Abstract
A hallmark of the immune system is the interplay among specialized cell types transitioning between resting and stimulated states. The gene regulatory landscape of this dynamic system has not been fully characterized in human cells. Here, we collected ATAC-seq and RNA-seq data under resting and stimulated conditions for up to 32 immune cell populations. Stimulation caused widespread chromatin remodeling, including response elements shared between stimulated B and T cells. Furthermore, several autoimmune traits showed significant heritability in stimulation-responsive elements from distinct cell types, highlighting the importance of these cell states in autoimmunity. Use of allele-specific read-mapping identified variants that alter chromatin accessibility in particular conditions, allowing us to observe evidence of function for a candidate causal variant that is undetected by existing large-scale studies in resting cells. Our results provide a resource of chromatin dynamics and highlight the need for characterization of effects of genetic variation in stimulated cells.
Editorial summary:
Analysis of gene expression and open chromatin regions in up to 32 immune cell populations under resting and stimulated conditions identifies widespread chromatin remodeling and shared response elements between stimulated B and T cells.
Introduction
Immune cells respond to stimuli with stereotyped transcriptional programs that enable specialized functions during an immune response. These programs, essential for immune homeostasis, are coordinated by precise interactions of transcription factors (TFs) that bind genomic sites to influence chromatin landscape and ultimately gene expression. Tight regulation of these programs is required for an appropriate immune response against cancer and infections, and the avoidance of autoimmunity. Genetic variation in regulatory regions that tune transcriptional programs can contribute to the risk of human autoimmune diseases.
Genome-wide association studies (GWAS) have identified hundreds of genetic variants that contribute to the risk of autoimmunity. Roughly 90% of these signals lie in non-coding regions, and thus presumably act by altering gene regulation, however most of these remain difficult to interpret1. Several studies have reported enrichment of variants linked to risk of immune-mediated disorders at key enhancers and cell-specific expression quantitative trait loci (eQTLs), suggesting potential mechanisms by which non-coding variants contribute to disease pathology1–7. Nonetheless, only a minority, perhaps 25%, of GWAS signals can currently be explained through known eQTLs8.
Several groups have shown that additional GWAS-eQTL overlap can remain hidden within stimulation-specific functional regions of immune cells1,9–13. Probing these response-specific functional regions can reveal previously undetected disease-associated mechanisms, emphasizing the unique role of stimulation to autoimmunity. For example, our group discovered a stimulation-response regulatory element which, when perturbed, resulted in a dysregulated immune response by delaying IL2RA expression in vivo14. This region harbors a fine-mapped GWAS variant linked to inflammatory bowel disease and type 1 diabetes15–17, thus connecting immune response to autoimmunity. However, these studies have been performed on few stimulated cell types. Thus, we lack a comprehensive view of the effects of stimulation on the chromatin landscape of diverse immune cell types, and the role of SNPs in these regions on autoimmune disease.
To address this gap, we developed a resource of chromatin accessibility and gene expression from 25 primary human immune cell types isolated from four human blood donors, in both resting and activated states. Additionally, to further understand chromatin structure in cells important for T cell development we included six thymocyte subsets and thymic epithelial cells (TECs) collected from fetal thymus samples.
Overall, we observed features of the chromatin landscape that depend upon cell lineage and response to stimulation. Notably, B and T cell subsets shared a significant proportion of these stimulation-responsive chromatin regions. Integrating these data with autoimmune disease GWAS, we found that stimulation-responsive chromatin regions explained significant trait heritability in multiple immune cell types, indicating distinct lineage contributions to autoimmunity. Finally, we leveraged stimulation-responsive chromatin elements and allele-specific imbalance of chromatin accessibility as a functional readout of variants from individual donors to identify new autoimmunity-related mechanisms. As proof-of-concept for the power of this approach, we found evidence of function for a candidate causal SNP that is associated with both rheumatoid arthritis and ulcerative colitis (rs6927172 - NC_00006.10:g. 138002175C>G), which putatively regulates the expression of TNFAIP3.
Results
An atlas of immune cells in resting and stimulated states
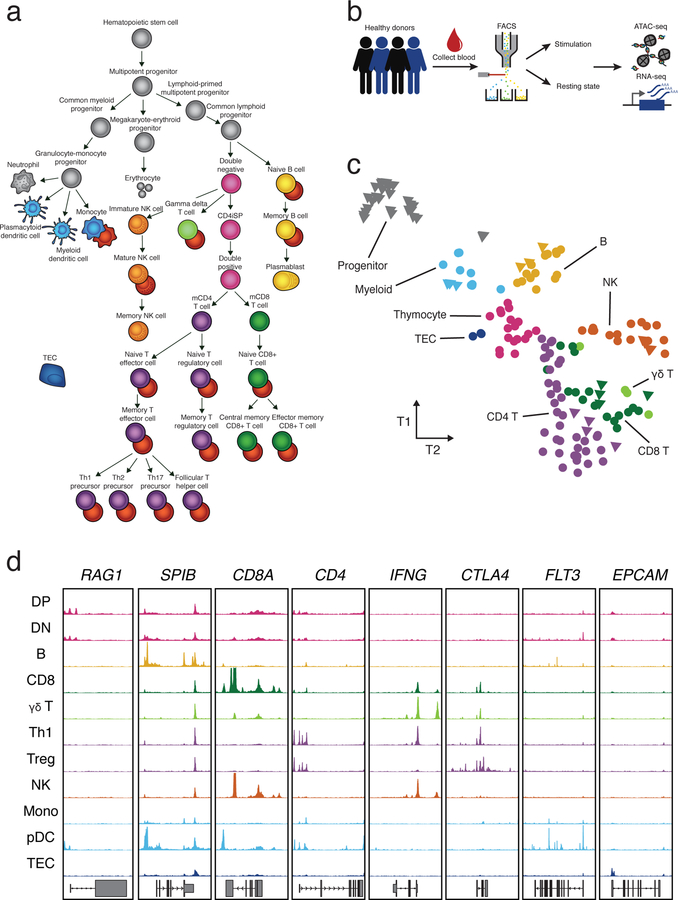
To identify regulatory elements underlying differentiation and stimulation responses in the human immune system, we generated a map of chromatin accessibility and gene expression in resting and stimulated immune cells (Fig. 1a). We isolated 25 specialized immune cell types by flow cytometry from peripheral blood of up to four healthy donors including different subsets of B cells, CD4+ T cells, CD8+ T cells, γδ T cells, monocytes, dendritic cells (DCs) and natural killer cells (NK) (Supplementary Fig. 1, Supplementary Table 1). Additionally, we collected fetal thymus samples from three donors and isolated a number of thymocyte subsets, including double positive (DP), double negative (DN), pre-T double negative (pre-T DN), immature single positive CD4+ (CD4+ iSP) and single positive mature CD4+ and CD8+ T cells, as well as thymic epithelial cells (TEC).
Fig. 1: Study workflow and tSNE of ATAC-seq data.
a, Illustration of isolated cell types, which include cell types that were previously published (gray) and resting (colored) and stimulated (red) immune cells obtained from this study. b, Schematic of sample processing pipeline. Immune cells were sorted by flow cytometry from up to four healthy donors, activated and subjected to ATAC-seq and RNA-seq. c, Exploratory tSNE of ATAC-seq chromatin accessibility from all cell types in a resting state. Each sample is colored by broad cell lineage. Samples for each cell type from different donors are plotted separately (counts can be found in Supplementary Table 1). Triangles represent previously published data19 and circles represent data generated in this study. d, Representative ATAC-seq profiles (y-axis = 0 to 400 RPKM) at several cell type-specific genes (see Methods). Cell type abbreviations listed in Supplementary Table 1.
We performed an assay for transposase-accessible chromatin using sequencing (ATAC-seq), which profiles chromatin-accessible regions as a sequencing depth read out18, and RNA-seq on isolated cell types. Details further verifying the quality of the samples can be found in the Supplementary Note. On average, across all resting cell type samples, we identified 36,810 accessible peaks (q-value < 0.05), controlling for the effects of read depth and sample quality.
Unsupervised clustering of ATAC-seq identified distinct chromatin signatures for different lineages (Fig. 1c, Supplementary Fig. 2c). For example, hematopoietic stem cell progenitors clustered separately from more differentiated immune cell types (data from Corces, et al.19). T cell subsets including CD4+, CD8+ and γδ T cells clustered closely together, indicating similar chromatin accessibility profiles for each of them under resting conditions. Circulating mature T cells could be distinguished from their single positive thymocyte precursors, suggesting further chromatin remodeling in the periphery. Finally, samples generally clustered as expected with published data from the same cell types (Fig. 1c).
The same major cell type clusters observed in the chromatin accessibility data were recapitulated when we clustered RNA-seq samples (Supplementary Fig. 2c,d). However, cell type clustering accuracy was higher when using chromatin accessibility than when using gene expression (mean HA-adjusted RAND index 0.19 and 0.12 respectively), consistent with previous analyses19.
