SUMMARY
Development and differentiation are associated with profound changes to histone modifications, yet their in vivo function remains incompletely understood. Here, we generated mouse models expressing inducible histone H3 lysine-to-methionine mutants, which globally inhibit methylation at specific sites. Mice expressing H3K36M developed severe anemia with arrested erythropoiesis, a marked hematopoietic stem cell defect, and rapid lethality. By contrast, mice expressing H3K9M survived up to a year and showed expansion of multipotent progenitors, aberrant lymphopoiesis and thrombocytosis. Additionally, some H3K9M mice succumbed to aggressive T cell leukemia/lymphoma while H3K36M mutants exhibited differentiation defects in testis and intestine. Mechanistically, H3K36M and H3K9M reduced H3K36 and H3K9 trimethylation patterns genome-wide and altered chromatin accessibility and gene expression landscapes. Strikingly, discontinuation of transgene expression largely restored differentiation programs. Our work shows that individual chromatin modifications are required at several specific stages of differentiation and introduces powerful tools to interrogate their roles in vivo.
INTRODUCTION
During development and tissue homeostasis, immature progenitor cells give rise to differentiated cells without altering genomic content. Post-translational histone modifications are thought to guide these processes by altering gene expression1,2. For example, H3K36me3 and H3K4me3 are associated with actively transcribed regions, while H3K9me3 and H3K27me3 correlate with repressed regions. However, the biological function of specific modifications is difficult to determine through mutational analyses3 as canonical histones are encoded by many genes in the mammalian genome4. As a result, the function of individual histone marks has been inferred from mutational analysis of histone modifying enzymes. Lysine methylation of histone H3 is regulated by multiple, redundant methyltransferases (KMT) and demethylases (KDMs). Since these enzymes also have non-histone substrates and act as scaffolds to recruit other factors, some KMT/KDM knockout phenotypes may be unrelated to the histone marks5–8. Hence, the direct biological impact of histone tail modifications remains incompletely understood.
Lysine-to-methionine (K-to-M) or lysine-to-isoleucine (K-to-I) point mutations on the variant histone, H3.3, suppress methylation levels at their respective sites9–14. Such mutations interfere with the active site of their cognate SET domain-containing KMTs9,10,12,15,16, resulting in a global decrease of individual histone marks. K-to-M mutations effectively function as hypomorphs, allowing the study of histone marks in cases where genetic disruption of the corresponding KMTs is lethal17. Of note, H3.3K36M mutations were identified in bone and soft-tissue sarcomas9–12,15,16, as well as head and neck cancer14,18. Moreover, H3.3K9M mutations in drosophila lead to defects in position effect variegation15,16. However, the impact of site-specific H3 methylation in untransformed, primary cells or mice remains incompletely explored. Here, we utilize K-to-M histone mutations in vivo and in vitro to modulate levels of histone methylation during complex biological processes, including tissue homeostasis, cellular differentiation and cancer.
RESULTS
Transgenic system to suppress H3K9 and H3K36 methylation
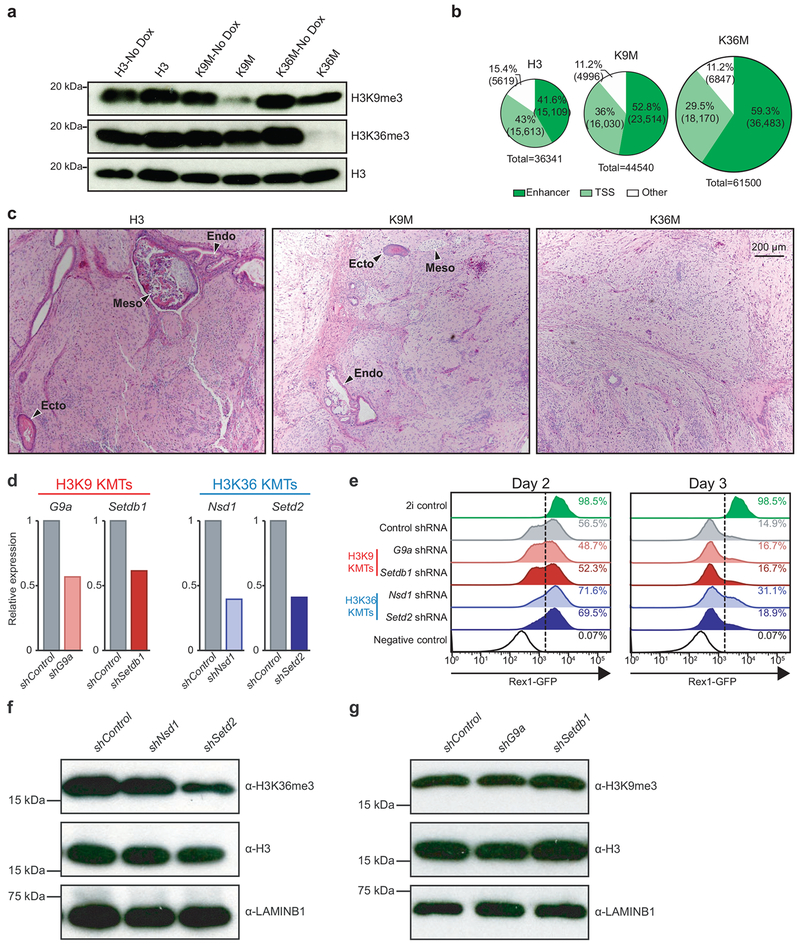
We targeted cDNAs encoding histone H3.3K9M, H3.3K36M, and wild-type H3.3 (hereafter referred to as H3K9M, H3K36M, and H3) into mouse embryonic stem (ES) cells using a site-specific, single-copy integration system19,20 (Fig. 1a). Our inducible system facilitated rapid and specific expression of the histone constructs following doxycycline (dox) administration (Fig. 1b and Extended Data 1a). Mutant histones partitioned with the nuclear fraction, suggesting that they were properly incorporated into chromatin (Fig. 1b). Consistent with previous reports, expression of H3K9M and H3K36M dramatically reduced the global levels of H3K9me3 and H3K36me310,14,16 (Fig. 1c). Dimethyl marks at both sites were also suppressed, albeit less appreciably, and H3K27me3 levels were slightly elevated with expression of H3K36M. Importantly, we observed no crosstalk between mutant histones (i.e., expression of H3K9M did not alter H3K36 methylation and vice versa) and expression of H3 had no effect on the methylation status of either residue (Fig. 1c and Extended Data 1a).
