRNA expression can occur independently of linear transcript expression8, and may be a gene’s most highly expressed isoform3,8,10,11. Brain circRNAs are also regulated during development5,10,12 and in response to neuronal excitation8. CircRNAs accumulate in aging mouse4 and fly3 brains, possibly due to their lack of free hydroxyl ends conferring resistance to exonucleases. Much is still unknown regarding circRNA biology; for example, it was only recently demonstrated that circRNAs can be translated in vivo13,14. Thus far, the most well-established role of circRNAs is in microRNA (miRNA) regulation via sequestration15, leading to loss of function.
Alzheimer disease (AD) is a progressive, neurodegenerative disorder and the most common cause of dementia, affecting millions worldwide16. AD is neuropathologically characterized by the accumulation of amyloid beta plaques and tau inclusions17,18 as well as widespread neuronal atrophy which results in dramatic cognitive impairment. Unfortunately, no effective preventative, palliative, or curative therapies currently exist for AD.
Previous studies investigating linear transcriptomic (mainly mRNA) differences in the context of AD have yielded insight into the pathogenic mechanisms underlying this disease as well as potential therapeutic targets19–22. Similar analyses for circRNAs remain outstanding, although a single circRNA that regulates specific microRNAs and synaptic function23 – CDR1-AS – has been reported to be downregulated in AD brains24. Here, we conduct a circular transcriptome-wide analysis of circRNA differential expression in AD cases and their correlation with clinical and neuropathological AD severity measures.
RESULTS
Study Design
Our study design included calling and quantifying circRNA counts in two independent RNA-seq datasets derived from neuropathologically-confirmed17,25 AD case and control brain tissues. In our discovery dataset, we generated 150nt paired-end, rRNA depleted, RNA-sequencing (RNA-seq) data from frozen parietal cortex tissue donated by 96 individuals (13 controls and 83 AD cases). These individuals were assessed at the Knight Alzheimer Disease Research Center (Knight ADRC) at Washington University School of Medicine and their demographic, clinical severity, and neuropathological information is presented in Supplementary Table 1. For replication, we leveraged an independent, publicly-available Advanced Medicine Partnership for AD: Mount Sinai Brain Bank (MSBB) dataset (syn3157743)26. In brief, the MSBB dataset includes 100nt single-end rRNA-depleted RNA-seq data derived from 195 samples (40 controls, 89 definite AD, 31 probable AD, and 35 possible AD) of inferior frontal gyrus tissue (Brodmann area (BM) 44) as well as data derived from three additional cortical regions (frontal pole (BM10), superior temporal gyrus (BM22), and parahippocampal gyrus (BM36)). Demographic, clinical severity, and neuropathological information for all individuals in the MSBB dataset, separated by cortical region, is presented in Supplementary Tables 2–5.
We used STAR software27 in chimeric read detection mode to align the reads from both RNA-seq datasets to the GENCODE28 annotated human reference genome (GRCh38). Chimeric reads were further processed and filtered using DCC software29 to identify backsplice junctions. Finally, we collapsed backsplice junction counts onto their linear gene of origin to generate a set of high-confidence circRNA counts for downstream analyses (ONLINE METHODS). Using this pipeline, we called 3,547 circRNAs in the discovery dataset and an average of 3,924 circRNAs in the four regions of the replication dataset (Supplementary Table 6). We focused replication analyses primarily on the BM44-derived data, as we observed the largest overlap between the circRNAs called in this region and the parietal dataset (Supplementary Figure 1), though analyses in the other cortical regions yielded similar results.
We performed circRNA differential expression analyses for neuropathological AD case-control status as well as correlation with AD quantitative traits: Braak score and clinical dementia rating at expiration/death (CDR) using DESeq2 software30. Braak score is a neuropathological measure of AD severity determined by the number and distribution of neurofibrillary tau tangles throughout the brain18. Braak scores range from 0 (absent, at most incidental tau tangles) to 6 (severe, extensive tau tangles in neocortical areas). CDR is a clinical measure of cognitive impairment with a range from 0 (no dementia) to 3 (severe dementia)31. These quantitative measures capture different aspects of the pathological mechanisms underlying AD and consequently are not perfectly correlated with each other nor AD case status (Supplementary Figure 2). Thus, we analyzed each trait separately, modeling the ordinal measures as continuous variables. We adjusted all analyses for post mortem interval (PMI), RNA quality as measured by median transcript integrity number (TIN)32, age at death (AOD), batch, sex, and genetic ancestry as represented by the first two principal components derived from genetic data (ONLINE METHODS). We extended our circRNA analyses to pre-symptomatic and autosomal dominant AD (Supplementary Tables 1–5) to investigate if circRNA expression changes occurred before symptom onset and whether these changes were restricted to sporadic AD. Finally, we investigated the AD-relevance and potential disease-influencing mechanisms of AD-associated circRNAs through relative importance, network co-expression, and microRNA binding site prediction analyses.
Discovery analysis to identify AD differentially expressed circRNAs
In the circular-transcriptome-wide discovery analysis (nCDR = 96, nBraak = 86, ncontrol = 13, nAD = 83), we identified 31 circRNAs significantly correlated with CDR passing a false discovery rate (FDR) of 0.05 (Supplementary Table 7). The most significantly correlated circRNA, was circHOMER1 (log2 fold change: −0.28 per unit of CDR, p-value: 8.22×10−12). circCDR1-AS (log2 fold change: 0.17 per unit of CDR, p-value: 3.18 × 10−02) was only nominally correlated with CDR, but, in contrast to the previous report24, we observed its expression to be upregulated with increasing dementia severity.
We also identified circRNAs significantly associated with the two other complementary AD traits: Braak score (nine circRNAs passed FDR, Supplementary Table 8) and neuropathological AD versus control status (nine circRNAs passed FDR; Supplementary Table 9). These analyses yielded both AD trait-specific associations as well as circRNAs that were consistently associated across all AD traits investigated. Three circRNAs passed FDR correction for all three analyses. For example, in addition to the CDR-association, circHOMER1 was also significantly associated with Braak score (p-value: 1.19×10−07) and AD versus control status (p-value: 2.76×10−06). In general, circRNAs associated with one AD trait were also, at least, nominally associated (p-value < 0.05) with the remaining two traits. We validated our RNA-seq findings for five circRNAs using an orthogonal qPCR approach with 13 discovery dataset RNA samples (ncontrol = 3, nPreSympAD = 3, nAD = 7). We demonstrate a strong correlation between RNA-seq-derived counts for the five circRNA transcripts and the GAPDH-normalized deltaCt values (median absolute correlation: 0.64, Supplementary Figure 3). Importantly, we also observe consistent direction of effect, thereby validating our RNA-seq results (Supplementary Figure 3). Altogether, we identified 37 circRNAs in the discovery analysis of the parietal cortex dataset that were significantly associated with at least one AD trait (Supplementary Figure 4).
Replication and meta-analysis of circRNA differential expression using an independent AD dataset
We performed replication analyses in the MSBB BM44 dataset (nCDR = 195, nBraak = 188, ncontrol = 40, nDefinite AD = 89). Twenty-seven of the 31 circRNAs that were correlated with CDR in the discovery dataset also showed, at minimum, a nominal p-value, with the same directions of effect and comparable effect sizes (effect size Pearson correlation: 0.97, p-value: 1.69×10−17, Supplementary Table 10). For example, we replicated decreasing circHOMER1 expression with increasing dementia severity (log2 fold change: −0.13 per unit of CDR, p-value: 2.27×10−09). A meta-analysis of the discovery and replication results revealed a total of 148 circRNAs that were significantly correlated with CDR after FDR correction (Supplementary Table 11), with 33 passing the stringent gene-based, Bonferroni multiple test correction of 5×10−06 (Table 1), including cirHOMER1 (p-value: 2.21×10−18) and circCDR1-AS (p-value: 2.83×10−08).
Table 1 |.