Identifying immune memory-associated accessible regions
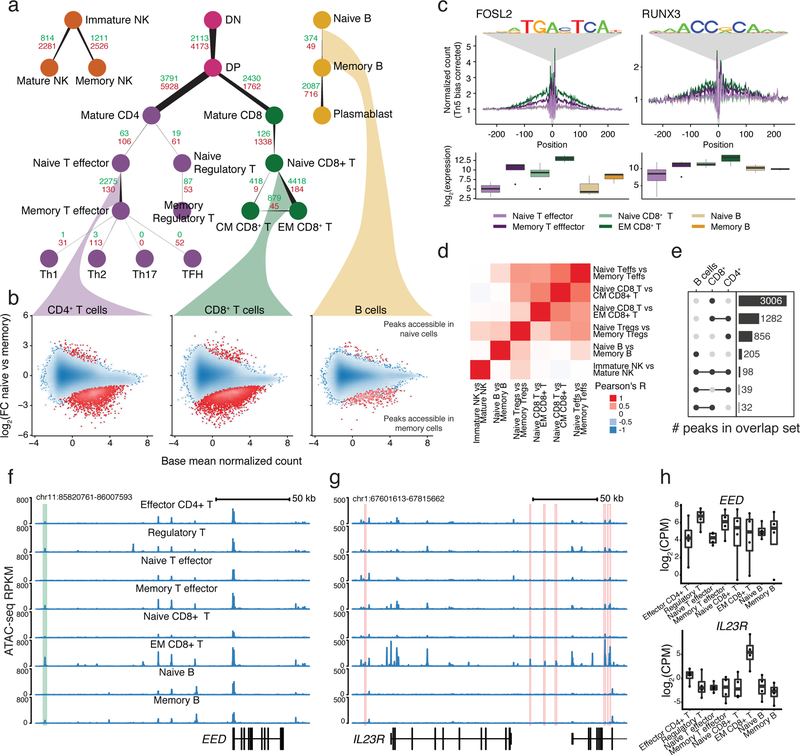
Taking advantage of the variety of cell types profiled, we quantified changes in chromatin accessibility and gene expression along paths of lymphoid cell differentiation in resting cell types (Fig. 2a; Supplementary Data 1 and 2).
Fig. 2: Identification of accessible regions associated with memory.
a, Number of differentially accessible regions when comparing cell subtypes to their progenitors. The count of regions that gain versus lose accessibility are labeled green and red, respectively. Edge starting and ending widths are proportional to the number of peaks losing and gaining accessibility. b, Example MA-plots comparing accessibility during the transition from naïve to memory states. Point density is shown as blue shading. The 0.1% of points in the least densely populated regions of the plot are shown as separate points. c, Transcription factor footprints showing genome-wide aggregate accessibility at PWM-predicted binding sites stratified by cell subset and with the distribution of normalized RNA expression of corresponding genes. d, Heatmap displaying Pearson’s R correlation between log2(FC) estimates of memory-associated chromatin changes between cell subsets. e, UpSet51 plot of the number of shared or unique regions that gain accessibility in memory formation in T cell and B cell lineages. f, ATAC-seq profile highlighting a region that increases in accessibility upon transition to memory that is shared among multiple cell types. g, ATAC-seq profile highlighting a region associated with effector memory CD8+ T cells that contains a GWAS variant linked with either Crohn’s disease, ankylosing spondylitis, or primary biliary cirrhosis. h, Boxplot (see Methods) of the gene expression of the genes highlighted in (f) and (g). All comparisons were performed on cells in a resting state and the number of samples used is listed in Supplementary Table 1.
Focusing on immune memory formation in different lineages, we found that B and T cells gained accessible chromatin regions as they mature from naïve to memory cells (Fig. 2a,b; Supplementary Note). To discover factors that potentially drive these chromatin changes, we scanned for transcription factor motifs within accessible regions genome-wide. We observed an enrichment of binding sites for known immune regulators such as JUN/FOS and RUNX3 (Fig. 2c), in addition to increased expression of these transcription factors in memory cells compared to naïve cells (Supplementary Fig. 4b)20.
Comparing memory-associated signatures of chromatin accessibility between different lineages, we observed concordant changes between different T cell subsets and a marginal overlap with the accessibility dynamics during B cell memory formation, quantified in terms of Pearson’s R correlation coefficients as well as overlap between sets of differentially accessible peaks (Fig. 2d,e).
In accordance with previous studies21–23, many genes which are dynamic during memory formation are associated with genomic regions that are putatively regulatory. For example, we found regions exhibiting increased accessibility in memory cells of the B and T lineage compared to naïve cells upstream of the chromatin regulator gene EED, which also correlated with increased expression in memory cells (Fig. 2f,h).
To highlight the importance of these regions for understanding autoimmunity, we integrated our estimates of memory-associated accessible regions and differentially expressed genes with a database of GWAS variants24. As an example, we found several regions that increase in accessibility around the IL23R gene in effector memory CD8+ T cells that contained variants associated with several autoimmune traits (Fig. 2g, Supplementary Fig. 4a). Moreover, IL23R was significantly upregulated in effector memory CD8+ T cells (Fig. 2h). Thus, dysregulation of immune memory-associated chromatin accessible regions represents a potentially important effect on autoimmunity.
Stimulation leads to large-scale chromatin changes
Previous studies have shown large-scale chromatin remodeling upon stimulation of individual cell types9,10,25,26. We set out to perform a comprehensive analysis of stimulation-dependent chromatin and gene expression changes across a wide range of cell types and lineages. To investigate these effects, we ex vivo stimulated the majority of the collected cell types and performed ATAC-seq and RNA-seq.
Our aim was to provide a strong stimulus to each cell type in order to measure chromatin dynamics of strongly activated cells. Subsets from distinct lineages were activated with distinct stimuli to induce biologically relevant responses: T cell subsets were stimulated by cross linking T cell receptor and co-stimulatory receptor (anti CD3/CD28 dynabeads)27–29; B cell subsets were stimulated with anti-human IgG/IgM and human IL-430–32; Monocytes were stimulated with lipopolysaccharide (LPS)9; and NK cells were stimulated with IL2, CD2 and CRTAM (NCR1) coated beads33. Stimuli exposure duration was chosen according to the corresponding literature and activation status was confirmed by inspecting surface expression of CD69 (Supplementary Fig. 7).
Overall, stimulation drives dramatic changes in chromatin landscapes of B and T cells. In contrast, we saw only limited effects in innate lineage cells (Supplementary Fig. 5a,b). Upon further inspection of the monocyte dataset, even though differentially expressed genes replicated results from Alasoo et al.10, we did not observe a robust stimulation response at the chromatin level or changes in CD69 surface expression (Supplementary Fig. 6b,c; 7). Therefore, we do not discuss the stimulated monocytes further.
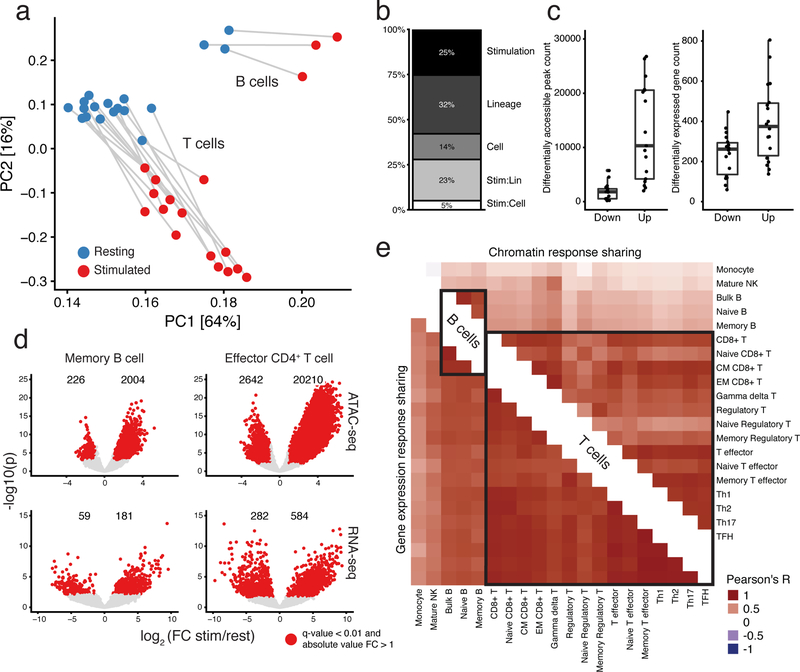
Stimulation was a major driver of sample clustering of B and T cells (Fig. 3a). The primary axes of variation in chromatin accessibility were associated with both stimulation and cell type. Moreover, stimulated samples moved in a similar direction, suggesting an underlying shared chromatin response to stimulation. Furthermore, many similar pathways were enriched for significant stimulation-associated genes among distinct subsets (Supplementary Fig. 8, Supplementary Table 1).