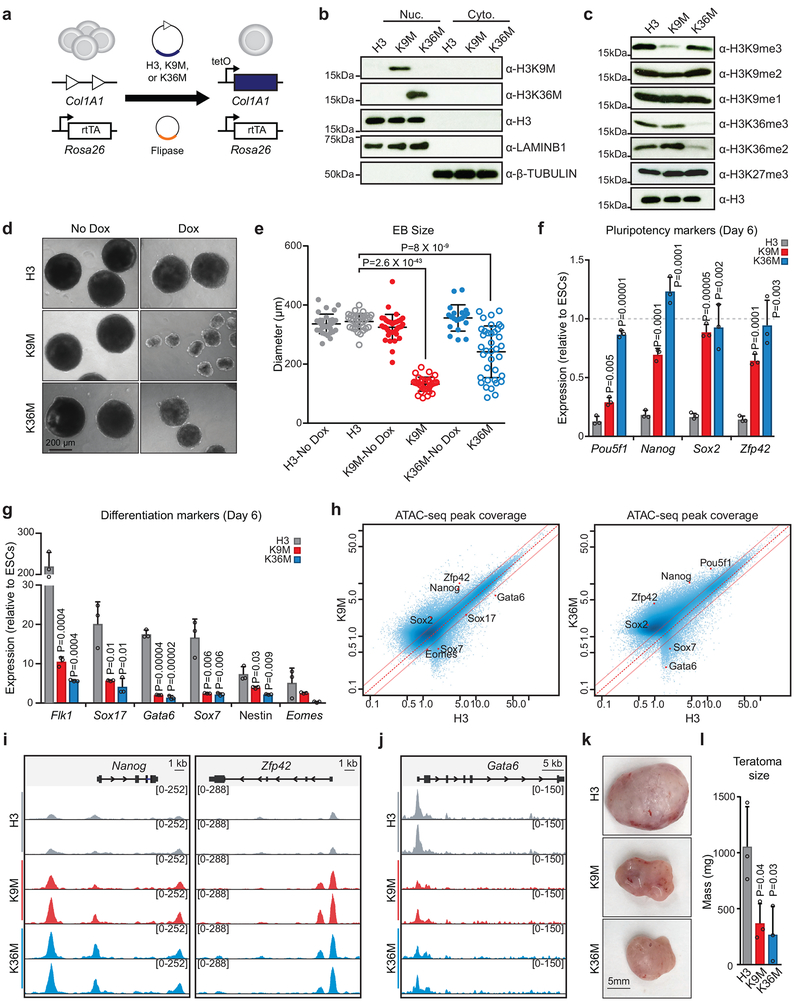
Fig. 1. Dox-inducible K-to-M histone mutants globally suppress site-specific histone methylation and impair differentiation of ES cells.
(a) A schematic of the strategy used to generate cells harboring inducible histone constructs. (b) Western blot analysis of nuclear (Nuc.) and cytoplasmic (Cyto.) fractions from ES cells expressing mutant histones. The H3 loading control is the same image as panel c. (c) Western blot analysis for the indicated histone modifications in ES cells expressing mutant histones. The H3 loading control is the same image as panel b. (d) Images of EBs at day 9 of induction with and without expression of H3K9M and H3K36M; scale bar=200 μm. (e) Quantification of EB diameters for each condition in technical replicate (H3, n=32; H3+dox, n=34; H3K9M, n=37; H3K9M+dox, n=33; H3K36M, n=20; H3K36M+dox, n=37). The center bar represents the mean and the whiskers represent the standard deviation of the mean. Statistical significance was determined using a two-tailed unpaired Student’s t-test. (f) qRT-PCR for pluripotency markers at day 6 of induction. Columns represent the mean and error bars represent standard deviation of the mean for n=3 independent experiments. Statistical significance was determined using a two-tailed unpaired Student’s t-test. (g) qRT-PCR for differentiation markers at day 6 of induction. Columns represent the mean and error bars represent standard deviation of the mean for n=3 independent experiments. Statistical significance was determined using a two-tailed unpaired Student’s t-test. (h) Scatter plots comparing ATAC-seq peak coverage for each mutant histone sample compared to H3 control. (i) Gene tracks showing ATAC-seq data for pluripotency genes. (j) Gene tracks showing ATAC-seq data for differentiation-associated genes. (k) Images of teratomas expressing H3K9M and H3K36M; scale bar=5 mm. (l) Quantification of teratoma mass for each condition in biological triplicate. Columns represent the mean and error bars represent standard deviation of the mean. N=3 teratomas. Statistical significance was determined using a two-tailed unpaired Student’s t-test. See source data for full membrane Western blot images. Data in b,c,d,k are representative of 3 independent experiments.
H3K9M and H3K36M expression impairs ES cell differentiation
To study the impact of our mutants on pluripotent stem cell differentiation, we generated embryoid bodies (EB). Expression of both H3K9M and H3K36M yielded significantly smaller EBs compared to the control (Fig. 1d,e), suggesting a defect in differentiation. Consistent with this observation, EBs expressing H3K9M and H3K36M retained expression of the pluripotency genes Oct4, Nanog, Sox2 and Zfp42 (Rex1) compared to control EBs (Fig. 1f). Moreover, both mutant EBs expressed markedly lower levels of the differentiation markers Flk1, Eomes, Gata6, Sox7, Sox17 and Nestin, relative to control EBs (Fig. 1g). ATAC-Seq analysis of these samples suggested that chromatin was more accessible overall in the mutant EBs, consistent with a less differentiated state (36,341 accessible regions in control; 44,540 in H3K9M; 61,500 in H3K36M; Extended Data 1b). For example, chromatin was more open in regions associated with crucial pluripotency genes that remained active in H3K9M- and H3K36M-expressing EBs (e.g., Pou5f1, Nanog and Zfp42) (Fig. 1h,i). Conversely, chromatin associated with differentiation markers (e.g., Gata6, Sox7) was closed in mutant EBs compared to control (Fig. 1h,j). In line with the observed defect in EB differentiation, cells expressing either H3K9M or H3K36M generated significantly smaller teratomas upon injection into the flanks of immunocompromised mice (Fig. 1k,l). Histological analysis revealed that control cells formed well-differentiated teratomas containing structures characteristic of all three germ layers (Extended Data1c). By contrast, H3K36M, and to a lesser extent, H3K9M produced poorly differentiated teratomas with large, undifferentiated areas and fewer differentiated structures (Extended Data 1c). Together, these data suggest that H3K9 and H3K36 methylation are crucial for exiting pluripotency as well as for timely induction of differentiation.
To test whether individual suppression of H3K9 and H3K36 KMTs phenocopy K-to-M mutations, we transduced Rex1-dGFP ES cells21 with shRNAs targeting H3K9-specific KMTs (G9a or Setdbl) or H3K36-specific KMTs (Nsd1 or Setd2) (Extended Data 1d), allowing for a sensitive, flow-cytometry-based readout of pluripotency exit. Indeed, suppression of H3K36 KMTs Nsd1 or Setd2 partially recapitulated the differentiation block observed with H3K36M expression at day 2 of EB formation, which coincided with a modest reduction in H3K36me3 levels (Extended Data 1e,f). These results are consistent with previous findings in Setd2 knockout ES cell lines22. By contrast, neither G9a nor Setdbl suppression impaired differentiation in our exit system. However, Western blot analysis revealed that knockdown of these enzymes was insufficient to reduce H3K9me3 levels (Extended Data 1g). Previous studies showed that disruption of H3K9 KMTs is compensated by alternative KMTs22,23, which likely explains the lack of an observable reduction in H3K9me3 following H3K9 KMT knockdown. Of note, shRNAs that more efficiently suppressed G9a and Setdbl led to widespread differentiation in ES cells under self-renewal conditions, precluding the use of more potent hairpins, which is in line with previous results24. These difficulties in interpreting KMT shRNA knockdown experiments thus underscore the power of our K-to-M mutations to study histone methylation.