Cortical circRNAs are significantly associated with AD case status, dementia severity, and neuropathological severity
| CDR - Discovery |
CDR - Replication |
Meta-Analysis |
||||||
|---|---|---|---|---|---|---|---|---|
| circRNA | Chr | log2FC | p-value | log2FC | p-value | CDR p-value | Braak p-value | AD Case p-value |
| circHOMER1 | 5 | −0.28 | 8.22×10−12 | −0.13 | 2.27×10−09 | 2.21×10−18 | 4.77×10−12 | 4.35×10−10 |
| circDOCK1 | 10 | 0.30 | 8.49×10−06 | 0.20 | 7.55×10−08 | 6.47×10−12 | 8.68×10−07 | 3.74×10−06 |
| circKCNN2 | 5 | −0.12 | 7.27×10−04 | −0.12 | 1.93×10−09 | 1.47×10−11 | 4.43×10−08 | 8.38×10−08 |
| circMAN2A1 | 5 | 0.23 | 2.46×10−04 | 0.17 | 2.92×10−07 | 5.59×10−10 | 1.25×10−06 | 3.75×10−09 |
| circST18 | 8 | 0.37 | 1.27×10−04 | 0.28 | 6.60×10−07 | 6.80×10−10 | 7.30×10−06 | 1.22×10−09 |
| circATRNL1 | 10 | −0.13 | 2.42×10−03 | −0.13 | 4.15×10−08 | 9.47×10−10 | 4.26×10−05 | 2.73×10−06 |
| circEXOSC1 | 10 | 0.14 | 3.66×10−02 | 0.18 | 8.13×10−09 | 7.92×10−09 | 6.22×10−05 | 1.27×10−06 |
| circICA1 | 7 | −0.16 | 7.40×10−05 | −0.11 | 2.33×10−05 | 1.77×10−08 | 3.43×10−02 | 2.08×10−06 |
| circFMN1 | 15 | −0.16 | 1.01×10−04 | −0.11 | 2.13×10−05 | 2.07×10−08 | 2.12×10−06 | 3.79×10−06 |
| circRTN4 | 2 | 0.14 | 8.36×10−03 | 0.13 | 2.72×10−07 | 2.18×10−08 | 6.96×10−08 | 4.81×10−09 |
| circCDR1-AS | 23 | 0.17 | 3.18×10−02 | 0.19 | 4.90×10−08 | 2.83×10−08 | 1.54×10−03 | 5.29×10−12 |
| circMAP7 | 6 | 0.17 | 1.83×10−05 | 0.10 | 1.66×10−04 | 5.51×10−08 | 1.07×10−06 | 5.41×10−08 |
| circTTLL7 | 1 | 0.18 | 2.59×10−03 | 0.16 | 3.42×10−06 | 6.18×10−08 | 1.22×10−06 | 1.07×10−07 |
| circFANCL | 2 | 0.21 | 9.12×10−03 | 0.15 | 9.88×10−07 | 7.65×10−08 | 1.75×10−03 | 1.11×10−03 |
| circEPB41L5 | 2 | −0.13 | 1.12×10−03 | −0.09 | 1.02×10−05 | 7.84×10−08 | 1.71×10−05 | 2.67×10−04 |
| circCORO1C | 12 | 0.12 | 7.19×10−04 | 0.11 | 2.20×10−05 | 1.14×10−07 | 7.97×10−06 | 2.45×10−07 |
| circDGKI | 7 | −0.12 | 3.86×10−02 | −0.14 | 2.42×10−07 | 1.41×10−07 | 3.78×10−03 | 1.05×10−03 |
| circKATNAL2 | 18 | −0.14 | 2.39×10−02 | −0.21 | 5.78×10−07 | 1.55×10−07 | 2.11×10−03 | 8.74×10−05 |
| circWDR78 | 1 | 0.14 | 5.84×10−04 | 0.11 | 3.59×10−05 | 1.57×10−07 | 2.62×10−04 | 2.95×10−05 |
| circADGRB3 | 6 | −0.07 | 1.10×10−02 | −0.07 | 2.20×10−06 | 1.94×10−07 | 5.97×10−03 | 1.47×10−03 |
| circPLEKHM3 | 2 | −0.19 | 6.13×10−06 | −0.10 | 1.00×10−03 | 2.32×10−07 | 3.77×10−04 | 4.13×10−06 |
| circERBIN | 5 | 0.25 | 1.34×10−03 | 0.17 | 2.92×10−05 | 2.67×10−07 | 2.42×10−04 | 1.20×10−05 |
| circPICALM | 11 | 0.07 | 1.29×10−02 | 0.08 | 4.63×10−06 | 4.54×10−07 | 3.12×10−06 | 3.35×10−08 |
| circRNASEH2B | 13 | 0.20 | 3.57×10−03 | 0.14 | 3.13×10−05 | 7.11×10−07 | 1.72×10−03 | 4.63×10−03 |
| circPDE4B | 1 | −0.13 | 5.84×10−03 | −0.11 | 1.98×10−05 | 7.47×10−07 | 1.94×10−03 | 5.33×10−05 |
| circPHC3 | 3 | 0.16 | 7.43×10−04 | 0.11 | 1.40×10−04 | 7.99×10−07 | 2.09×10−02 | 1.01×10−02 |
| circFAT3 | 11 | −0.23 | 4.75×10−03 | −0.21 | 3.11×10−05 | 9.31×10−07 | 8.21×10−03 | 2.04×10−04 |
| circMLIP | 6 | −0.08 | 5.75×10−02 | −0.10 | 3.41×10−06 | 2.24×10−06 | 7.22×10−06 | 2.71×10−07 |
| circLPAR1 | 9 | 0.17 | 2.17×10−02 | 0.20 | 1.72×10−05 | 2.68×10−06 | 1.49×10−03 | 4.58×10−06 |
| circSLAIN2 | 4 | 0.14 | 5.25×10−04 | 0.12 | 5.62×10−04 | 2.70×10−06 | 2.51×10−02 | 2.63×10−05 |
| circSPHKAP | 2 | −0.39 | 1.48×10−03 | −0.27 | 3.16×10−04 | 3.32×10−06 | 2.88×10−02 | 2.44×10−01 |
| circYY1AP1 | 1 | 0.20 | 4.47×10−04 | 0.11 | 9.71×10−04 | 4.40×10−06 | 1.83×10−04 | 1.15×10−03 |
| circDNAJC6 | 1 | 0.16 | 6.63×10−03 | 0.11 | 1.27×10−04 | 4.99×10−06 | 2.04×10−05 | 8.21×10−06 |
circRNA association with AD traits in the discovery Knight ADRC parietal dataset, replication MSBB Brodmann Area 44 (BM44) dataset, and meta-analyses. Presented are the log2 fold changes (log2FC) and p-values generated via a Wald-log test for the discovery (nCDR = 96) and replication (nCDR = 195) analyses and the inverse/Stouffer’s method combined p-values for the meta-analyses. Discovery and replication analyses were adjusted for post-mortem interval, RNA quality (median transcript integrity number), age at death, batch, sex, and genetic ancestry (principal components 1–2). Braak, Braak score; CDR, clinical dementia rating at expiration/death; Chr, chromosome.
Similarly, five of the nine circRNAs that were correlated with Braak score in the discovery dataset replicated in the MSBB dataset (effect size Pearson correlation: 0.99, p-value: 9.29×10−06, Supplementary Table 12). A total of 33 circRNAs were significantly associated with Braak score after FDR correction in the meta-analysis (Supplementary Table 13). Finally, five of nine circRNAs associated with AD case-control status replicated in the MSBB dataset (effect size Pearson correlation: 0.99, p-value: 6.12×10−05, Supplementary Table 14) and 75 circRNAs associated with AD case-control status after FDR correction (Supplementary Table 15) in the meta-analysis.
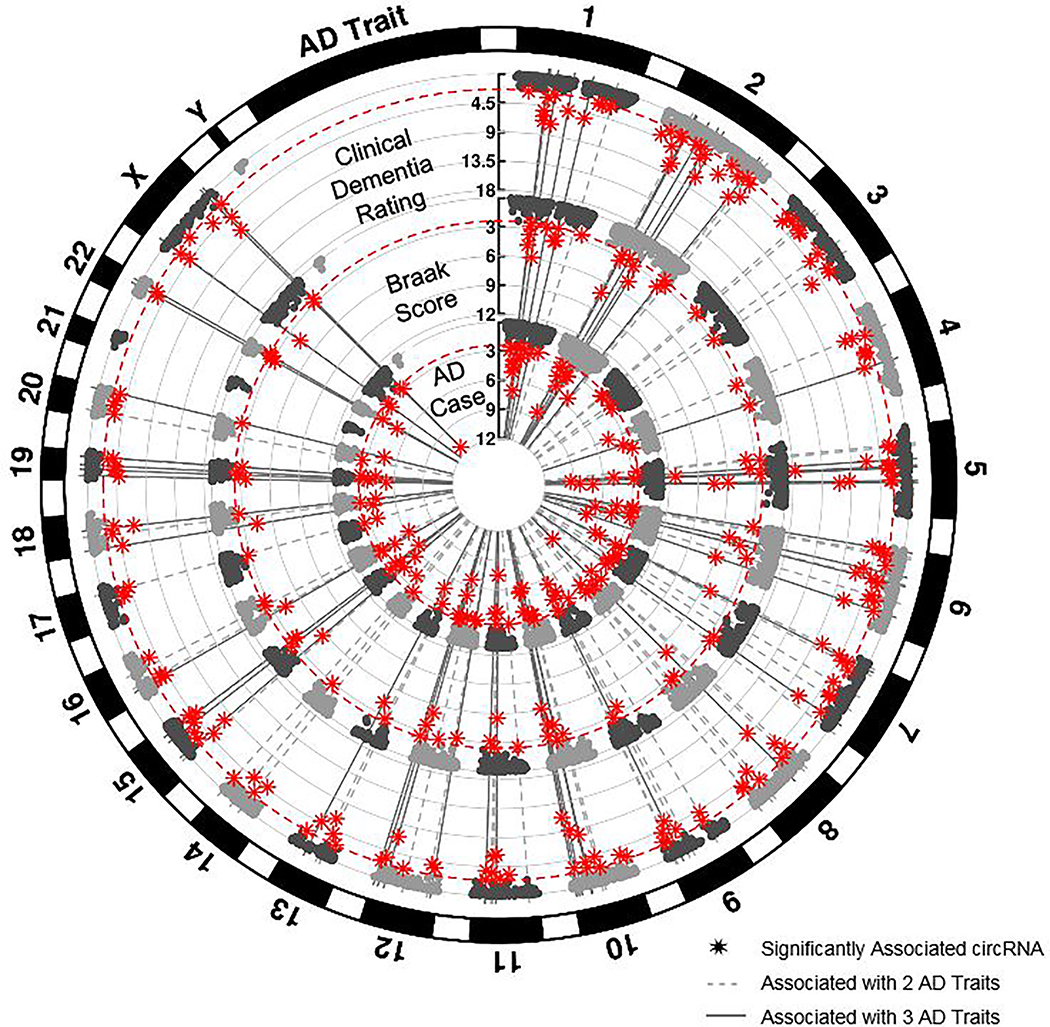
Overall, we identified 164 circRNAs that were significantly associated with at least one AD trait in the meta-analyses (Figure 1). Twenty-eight of these circRNAs, including circHOMER1 and circCORO1C, were significantly associated with all three traits investigated (Supplementary Figure 5). Nine cross-trait circRNA-associations had p-values passing the gene-based stringent threshold of 5×10−06 (Table 1). Altogether, these results support a consistent, replicable, and highly significant association between changes in circRNA expression and AD traits.
Figure 1: Cortical circRNAs are associated with AD traits.
Each circular Manhattan plot presents the results from a meta-analysis of circRNA AD-association results from discovery (parietal cortex) and replication (inferior frontal gyrus (Brodmann Area 44)) datasets. In order from outermost to innermost circular plot, the AD traits include: clinical dementia rating at expiration/death (CDR), Braak neuropathological severity score, and AD case-control status (AD case). Study-wide significance threshold is based on a false discovery rate of 0.05 and depicted by the red, dashed line. circRNAs that passed this threshold are displayed with star symbols. Lines extending through all three plots identify circRNAs that are significantly associated with multiple AD traits – dotted line: 2 traits; solid line: 3 traits.
AD-associated changes in circRNA expression demonstrate independence from AD-associated changes in their cognate linear mRNAs and AD-associated changes in estimated brain cell-type proportions
Circular and their cognate linear mRNAs can demonstrate independent expression8, but some level of correlation is expected given the shared genomic origin and biogenesis machinery. This correlation is also technically biased because the majority of RNA-seq reads covering a circRNA transcript will not contain the circRNA-defining backsplice junction and thus be incorrectly counted as originating from a linear mRNA rather than a circRNA transcript. For example, linear forms of circCDR1-AS are expressed at such low levels33 that they have been historically undetectable23,33. However, we observe ‘linear’ CDR1-AS counts in our linear mRNA quantification, consistent with the technical bias. This artifact is expected to bias circRNA AD-associations to the null when the relatively less abundant circRNAs are included together in the same regression models as their cognate linear mRNAs. Nevertheless, we demonstrate that a majority of CDR-associated changes in circRNA expression are independent from CDR-associated changes in their cognate linear mRNAs using this regression-based approach.
In the meta-analysis of the discovery and replication linear and circRNA combined regression results, we observe that 109 of 146 circRNAs retain a significant association (p-value < 0.05, Supplementary Table 16) with CDR, for example circHOMER1 (p-value: 3.11×10−06) or circDOCK1 (p-value: 1.65×10−05), demonstrating an independent association. In addition, 62 CDR-associated circRNAs had association p-values less than the association p-values of their cognate linear mRNA and 78 CDR-associated circRNAs explained as much or more of the variation in CDR compared to their cognate linear mRNAs (Supplementary Table 16). In a separate analysis, we employ the same regression-based approach to demonstrate that most (106 of 148, Supplementary Table 17) CDR-associated circRNAs - for example circHOMER1 (p-value: 8.15×10−13) or circDOCK1 (p-value: 1.03×10−05) - are similarly independent of AD-associated neuronal and other estimated brain cell-type proportion changes34 (Supplementary Results). Together, these results demonstrate that the majority of AD-circRNA associations are independent of AD-associated changes in linear mRNA or brain cell-type proportions.