Fig. 3: Stimulation induces large-scale changes in chromatin and gene expression in B and T cells.
a, Principal component analysis (PCA) of ATAC-seq read counts of B and T cell subsets (excluding plasmablast cells), which were merged from multiple donors. Analysis based on the top 100k most variable peaks. b, Explained proportion of variation in chromatin accessibility of the samples in (a) with at least 3 biological replicates, explained by biological factors of interest (see Methods). The interaction effect for lineage is labeled Stim:Lin and the interaction effect for cell type is labeled Stim:Cell. c, Counts of significant differentially accessible chromatin regions (left) and expressed genes (right) identified for B and T cells during stimulation (see Methods). Naïve regulatory T cells were excluded due to a lack of power because we had too few biological samples passing QC. d, Volcano plots showing stimulation effects for ATAC-seq (top) and RNA-seq (bottom) for memory B (left) and effector CD4+ T (right) cells. e, Displayed is the Pearson’s R correlation between samples from stimulation-response chromatin (top-right triangle) and gene expression (bottom-left triangle) effects, at sites or genes with a significant stimulation response in at least one of the two cell types in the comparison. All estimates are from at least three biological replicates, except the Naïve Regulatory T cells, which had two. Counts of the number of samples included and overlapping significant stimulation-associated peaks between cell subsets can be found in Supplementary Table 1.
To quantify these observations, we used a random effects model to estimate the proportion of biological variance of chromatin accessibility explained by stimulation condition, lineage (CD4+, CD8+, and B) and cell subset (e.g. naïve, memory and T helper cells; Methods). We found that stimulation, regardless of lineage or cell type, accounted for roughly a quarter of the explained chromatin variation (Fig. 3b). In fact, the chromatin differences due to stimulation were nearly as large as differences between cell lineages (25% versus 32%, respectively). Additionally, lineage-dependent stimulation changes explained a significantly greater amount of chromatin variation than cell subset-specific stimulation. In summary, broadly shared simulation and lineage-specific stimulation effects drove a large proportion of observed chromatin variation.
One major effect of stimulation was a marked increase in the number of accessible sites and in widespread up-regulation of gene expression (Fig. 3c). Specifically, we found 30,224 additional peaks in stimulated cell types compared to resting state (Supplementary Fig. 5c). Testing for differential accessibility, we found, on average, that 12,119 peaks were significantly more accessible, versus 1,722 peaks that were significantly less accessible following stimulation (Fig. 3c, left panel; Supplementary Data 3). We observed a similar pattern in gene expression where, per sample, an average of 394 genes were up-regulated compared to 237 down-regulated genes in stimulated cell types relative to their resting cell state (Fig. 3c, right panel; Supplementary Data 4).
For example, stimulation of memory B cells leads to considerable increases of accessibility and expression. At the same time, only 226 peaks become less accessible and 59 genes are down-regulated following stimulation. We found a similar trend within subsets of the T cell lineage (Fig. 3d).
Finally, given that both B and T cell subsets have highly distinct functions during an immune response, we were interested in quantifying the strength of shared effects of stimulation on chromatin remodeling. Therefore, we computed Pearson’s R correlation of effects estimated from stimulation-induced changes in chromatin accessibility and gene expression between each pair of cell types as a summary statistic of the shared effect of stimulation (Fig. 3e).
We observed large proportions of shared stimulation-induced chromatin accessible regions between CD4+, CD8+ and γδ T cells (mean R = 0.74) upon activation. Similarly, we observed strong sharing among B cell subsets (mean R = 0.82). Furthermore, despite the divergent mechanisms of activation in B and T cells, we identified a significant level of sharing globally between stimulation-response in B and T cells (mean R = 0.37), indicating that some stimulation-induced regulatory networks are utilized by both B and T cells. However, while stimulation induces both unique and shared chromatin responses, the stimulation-induced transcriptional signatures are broadly shared among cell types (mean R = 0.77). We further characterize transcription factors driving stimulation chromatin effects and their connection to gene expression in the Supplementary Note.
Context-specific allelic imbalance
In addition to identifying stimulation-associated chromatin regions, we were interested in characterizing genetic variants that alter context-specific chromatin regulation. Due to the small number of individuals included in this study, and thus insufficient power to call chromatin accessibility quantitative trait loci (caQTLs), we used the observed allelic imbalance of ATAC-seq reads that map to heterozygous SNPs to identify significant sites of allele-specific chromatin accessibility (ASC). The hypothesis is that a heterozygous variant may cause local, allele-specific changes in chromatin accessibility, for example by disrupting local transcription factor binding. Such events result in an imbalance in the number of ATAC-seq reads overlapping each allele34–38. After read-filtering to account for mapping biases39, we computed binomial test p-values and identified 607 significant ASC sites, on average, per sample, and a total of 10,780 sites overall (q-value < 0.1, see Methods, Supplementary Table 1).
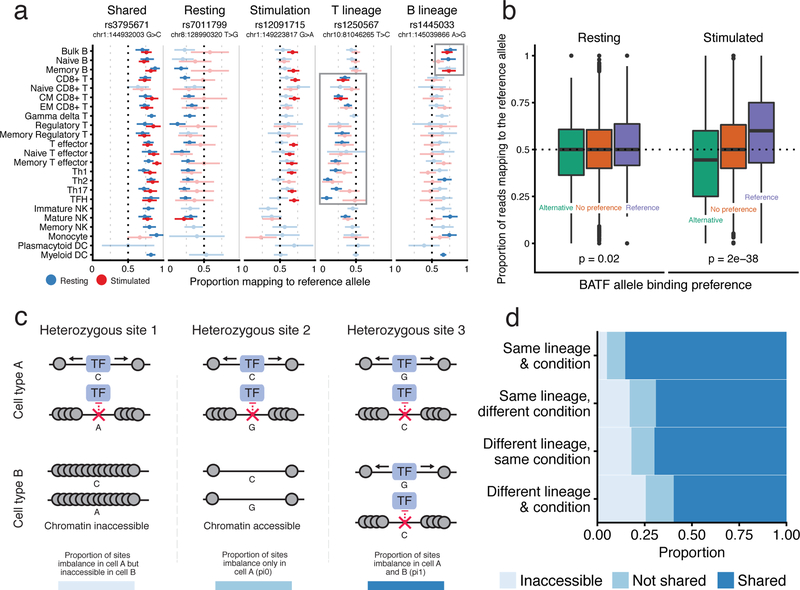
Because we collected many cell samples from each donor, we could test for allelic imbalance of chromatin accessibility at an individual heterozygous site across cell and condition-specific contexts. For instance, we observed a heterozygous variant, rs3795671, that resulted in increased chromatin accessibility at the reference allele across resting and stimulated samples (Fig. 4a). Other variants exhibited allelic imbalance in a subset of cell type states; these include rs7011799, rs12091715, rs1250567, and rs1445033, which are associated with chromatin accessibility within resting, stimulated, T, and B lineage contexts, respectively.
Fig. 4: Observed allelic imbalance in chromatin accessibility data.
a, Examples of allele-specific chromatin accessibility imbalance shared across various groups. For each heterozygous site, we display the proportion of reads mapping to the reference allele (x-axis) for cell samples (y-axis). Error bars represent 95% confidence intervals computed from read depth. Samples without significant imbalance are lightly shaded. We excluded samples from the visualization with fewer than four reads at the specific variant. b, Heterozygous sites were grouped into three bins based on the PWM-predicted BATF binding affinity: preference for the alternative, no preference, preference for the reference allele. The y-axis represents the aggregate proportion of reads mapping to the reference allele for these groups in Th1 precursor cells under resting (left) and stimulated (right) conditions. Sites with fewer than four reads were excluded. c, Scenarios of allele-specific chromatin accessibility imbalance in two different cell types or conditions for the same donor. d, Proportion plot displaying the estimated average proportions for each case from (c), stratified by whether the two samples were of the same lineage and condition. Innate cells were excluded from this analysis. All plots are for Donor 1, who had the highest sequencing depth, although similar trends were found in the other donors (Supplementary Fig. 9b,c). Read counts for (a) and sample sizes for (b) and (d) can be found in Supplementary Table 1.
Next, we leveraged these data to verify transcription factors that regulate chromatin in specific contexts, and thus drive context-specific allelic imbalance. As described in the Supplementary Note, motif enrichment analysis (Supplementary Fig. 5d) suggested that BATF (or perhaps another transcription factor from the AP-1 family, which can have similar position weight matrices (PWMs)), among others, is an important regulator of chromatin in stimulated cells26. We therefore hypothesized that sites that affect BATF binding would result in ASC within stimulated samples. As a proxy for true BATF binding, for each heterozygous site within a BATF binding motif, we computed the relative binding affinity for the reference and the alternative allele using the BATF PWM. Upon stimulation, alleles that were predicted to increase BATF binding affinity were associated with increased accessibility. However, there was no such effect in resting cells, indicating that BATF does not impact chromatin accessibility in the resting state (Fig. 4b). We observed a similar preferential binding effect for other stimulation-associated transcription factors, including JUNB, FOSL1 and BACH1 (Supplementary Fig. 9a). Thus, the PWMs of these factors can predict sequence dependent changes of stimulation-specific effects on chromatin accessibility. However, because of correlation between PWMs of distinct transcription factors, further study will be required to link these signals definitively to specific transcription factors.