Expression of H3K9M and H3K36M perturbs tissue homeostasis
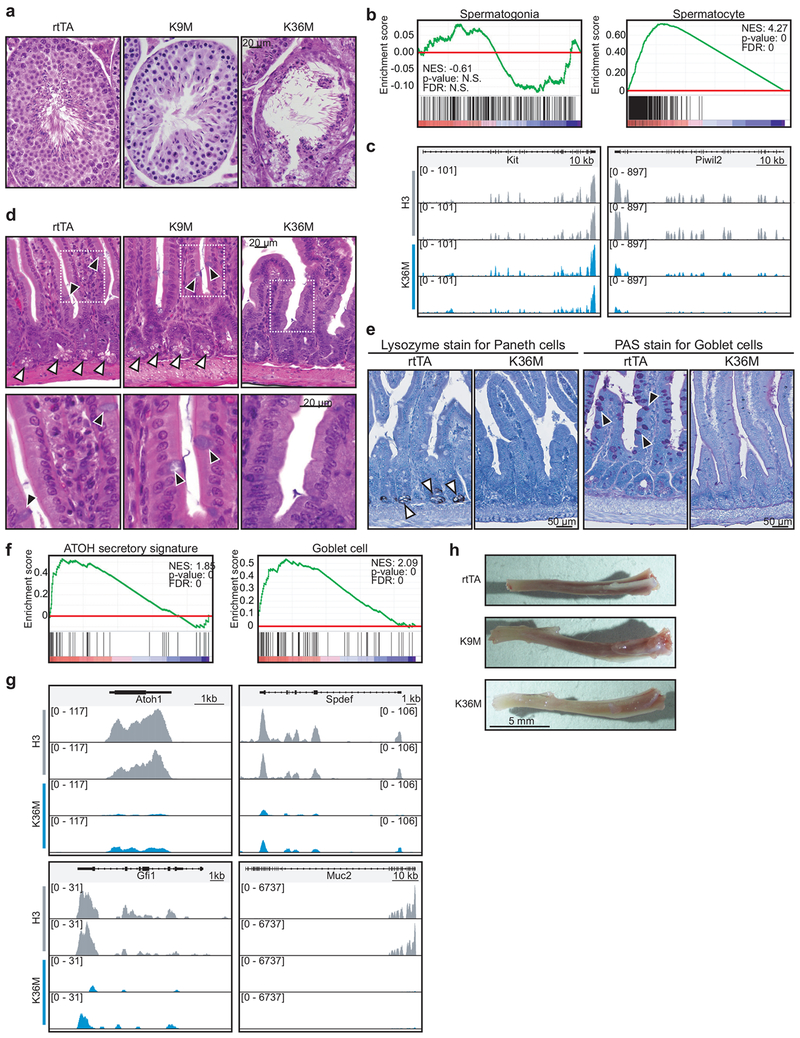
We next produced transgenic mice from our targeted ES cells and induced experimental and control animals with dox at 6–8 weeks of age (Fig. 2a). Strikingly, H3K36M mice developed poor body condition and became moribund within 4–7 weeks of induction (Fig. 2b). Upon postmortem examination, male H3K36M mice exhibited testicular atrophy with deterioration of the seminiferous tubule and concomitant loss of mature testis cells (Extended Data 2a). RNA-seq analysis of testes after 4 weeks of dox treatment suggested that the transcriptional program associated with undifferentiated spermatogonia is unaffected in H3K36M mice based on gene set enrichment analysis (GSEA) and spermatogonia marker genes (e.g., Stra8, Kit, Prdm9), (Extended Data 2b,c; left panels). However, we found a marked depletion of the more mature spermatocyte-specific program (e.g. Piwil2, Meioc, Hormadl) (Extended Data 2b,c; right panels). These results point to a specific role for H3K36 methylation between the spermatogonia and spermatocyte stages of differentiation, resembling the phenotype of Setd2 knockout mice25.
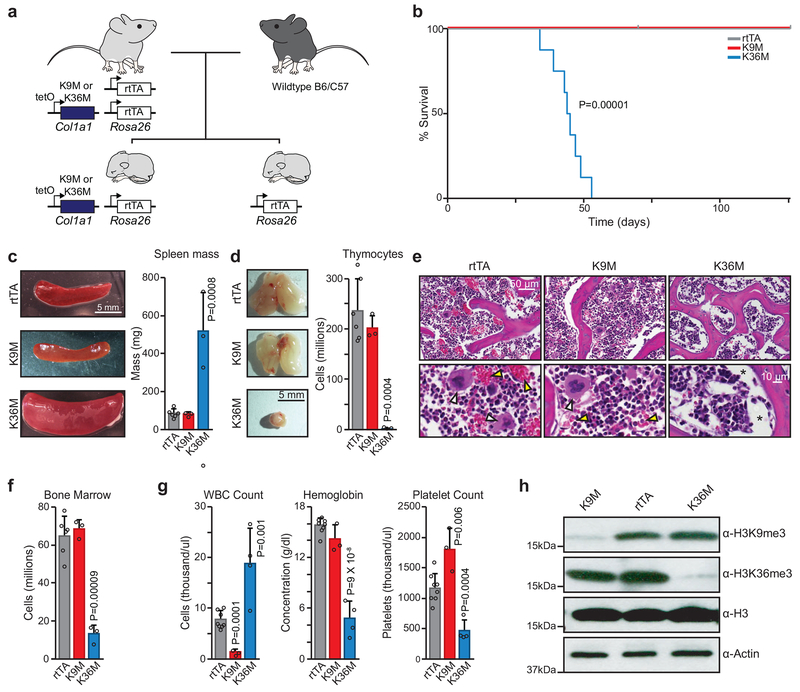
Fig. 2. In vivo expression of H3K36M leads to rapid death with severe anemia and organ abnormalities.
(a) A schematic of the strategy used to generate transgenic mice with littermate controls. (b) A survival curve for transgenic mice following doxycycline administration (rtTA, n=10 mice; H3K9M, n=3; H3K36M, n=8). Statistical significance was determined using a two sided Log-rank (Mantel-Cox) test. (c) Representative images of spleens taken from mice induced for 4 weeks and quantified by mass. Scale bar=5 mm. Columns represent the mean and error bars represent standard deviation of the mean for biological replicates (rtTA, n=6; H3K9M, n=3; H3K36M, n=3). Statistical significance was determined using a two-tailed unpaired Student’s t-test. (d) Representative thymus images taken from mice induced for 4 weeks with total thymocytes quantified by cell count. Scale bar=5 mm. Columns represent the mean and error bars represent standard deviation of the mean for biological replicates (rtTA, n=6; H3K9M, n=3; H3K36M, n=3). Statistical significance was determined using a two-tailed unpaired Student’s t-test. (e) Histological analysis of bone from mice induced for 4 weeks. Scale bar=50 μm (upper panels) and 10 μm (lower panels). White arrowheads indicate megakaryocytes, yellow arrowheads indicate mature erythroid cells; asterisks indicate areas with low cellularity. (f) Bone marrow cellularity quantified by cell count. Error bars represent standard deviation of the mean for biological replicates (rtTA, n=6; H3K9M, n=3; H3K36M, n=3). Statistical significance was determined using a two-tailed unpaired Student’s t-test. (g) CBC data for white blood cells, hemoglobin, and platelets sampled at 6 weeks (as H3K36M mice became moribund). Error bars represent standard deviation of the mean for biological replicates (rtTA, n=8; H3K9M, n=3; H3K36M, n=4). Statistical significance was determined using a two-tailed unpaired Student’s t-test. (h) Western blot analysis for tri-methyl H3K9 and H3K36 in LKS cells sorted from transgenic mice following one week of doxycycline administration. See source data for full membrane Western blot images. Data in e,h are representative of 3 independent experiments.