AD-associated changes in circRNA expression are consistent across cortical regions
The MSBB dataset also includes RNA-seq data derived from three additional brain cortical regions: BM10 (Supplementary Table 2), BM22 (Supplementary Table 3), and BM36 (Supplementary Table 4). To determine if AD-associated changes in circRNA expression were consistent across the cortex, we performed circular-transcriptome-wide analyses in these additional datasets. As before, we investigated for circRNA correlation with CDR (Supplementary Tables 18–20) and Braak score (Supplementary Tables 21–23), and association with AD case-control status (Supplementary Tables 24–26). We performed three sets of meta-analyses with the parietal discovery results, one for each of the additional cortical regions: BM10 (Supplementary Tables 27–29), BM22 (Supplementary Tables 30–32), and BM36 (Supplementary Tables 33–35). We then compared these results with the BM44 meta-analysis results to identify consistent AD-associated circRNA expression changes.
We identified 23 circRNAs that were significantly associated with CDR in all four meta-analyses, with comparable effect sizes and the same directions of effect (overlap p-value: 1.60×10−94, Supplementary Figure 6A). Similarly, we identified 14 circRNAs that were significantly associated with Braak score (overlap p-value: 1.38×10−70, Supplementary Figure 6B) and five that were significantly associated with AD case status (overlap p-value: 3.90×10−26, Supplementary Figure 6C) with consistent directions of effect in all four meta-analyses. Three circRNAs: circHOMER1, circKCNN2, and circMAN2A1, were significantly associated with all three AD traits in all four meta-analyses. Eleven circRNAs were associated with the two quantitative AD traits in all four meta-analyses: circDGKB, circDNAJC6, circDOCK1, circERBIN, circFMN1, circHOMER1, circKCNN2, circMAN2A1, circMAP7, circSLAIN1, and circST18.
The MSBB dataset includes an additional measure of neuropathological severity, mean number of amyloid plaques. Results for circRNA correlation with mean number of plaques were consistent with the other traits in all MSBB cortical regions (Supplementary Tables 36–39, Supplementary Figure 7 and Supplementary Results). Together, these results suggest that expression changes in some circRNAs are a consistent phenomenon across cortical regions in the context of AD.
Evidence supporting circRNA differential expression in pre-symptomatic AD
We investigated for early AD-related changes in circRNA expression in a small number (nDiscovery = 6 and nReplication = 6) of individuals with pre-symptomatic AD – i.e., neuropathological evidence of AD but, at most, very mild dementia (CDR <= 0.5).
We first compared circRNA expression between pre-symptomatic AD (PreSympAD) versus controls (control nDiscovery = 13, control nReplication = 40) in each dataset individually, but failed to detect significant circRNA differential expression. Nevertheless, we did identify several nominal associations with directions and magnitudes of effect (log2 fold change) consistent with those observed in complementary analyses identifying circRNA differential expression between symptomatic (CDR >= 1) individuals with AD neuropathology (SympAD) versus controls in the BM44 dataset (nSympAD = 137 Supplementary Tables 40), but not in the smaller parietal dataset (nSympAD = 77, Supplementary Table 41).
These results suggested that changes in circRNA expression occur in PreSympAD, but we had too few individuals to detect this on a transcriptome-wide basis. If this hypothesis is correct, then the effect size correlation between nominally PreSympAD-associated circRNAs and significantly SympAD-associated circRNAs should be stronger for the SympAD-associated circRNAs compared to the background, non-SympAD-associated circRNAs. Thus we generated bootstrapped confidence intervals35 for the Pearson correlation between effect sizes.
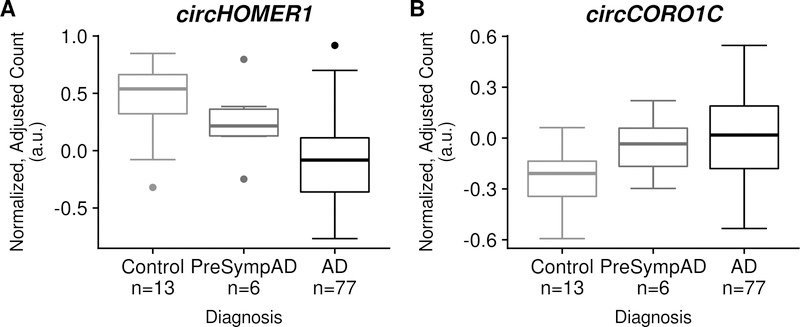
We observed that the bootstrapped effect size correlation coefficient distribution for the SympAD-associated circRNAs was significantly higher than the background distribution in both the parietal discovery (14 SympAD-associated circRNAs, effect size correlation: 0.67 [0.43, 0.90] versus 713 background circRNAs, effect size correlation: 0.21 [0.14, 0.29], p-value: < 2.2×10−16; Figure 2) and the BM44 replication (100 SympAD-associated circRNAs, effect size correlation: 0.78 [0.68, 0.85] versus 1544 background circRNAs, effect size correlation: 0.36 [0.31, 0.41], p-value < 2.2×10−16) datasets (Supplementary Table 42).
Figure 2: Changes in cortical circRNA expression tracks with AD clinical severity.
Presented are boxplots of library-size normalized, differential expression covariate-adjusted counts for two AD-associated circRNAs: circHOMER1 and circCORO1C in the Knight ADRC parietal dataset. AD: Alzheimer disease. PreSympAD (Pre-symptomatic AD: neuropathological evidence of AD but, at most, very mild dementia (Clinical dementia rating <= 0.5). Box plot elements: center line (median), box (first and third quartiles), whiskers (quartile ± 1.5×interquartile range), dots (outlier points as defined by falling outside of whiskers).
When we extended these analyses to the three other cortical regions of the MSBB dataset (Supplementary Tables 43–45), we also observed evidence for pre-symptomatic changes in circRNA expression (Supplementary Table 42, p-values: < 2.2×10−16). The SympAD-associated circRNA effect size correlation distribution width varied by cortical region (Supplementary Table 42): BM44 ~ BM36 < BM22 < parietal cortex < BM10, in a sequence reminiscent of the observed spatiotemporal progression of AD pathology within the cortex18,36. Together, these results support early changes in circRNA expression in multiple cortical regions in PreSympAD.
Changes in circRNA expression are more severe in individuals with autosomal dominant AD
Autosomal dominant AD (ADAD) is an early-onset form of AD caused by pathogenic mutations in APP, PSEN1, or PSEN237. We investigated whether changes in circRNA expression also occur in the context of ADAD by generating parietal cortex-derived RNA-seq data from 21 brains donated by individuals with ADAD who were enrolled in the Dominantly Inherited Alzheimer Network (DIAN) study. ADAD participant demographic, clinical, and neuropathological data is presented in Supplementary Table 1. We generated the ADAD RNA-seq data at the same time as the discovery RNA-seq data and called and filtered circRNAs in both datasets simultaneously.
In a circular-transcriptome-wide analysis of circRNA differential expression between ADAD (n=21) and discovery dataset controls (n=13), we identified 236 ADAD-associated circRNAs that were significant under the FDR threshold (Supplementary Table 46). These included almost all (8/9) AD case-control status-associated circRNAs identified in the discovery analysis, with consistent direction of effect (Supplementary Figure 8). However, the magnitudes of effect were greater in the ADAD versus control analysis (e.g. circHOMER1: AD versus control, log2 fold-change: −0.64; ADAD versus control log2 fold-change: −0.95).
To investigate whether the larger effect size was due to the greater pathological severity in the ADAD brains (Supplementary Table 1), we performed a Braak score-adjusted circRNA differential expression analysis between ADAD and discovery dataset AD (samples with available Braak score: nADAD = 17, nAD = 73). We identified 77 significantly differentially expressed circRNAs (Supplementary Table 47) and 59/77 of these were identified in the ADAD versus controls analysis (Supplementary Figure 8). As before, these 59 differentially expressed circRNAs had consistent directions of effect, and the majority (56/59) had greater magnitudes of effect when comparing controls versus AD versus ADAD. Altogether, these results demonstrate that changes in circRNA expression also occur in the context of ADAD and are more severe in magnitude, even when adjusting for neuropathological severity.
AD-associated circRNAs explain more of the variation in AD quantitative measures than number of APOE4 alleles or estimated neuronal proportion
We performed relative importance analyses38 to assess the contribution of circRNA expression to the variation in AD quantitative traits: CDR and Braak score compared to two known contributors: number of APOE4 alleles (APOE4) – the most common genetic risk factor for AD16 – and the estimated proportion of neurons (EstNeuron)34.
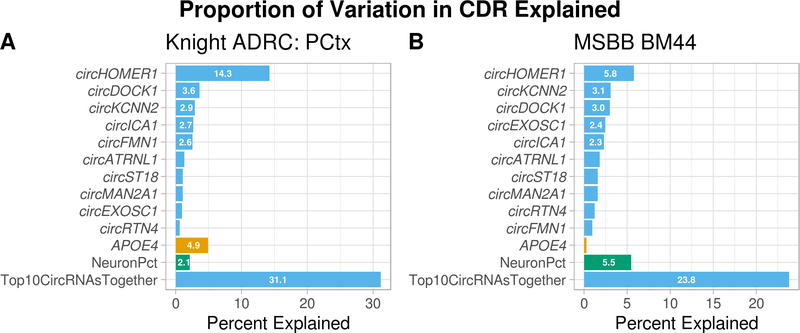
We selected the meta-analysis top 10 most significantly CDR-associated circRNAs for the proportion of variation explained analyses. In the discovery dataset (nCDR = 96), these circRNAs - included in the same multivariate model as APOE4 and EstNeuron – explained a total of 31.1% of the observed variation in CDR (Figure 3A and Supplementary Table 48). Our BM44 replication dataset (nCDR = 195) results with the same circRNAs were consistent, with the circRNAs explaining a total of 23.8% to the variation in CDR (Figure 3B, Supplementary Table 49). In both the discovery and replication datasets, we observed some circRNAs individually, and the top 10 circRNAs together, to explain more of the variation in CDR compared to APOE4 and EstNeuron (Figure 3A–B). We observed the same pattern when assessing the relative contribution of circRNAs to the observed variation in Braak score (Supplementary Figure 9 and Supplementary Tables 50–51) and when analyzing the other MSBB tissues for contribution of circRNAs to variation in CDR (Supplementary Tables 52–54), Braak score (Supplementary Tables 55–57), and mean number of plaques (Supplementary Tables 58–61 and Supplementary results). Finally, we also observed that circRNAs explain more of the variation in Braak score in individuals with ADAD than APOE4 and EstNeuron (Supplementary Table 62 and Supplementary Results).