Intrigued by the stimulation-specific effect of the BATF-associated PWM, we wanted to leverage our ASCs to determine the prevalence of context-specific chromatin regulation. We propose three possible models of chromatin effects at a significant ASC site, in pairs of cell types or conditions (Fig. 4c, from left to right). The variant can: 1) differentially affect chromatin in cell type A, while the region is inaccessible in cell type B; 2) differentially affect chromatin in cell type A, but not in cell type B, even though the region is accessible therein, or; 3) differentially affect chromatin in both cell types. The relative fractions of case 2 versus case 3 were estimated using a method that accounts for incomplete power in each statistical test40.
We quantified the proportion of ASCs that fit each possible model and observed that most ASCs have shared effects on chromatin regulation (Methods). To further investigate changes in the prevalence of shared regulation across differences in cell state, we varied whether the two compared samples were of the same lineage and condition. For the purpose of this analysis, we grouped all “stimulated” samples.
Notably, most effects on allelic imbalance are shared regardless of lineage or condition (Fig. 4d). In contrast, the proportion of accessible sites covaried more strongly with differences in lineage and condition between the two samples. These observations suggest that changes in cellular function due to chromatin remodeling occur primarily through changes in chromatin accessibility, rather than regulation of already accessible chromatin. Moreover, these trends held when the analysis was performed in the other donors (Supplementary Fig. 9b,c). The BATF analysis in Fig. 4b indicates that at least some sites containing BATF motifs are likely differentially regulated upon stimulation, however this scenario represents a small minority of all ASC sites, which corroborates findings from previous studies on caQTLs41.
GWAS enrichment in immune-specific accessible regions
Understanding the molecular mechanisms behind autoimmune disease risk variants requires the identification of disease-relevant cell types and states. With chromatin accessibility information from a large number of resting and activated immune cell types, we can identify cell types and conditions enriched for variants associated with autoimmune disease. For example, the inclusion of thymocytes and progenitor cells allow us to examine the possibility that autoimmunity risk variants affect enhancers that are selectively accessible during early differentiation.
Using publicly available summary statistics for nine autoimmune disorders and four non-immune traits (as controls), we identified trait-relevant functional annotations using the partitioning heritability functionality of LD Score regression2. This allowed us to compute the proportion of heritability explained by SNPs in open chromatin for a particular cell type. This quantity, divided by the overall proportion of open chromatin SNPs in that cell type, will be referred to as the enrichment of heritability (Supplementary Table 1).
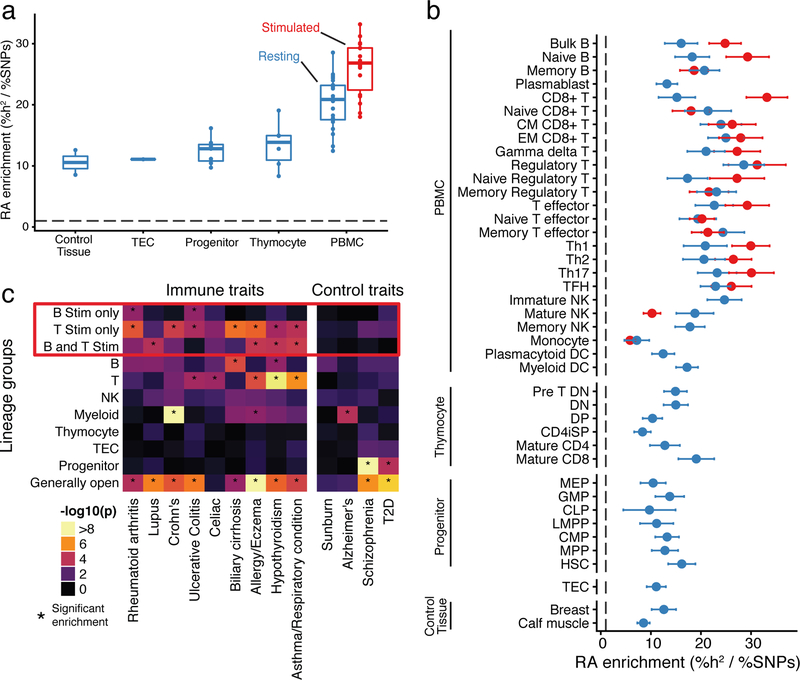
We observed greater enrichment of heritability in differentiated immune cells compared to progenitor cells and non-immune tissues for most autoimmune traits (Fig. 5a; Supplementary Fig. 5a,b). Specifically for rheumatoid arthritis (RA) we saw about ten-fold enrichment of heritability in open chromatin from control tissues and progenitors compared to the genome-wide background (Fig. 5a), and a median 23-fold enrichment in open chromatin of adult immune cell types. Heritability enrichment in non-immune tissues compared to the genome-wide background is likely due to the fact that many open chromatin regions are shared among diverse cell types. Thus, many of the SNPs that affect disease risk through immune cell functions are also located in open chromatin in many other tissues42.
Fig. 5: GWAS analysis of accessible regions.
a, Rheumatoid arthritis heritability enrichment is aggregated by differentiation and condition. Stimulated innate cells were excluded from this visualization. The dashed line represents a baseline proportion of disease heritability across all SNPs. b, Enrichment of rheumatoid arthritis (RA) heritability (x-axis) in open chromatin regions for resting (blue) and stimulated (red) samples (y-axis). Error bars indicate one standard deviation in each direction. The dashed line represents a baseline proportion of disease heritability across all SNPs. c, We grouped peaks into disjoint clusters based on their patterns of accessibility across cell types (x-axis). Then, we used partitioning heritability functionality of LDScore regression to estimate enrichments of trait signal (x-axis) in these peak clusters (y-axis). We highlighted groups of peaks related to stimulation with a red box, and asterisks (*) indicate significant enrichment of trait heritability (Bonferroni adjusted p < 0.05). Sample sizes for panels a, b and c can be found in Supplementary Table 1.
Notably, heritability enrichment was particularly strong in stimulated immune cells, compared to their resting counterparts (Fig. 5b). This signal was spread across diverse cell types, including both B and T cell lineages, and did not implicate particular immune subsets as drivers of rheumatoid arthritis. We wondered whether this relatively diffuse signal arises because multiple cell types play causal roles in rheumatoid arthritis, or simply because the broad sharing of stimulation peaks makes it difficult to identify a critical cell type.
To investigate this further, we grouped the open chromatin peaks into 11 disjoint clusters based on their profiles of accessibility across cell types and conditions (Methods, Supplementary Fig. 10c). For example, we defined a cluster of peaks that are open only in stimulated B cells, and a cluster of peaks that are open only in stimulated T cells. We used these peak groupings to test which clusters are enriched for autoimmune trait heritability (Fig. 5c).
Overall, we observed strong enrichment of heritability in accessible regions from multiple peak groups, especially among the stimulation-related peaks. No single peak group could explain the entirety of the signal. Since these groups are disjointed, this observation implies that multiple separate immune components contribute to heritability. In the case of rheumatoid arthritis, we identified enriched heritability in stimulation peaks specific to B lineage cells, and in stimulation regions specific to T lineage cells, thus implicating a role for both stimulated B and T cells. Previous studies have separately identified B19,43 and T cells1,4 in driving the development of rheumatoid arthritis, although our analysis would suggest that both contribute to autoimmunity. In addition to signals from cell type-specific clusters, all traits had significant enrichment within loci that were broadly accessible, highlighting the contribution of broadly open “housekeeping” peaks to heritability.
Taken together, these results suggest that stimulated cells from both B and T lineages likely contribute to autoimmunity. Furthermore, it remains difficult to implicate more narrowly-defined subsets through this type of analysis, due to the extensive sharing of open chromatin among closely related subsets.