In the intestine, PAS+ goblet and Lysozyme+ Paneth cells were absent in H3K36M mice while the typical crypt-villus structure of the epithelium was preserved, suggesting defects in secretory lineage commitment (Extended Data 2d,e). Indeed, RNA-seq analysis demonstrated that H3K36M intestinal cells were depleted for secretory lineage-associated transcriptional programs (Extended Data 2f; left panel). Key regulators of secretory differentiation such as Atoh1 and Gfi1 were strongly reduced (Extended Data 2g; left panels). Likewise, mRNA expression characteristic of goblet cells was disrupted in the intestine of mice expressing H3K36M, with a severe reduction in transcript levels for the goblet cell markers Spdef and Muc2 (Extended Data 2f,g; right panels). Thus, H3K36M disrupts distinct differentiation programs in the intestine.
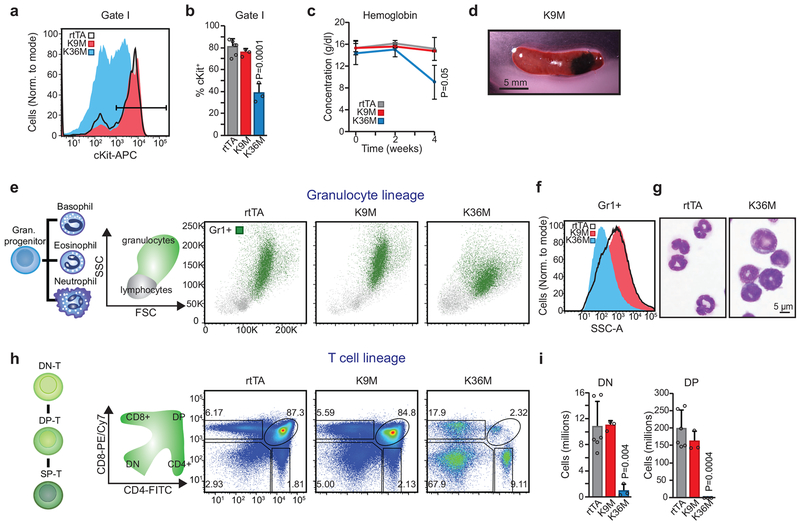
Strikingly, H3K36M mice also exhibited splenomegaly, thymic atrophy, and lack of red marrow in leg bones (Fig. 2c,d and Extended Data 2h). Histological sections and cell counts revealed markedly decreased bone marrow cellularity in H3K36M mice, with a complete absence of megakaryocytes and few terminally differentiated erythroid cells compared to both H3K9M mice and rtTA littermate controls (Fig. 2e,f). Complete blood counts (CBCs) pointed to a significant increase in white blood cell numbers (predominantly from circulating early erythroid cells), reduced platelet numbers, and severely reduced hemoglobin levels in H3K36M-expressing mice (Fig. 2g). By contrast, H3K9M mice displayed partially reciprocal abnormalities, including a 4-fold decrease in white blood cells and a 1.5-fold increase in platelets (Fig. 2g). We confirmed strong reduction of H3K9me3 and H3K36me3 levels in H3K9M and H3K36M progenitor cells (lineage−, c-Kit+, Sca1−), respectively, using Western blot analysis (Fig. 2h). However, levels of total H3 were not appreciably elevated with dox administration, suggesting that the mutant transgenes are expressed at physiological levels. Together, these results point to widespread and distinct functions of H3K9 and H3K36 methylation in tissue maintenance and blood cell homeostasis.
K-to-M mutations influence cell fate at discrete points during hematopoiesis
To refine our hematopoietic analysis, we first examined the myeloid lineages after 4 weeks of mutant histone induction, before mice were overtly sick. Consistent with the severe anemia in H3K36M mice, we observed a dramatic increase in early erythroid cells, such as progenitors (CFUEs) and proerythroblasts with a concomitant decrease in more mature stages, specifically Ter119+ erythroblasts and mature enucleated erythrocytes (Fig. 3a,b, see Gate I). Notably, the immature erythroid cells expressed abnormally low levels of the progenitor marker c-Kit (Extended Data 3a,b), suggesting that they were not simply expanded progenitors. Indeed, May-Grünwald-Giemsa (MGG) staining revealed that the abnormal cells exhibit morphologies characteristic of both immature progenitors (pro-erythroblasts) and more mature precursors (erythroblasts), despite the absence of the erythroblast marker, Ter119 (Fig. 3c). Critically, hemoglobin levels were significantly lower by 4 weeks of H3K36M induction and dropped precipitously by death at 5–6 weeks of induction, confirming a severe differentiation defect (Fig. 2g and Extended Data 3c). H3K9M mice presented no overt defects in erythropoiesis (Fig. 3a–c and Extended Data 3a–c). Collectively, these data demonstrate that global loss of H3K36 methylation but not H3K9 methylation results in a robust block in erythroid differentiation, yet preserves early, albeit abnormal and ineffective, erythropoiesis.
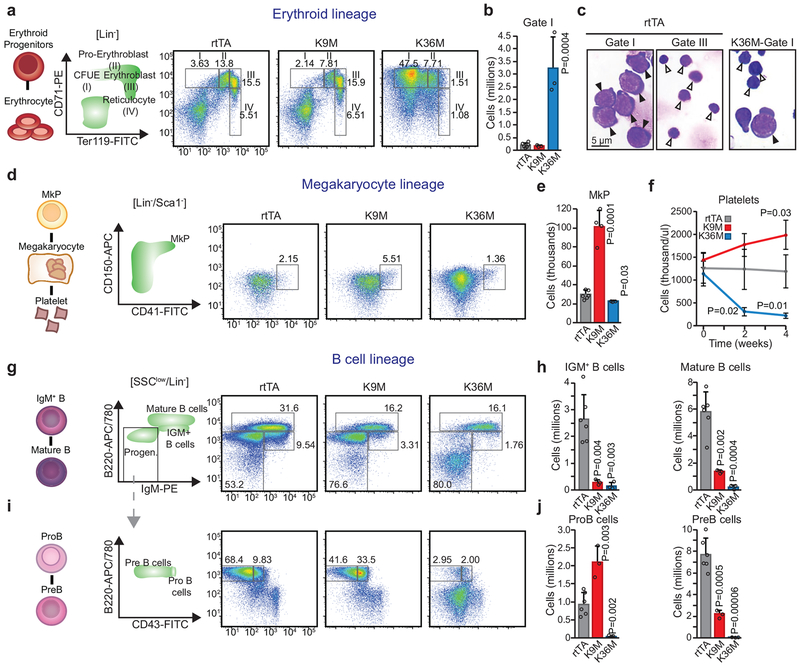
Fig. 3. In vivo expression of H3K36M dramatically alters myeloid and lymphoid differentiation.