Figure 3: AD-associated circRNAs explain more of the observed variation in clinical dementia rating compared to number of APOE4 alleles or the estimated proportion of neurons.
Percent of variation in clinical dementia rating (CDR) explained by the top 10, most meta-analysis significant CDR-associated circRNAs compared to two known contributors number of APOE4 alleles – the most common genetic risk factor for AD – and the estimated proportion of neurons. Knight ADRC: PCtx – parietal discovery dataset (nCDR = 96); MSBB BM44 – inferior frontal gyrus replication dataset (nCDR = 195).
In addition to the proportion of variation analyses, we also compared the AD predictive ability of the same meta-analysis 10 most significant CDR-associated circRNAs to the AD predictive ability of baseline models that include number of APOE4 alleles and the differential expression covariates. Consistent with the relative importance analyses, we found that circRNAs alone provided similar or greater predictive value compared with the baseline genetic-demographic models, and even improved the predictive ability when combined with the baseline genetic-demographic data (Supplementary Table 63, Supplementary Figure 10, and Supplementary Results). Altogether, these results demonstrate that circRNA expression is strongly associated with AD quantitative traits and contributes significantly to the variation in these AD severity measures.
Differentially expressed circRNAs co-express with AD-relevant genes and pathways
Analyzing circRNA co-expression with linear transcripts provides an opportunity to infer the biological and pathological relevance of circRNAs. We computed co-expression networks in the discovery parietal dataset (Supplementary Tables 64–65) as well as in each of the cortical regions of the MSBB dataset: BM10 (Supplementary Tables 66–67), BM22 (Supplementary Tables 68–69), BM36 (Supplementary Tables 70–71), and BM44 (Supplementary Tables 72–73) based on Spearman correlation using MEGENA software39. We further calculated the correlation between the eigengenes40 of these networks and CDR.
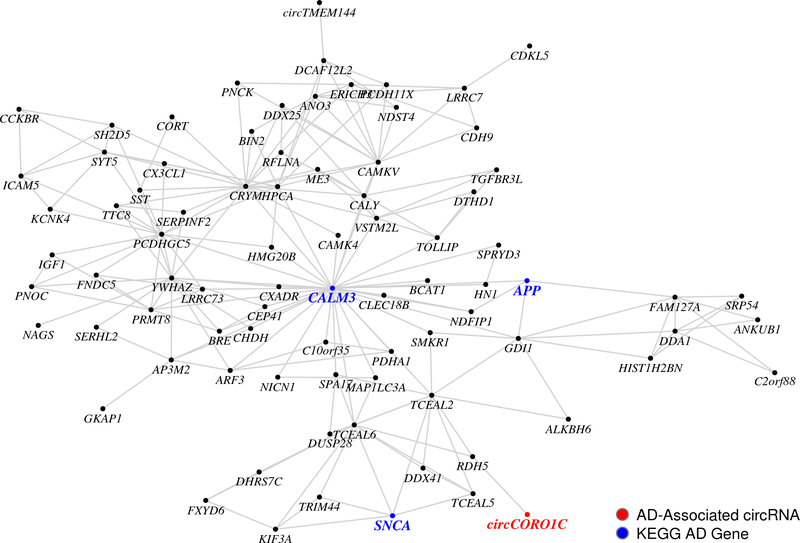
In the parietal dataset, we identified 49 hierarchical co-expression modules that were significantly correlated with CDR (Supplementary Table 64) and contained at least one AD-associated circRNA (Supplementary Table 65). Similarly, in the MSBB BM44 dataset, we identified 20 hierarchical co-expression modules that significantly correlated with CDR (Supplementary Table 72) and contained at least one AD-associated circRNA (Supplementary Table 73). CircHOMER1 expressed in module c1_16 (module correlation with CDR, p-value: 5.94×10−04) in the parietal dataset. This module included linear transcripts that are significantly enriched for AD pathways (KEGG Alzheimer’s Disease, 66/156 genes, adjusted p-value: 1.07×10−15) and oxidative phosphorylation-related genes (KEGG Oxidative Phosphorylation, 58/115 genes, adjusted p-value: 2.76×10−18). Similarly, the AD-associated circRNA, circCORO1C, co-expressed in BM44 dataset module c1_46 (module correlation with CDR, p-value: 1.52×10−07), which also included the AD genes APP and SNCA (Figure 4).
Figure 4: AD-associated circRNAs co-express with AD-relevant genes.
Spearman correlation-based network co-expression module c1_46 (module association with clinical dementia rating (CDR), p-value: 1.52×10−07) in the MSBB BM44 dataset (n = 195). Module association with CDR was determined from a multivariate linear regression with module eigengene and differential expression covariates. Significance of the module eigengene association with CDR was determined using a two-tailed t-test. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Our MEGENA results in the other cortical regions of the MSBB dataset were consistent with AD-associated circRNAs co-expressing with AD-related genes and pathways. For example, we observed APP co-expressing with several AD-associated circRNAs (Supplementary Table 69) in the BM22 module c1_14 (module correlation with CDR, p-value: 2.39×10−06). Altogether, these results suggest an important role for circRNAs in AD.
AD-associated circRNAs contain binding sites for microRNAs that potentially regulate AD-associated pathways and genes
The functional consequences of circRNA expression is an area of active research. While recent studies have demonstrated that circRNAs can regulate transcription2 and even be translated13,14, their most well-characterized function is in miRNA regulation via sequestration2,15. For example, circCDR1-AS contains over 70 binding sites for miR-72,23 and reducing circCDR1-AS expression results in the downregulation of miR-7 target mRNAs2,5,15. However, even a single miRNA binding site on a circRNA appears sufficient to regulate miRNA function41.
To identify miRNAs potentially regulated by AD-associated circRNAs, we utilized TargetScan70 software42 to predict miRNA binding sites in circRNA sequences (Supplementary Tables 74–75). We replicated the previously reported finding of over 70 miR-7 predicted binding sites in the circCDR1-AS sequence (Supplementary Table 74) and predicted binding sites for several intriguing miRNAs in the other AD-associated circRNAs. CircATRNL1 contained 18 predicted binding sites for miR-136 (Supplementary Tables 74–75), an miRNA whose increased expression triggers apoptosis in glioma cells43. circHOMER1 contained 5 predicted binding sites for miR-651 (Supplementary Tables 74–75), which is an miRNA predicted to target the AD-related genes PSEN1 and PSEN242. Finally, circCORO1C which we identified as co-expressing with the AD-related genes APP and SNCA (Supplementary Table 73) contains two predicted binding sites for miR-105 (Supplementary Table 74), which is an miRNA predicted to target APP and SNCA42. While these bioinformatics results require functional validation in future studies, they suggest that some AD-associated circRNAs may exert functional effects through miRNA regulation.
DISCUSSION
Transcriptional regulation underlies the complexity of the human nervous system, and its misregulation can contribute to disease44. Indeed, several studies focused on the linear transcriptome have identified co-expression networks and changes in splicing associated with AD status19–22. Here, we provide insight into the AD-associated circular transcriptome.
Using two large and independent brain-derived RNA-seq datasets, we establish that changes in specific circRNAs are a replicable and highly significant phenomenon in AD. We demonstrate that circRNA expression levels are robustly correlated with both neuropathological and clinical measures of AD severity, suggesting an important role in the disease (Table 1). This role is further supported by evidence for changes in circRNA expression in pre-symptomatic AD. The pathological processes underlying AD follow a well characterized spatiotemporal progression18 which begins decades before symptom onset. Thus, changes in circRNA expression during the pre-symptomatic stage, which we observe to occur in a sequence consistent with the known spatiotemporal progression, may directly contribute to disease rather than being merely correlated. Our finding that the effect sizes of changes in circRNA expression were greater in individuals with the genetically-driven ADAD compared to sporadic AD, even after adjusting for neuropathological severity, also argues against AD-associated circRNAs being merely correlated with disease. This important role is also supported by our network analyses, which demonstrate that AD-associated circRNAs co-express with genes known to be part of AD causal pathways.
We identify 164 AD-associated circRNAs on meta-analysis and perform network co-expression and microRNA binding site prediction analyses to infer biological context and facilitate the interpretation of our results. For example, circHOMER1, which was significantly associated with all three AD traits, co-expressed with linear genes involved in AD and oxidative phosphorylation, perhaps suggesting a role for this circRNA in brain hypometabolism associated with AD45–47. Brain hypometabolism has also been demonstrated in PSEN1 mutation-driven ADAD48,49 and circHOMER1 contains multiple predicted bindings sites for miR-651, an miRNA predicted to target PSEN1 and PSEN242. Similarly, we identified circCORO1C to co-express with the AD-related genes APP and SNCA and further identified the presence of multiple predicted miR-105 binding sites in circCORO1C. MiR-105 is predicted to target both APP and SNCA42, suggesting that the co-expression we observe may be mediated through this microRNA. Importantly, if this and other AD-associated circRNAs exert functional effects through miRNA regulation, then subtle changes in circRNA expression may have major impacts on downstream gene expression.
Our identification of high-confidence circRNA expression is technically limited by the high depth of sequencing and large number of samples required to generate sufficient reads for calling and stringently filtering backsplice junctions. In addition, circRNAs can only be called in ribosomal RNA (rRNA)-depleted RNA-seq datasets which are currently uncommon. Our results support the generation of additional AD and control brain rRNA-depleted RNA-seq datasets. As these datasets become available, it will be important to confirm our findings. In particular, our ADAD analyses should be replicated with age-matched controls and our PreSympAD findings should be replicated in a larger dataset as these are both limitations of our current study. Another limitation of our study is the fact that our independent replication dataset is derived from a different cortical region than our discovery dataset. Nevertheless, analyzing RNA-seq data from four different cortical regions in the MSBB replication dataset allowed us to observe changes in circRNA expression as a consistent phenomenon across the cortex in a sequence following the known spatiotemporal progression of AD.
Our sensitivity analyses demonstrate that the majority of circRNA AD-associations are independent of cognate linear mRNA or cell-type proportion changes associated with AD – despite the inherent technical (linear) or biological (cell-type proportion) correlation. Nevertheless, the linear-circular technical correlation limits the interpretation of co-expression modules that include both AD-associated circRNAs and their cognate linear mRNAs. In addition, some AD-associated circRNAs may not be independent of their AD-linear mRNA-associations, but as the biological functions of circRNAs are different, these AD-associated circRNAs may still be pathologically relevant. Finally, we observe instances where circRNAs rather than their cognate linear mRNAs appear to be driving the association with AD. Consequently, circRNA analyses should be conducted alongside traditional linear mRNA analyses to test for this possibility in other rRNA-depleted RNA-seq datasets.