Stimulation-specific data increase GWAS and eQTL overlap
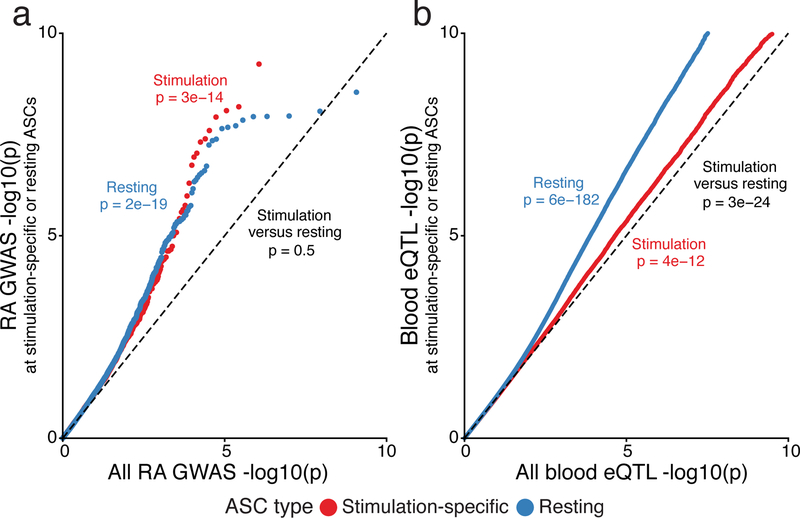
One important approach for linking noncoding GWAS hits to the genes they regulate is through eQTL mapping. However, only a small proportion of significant GWAS sites have been successfully linked to an eQTL. One hypothesis for the limited overlap is that many eQTLs may be context-specific and thus remain unidentified if not all of the relevant cell types or conditions have been studied44. Since we found a strong signal of autoimmune trait risk variants located within stimulation-specific chromatin accessibility peaks, we sought to investigate whether these variants would have been identified as candidate regulatory variants in a standard tissue-based eQTL study. We present results primarily from rheumatoid arthritis, because the heritability of this disorder was highly enriched in the stimulation-specific accessible regions in our previous analysis.
For this analysis, we partitioned the set of allelic imbalance sites into two groups: ASC sites that were identified in resting samples, and ASC sites that were only identified in stimulated samples (either B or T cells; nominal p < 0.10). In this analysis, sites that were present in both resting and stimulated cells were included in the resting set, as those could be identified even without using stimulated cell data. Since variants that affect chromatin accessibility are strongly associated with regulation of gene expression34, the former set of ASCs likely contain sites that regulate gene expression in the resting cell state, while the latter contain sites associated with stimulation-specific gene regulation.
Both sets of SNPs exhibited highly significant enrichment of rheumatoid arthritis GWAS signal relative to control SNPs (p = 2 × 10−19 (resting), and p = 3 × 10−14 (stimulation only); Fig. 6a). However, the stimulation-specific ASCs would not have been identified in resting samples. Moreover, we compared these two sets to all variants for which an ASC p-value was computed and similar trends were observed (Supplementary Fig. 11a). Thus, stimulation-specific chromatin is as important, in terms of disease risk enrichment, as resting-specific chromatin from immune cells.
Fig. 6: GWAS and expression quantitative trait loci (eQTL) enrichment in sites of allele-specific chromatin.
a, Comparison of rheumatoid arthritis GWAS enrichment within the set of SNPs that regulate chromatin accessibility, in either B or T cells, under stimulation (red) or resting (blue) conditions. b, Comparison of eQTL signal in the same two sets of variants from (a), using eQTL data from GTEx v7. For both plots the x-axis reflects an empirical distribution of p-values. Sample sizes can be found in Supplementary Table 1. We computed p-values with a two-sided Mann–Whitney U text.
Thus far there have been many studies of blood eQTLs (PBMCs) encompassing many thousands of samples45. However, since the proportion of stimulated cells in bulk tissues is relatively low, we hypothesized that sites that affect stimulation-specific chromatin accessibility may not be detected as eQTLs using whole blood or resting immune cells. To test this hypothesis, we determined the enrichment of blood eQTL signal from GTEx in the two previously described sets of resting and stimulation-specific ASC sites. We observed a strong enrichment (p = 3 × 10−24) of blood eQTL signal within the resting state-specific set of ASC variants compared to the stimulation-specific set (Fig. 6b). Furthermore, a similar trend was observed when using eQTLs from naïve T cells (Supplementary Fig. 11b)7. These observations indicate a clear need for large-scale eQTL mapping in stimulated immune cells and argue that the low overlap of autoimmune GWAS and eQTL data8 is at least partly driven by the current lack of such data.
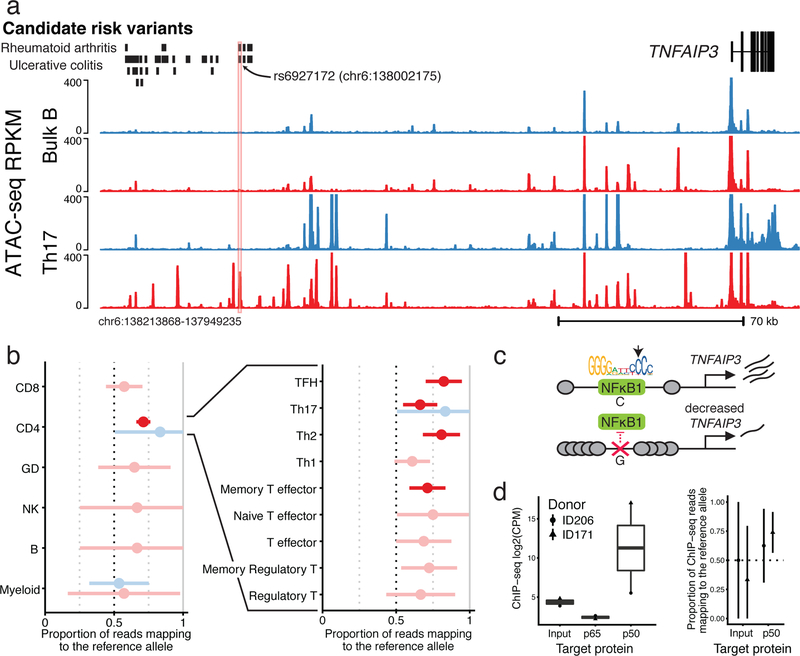
These observations suggest that the inclusion of data from stimulated cells could help fine-map previously unexplained causal mutations. To identify such examples, we intersected a database of fine-mapped candidate causal GWAS SNPs1 with a set of stimulation-specific allelic imbalance heterozygous SNPs and PWM-disruption scores for TFs (Supplementary Data 5).
Using these data, we identified a T cell lineage-specific stimulation peak within an intergenic region upstream of the TNFAIP3 locus (encoding A20 protein). Moreover, this region contains several variants that are included in a credible set of regions identified for rheumatoid arthritis and ulcerative colitis. For rheumatoid arthritis, only rs6927172 falls within a stimulation-specific chromatin accessible peak (Fig. 7a). For ulcerative colitis, in addition to rs6927172, three additional variants fall within another nearby stimulation-responsive peak, yet this second peak does not demonstrate as strong a response as the first peak (Supplementary Fig. 13a). Notably, rs6927172 was associated with altered chromatin accessibility in stimulated CD4+ T cells across multiple donors, suggesting a potential role of the SNP in regulating chromatin remodeling (Fig. 7b).
Fig. 7: Identifying rs6927172 as a stimulation-specific chromatin regulator in a complex autoimmune GWAS region.
a, Chromatin accessibility profile for stimulated (red) and resting (blue), bulk B (top) and Th17 (bottom) cells, around variant rs6927172. This region contains significant GWAS signals for ulcerative colitis and rheumatoid arthritis, but the causal variant(s) have not been determined (credible set indicated). We include a trackplot with all samples in Supplementary Fig. 12. b, Allele-specific ATAC-seq reads at rs6927172 in the three heterozygous donors (the fourth was not heterozygous at this site). Displayed is the proportion of reads mapping to the reference allele. Error bars represent 98% confidence intervals and were computed from read depth. Significant (p < 0.01) allelic imbalance associations are colored (see Methods). Exact read counts for these sites can be found in Supplementary Table 1. c, A proposed negative feedback model of gene regulation linking NFKB1 toTFNAIP3. We included the canonical PWM for the p50 subunit of NFKB1, as downloaded from the Jaspar TF motif database. The heterozygous allele disrupts the nucleotide indicated by the arrow. d, ChIP-seq read count for the input genomic DNA control, and p50 and p65 subunits of NFKB1 (left, n = 2, read counts found in Supplementary Table 1). Allelic-imbalance of ChIP-seq reads mapping to rs6927172 (right).
Indeed, the cytosine to guanine mutation leads to a large change in the PWM-predicted binding affinity of the p50 subunit of NFKB1 (Fig. 7c, Supplementary Fig. 13b; see Supplementary Fig. 13c for expression levels). To validate this observation, we performed ChIP-seq for the p65 and p50 subunits of NFKB1 from stimulated CD4+ T cells isolated from two healthy donors who are heterozygous for rs6927172. We observed an increased count of p50 ChIP-seq reads that mapped to the region containing our SNP of interest in one of the two donors. As predicted, we detected significant allelic-imbalance for p50 in the same donor (Fig. 7d).
As NFKB1 has been shown to regulate TNFAIP3 expression46,47, this result suggests that rs6927172 may contribute to the pathology of autoimmune diseases by disrupting binding of NFKB1 in stimulated T cells to the TNFAIP3 locus and therefore inhibit gene expression (Fig. 7c).