(a) Flow cytometry analysis for erythroid cells. Frequencies for each cell type are indicated as a percentage of the parent gate. (b) Quantification of erythroid progenitors. Columns represent the mean and error bars represent standard deviation of the mean for biological replicates (rtTA, n=6; H3K9M, n=3; H3K36M, n=3). Statistical significance was determined using a twotailed unpaired Student’s t-test. (c) Cytospin images of erythroid progenitors sorted from H3K36M and rtTA mice. Black arrowheads indicate proerythroblasts, white arrowheads indicate erythroblasts; scale bar=5 pm. (d) Flow cytometry analysis for megakaryocyte progenitors (MkPs). Frequencies for each cell type are indicated as a percentage of the parent gate. (e) Quantification of MkPs. Columns represent the mean and error bars represent standard deviation of the mean for biological replicates (rtTA, n=6; H3K9M, n=4; H3K36M, n=3). Statistical significance was determined using a two-tailed unpaired Student’s t-test. (f) Time course analysis for platelets assessed by CBC. Columns represent the mean and error bars represent standard deviation of the mean for biological replicates (rtTA, n=5; H3K9M, n=4; H3K36M, n=3). Statistical significance was determined using the Holm-Sidak method. (g) Flow cytometry analysis for B cells. Frequencies for each cell type are indicated as a percentage of the parent gate. (h) Quantification of mature B cells and IGM+ B cells. Columns represent the mean and error bars represent standard deviation of the mean for biological replicates (rtTA, n=6; H3K9M, n=3; H3K36M, n=3). Statistical significance was determined using a two-tailed unpaired Student’s t-test. (i) Flow cytometry analysis for B cell progenitors. Frequencies for each cell type are indicated as a percentage of the parent gate. (j) Quantification of ProB cells and PreB cells. Columns represent the mean and error bars represent standard deviation of the mean for biological replicates (rtTA, n=6; H3K9M, n=3; H3K36M, n=3). Statistical significance was determined using a two-tailed unpaired Student’s t-test. Data in a,c,d,g,i are representative of 3 independent experiments.
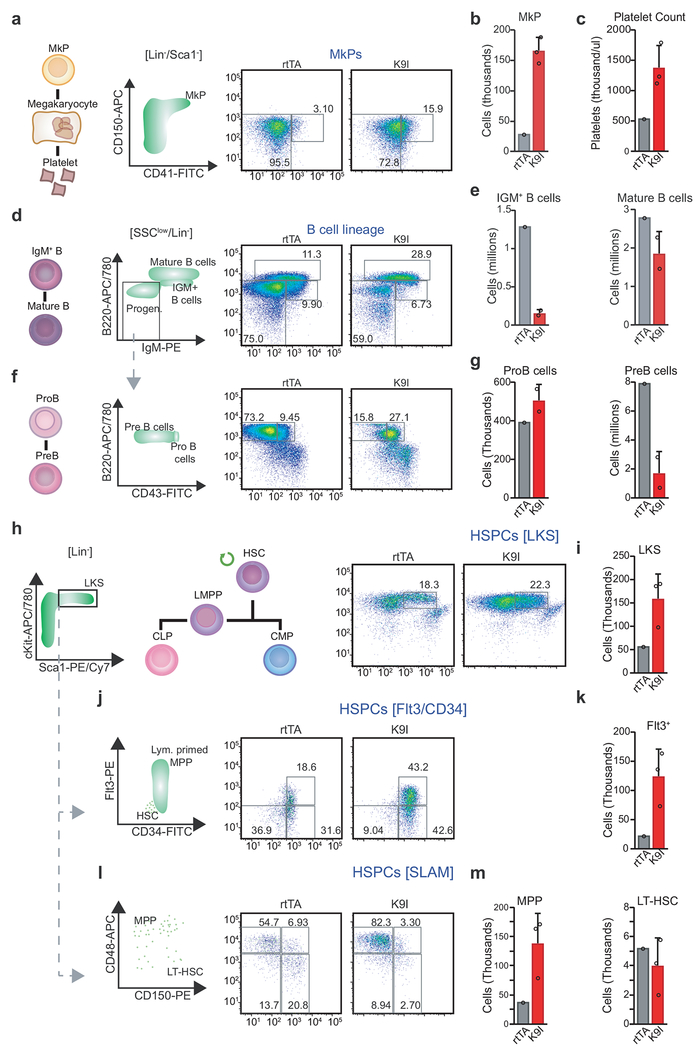
Considering the abnormal platelet counts observed with both mutants (Fig. 2g), we next quantified megakaryocyte progenitors (MkPs) by flow cytometry following 4 weeks of dox induction. MkPs in H3K9M mice increased nearly 3-fold while H3K36M expression caused a subtle but significant decrease in MkP numbers (Fig. 3d,e). Serial CBCs revealed that H3K36M and H3K9M expression had reciprocal effects on platelet numbers as early as 2 weeks after induction (Fig. 3f). Moreover, 2 of 3 examined H3K9M mice suffered spleen infarction (Extended Data 3d), which may result from the increased platelet numbers and/or altered platelet function. Thus, reduced H3K36/H3K9 methylation has opposite effects on the megakaryocyte lineage.
During flow cytometric analysis of H3K36M mice, we noticed a decrease of cells exhibiting high side scatter (SSC), a measure of granularity that identifies mature granulocytes. Accordingly, bone marrow cells positive for the pangranulocyte marker Gr1 exhibited reduced SSC compared with both control and H3K9M cells (Extended Data 3e,f). Moreover, sorted granulocytes from H3K36M animals appeared underdeveloped after MGG staining with thicker, less segmented nuclei and more immature (blue) cytoplasm compared to control cells (Extended Data 3g). We conclude that H3K36M expression impedes granulocyte maturation without impacting early granulocyte specification.
Strikingly, mice expressing either H3K9M or H3K36M exhibited reduced B cell numbers (Fig. 3g,h). Analysis of B cell progenitors revealed that H3K9M induction led to an accumulation of ProB cells at the expense of PreB cells, indicating a specific block during the ProB-to-PreB stage of differentiation (Fig. 3i,j). By contrast, mice expressing H3K36M were nearly devoid of early B cells, suggesting an earlier block in lymphoid differentiation. Supporting this notion, H3K36M mice exhibited thymic atrophy (Fig. 2D) with a severe reduction in CD4/CD8 double-positive T cells compared to H3K9M mice (Extended Data 3h,i). These data show that H3K36 methylation levels affect early lymphoid specification, while H3K9 methylation is required for the transition of more committed progenitor states.