Future studies to better understand and functionally characterize AD-associated circRNAs may yield novel quantitative trait loci or even biomarkers and therapeutic targets, as has been recently demonstrated for acute ischemic stroke41. We observed circRNA expression to yield strong predictive ability for AD case status, even in the absence of demographic or APOE4 risk factor data. This observation coupled with the relative stability of circRNAs in biofluids like CSF and plasma7 and their enrichment in exosomes50 suggests that circRNAs will likely have utility as peripheral biomarkers of pre-symptomatic and symptomatic AD and potentially other neurodegenerative diseases.
Ethics approval and consent to participate
All research participants contributing clinical, genetic, or tissue samples for analysis in this study provided written informed consent, subject to oversight by the Washington University in St. Louis or Mount Sinai School of Medicine institutional review boards. All studies conducted using Knight ADRC (201105102) and DIAN (201106339) data were approved by the Washington University Human Research Protection Office and written informed consent was obtained from each participant.
ONLINE METHODS
Code Availability
A description of how all software has been run for this study, including relevant command flags, is included in the Online Methods. In addition, the code used for analysis is provided in the included Supplementary Software.
RNA-sequencing
Discovery (Knight ADRC) and Autosomal Dominant AD (DIAN) datasets
We generated 151 nucleotide (nt), paired-end, rRNA depleted RNA-sequencing (RNA-seq) data from frozen brain parietal cortex tissue. The frozen brain tissues were donated by participants in either the prospective Knight Alzheimer’s Disease Research Center (Knight ADRC) Memory and Aging Project study at Washington University School of Medicine or the Dominantly Inherited Alzheimer’s Network (DIAN) study. All participants consented to brain donation and neuropathological analysis. We first disrupted the frozen cortical tissues using a TissueLyser LT and purified the RNA from this disrupted tissue using RNeasy Mini Kits. (Qiagen, Hilden, Germany). We calculated the RNA Integrity Number (RIN) using a RNA 6000 Pico assay on a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, USA). We also quantified the extracted RNA using the Quant-iT RNA assay (Invitrogen, Carlsbad, USA) on a Qubit Fluorometer (Fisher Scientific, Waltham, USA). Prior to library construction, we introduced External RNA Controls Consortium (ERCC)51 RNA Spike-In Mix (Invitrogen, Carlsbad, USA). rRNA depleted cDNA libraries were prepared using a TruSeq Stranded Total RNA Sample Prep with Ribo-Zero Gold kit (Illumina, San Diego, USA) and sequenced on an Illumina HiSeq 4000 at the McDonnell Genome Institute at Washington University in St. Louis. All samples were randomly assigned to a sequencing pool prior to sequencing and RNA extraction and sequencing library preparation were performed blind to neuropathological case-control status. The average number of raw sequencing reads per individual was 58,094,683 (Supplementary Table 6).
Replication Dataset (MSBB)
We downloaded publicly available RNA-seq data from the Synapse portal (syn3157743, accessed May 2018) from the Advanced Medicine Partnership for AD: Mount Sinai Brain Bank (MSBB) dataset. In short, this dataset was generated by sequencing RNA derived from four different cortical regions: frontal pole (Brodmann area (BM) 10), superior temporal gyrus (BM22), parahippocampal gyrus (BM36) and inferior frontal gyrus tissue (BM44) from 301 individuals. rRNAs was depleted using the Ribo-Zero rRNA Removal Kit (Human/Mouse/Rat) (Illumina, San Diego, USA). Sequencing libraries were prepared using TruSeq RNA Sample Preparation kit v2. From these libraries, rRNA-depleted 101nt single-end, and non-stranded RNA-seq data was generated via an Illumina HiSeq 2500 (Illumina, San Diego, USA)26. The average number of raw sequencing reads per individual was 35,062,514.
Alzheimer Disease Traits
In this study, we investigated differential expression and correlation of circular RNA (circRNA) expression in human cortical tissues with Alzheimer Disease (AD) case-control status, autosomal dominant Alzheimer Disease (ADAD) case-control status, and two AD quantitative traits: clinical dementia rating at expiration/death (CDR) and Braak score.
Case-control status was determined by post-mortem, neuropathological analysis of study participant brains following CERAD17 and/or Khachaturian25 criteria. ADAD status was determined via pre-mortem sequencing of APP, PSEN1, and PSEN2 genes to identify established, pathogenic mutations37. CDR is a clinical measure of cognitive impairment with a range from 0 (no dementia) to 3 (severe dementia)31. Braak score is a neuropathological measure of AD severity, as determined by the number and distribution of neurofibrillary tau tangles through the brain18. Braak scores range from 0 (absent, at most incidental tau tangles) to 6 (severe, extensive tau tangles in neocortical areas). Importantly, the neuropathological diagnoses available are based on criteria that require the presence of “neuritic” or “senile” plaques and thus individuals with neurofibrillary tau tangles but without plaques may still be considered controls. We identified a subset of the AD brains that were from individuals with pre-symptomatic or pre-clinical AD. These individuals did not have clinically significant dementia (clinical dementia rating <= 0.5, at most, very mild dementia) but their brains had evidence of AD neuropathological changes. Finally, the MSBB dataset included an additional AD neuropathological quantitative trait, mean amyloid plaque number.
Phenotype Processing
Discovery Dataset: Knight ADRC
We generated genetic ancestry covariates through principal components analysis via PLINK v1.9 software52 using previously generated GWAS data. In brief, we merged genetic microarray data from the Knight ADRC study participants with the HapMap reference panel53, filtered to only include variants with a mean allele frequency greater than 5% and a genotype rate greater than 95%, pruned to only include those variants that were not in linkage disequilibrium, and used the –pca command. We used the first two principal components to represent genetic ancestry for downstream analyses. We only included parietal cortex-derived samples for differential expression, correlation, and meta- analyses from individuals for whom all differential expression analysis covariates (post mortem interval (PMI), median transcript integrity number32 (TIN) – a measure of RNA quality, age at death (AOD), batch, sex, and genetic ancestry covariates) were available.
We excluded samples from individuals who were neuropathologically classified as controls but had mild or worse dementia (CDR >= 1), i.e. demented controls, as their dementias can be expected to have non-AD etiologies.
We excluded four samples as their circular transcriptomic profiles, as measured by the first two transcriptomic principal components, were outliers compared to the distribution of other parietal region samples.
Replication Dataset: MSBB
We downloaded additional data from the MSBB replication dataset, including clinical phenotype and RNA-seq covariates (syn12178045), whole genome sequencing (WGS) data (syn10901600), and quality control remapping data (syn12178045) from the Synapse portal (accessed, May 2018). We processed this data as follows:
Age at death (AOD) listed as ‘90+’ was reassigned as ‘90’ in order to make the variable quantitative.
Post mortem interval (PMI) was adjusted from minutes to hours in order to match the discovery dataset scale.
Number of APOE4 alleles was inferred using the WGS data based on the SNP: rs429358. After confirming that there existed a high concordance between the non-missing number of APOE4 alleles provided in the clinical covariates file and this inferred number, we used the inferred number of alleles for all downstream analyses as to increase the number of individuals with this data.
We generated genetic ancestry covariates from the MSBB WGS data through principal components analysis via PLINK v1.9 software, as with the discovery dataset.
We assigned missing batch and RIN information to files that had been resequenced using information from the original sequencing run, matching the two files on the basis of a common barcode.
Between the originally sequenced and resequenced sample, we selected the RNA-seq data with a greater number of mapped reads.
We excluded individuals and reassigned sample-swap IDs on the basis of information provided in the quality control remapping data (syn12178047) file.
We excluded samples from individuals who were neuropathologically classified as controls but had mild or worse dementia (CDR >= 1), i.e. demented controls, as their dementias can be expected to have non-AD etiologies.
We excluded five samples as their circular transcriptomic profiles, as measured by the first two transcriptomic principal components, were outliers compared to the distribution of other samples from that same cortical region.
We only included samples for differential expression, correlation, and meta-analyses from individuals for whom data for all differential expression analysis covariates (post mortem interval (PMI), median TIN, age at death (AOD), batch, sex, and genetic ancestry covariates) were available.
RNA-seq Data Processing and Alignment
In order to increase detection power, we processed and aligned RNA-seq data derived from all available samples in each dataset, not just those from samples that met inclusion criteria for downstream analyses. All RNA-seq data processing and alignment was performed blind to neuropathological case-control status.
We aligned raw sequencing reads from the discovery RNA-seq dataset to the primary assembly of the human reference genome, GRCh38, using STAR v2.5.3a27 in chimeric alignment mode using parameters suggested by the documentation of the circRNA calling software, DCC29. We first prepared an alignment index with an overhang splice junction database overhang of 150 (--sjdbOverhang 150) using the GENCODE v2628 comprehensive gene annotation. We then aligned each mate pair individually and together, for a total of 3 alignments per sample, using the following parameters:
--outSJfilterOverhangMin 15 15 15 15 --alignSJoverhangMin 15 --alignSJDBoverhangMin 15 --seedSearchStartLmax 30 --outFilterMultimapNmax 20 --outFilterScoreMin 1 --outFilterMatchNmin 1 --outFilterMismatchNmax 2 --chimSegmentMin 15 --chimScoreMin 15 --chimScoreSeparation 10 --chimJunctionOverhangMin 15
The replication MSBB RNA-seq dataset was provided as aligned and unmapped files and thus required additional processing prior to alignment. After downloading aligned and unmapped files for each sample from the Synapse web portal (syn3157743), we used Picard tools’ RevertSam, FastqToSam, and MergeSamFiles (http://broadinstitute.github.io/picard/) functions to generate raw, unaligned files. We aligned these generated files as above using STAR v2.5.3a but with an alignment index suitable for 101n reads (--sjdbOverhang 100) and only once per sample due to its single-ended nature.
For all alignments, we soft-clipped any adapter sequence from the reads based on the generic Illumina adapter sequence.
Calling circRNA-defining backsplices
We used DCC software v0.4.429 to detect, annotate, quantify, filter, and call circRNA-defining backsplices from the chimeric junctions identified during STAR alignment. We performed additional filtering following DCC software documentation: backsplice junctions were excluded if they were located in repetitive regions of the genome (as defined in the UCSC Genome Browser: RepeatMasker and Simple Repeats tables), spanned multiple gene annotations, or were located in the mitochondrial chromosome. When analyzing paired-end data, DCC software takes into account chimeric junctions identified in both mates individually and together to improve sensitivity. DCC software can also assign the circRNA strand of origin based on sequence if it is provided with non-stranded data.