Discussion
While increasing evidence points to an important role of stimulation response in autoimmune dysregulation, little work has been done to characterize shared and cell subset-specific stimulation effects across multiple differentiation lineages. Therefore, we developed a map of stimulation-responsive chromatin accessibility and transcription in a diverse set of immune cell populations.
To study the impact of genetic variation on the chromatin landscape, we used allele-specific measurement of open chromatin to identify genotype-dependent sites (ASC). While the majority of ASC are shared between closely related cell types, only 60% of ASC are shared for comparisons between lineages and between stimulated and resting cells. Most non-sharing of ASC is due to differential accessibility between cell types/conditions, while a smaller fraction is due to differential regulation of shared open sites as exemplified by BATF activity in stimulated cells.
Several past studies have mapped GWAS signals to cell type-specific functional elements, allowing the prediction of the cell types that may be involved in disease pathology1–6,48. We observed significant autoimmune trait heritability within accessible regions of distinct stimulated cell types, suggesting contributions of multiple stimulation-response components to autoimmunity. Moreover, we showed that genetic variation associated with stimulation-specific gene regulation is significantly underrepresented in existing large-scale eQTL datasets from whole blood PBMCs. As a concrete example of this point we identified rs6927172, which was associated with CD4+ T cell stimulation-specific changes in chromatin accessibility near TNFAIP3 and affects the predicted binding of the p50 subunit of NFKB1. This region has been associated with several autoimmune phenotypes, such as variable patient responses to anti-TNF treatment49, ulcerative colitis15, and rheumatoid arthritis50. We further propose a model by which this variant affects autoimmunity in the Supplementary Note.
Taken together, these data provide insights into the interplay of specific loci and regulatory networks involved in the human immune system. Additionally, we present a powerful platform to map genetic variants to cell and context-specific functional regions genome-wide, and therefore, to their context-specific effects on disease phenotypes.
Methods
Data collection
Sample collection and processing
This study was approved by the UCSF Committee on Human Research and written consent was obtained from all donors. Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood (~450 ml) using Ficoll-Plaque Plus (GE Healthcare, IL, USA) centrifugation. Bulk population of CD4+ T cells, B cells and monocytes were positively enriched, whereas pan T cells, NK cells and dendritic cells were negatively enriched using magnetic beads prior to sorting (STEMCELL, Canada). Different cell subsets were sorted on a FACS Aria flow cytometer (BD Biosciences, CA, USA) up to >95% purity (Supplementary Table 1). Sorted cells were washed once in PBS, cryopreserved in Bambanker freezing media (LYMPHOTEC Inc, Japan) for ATAC experiments and in TriReagent (Sigma-Aldrich, MO, USA) for RNA experiments. Cells frozen in Bambanker freezing media were stored in liquid nitrogen until ready to use. Cells frozen in TriReagent were stored at −80°C until further use.
Ex vivo activation
Freshly sorted cells were cultured in RPMI-1640 medium supplemented with glutamine, sodium pyruvate, penicillin, streptomycin, non-essential amino acid and 10% FCS (Sigma-Aldrich, MO, USA). T lymphocytes were stimulated for 24 hours with anti-human CD3/CD28 dynabeads (Thermo Fisher Scientific, MA, USA) at a 1:1 cell to bead ratio and human IL-2 (UCSF Pharmacy, 300 unit/ml for regulatory T cells, 50 unit/ml for other T lymphocytes). B lymphocytes were activated for 24 hours with 10 μg/ml F(ab)’2 anti-human IgG/IgM (Affymetrix, CA, USA) and 20 ng/ml human IL-4 (Cell Sciences, MA, USA). NK cells were activated using 2 conditions: 1) with the NK Cell Activation/Expansion Kit as per manufacturer’s instructions for 48 hours (Milteny, Germany) and 2) with 500 unit human IL-2 for 24 hours. Monocytes were stimulated with 100 ng/ml and 1μg/ml LPS (Sigma-Aldrich, MO, USA) for 6 hours and 24 hours, respectively. The detailed activation conditions are listed in Supplementary Table 1). After stimulation, cells were washed once with PBS, cryopreserved in Bambanker freezing media and TriReagents, and stored in liquid nitrogen until ready to use.
RNA-seq library preparation
Cells frozen in TriReagent were thawed at room temperature for 10 min. Technical replicates were done for each cell aliquot. One hundred μl chloroform was added to 500 μl of sample, mixed, incubated at room temperature for 10 min and centrifuged at 12,000 g for 10 min at 4°C. The aqueous phase was transferred to a new tube and an equal volume of 100% ethanol was added. For further RNA extraction the Direct-zol RNA MicroPrep kit (Zymo Research, CA, USA) was used. Samples were mixed thoroughly before transferring onto a Zymo-Spin IC Column and RNA was extracted according to the manufacturer’s protocol starting at step 2. RNA sequencing library preparation was based on the Smart-seq2 protocol52. For oligo dT annealing, 2 μl RNA was mixed with 1 μl recombinant RNase inhibitor (Takara Bio USA, CA, USA), 1 μl of 10 μM oligo dT (Integrated DNA Technologies, [IDT], IA, USA) and 1 μl of 10 mM dNTPs (New England Biolabs, MA, USA), and incubated at 72°C for 3 min. Reverse transcription and template switching were done in 1x first strand synthesis buffer (Thermo Fisher Scientific, MA, USA), 100 U SuperScript II reverse transcriptase (Thermo Fisher Scientific), 10 U RNase inhibitor (Takara Bio USA), 5 mM DTT, 1 M Betaine (Sigma-Aldrich, MO, USA), 6 mM MgCl2, and 1 μM TSO (IDT). The mixture was incubated at 42°C for 90 min, 10 cycles of 50°C for 2 min and 42°C for 2 min, followed by a final incubation of 15 min at 70°C. Transcribed RNA was amplified in 1x KAPA HiFi HotStart ReadyMix (Kapa Biosystems, MA, USA), 0.1 μM IS primer and 0.6 ×Sybr Green (Thermo Fisher Scientific) as follows: 98°C for 3 min, 18 cycles of 98°C for 20 seconds, 67°C for 15 seconds, followed by a final extension at 72°C for 5 min. Samples were purified using AMPure XP beads (Beckman Coulter, CA, USA) in a 1:1 ratio. Tagmentation of amplified cDNA was carried out using the Nextera XT DNA library kit (Illumina, CA, USA). In a 20 μl reaction, 150–200 pg cDNA was added to 1x TD buffer (Illumina) and 5 μl of ATM (Illumina). The mixture was incubated at 55°C for 5 min. Five μl of NT buffer was added to the mixture to stop the fragmentation reaction and incubated for 5 min at room temperature. Tagmented libraries were further amplified in a total reaction volume of 50 μl by adding 15 μl of NPM (Illumina) and 1.25 μM of each, i7 and i5, custom Nextera barcoded PCR primers53 for 3 min at 72°C, 30 seconds at 95°C, and 12 cycles of 10 seconds at 95°C, 30 seconds at 55°C, and 30 seconds at 72°C. Amplified libraries were purified using AMPure XP beads (Beckman Coulter) using a 1:1 ratio.
ATAC-seq library preparation
ATAC-seq libraries were prepared following the Fast-ATAC protocol19. In detail, frozen cells were thawed in a water bath, resuspended in 2 ml media and centrifuged at 500 g for 5 min. Pelleted cells were resuspended in 1 ml and counted using a hemocytometer. For the ATAC reaction, we took aliquots of 10,000 live cells, unless limited by cell number. Technical replicates were done for all samples. Cells were spun down at 4°C at 500 g for 5 min, washed once in cold PBS and resuspended in the Tn5 reaction buffer (1x TD buffer [Illumina, CA, USA], 50 μl TDE1 Tn5 transposase [Illumina] per ml Tn5 reaction buffer and 0.01% Digitonin). 5,000–10,000 cells were transposed in 50 μl of reaction buffer. The reaction volume of samples containing fewer than 5,000 cells was linearly scaled to the number of cells present whereby, for example, 4,000 cells were done in a 40 μl reaction and 2,500 cells were done in a 25 μl reaction. The Tn5 reaction mix was incubated at 37°C for 30 min at 300 rpm. Transposed samples were purified using MinElute PCR purification columns according to the manufacturer’s protocol (Qiagen, Germany). Purified samples were amplified and indexed using custom Nextera barcoded PCR primers (Supplementary Table 1) as described in Buenrostro, et al.18. Amplified libraries were purified using MinElute PCR purification columns.