H3K9M and H3K36M have distinct effects on HSPC maintenance
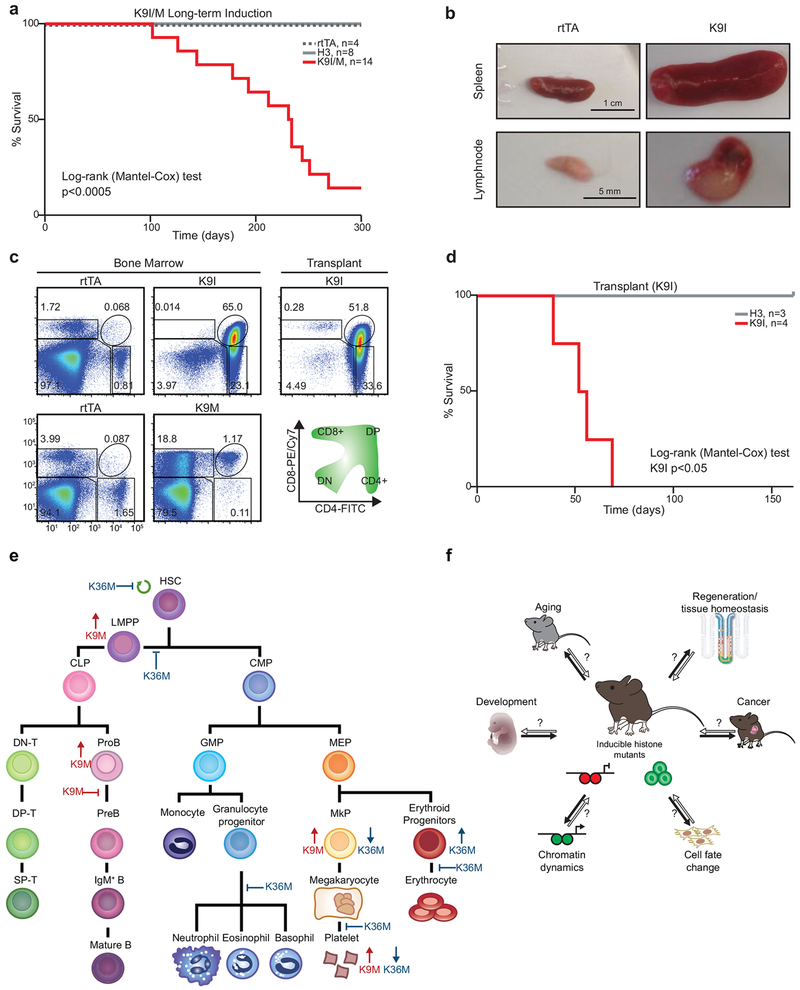
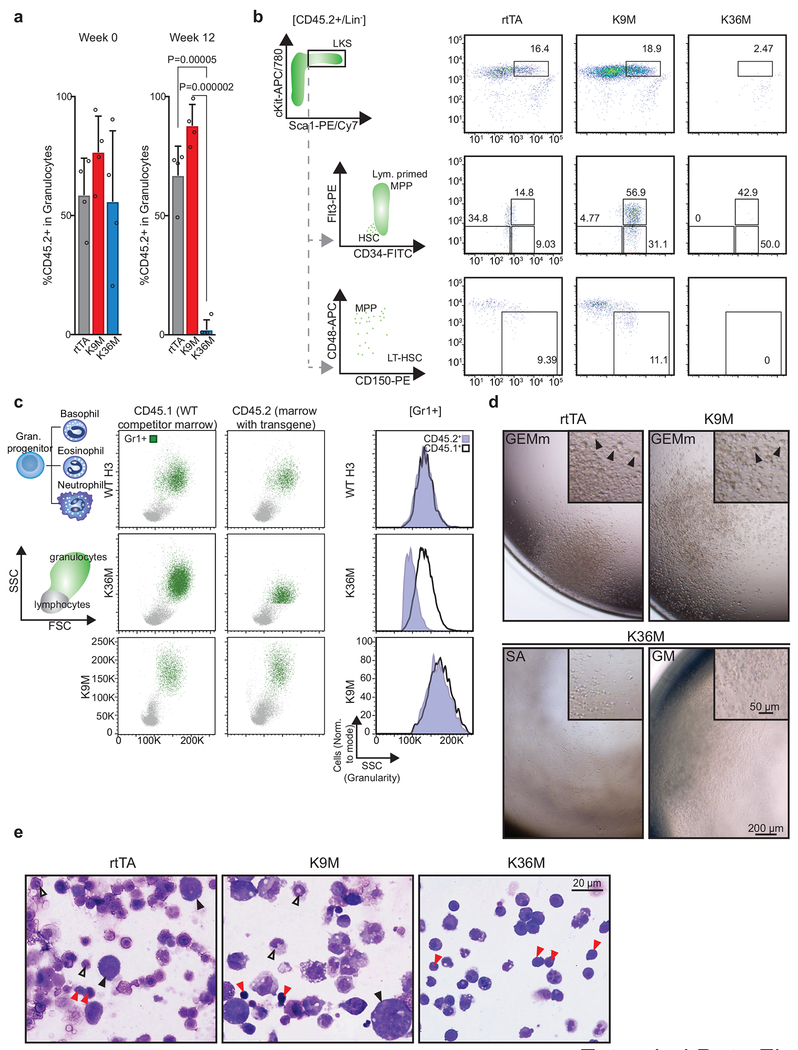
Within the bone marrow, the lineage−, cKit+, and Sca1+ (LKS) population contains all long-term HSCs (LT-HSCs) as well as short term HSCs (ST-HSCs), and multipotent progenitors (MPPs). H3K9M induction led to a robust increase in the number of LKS cells (Fig. 4a,b), suggesting that H3K9 methylation limits their expansion. More detailed analysis using Flt3 and SLAM markers suggested that the increase in LKS cells was due to increased abundances of ST-HSCs and MPPs (Fig. 4c–f), while LT-HSCs were not significantly altered (Fig. 4f, right panel). Consistent with this notion, a large proportion of the LKS population in H3K9M mice expressed high levels of Flt3, which marks lymphoid-primed multipotent progenitors (LMPPs; Fig. 4c,d)26,27 In stark contrast, expression of H3K36M reduced the number of LKS cells, LT-HSCs, and LMPPs (Fig. 4a–f), suggesting a potent role of H3K36 methylation during very early lymphoid differentiation as well as stem cell maintenance, consistent with a recent report that implicates Setd2 in HSPC self-renewal28. Together, these data show that H3K36 and H3K9 methylation have profound, partially opposing effects on early HSPC populations.
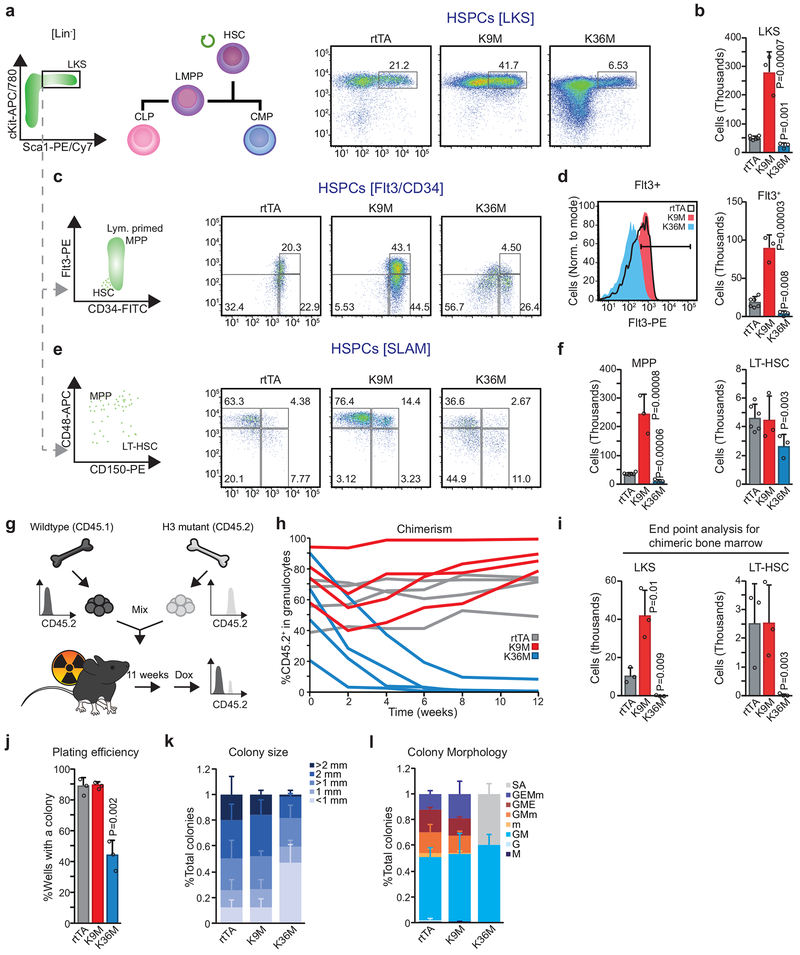
Fig. 4. Expression of H3K36M and H3K9M have opposing, cell autonomous effects on HSPC function.