For the discovery dataset, we ran DCC in paired-end, stranded mode with the following parameters:
-D -R GRCh38_Repeats_simpleRepeats_RepeatMasker.gtf -an gencode.v26.primary_assembly.annotation.gtf -Pi -F -M -Nr 1 1 -fg -G -A GRCh38.primary_assembly.genome.fa
For the replication dataset, we ran DCC in single-end, non-stranded mode with the following parameters:
-D -N -R GRCh38_Repeats_simpleRepeats_RepeatMasker.gtf -an gencode.v26.primary_assembly.annotation.gtf -F -M -Nr 1 1 -fg -G -A GRCh38.primary_assembly.genome.fa
We also called backsplices using an additional software package, circRNA_finder3, observing an average Pearson correlation of 0.99 between the counts called by the two methods. Similar to DCC, circRNA_finder calls backsplices from the chimeric junctions identified via STAR, but does not have parameters to adjust for type of RNA-seq data. Due to this limitation, the DCC-called backsplices were retained for downstream analyses. Backsplice calling was performed blind to neuropathological case-control status.
Filtering and collapsing annotated backsplices to identify high-confidence circRNAs
circRNAs are detected in RNA-seq data by calling backsplices from chimeric junctions. Such junctions can form artifactually during library preparation via a template switching process54. As these artifactual junctions are formed randomly, filtering called backsplices by the number of samples in which they are observed as well as the minimum ratio of linearly-aligning versus chimerically-aligning reads (circ:linear ratio) at each backsplice junction allows for the selection of a high-confidence set of backsplices. In order to empirically determine the number of samples and circ:linear ratio filtering thresholds, we called artifactual backsplices identified in spiked-in linear (External RNA Controls Consortium) ERCC51 RNAs from our discovery dataset. As these spike-in RNAs are linear, backsplices identified in ERCC sequences are expected to arise artifactually during the library preparation. As before, we aligned the raw sequencing reads using STAR v2.5.3a using the same parameters as the discovery dataset but used the ERCC92 fasta and gtf files (Invitrogen, Carlsbad, USA) rather than the human reference genome files, in order to identify the artifactual junctions. We also used DCC in stranded, paired-end mode, but without filtering for human genome annotations. As expected, we were able to detect artifactual backsplices in the ERCC spike-in RNA (Supplementary Table 6 and Supplementary Figure 11). Based on this data, we selected a highly conservative threshold of being observed in at least 3 samples and having a minimum circ:linear ratio of 0.1 for inclusion in downstream analyses.
In our discovery, parietal cortex dataset, the majority (5,090/7,450) of the backsplice junctions we identified using this calling and filtering approach have been previously identified using a different calling algorithm in an independent analysis of healthy parietal cortex tissue10,55. After identifying high-confidence backsplice junctions, we collapsed each of them on to its annotated linear gene of origin / cognate linear mRNA for downstream differential expression and correlation analyses. Backsplices without a linear gene of origin annotation were excluded from the analysis. For the MSBB replication dataset circRNA calls - which are derived from non-stranded data - we updated the strand and linear gene of origin annotation to match that of the stranded parietal dataset, but only if the backsplice calls had the same chromosome, start, and end positions.
Overall, we called a total of 3,547 well-supported circRNAs in the discovery dataset and 4,330 in the larger replication dataset. There were 3,146 well-supported circRNAs common to both the discovery and replication datasets. We visualized the overlap between the circRNAs called in each dataset using the Venn tool at: http://bioinformatics.psb.ugent.be/webtools/Venn/ (Supplementary Figure 1). All circRNA identification was performed blind to neuropathological case-control status.
Calling linear transcripts
We called linear transcripts using Salmon software v0.8.256 in quasi-mapping-based alignment mode. In short, we generated a quasi-mapping index using the primary assembly of the human reference genome, GRCh38, and the GENCODE v2628 comprehensive gene annotation. We then quantified the linear transcript expression from the raw, unaligned RNA-seq files for both the discovery and replication datasets using the default Salmon pipeline parameters. All linear transcript calling was performed blind to neuropathological case-control status.
Measuring Transcript Integrity Number
Transcript integrity number (TIN) is measure of RNA quality that is derived from the sequencing data and directly measures the degradation of mRNA32. The median TIN score for each sample has been demonstrated to have robust concordance with the RNA integrity number (RIN) – a commonly used measure of mRNA integrity based on ribosomal RNA amounts - in multiple independent RNA-seq datasets. We calculated TIN for representative, protein-coding transcripts in each sample using the RSeQC software v2.6.457 in order to provide a consistent quality control covariate for our differential expression and correlation analyses. In brief, we utilized STAR-aligned RNA-seq data and the representative (annotated as “basic”) protein-coding transcript annotations in GENCODE v26 to calculate median TIN for each sample in the discovery and replication datasets (Supplementary Table 6).
Differential expression and correlation analyses
We performed differential expression and correlation analyses between the sets of high-confidence cortical circRNA counts and AD traits using the negative binominal family logistic regression and two-tailed statistical Wald test capabilities of DESeq2 v.1.18.130. Our analysis approach follows previously published studies that include analyses of circRNA differential expression8,10. In general, differential expression analyses assume that the background distribution of RNA expression to be equivalent between samples with observed differences being attributable to adjustable technical differences (such as sequencing depth / library size or RNA quality), adjustable biological differences (such as sex or age of death), or finally due to biological traits of interest (such as disease status or severity). Our DESeq2 analysis approach takes all these factors into account. Prior to performing the logistic regression and Wald test, circRNA counts for each sample were normalized on the basis of sequencing depth / library size-derived size factor, estimated using circRNA counts from all samples derived from the same cortical region. Following this normalization, the samples were subsetted as to only include samples for which complete information – including differential expression covariate data - was available for the particular AD trait under investigation. For example, Braak score was only available for 86/96 participants in the discovery dataset and thus the sample size for discovery Braak score circRNA correlation analysis was 86. We performed all differential expression and correlation analyses with these subsets, and, in general, adjusted for the following covariates: post mortem interval (PMI), median TIN, age at death (AOD), batch, sex, and genetic ancestry - represented by the first two principal components derived from genetic data. Importantly, restricting the discovery analysis to only individuals of European genetic ancestry, i.e. dropping the 6 black individuals (5 AD cases and 1 control), yielded consistent results (effect size, Pearson correlation for CDR-associated circRNAs in the European-only vs. original discovery analysis: 0.94). We did not adjust the analyses that included ADAD samples for AOD. ADAD is early-onset37 and ADAD brains were donated by individuals who had a younger AOD compared to both control and AD participants (Supplementary Table 1), rendering AOD collinear with status. In addition, as GWAS data to calculate genetic ancestry covariates was unavailable for ADAD samples, we substituted self-reported ethnicity for genetic ancestry covariates in all analyses that included ADAD samples. We restricted analyses to only include samples for which complete information for all included differential expression covariates was available. We set a statistical significance false discovery rate (FDR) threshold of 0.05 and present uncorrected p-values, noting if they pass FDR correction. DESeq2 software automatically filters out circRNAs with low expression prior to statistical analyses.
In our discovery and ADAD datasets, we used this approach to investigate for cortical circRNAs that are significantly differentially expressed between AD versus controls and ADAD versus controls. We also investigated for cortical circRNAs that are significantly differentially expressed between ADAD versus AD, adjusting for neuropathological severity as measured by Braak score. We investigated for cortical circRNAs that were significantly correlated with CDR and similarly, investigated for circRNAs that were significantly correlated with Braak score in the discovery dataset samples for which these AD traits were available. To replicate these findings, we performed similar analyses in the MSBB datasets. We selected BM44 to be our primary replication dataset, but performed the analyses in all cortical regions separately. We investigated for differential cortical circRNA expression between definite AD versus control status, significant correlation between CDR and cortical circRNA expression, and significant correlation between Braak score and cortical circRNA expression. Finally, we performed analyses to investigate for significant correlations between circRNAs and mean number of plaques in the MSBB dataset. With the exception of invalid statistical models due to collinearity between the quality control metric and the particular AD trait under investigation, substituting median TIN or RIN quality control metrics, yielded similar differential expression and correlation results. For example, the effect size Pearson correlation for the 31 discovery analysis CDR-associated circRNAs obtained after substituting RIN for TIN is 0.99 (p-value: 5.43×10−26).
Validating RNA-seq Counts and Direction of Effect via Quantitative PCR
We designed divergent primers to the backsplice junction of circHOMER1 (Forward 5’- TTTGGAAGACATGAGCTCGA −3’; Reverse 5’- AAGGGCTGAACCAACTCAGA −3’), circKCNN2 (Forward 5’- GACTGTCCGAGCTTGTGAAA −3’; Reverse 5’- GGCCGTCCATGTGAATGTAT −3’), circMAN2A1 (Forward 5’- TGAAAGAAGACTCACGGAGGA −3’; Reverse 5’- TAGCAAACGCTCCAAATGGT −3’), circICA1 (Forward 5’- TTGATGATTTGGGGAGAAGG −3’; Reverse 5’- TGGATGAAGGACGTGTCTCA −3’), circFMN1 (Forward 5’- GGTGGCTATGCAGAGAAAGC −3’; Reverse 5’- CAGGGAAGACCACAGCTGAG −3’), circRNA transcripts based on circRNA fasta sequences extracted via the getcircfasta.py script provided with DCC software29. Divergent primers face outwards - as opposed to inward facing, typical primers – and as a result they will only produce a PCR product if there exists a backsplice junction formed via circularization of a transcript or rarely by tandem exon duplication1. We confirmed that these primers were divergent through in silico PCR (https://genome.ucsc.edu/cgi-bin/hgPcr) and confirmed that the amplication efficiency of each divergent primer pair was suitable for quantitative PCR (qPCR). We then selected 13 parietal cortex-derived RNA samples from individuals in the discovery study (3 controls, 3 PreSympAD, and 7 AD) to generate GAPDH-normalized (Forward 5’- TGCACCACCAACTGCTTAGC −3’; Reverse 5’- GCCATGGACTGTGGTCATGAG −3’) expression values to correlate with our RNA-seq-derived counts. We generated cDNA from the RNA samples using SuperScript VILO cDNA synthesis kit (Invitrogen) following the manufacturer’s recommended protocol. With this cDNA, we performed the qPCR experiment using PowerUp SYBR Green Master Mix (Applied Biosystems) on a QuantStudio 12K Flex Real-Time PCR System. We calculated the relative expression following the standard DeltaDeltaCt method. In brief, we averaged the triplicate readings of Ct for each primer pair and subtracted the average linear GAPDH Ct from the average circRNA Ct to calculate DeltaCt. We further calculated the DeltaDeltaCt of each circRNA by subtracting the average control (n=3) DeltaCt for each primer from the DeltaCts. Finally, we generated relative expression using the following formula: Relative Expression = 2−ΔΔCt.
Meta-analyses and Overlap calculations
We performed meta-analyses of the cortical circRNA differential expression and correlation discovery and replication results using the metaRNA-seq R package v1.0.2. We chose to combine the p-values of the circRNAs common to both replication and discovery results using the inverse/Stouffer method due to the differences in sample size between the datasets. As before, we set a statistical significance threshold and false discovery rate (FDR) threshold of 0.05 and present uncorrected p-values, noting if they pass FDR correction. We visualized the results of our meta-analyses using the CMplot R package v3.3.1.