DNA sequencing
To circumvent index hopping, subpools of samples were created, ran over a 2.5% Agarose gel and size selected, excluding high molecular weight DNA, free PCR primers, and primer dimers. Samples were purified using a MinElute Gel Purification kit according to manufacturer’s instructions (Qiagen). Prior to sequencing, samples were amplified in 1x NEBNext Master Mix (New England BioLabs, MA, USA), 1.25 μM oligo C (Illumina P5) and 1.25 μM oligo D (Illumina P7) using the following cycle conditions: 30 seconds at 98°C followed by 3–4 cycles of 10 seconds at 98°C, 30 seconds at 63°C and 1 min at 72°C. All samples were purified using MinElute PCR purification columns (Qiagen). Samples run on the NovaSeq 6000 were additionally purified using 1x AMPure XP beads (Beckman Coulter, CA, USA) to remove free oligo C and D primers. ATAC-seq samples were sequenced on a HiSeq 4000 in paired-end 76 bp cycle mode or on a NextSeq 500, without prior gel clean-up, in paired-end 76 bp cycle, high-output mode. RNA-seq (cDNA) libraries were either sequenced on a NovaSeq using a S2 flow cell in paired-end 100 bp cycle mode or on a HiSeq 4000 in paired-end 76 bp cycle mode.
Genotyping
Three hundred μl of donor blood (donors: 1001,1002, 1003 and 1004) was used to extract genomic DNA using DNeasy Blood and Tissue Kits (Qiagen, Germany) as per manufacturer’s protocol. We genotyped 958,497 markers using Infinium OmniExpressExome-8 v1.4 kit (Illumina, CA, USA). We phased and imputed all biallelic common variants (MAF > 1%) using the Michigan imputation server that ran minimac3 with 1000G Phase 3 v5 as the reference genome and Eagle v2.354. All genotypes with an imputed heterozygous probability of 0.9 or greater were included in downstream allele-specific chromatin accessibility analysis.
Additional details regarding data collection can be found in the Supplementary Note.
Data analysis
RNA-seq
We trimmed remaining transposase adapters with cutadapt version 1.13 with a -minimum-length of 20 and an -overlap of 5 in paired-end mode55. RNA-seq reads were pseudoaligned using Kallisto version 0.43.056. The Kallisto index was made with default parameters and the gencode v25lift37 fasta file57 and was run in quant mode with default parameters. Following pseudoalignment we computed gene abundances using tximport version 1.2.058. We excluded donor cell type samples with fewer than one million total reads from all technical replicates or that were extreme PCA outliers. Discarded cell types include thymocyte subsets and TEC cells.
ATAC-seq
We trimmed remaining transposase adapters with cutadapt version 1.13 with a -minimum-length of 20 and an -overlap of 5 in paired-end mode55. We aligned ATAC-seq reads using bowtie2 version 2.2.9 with default parameters and a maximum paired-end insert distance of 2kbp. The bowtie2 index was constructed with the default parameters for the hg19 reference genome. We filtered out reads that mapped to chrM and used samtools version 1.4 to filter out reads with MAPQ < 30 and with the flags ‘-F 1804’ and ‘-f 2’. Additionally, duplicate reads were discarded using picard version 1.134 (http://broadinstitute.github.io/picard/). We calculated percentage aligning to mitochondria, percentage within blacklist regions59, and enrichment of reads at TSS regions relative to 2kb away using the RefSeq gene annotation (referred to elsewhere as the TSS enrichment). Chromatin accessibility peaks were identified with MACS2 version 2.1.1 under default parameters and ‘--nomodel --nolambda --keep-dup all --call-summits’. The count of absolute peaks per cell type refers to the number of peak regions reported in the ‘narrowPeak’ file (peaks with multiple summits are only counted once). We reported the peak count estimate after linearly adjusting for estimated confounding effects from sample read depth, and TSS enrichment (a proxy for sample quality). A consensus set of peaks was defined by merging overlapping (1bp or more) peaks identified in at least two samples across all samples. This set of peaks had a median peak length of 346 bps following removal of peaks that were greater than 3 kb or non-autosomal. We then used the ‘get_count’ function from the nucleoATAC python package to count the number of fragments within the consensus peak set across all samples60. We excluded donor samples with fewer than 5 million reads from all technical replicates that passed quality filters, TSS enrichment score less than 4, more than 0.5% of reads mapping to a known set of region blacklist59, more than 25% of reads mapping to the mitochondria, or that were extreme PCA outliers. We used deeptools version 2.5.1 to generate bigwig tracks with a genomic bin size of 10 bp, RPKM normalization and the ‘—extendReads’ parameter.
Differentially expressed genes
For this analysis, we included only unique protein coding genes from gencode v25. Additionally, we eliminated all genes that had below ten counts per million (CPM) in at least two biological replicate samples, resulting in 13,512 genes tested. We used the TMM normalization method to compute scale factors for each sample, and voom to compute weights capturing the relationship between gene expression mean and variance. For each cell type, we used limma to estimate the log2 fold change (log2FC) of gene expression for each gene upon stimulation. We included donor in the design matrix to correct for donor-specific effects. We considered a gene differentially expressed if the limma-reported Benjamini-Hochberg (BH) corrected p-value61, which we refer to as a q-value, was less than 0.01 and the absolute log2FC was greater than 1.
Differentially accessible chromatin regions
Starting with the consensus peak set we eliminated all peaks with fewer than one CPM in at least two biological replicate samples, resulting in 671,448 tested peaks. We used the TMM method to compute scale factors for each sample, and voom to compute weights capturing the relationship between mean and variance chromatin accessibility across samples. For each cell type we used limma to estimate the log2FC of each chromatin accessibility region upon stimulation, while controlling for donor and the enrichment of reads at transcription start sites (the latter serves as a proxy for sample quality). We considered a region a significant differentially accessible chromatin peak if the limma-reported BH corrected p-value was less than 0.01 and the estimated absolute log2FC from the resting to stimulation condition was greater than 1. We initially considered different stimulation conditions of NK and monocyte samples separately. However, estimated log2FC stimulation effects were nearly identical among significant stimulation peaks when the conditions were pooled or unpooled (adjusted R2 = 0.88 for Mature NK cells and R2 = 0.99 for Monocytes). Thus, we reported summary statistics from the pooled differential accessibility analysis.
Quantifying the amount of shared stimulation response
We aimed to quantify sharing of simulation effects between two cell types, denoted A and B. To do this we first determined the set of differentially accessible peaks (q<0.01 and |log2FC|>1.0) from cell type A (using limma-voom, see Differentially accessible chromatin regions above). For this set of peaks, we computed Pearson correlation (with the ‘cor’ function in R) between stimulation effects (log2FC) in cell type A and B. This analysis results in asymmetric sharing estimates conditioning on significant peaks from cell type A or B. We found that the asymmetric estimates were broadly consistent, so we report the mean correlation weighted by the number of significant peaks in each cell type (individual asymmetric correlation estimates can be found in Supplementary Table 1).
Allele-specific chromatin accessibility
We collected all aligned reads that overlap heterozygous sites. Initially, we used the set of heterozygous sites identified with ‘HaplotypeCaller’ from GATK version 3.7 with ‘—minPruning 10’ and ‘-stand_call_conf 20’ run on a donor-specific bam file including all samples from the donor62. The final set of heterozygous sites that we used for analysis were the intersection of the GATK and genotyped sites passing filters (see Genotyping above). Next, we passed all aligned reads that overlap heterozygous sites through WASP filtering39. Briefly, the read is remapped with the SNP allele flipped (we only examine biallelic sites) and is only retained if it maps to the same location. This filtering approach effectively eliminates reference genome biases enabling unbiased estimation of the proportion of reads mapping to the reference allele. At each heterozygous site we counted the number of WASP filtered reads mapping to the reference and the alternative allele. We computed a p-value per heterozygous site using a binomial test, and for each sample corrected for multiple hypotheses by computing BH q-values. P-values were converted to BH q-values using the ‘p.adjust’ function in R.
Estimating the proportion of shared allele-specific imbalance effects
We first computed the posterior mean and variance of the proportion of reads mapping to the reference allele, assuming a uniform prior on the proportion and a binomial likelihood. The posterior under this model is Beta(r + 1, a + 1), where r and a are the numbers of reads mapping to the reference and alternative allele respectively. At each heterozygous site, we defined the effect as the difference between the proportion of reads mapping to the reference and the expectation under the null of no allele-specific effect, i.e., 50%.
We aimed to estimate the proportion of shared imbalance effects between two cell types, denoted A and B. To do this we first determined the set of significant ASC sites (q-value < 0.01) from cell type A (see Allele-specific chromatin accessibility above). We collected effects and variances for these ASC sites in cell type B, and then used ashR40 to estimate the proportion of these effects that are nonzero. While comparable methods such as Storey’s pi1 only uses p-values63, ashR leverages both effect size and standard error estimates to improve power. We reported the average sharing estimates, however individual values can be found in Supplementary Table 1.
GWAS and eQTL enrichments
To identify genomic annotations enriched for genetic trait heritability we used LD Score regression version 1.0.0 under the partitioning heritability mode with default parameters. We excluded SNPs from the MHC region for this analysis. To assess enrichment of rheumatoid arthritis signal in ASC sets, we compared against the distribution of the p-values for the rheumatoid arthritis GWAS genome-wide (the same summary statistics that were used in the LD Score analysis). To assess enrichment of blood eQTL signal in ASC sets, we compared against the distribution of p-values from all variant-gene pairs available.