(a) Flow cytometry analysis of LKS cells. Frequencies as a percentage of parent gate. (b) Quantification of LKS cells. Columns represent mean and error bars represent standard deviation of mean (biological replicates, rtTA, n=6; H3K9M, n=3; H3K36M, n=3, two-tailed unpaired Student’s t-test). (c) Analysis of stem cell populations, gated on LKS cells. Frequencies shown as a percentage of parent gate. (d) Flt3+ distribution within LKS cells (left panel) with quantification of LMPPs (Right panel), defined as Flt3high cells (top 25% of Flt3+ cells set on rtTA control). Columns represent means and error bars standard deviations (biological replicates, rtTA, n=6; H3K9M, n=3; H3K36M, n=3, two-tailed unpaired Student’s t-test). (e) Analysis of SLAM markers, gated on LKS cells. Frequencies indicated as a percentage of the parent gate. (f) Quantification of MPPs (left panel) and long-term HSCs (right panel). Columns represent the mean +/− standard deviation (biological replicates, rtTA, n=6; H3K9M, n=3; H3K36M, n=3, two-tailed unpaired Student’s t-test). (g) Scheme of competitive bone marrow transplant assay. (h) Time course of the contribution of mutant or rtTA control bone marrow in a competitive bone marrow transplant assay. (i) Quantification for donor-derived, CD45.2+ LKS cells and long-term hematopoietic stem cells at conclusion of competitive bone marrow transplant assays. Columns represent the mean +/− standard deviation (n=3, biological replicates, two-tailed unpaired Student’s t-test). (j) Quantification of plating efficiency for methylcellulose assays. Columns represent the mean +/− standard deviation (rtTA, n=3; H3K9M, n=3; H3K36M, n=3, biological replicates, two-tailed unpaired Student’s t-test). (k) Quantification of colony size from methycellulose assays. Columns represent the means and error bars represent standard deviations (rtTA, n=3; H3K9M, n=3; H3K36M, n=3, biological replicates). (l) Quantification of colony morphology from methycellulose assays. SA=sparse, atypical; G=granulocyte; M=monocyte; E=erythrocyte; m=megakaryocyte. Columns represent the mean +/− standard deviation (rtTA, n=3; H3K9M, n=3; H3K36M, n=3, biological replicates). Data in a,c,e are representative of 3 independent experiments.
To study the mutants’ intrinsic role in HSCs, we analyzed hematopoiesis after competitive bone marrow transplantation into mice with an unperturbed microenvironment (Fig. 4g). Recipients were induced with dox 11 weeks after transplantation to ensure complete engraftment (Fig. 4h). Strikingly, H3K36M expressing granulocytes (CD45.2+, mutant donor-derived) progressively diminished compared to rtTA control granulocytes and became undetectable by 12 weeks (Fig. 4h and Extended Data 4a). By contrast, the contribution from H3K9M expressing granulocytes remained robust over this time period (Fig. 4h and Extended Data 4a). To confirm that these effects reflected impaired HSCs, we examined the levels of donor-derived HSPCs via flow cytometry at the conclusion of the time course. Indeed, H3K36M donor-derived HSCs were absent after 12 weeks of induction (Fig. 4i and Extended Data 4b). Moreover, granulocytes exhibited an abnormal SSC profile akin to H3K36M transgenic mice, indicating that the observed granulocyte defect is blood cell intrinsic (Extended Data 4c). Similar to transgenic mice, the pool of H3K9M-expressing LKS cells and LMPPs was expanded while the number of LT-HSCs was not significantly altered in H3K9M bone marrow recipients, confirming that these phenotypes are also cell-intrinsic (Fig. 4i and Extended Data 4b). These results demonstrate that both H3K36M and H3K9M induction cause cell-autonomous early hematopoietic defects.
To study the HSC defect at a single-cell level in vitro, we induced mutant and control mice for 4 weeks and sorted single LT-HSCs into 96-well plates in the presence of a replete cytokine cocktail and dox. H3K36M expressing LT-HSCs showed markedly diminished plating efficiency and generated smaller colonies compared to both control and H3K9M cells (Fig. 4j,k). Scoring for morphology, we observed severe lineage restriction in colonies derived from H3K36M expressing LT-HSCs (Fig. 4l). Specifically, no erythroid differentiation was appreciable by light microscopy after H3K36M induction, although cells with morphology characteristic of arrested, early erythroid lineage differentiation were detected after MGG staining (Extended Data 4d,e). These findings recapitulate the abnormalities observed in the bone marrow and demonstrate that the erythroid differentiation defect is lineage-intrinsic. Moreover, no megakaryocytes were detected in methylcellulose assays after H3K36M induction, as observed in mice (Fig. 4l and Extended Data 4d,e). By contrast, colonies from HSCs expressing H3K9M comprised all differentiated cell types at expected frequencies (Fig. 4l and Extended Data 4d,e). Collectively, these in vitro results corroborate our conclusion that H3K36M but not H3K9M induction profoundly perturbs HSC proliferation and differentiation.