We visualized overlap between meta-analysis results using the VennDiagram R package v1.6.20 and calculated significance of overlap using the SuperExactTest R package V 1.0.0, which reports one-tailed p-values58.
Independence of Circular versus Cognate Linear RNA AD-Associations or AD-Associated Changes in Estimated Brain Cell-type Proportions via Regression-Based Analyses
To demonstrate the independence of circular versus their cognate linear mRNA AD-associations, we included library-size normalized counts for the CDR-associated circRNAs and their cognate linear mRNAs in the same regression models predicting CDR. The regression models also included the differential expression covariates: PMI, median TIN, AOD, batch, sex, and genetic ancestry. Given the fact that circRNA expression levels are lower than their cognate linear mRNA expression levels, and the majority of RNA-sequencing reads covering a circRNA will not include the backsplice (thereby inflating the cognate linear mRNA counts); we consider circRNAs to demonstrate an independent association with CDR if they retain a significant (p-value < 0.05) association in the combined regression model. We perform these regression analyses for the CDR-associated circRNAs in both the discovery and replication datasets and combine the results using a fixed effects meta-analysis. In addition, we calculate the proportion of variation in CDR explained by circRNAs versus their cognate linear mRNAs38 and present the average proportion of variation explained in the two datasets. Two of 148 meta-analysis, CDR-associated circRNAs did not have a cognate linear RNA and were excluded from these analyses.
We demonstrated the independence of circRNA AD-associations from AD-associated changes in brain cell-type proportions using a similar regression-based approach. We included library-size normalized counts for the CDR-associated circRNAs and computationally-deconvoluted34 estimated proportions of neurons, oligodendrocytes, and microglia. We did not include the deconvoluted estimated astrocyte proportion to avoid multicollinearity and also because we have previously reported that astrocyte and neuron estimated proportions are strongly inversely correlated34. AD-associated circRNAs that retained a significant (p-value < 0.05) association in these combined models are considered independent. We perform these regression analyses for all 148 CDR-associated circRNAs in both the discovery and replication datasets and combine the results using a fixed effects meta-analysis.
Pre-symptomatic AD Bootstrapped Correlation Coefficient Analyses
In our discovery and replication datasets, a small number of individuals with pre-symptomatic AD (PreSympAD) – i.e., neuropathological evidence of AD but, at most, very mild dementia (CDR <= 0.5) were included. We investigated if changes in expression in the PreSympAD brains were similar to the changes observed in symptomatic AD (SympAD) – i.e., neuropathological evidence of AD and dementia (CDR >= 1).
We first performed a cortical circRNA differential expression analysis between SympAD versus controls and then between PreSympAD versus controls, using the same methods as described above. Then, for all circRNAs that were not automatically filtered out by DESeq2 due to low expression, we calculated the correlation between the log2 fold change (log2FC, effect size) observed in the PreSympAD analysis and the log2FC observed in the SympAD analysis. If the SympAD versus control brain differentially expressed circRNAs demonstrate similar changes in expression in the PreSympAD, we expect the correlation between the log2FC values for these circRNAs to be stronger than those from the non-significant, background circRNAs. We tested this by performing 10,000 bootstrap simulations to identify a bias corrected and accelerated35 95% confidence interval for the two log2FC correlation coefficients – one for the SympAD-associated circRNAs and the other for the non-significant, background circRNAs. We generated p-values for the significantly associated distribution being higher than the background distribution using a one-tailed Kolmogorov–Smirnov test. We performed this analysis in the discovery dataset and in all cortical regions in the replication dataset to assess for regional differences in circRNA expression changes in PreSympAD. Bootstrap correlation coefficients and confidence intervals were generated using the boot R package V1.3–20.
Receiver Operating Characteristic (ROC) curve and Area under the curve (AUC) analyses
To evaluate the predictive ability of AD-associated circRNAs, we calculated logistic regression models predicting AD case status in both the discovery and replication datasets. We subsetted each dataset as to only include definite AD cases and controls and calculated three models. The first model (base) included the following as covariates: PMI, median TIN, AOD, batch, sex, genetic ancestry, and number of APOE4 alleles. The second model (circ) included the top 10 most significantly CDR-associated circRNAs from the meta-analysis. The third model (base+circ) combined the variables of the first two models together. We calculated ROC curves and AUCs using the R package pROC V1.12.1.
Relative importance analyses
The number of APOE4 alleles – the most common genetic risk factor for AD16 – and the estimated proportion of neurons34 are known to contribute to the observed variation in AD quantitative traits like CDR and Braak score. We assessed the relative importance of circRNA expression compared to these known contributors using the relaimpo R package, v2.2.338. To do this, we first selected the library-size normalized counts of the top 10 most significant AD-trait associated circRNAs and adjusted them for the same covariates used in the differential expression analyses: PMI, median TIN, AOD, batch, sex, and genetic ancestry. We then included these normalized, adjusted counts, first individually and then together in a multivariate model, with number of APOE4 alleles and estimated neuronal proportion in the same linear regression model predicting either CDR or Braak score, or mean number of plaques (only available in the replication MSBB dataset). We assessed the relative contribution of each of the model variables to the variation in the predicted AD quantitative trait using the lmg method of the relaimpo package. Thus, we measured the contribution of each of the top 10 most meta-analysis significant circRNAs compared to number of APOE4 alleles and estimated neuronal proportion both individually and when included together in the same model. We conducted these analyses in both the discovery dataset as well as all 4 cortical regions in the replication dataset, selecting the top 10 most meta-analysis significant circRNAs from each region-specific meta-analysis.
Network Co-expression Analyses
We computed circRNA and protein-coding linear transcript co-expression networks from AD and control samples in order to infer the biological and pathological relevance of circRNAs based on the linear transcripts they co-expressed with. We first adjusted library size-normalized, circRNA and linear transcript counts, from the same samples, for the differential-expression analyses covariates – PMI, median TIN, AOD, batch, sex, and genetic ancestry – and then combined them together. We included all circRNAs and the top 10,000 most variable protein-coding linear transcripts to reduce computational burden. We computed gene co-expression networks from these combined counts based on Spearman correlation using multiscale embedded gene co-expression network analysis (MEGENA, v1.3.639). Briefly, this method leverages planar maximally filtered graph techniques to identify compact gene expression networks and has been independently demonstrated to have high module conservation with, and to identify more modules than the older WGCNA method59. Importantly, this method identifies hierarchical networks with submodules existing within larger parent modules, when possible. As such the same linear transcript or circRNA may be assigned to multiple modules. Following module identification, we calculated each modules’ eigengene using the WGCNA R package v1.6340. To identify significant associations between modules and CDR, we performed two-tailed, p-value generating regression analyses between the module eigengenes and CDR adjusting for the differential expression covariates. Significance of the module eigengene association with CDR was determined using a two-tailed t-test. We identified significant gene enrichment and pathway associations for each module by extracting the linear transcript module members and processing them through the FUMA software’s hypergeometric – one-tailed – test60, with protein coding genes as the background gene list. Finally, we visualized networks using the igraph R package v1.2.1.
MicroRNA Binding Site Prediction
We generated a fasta file of circRNA sequences using the getcircfasta.py script provided with DCC software29. We predicted microRNA (miRNA) binding sites in these circRNA sequences using the targetscan_70.pl script provided with the TargetScan70 database42, March 2018 release. When multiple isoforms of the same circRNA were predicted to have different number of binding sites for the same miRNA, we selected the greatest number of predicted binding sites to present at the gene-level. We identified predicted targets of miRNA regulation from the March 2018 release of the TargetScanHuman database42.
Statistical Analysis
We tested for differential expression of circRNAs using DESeq2 v.1.18.130 to perform negative binominal family logistic regressions and a two-tailed Wald test to determine significance. We tested for circRNA association effect size correlations using Pearson correlation with significance determined by a two-tailed t-test. We demonstrated the independence of circRNA AD-associations from AD-associated changes in cognate linear mRNAs or AD-associated changes in estimated brain cell type proportions using linear regression analyses with significance determined by two-tailed t-tests. We calculated one-tailed p-values for the significance of overlap between different sets of differentially expressed circRNAs using the SuperExactTest R package V 1.0.058. We calculated whether bootstrapped effect size correlation distributions between SympAD-associated circRNAs was greater than the background distribution using a one-tailed Kolmogorov-Smirnov test. We calculated the proportion of variation in quantitative AD traits explained by circRNAs and other contributors using linear regression followed by relative importance analysis done using the relaimpo R package, v2.2.338. We generated circRNA and linear mRNA co-expression network modules based on Spearmann correlation using MEGENA, v1.3.639. We calculated module eigengenes and determined their association with CDR using linear regression with significance determined by two-tailed t-tests. Co-expression module enrichment for AD-related pathways was determined using a one-tailed hypergeometric test performed by FUMA software60. For parametric tests, data distribution was assumed to be normal but this was not formally tested. All statistical analysis was done using R statistical software61.
A Life Sciences Reporting Summary for our manuscript is available.
Data Availability
Knight ADRC dataset - NG00083 (https://www.niagads.org/datasets/ng00083)
Sequencing information derived from ADAD samples is protected and requires additional authorization from DIAN for access.
Mount Sinai Brain Bank, replication dataset: https://www.synapse.org/#!Synapse:syn3159438
Supplementary Material
Acknowledgements
We thank all the participants and their families, as well as the many institutions and their staff.
Funding: This work was supported by grants from the National Institutes of Health (R01AG044546, P01AG003991, RF1AG053303, R01AG058501, U01AG058922, RF1AG058501 and R01AG057777), the Alzheimer Association (NIRG-11-200110, BAND-14-338165, AARG-16-441560 and BFG-15-362540), NIH AG046374 (CMK), Tau Consortium (CMK), K23 AG049087 (JPC).
The recruitment and clinical characterization of research participants at Washington University were supported by NIH P50 AG05681, P01 AG03991, and P01 AG026276.
This work was supported by access to equipment made possible by the Hope Center for Neurological Disorders, and the Departments of Neurology and Psychiatry at Washington University School of Medicine.
The results published here are in part based on data obtained from the AMP-AD Knowledge Portal accessed via the cited accession numbers.
MSBB: These data were generated from postmortem brain tissue collected through the Mount Sinai VA Medical Center Brain Bank and were provided by Eric Schadt from Mount Sinai School of Medicine.
DIAN: Data collection and sharing for this project was supported by The Dominantly Inherited Alzheimer’s Network (DIAN, UF1AG032438) funded by the National Institute on Aging (NIA), the German Center for Neurodegenerative Diseases (DZNE), Raul Carrea Institute for Neurological Research (FLENI), Partial support by the Research and Development Grants for Dementia from Japan Agency for Medical Research and Development, AMED, and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI). This manuscript has been reviewed by DIAN Study investigators for scientific content and consistency of data interpretation with previous DIAN Study publications. We acknowledge the altruism of the participants and their families and contributions of the DIAN research and support staff at each of the participating sites for their contributions to this study.