Additional details regarding data analysis can be found in the Supplementary Note and the Life Sciences Reporting Summary.
Supplementary Material
Acknowledgements
We thank D. Yao for helping process samples, Chun Jimmie Ye and members of the Greenleaf, Marson, and Pritchard laboratories for helpful conversations and manuscript feedback. We relied on the Flow Cytometry Core at UCSF, which was supported by the Diabetes Research Center grants NIH P30 DK063720 and 1S10OD021822–01, and sequencing data generated by the Stanford Functional Genomics Facility on an Illumina HiSeq 4000 that was purchased with funds from NIH under award number S10OD018220. Sequencing that was generated on an Illumina NovaSeq was supported by the Chan Zuckerberg Biohub. Some of the computing for this project was performed on the Sherlock cluster. Sequencing of ChIP-seq libraries was supported by UCSF Center for Advanced Technology (CAT). We would like to thank Stanford University and the Stanford Research Computing Center for providing computational resources and support that contributed to these research results. Support for D.C. was provided by NLM Training Grant Number T15LM007033. A. Mezger is supported by the Swedish Research Council (grant 2015–06403). F.B. was supported by the Care-for-Rare Foundation and NIH/NIGMS funding for the HIV Accessory & Regulatory Complexes (HARC) Center (P50 GM082250; A. Marson). This work was supported by NIH grants (1R01HG008140 – J.P. serves as PI with subcontract to A. Marson and L.A.C. at UCSF, P30AR070155 to L.A.C., DP3DK111914–01 to A. Marson, P50HG007735 to W.J.G., UM1HG009442 to W.J.G, U19AI057266 to W.J.G.), the Howard Hughes Medical Institute (J.K.P.), the Rheumatology Research Foundation (L.A.C.), the UCSF-Stanford Arthritis Center of Excellence (L.A.C.)(supported in part by the Arthritis Foundation), the Rita Allen Foundation (W.J.G.), the Human Frontiers Science Program grant RGY006S (W.J.G.), the Burroughs Wellcome Fund (A. Marson), and the National Multiple Sclerosis Society (A. Marson; CA 1074-A-21). Both W.J.G and A. Marson are investigators at the Chan Zuckerberg Biohub. Additionally, A. Marson holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund, has received funding from the Innovative Genomics Institute (IGI) and is a member of the Parker Institute for Cancer Immunotherapy (PICI).
Competing interests
Stanford University has filed a provisional patent application on the methods described, and W.J.G. is named as an inventor. W.J.G. is a cofounder of Epinomics and consultant for 10x Genomics and Guardant Health. A.Marson is a co-founder of Arsenal Biosciences and Spotlight Therapeutics. A.Marson serves as on the scientific advisory board of PACT Pharma, is an advisor to Trizell, and was a former advisor to Juno Therapeutics. The Marson Laboratory has received sponsored research support from Juno Therapeutics, Epinomics, Sanofi and a gift from Gilead.
Footnotes
Data Availability Statement
Our GEO RNA-seq data repository: GSE118165
Our GEO ATAC-seq data repository: GSE118189
Our GEO ChIP-seq data repository: GSE126505
Progenitor data GEO accession: GSE74912
Additional supplementary information: http://web.stanford.edu/group/pritchardlab/dataArchive.html
References
- 1.Farh KK et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–43 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47, 1228–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pickrell JK Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am J Hum Genet 94, 559–73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu X et al. Integrating autoimmune risk loci with gene-expression data identifies specific pathogenic immune cell subsets. Am J Hum Genet 89, 496–506 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maurano MT et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trynka G et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet 45, 124–30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L et al. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell 167, 1398–1414 e24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun S et al. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nat Genet 49, 600–605 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim-Hellmuth S et al. Genetic regulatory effects modified by immune activation contribute to autoimmune disease associations. Nat Commun 8, 266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alasoo K et al. Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat Genet 50, 424–431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fairfax BP et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343, 1246949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee MN et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science 343, 1246980 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye CJ et al. Intersection of population variation and autoimmunity genetics in human T cell activation. Science 345, 1254665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simeonov DR et al. Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature 549, 111–115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang H et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547, 173–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onengut-Gumuscu S et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet 47, 381–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Lange KM et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet 49, 256–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buenrostro JD, Giresi PG, Zaba LC, Chang HY & Greenleaf WJ Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corces MR et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 48, 1193–203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moskowitz DM et al. Epigenomics of human CD8 T cell differentiation and aging. Sci Immunol 2(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Veeken J et al. Memory of Inflammation in Regulatory T Cells. Cell 166, 977–990 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He B et al. CD8(+) T Cells Utilize Highly Dynamic Enhancer Repertoires and Regulatory Circuitry in Response to Infections. Immunity 45, 1341–1354 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu B et al. Epigenetic landscapes reveal transcription factors that regulate CD8(+) T cell differentiation. Nat Immunol 18, 573–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leslie R, O’Donnell CJ & Johnson AD GRASP: analysis of genotype-phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics 30, i185–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ostuni R et al. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–71 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Gate RE et al. Genetic determinants of co-accessible chromatin regions in activated T cells across humans. Nat Genet (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hess K et al. Kinetic assessment of general gene expression changes during human naive CD4+ T cell activation. Int Immunol 16, 1711–21 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Diehn M et al. Genomic expression programs and the integration of the CD28 costimulatory signal in T cell activation. Proc Natl Acad Sci U S A 99, 11796–801 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trickett A & Kwan YL T cell stimulation and expansion using anti-CD3/CD28 beads. J Immunol Methods 275, 251–5 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Wortis HH, Teutsch M, Higer M, Zheng J & Parker DC B-cell activation by crosslinking of surface IgM or ligation of CD40 involves alternative signal pathways and results in different B-cell phenotypes. Proc Natl Acad Sci U S A 92, 3348–52 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Belle K et al. Comparative In Vitro Immune Stimulation Analysis of Primary Human B Cells and B Cell Lines. J Immunol Res 2016, 5281823 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodgkin PD, Go NF, Cupp JE & Howard M Interleukin-4 enhances anti-IgM stimulation of B cells by improving cell viability and by increasing the sensitivity of B cells to the anti-IgM signal. Cell Immunol 134, 14–30 (1991). [DOI] [PubMed] [Google Scholar]
- 33.Rieckmann JC et al. Social network architecture of human immune cells unveiled by quantitative proteomics. Nat Immunol 18, 583–593 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Degner JF et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 482, 390–4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kilpinen H et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science 342, 744–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kasowski M et al. Extensive variation in chromatin states across humans. Science 342, 750–2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McVicker G et al. Identification of genetic variants that affect histone modifications in human cells. Science 342, 747–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neph S et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature 489, 83–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van de Geijn B, McVicker G, Gilad Y & Pritchard JK WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat Methods 12, 1061–3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stephens M False discovery rates: a new deal. Biostatistics 18, 275–294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banovich NE et al. Impact of regulatory variation across human iPSCs and differentiated cells. Genome Res 28, 122–131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyle EA, Li YI & Pritchard JK An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 169, 1177–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsh AM et al. Integrative genomic deconvolution of rheumatoid arthritis GWAS loci into gene and cell type associations. Genome Biol 17, 79 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Consortium GTEx. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westra HJ et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45, 1238–1243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krikos A, Laherty CD & Dixit VM Transcriptional activation of the tumor necrosis factor alpha-inducible zinc finger protein, A20, is mediated by kappa B elements. J Biol Chem 267, 17971–6 (1992). [PubMed] [Google Scholar]
- 47.Housley WJ et al. Genetic variants associated with autoimmunity drive NFkappaB signaling and responses to inflammatory stimuli. Sci Transl Med 7, 291ra93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calderon D et al. Inferring Relevant Cell Types for Complex Traits by Using Single-Cell Gene Expression. Am J Hum Genet 101, 686–699 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bank S et al. Associations between functional polymorphisms in the NFκB signaling pathway and response to anti-TNF treatment in Danish patients with inflammatory bowel disease. The Pharmacogenomics Journal 14, 526–534 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Thomson W et al. Rheumatoid arthritis association at 6q23. Nat Genet 39, 1431–3 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lex A, Gehlenborg N, Strobelt H, Vuillemot R & Pfister H UpSet: Visualization of Intersecting Sets. IEEE Trans Vis Comput Graph 20, 1983–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Picelli S et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9, 171–81 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Buenrostro JD et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das S et al. Next-generation genotype imputation service and methods. Nat Genet 48, 1284–1287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal 1, 10–12 (2011). [Google Scholar]
- 56.Bray NL, Pimentel H, Melsted P & Pachter L Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–7 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Harrow J et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22, 1760–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soneson C, Love MI & Robinson MD Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schep AN et al. Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res 25, 1757–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hochberg YBY Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B 57, 289–300 (1995). [Google Scholar]
- 62.McKenna A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Storey JD & Tibshirani R Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100, 9440–5 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.