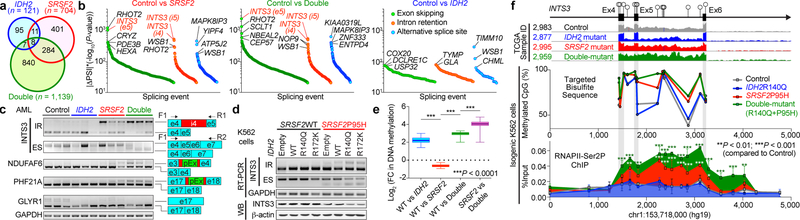
Fig. 3 |. Collaborative effects of mutant IDH2 and SRSF2 on aberrant splicing.
a, Venn diagram showing numbers of differentially spliced events from TCGA AML samples. b, Differentially spliced events (|ΔPSI| > 10% and P < 0.01) in indicated genotype are ranked by y-axis ((|ΔPSI × (−Log10(P-value)) and class of event (e5: exon 5; i4/5: intron 4/5) (PSI and P-values adjusted for multiple comparisons were calculated using PSI-Sigma25). c, Representative RT-PCR results of aberrantly spliced transcripts in AML patient samples (pEx: exon with premature stop-codon; n = 3 patients per genotype; three technical replicates with similar results). d, RT-PCR and WB of INTS3 in isogenic K562 cells (representative images from three biologically independent experiments with similar results). e, Mean log2 fold-change in DNA cytosine methylation (y-axis) at regions of genomic DNA encoding mRNA which undergo differential splicing (x-axis). DNA methylation levels were determined by eRRBS (n = 3 per genotype; the mean represented by the line inside the box and the box expands from the 25th to 75th percentiles with whiskers drawn down to the 2.5 and 97.5 percentiles; one-way ANOVA with Tukey’s multiple comparison test; ***P < 2.2e-16). f, Diagram of the genomic locus of INTS3 around exons 4–6 with CpG dinucleotides, representative RNA-seq from four AML patients, targeted bisulfite sequencing (n = 1 per genotype), and results of anti-RNAPII-Ser2P ChIP-walking experiments (n = 3; the mean ± s.d.; two-way ANOVA with Tukey’s multiple comparison test). *P < 0.05; **P < 0.01; ***P < 0.001.