Footnotes
DIAN Consortium Members: Ricardo Allegri12, Fatima Amtashar13, Tammie Benzinger13, Sarah Berman14, Courtney Bodge15, Susan Brandon13, William (Bill) Brooks16, Jill Buck17, Virginia Buckles13, Sochenda Chea18, Patricio Chrem12, Helena Chui19, Jake Cinco20, Jack Clifford18, Mirelle D’Mello16, Tamara Donahue13, Jane Douglas20, Noelia Edigo12, Nilufer Erekin-Taner18, Anne Fagan13, Marty Farlow17, Angela Farrar13, Howard Feldman21, Gigi Flynn13, Nick Fox20, Erin Franklin13, Hisako Fujii22, Cortaiga Gant13, Samantha Gardener23, Bernardino Ghetti17, Alison Goate24, Jill Goldman25, Brian Gordon13, Julia Gray13, Jenny Gurney13, Jason Hassenstab13, Mie Hirohara26, David Holtzman13, Russ Hornbeck13, Siri Houeland DiBari27, Takeshi Ikeuchi28, Snezana Ikonomovic14, Gina Jerome13, Mathias Jucker29, Kensaku Kasuga28, Takeshi Kawarabayashi26, William (Bill) Klunk14, Robert Koeppe30, Elke Kuder-Buletta29, Christoph Laske29, Johannes Levin27, Daniel Marcus13, Ralph Martins23, Neal Scott Mason32, Denise Maue-Dreyfus13, Eric McDade13, Lucy Montoya19, Hiroshi Mori22, Akem Nagamatsu33, Katie Neimeyer25, James Noble25, Joanne Norton13, Richard Perrin13, Marc Raichle13, John Ringman19, Jee Hoon Roh31, Peter Schofield16, Hiroyuki Shimada22, Tomoyo Shiroto26, Mikio Shoji26, Wendy Sigurdson13, Hamid Sohrabi23, Paige Sparks36, Kazushi Suzuki33, Laura Swisher13, Kevin Taddei23, Jen Wang24, Peter Wang13, Mike Weiner37, Mary Wolfsberger13, Chengjie Xiong13, Xiong Xu13
12FLENI Institute of Neurological Research (Fundacion para la Lucha contra las Enfermedades Neurologicas de la Infancia)
13Washington University in St. Louis School of Medicine
14University of Pittsburgh
15Brown University-Butler Hospital
16Neuroscience Research Australia
17Indiana University
18Mayo Clinic Jacksonville
19University of Southern California
20University College London
21University of California San Diego
22Osaka City University
23Edith Cowan University, Perth
24Icahn School of Medicine at Mount Sinai
25Columbia University
26Hirosaki University
27German Center for Neurodegenerative Diseases (DZNE) Munich
28Niigata University
29German Center for Neurodegnerative Diseases (DZNE) Tubingen
30University of Michigan
31Asan Medical Center
32University of Pittsburgh Medical Center
33Tokyo University
36Brigham and Women’s Hospital-Massachusetts
37University of California San Francisco
Competing interests: CC receives research support from: Biogen, EISAI, Alector and Parabon. The funders of the study had no role in the collection, analysis, or interpretation of data; in the writing of the report; or in the decision to submit the paper for publication. CC is a member of the advisory board of Vivid genetics, Halia Therapeutics and ADx Healthcare.
Accession codes:
Knight ADRC Parietal Cortex Dataset – NG00083 (https://www.niagads.org/datasets/ng00083)
MSBB Dataset - syn3159438 (https://www.synapse.org/#!Synapse:syn3159438)
References
- 1.Barrett SP & Salzman J Circular RNAs: analysis, expression and potential functions. Development 143, 1838–1847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X, Yang L & Chen L-L The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 71, 428–442 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Westholm JO et al. Genome-wide Analysis of Drosophila Circular RNAs Reveals Their Structural and Sequence Properties and Age-Dependent Neural Accumulation. Cell Rep. 9, 1966–1980 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gruner H, Cortés-López M, Cooper DA, Bauer M & Miura P CircRNA accumulation in the aging mouse brain. Sci. Rep 6, 38907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Memczak S et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Salzman J, Gawad C, Wang PL, Lacayo N & Brown PO Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One 7, e30733 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maass PG et al. A map of human circular RNAs in clinically relevant tissues. J. Mol. Med 1–11 (2017). doi: 10.1007/s00109-017-1582-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.You X et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci 18, 603–610 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashwal-Fluss R et al. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56, 55–66 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Rybak-Wolf A et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 58, 870–885 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Liang D et al. The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Mol. Cell 0, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venø MT et al. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 16, 245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Legnini I et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 66, 22–37.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pamudurti NR et al. Translation of CircRNAs. Mol. Cell 66, 9–21.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen TB et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Scheltens P et al. Alzheimer’s disease. The Lancet 388, 505–517 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Mirra SS et al. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology 41, 479–486 (1991). [DOI] [PubMed] [Google Scholar]
- 18.Braak H, Alafuzoff I, Arzberger T, Kretzschmar H & Del Tredici K Staging of Alzheimer disease-associated neurofibrillary pathology using paraffin sections and immunocytochemistry. Acta Neuropathol. (Berl.) 112, 389–404 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang B et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karch CM et al. Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PloS One 7, e50976 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raj T et al. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat. Genet 50, 1584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verheijen J & Sleegers K Understanding Alzheimer Disease at the Interface between Genetics and Transcriptomics. Trends Genet. TIG 34, 434–447 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Piwecka M et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357, eaam8526 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Lukiw WJ Circular RNA (circRNA) in Alzheimer’s disease (AD). Front. Genet 4, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khachaturian ZS Diagnosis of Alzheimer’s disease. Arch. Neurol 42, 1097–1105 (1985). [DOI] [PubMed] [Google Scholar]
- 26.Wang M et al. The Mount Sinai cohort of large-scale genomic, transcriptomic and proteomic data in Alzheimer’s disease. Sci. Data 5, 180185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinforma. Oxf. Engl 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrow J et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng J, Metge F & Dieterich C Specific identification and quantification of circular RNAs from sequencing data. Bioinforma. Oxf. Engl 32, 1094–1096 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris JC The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology 43, 2412–2414 (1993). [DOI] [PubMed] [Google Scholar]
- 32.Wang L et al. Measure transcript integrity using RNA-seq data. BMC Bioinformatics 17, 58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrett SP, Parker KR, Horn C, Mata M & Salzman J ciRS-7 exonic sequence is embedded in a long non-coding RNA locus. PLoS Genet. 13, e1007114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z et al. Genetic variants associated with Alzheimer’s disease confer different cerebral cortex cell-type population structure. Genome Med. 10, 43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carpenter J & Bithell J Bootstrap confidence intervals: when, which, what? A practical guide for medical statisticians. Stat. Med 19, 1141–1164 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Schroeter ML et al. Executive deficits are related to the inferior frontal junction in early dementia. Brain 135, 201–215 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bateman RJ et al. Autosomal-dominant Alzheimer’s disease: a review and proposal for the prevention of Alzheimer’s disease. Alzheimers Res. Ther 3, 1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Relative Importance for Linear Regression in R: The Package relaimpo | Groemping | Journal of Statistical Software. doi: 10.18637/jss.v017.i01 [DOI] [Google Scholar]
- 39.Song W-M & Zhang B Multiscale Embedded Gene Co-expression Network Analysis. PLOS Comput. Biol 11, e1004574 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langfelder P & Horvath S WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai Y et al. Circular RNA DLGAP4 Ameliorates Ischemic Stroke Outcomes by Targeting miR-143 to Regulate Endothelial-Mesenchymal Transition Associated with Blood-Brain Barrier Integrity. J. Neurosci. Off. J. Soc. Neurosci 38, 32–50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agarwal V, Bell GW, Nam J-W & Bartel DP Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y et al. MiR-136 promotes apoptosis of glioma cells by targeting AEG-1 and Bcl-2. FEBS Lett. 586, 3608–3612 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Lee TI & Young RA Transcriptional Regulation and Its Misregulation in Disease. Cell 152, 1237–1251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kljajevic V, Grothe MJ, Ewers M & Teipel S Distinct pattern of hypometabolism and atrophy in preclinical and predementia Alzheimer’s disease. Neurobiol. Aging 35, 1973–1981 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Liang WS et al. Alzheimer’s disease is associated with reduced expression of energy metabolism genes in posterior cingulate neurons. Proc. Natl. Acad. Sci 105, 4441–4446 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perkins M et al. Altered Energy Metabolism Pathways in the Posterior Cingulate in Young Adult Apolipoprotein E ɛ4 Carriers. J. Alzheimers Dis 53, 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith R et al. Posterior Accumulation of Tau and Concordant Hypometabolism in an Early-Onset Alzheimer’s Disease Patient with Presenilin-1 Mutation. J. Alzheimers Dis. JAD 51, 339–343 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Mosconi L et al. Hypometabolism exceeds atrophy in presymptomatic early-onset familial Alzheimer’s disease. J. Nucl. Med. Off. Publ. Soc. Nucl. Med 47, 1778–1786 (2006). [PubMed] [Google Scholar]
- 50.Li Y et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25, 981–984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-only - REFERENCES
- 51.Pine PS et al. Evaluation of the External RNA Controls Consortium (ERCC) reference material using a modified Latin square design. BMC Biotechnol. 16, 54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gibbs RA et al. The International HapMap Project. Nature 426, 789–796 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Tang C et al. Template switching causes artificial junction formation and false identification of circular RNAs. bioRxiv 259556 (2018). doi: 10.1101/259556 [DOI] [Google Scholar]
- 55.Glažar P, Papavasileiou P & Rajewsky N circBase: a database for circular RNAs. RNA (2014). doi: 10.1261/rna.043687.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patro R, Duggal G, Love MI, Irizarry RA & Kingsford C Salmon: fast and bias-aware quantification of transcript expression using dual-phase inference. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L, Wang S & Li W RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Wang M, Zhao Y & Zhang B Efficient Test and Visualization of Multi-Set Intersections. Sci. Rep 5, 16923 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chella Krishnan K et al. Integration of Multi-omics Data from Mouse Diversity Panel Highlights Mitochondrial Dysfunction in Non-alcoholic Fatty Liver Disease. Cell Syst. 6, 103–115.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watanabe K, Taskesen E, van Bochoven, A. & Posthuma D Functional mapping and annotation of genetic associations with FUMA. Nat. Commun 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/ (2018). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Knight ADRC dataset - NG00083 (https://www.niagads.org/datasets/ng00083)
Sequencing information derived from ADAD samples is protected and requires additional authorization from DIAN for access.
Mount Sinai Brain Bank, replication dataset: https://www.synapse.org/#!Synapse:syn3159438