H3K9M and H3K36M expression elicit aberrant transcriptional programs
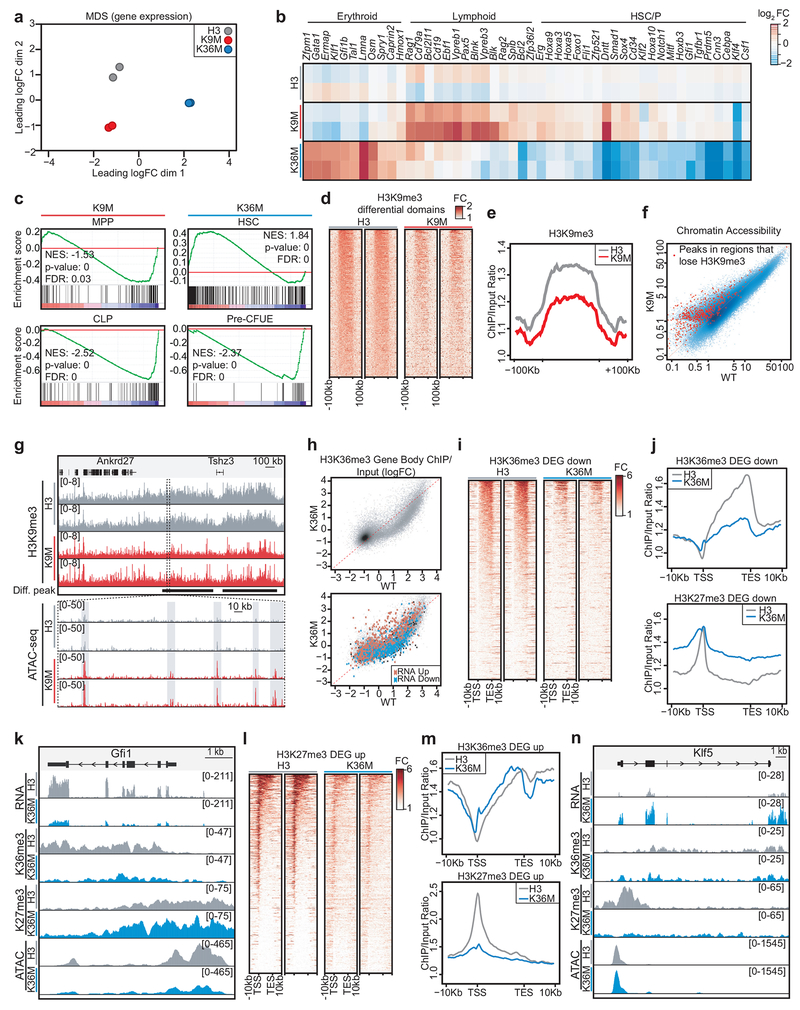
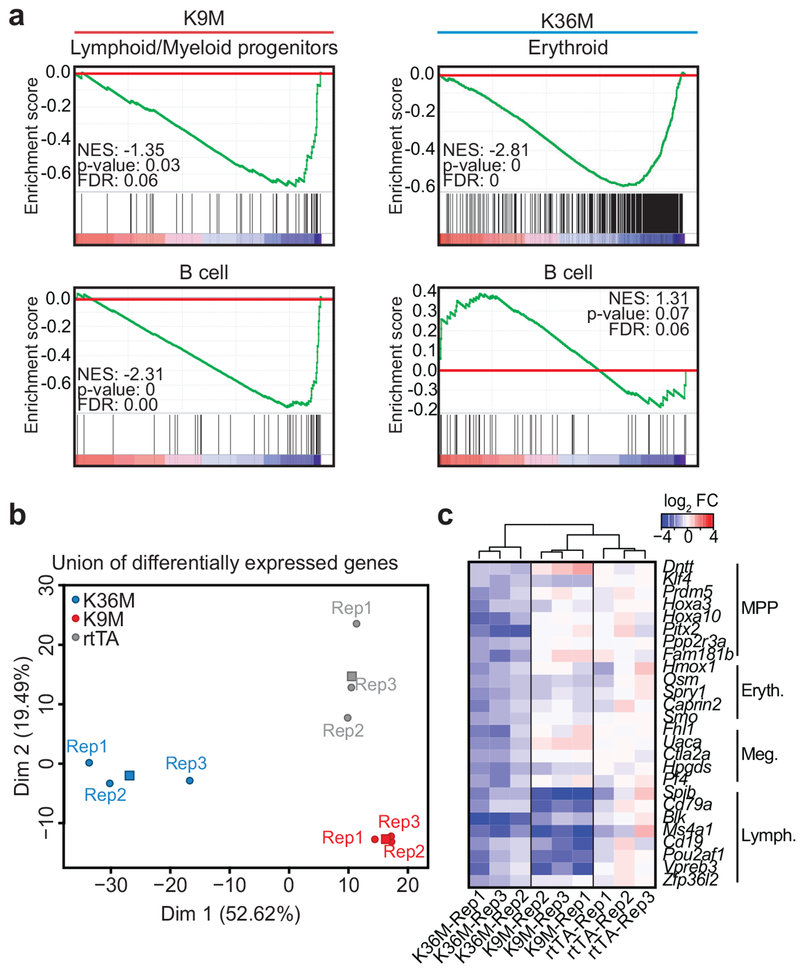
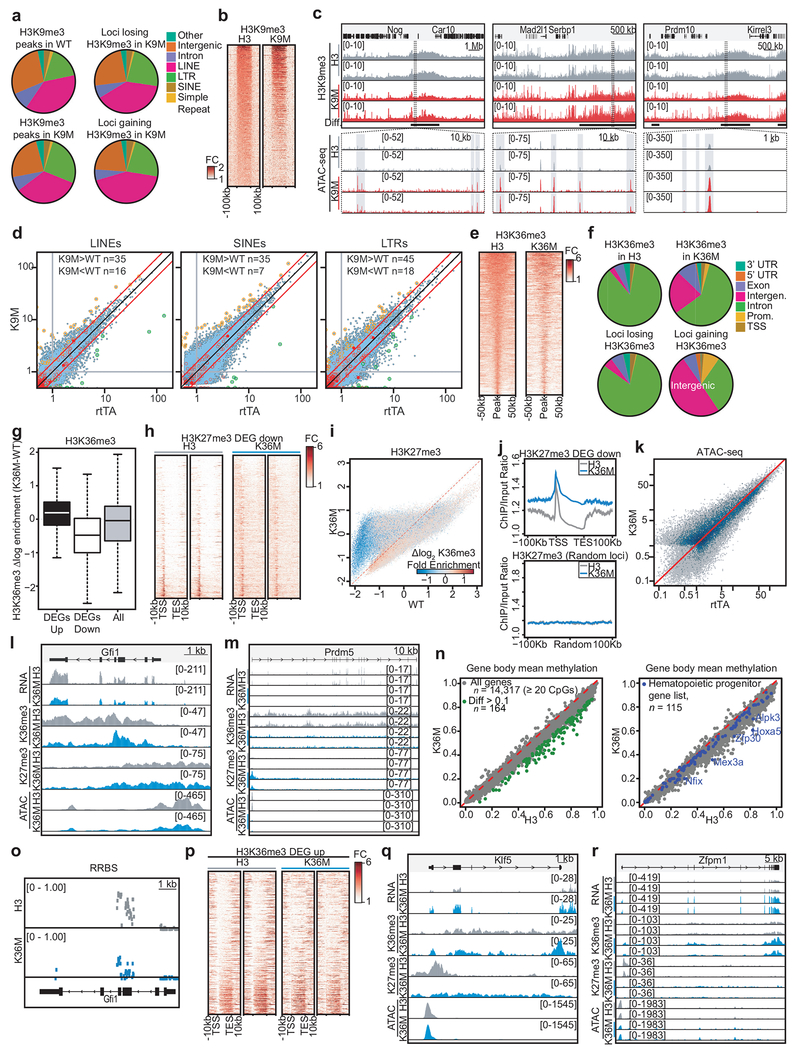
To investigate the molecular mechanisms by which our mutants impact cell fate, we analyzed gene expression patterns in lineage−, cKit+ HSPCs using RNA-seq following 4 weeks of dox induction. In general, gene expression changes following H3K9M induction were subtle, while induction of H3K36M led to robust expression differences. Compared to control, 441 and 2,306 genes were differentially expressed (2-fold or greater change in expression, FDR<0.05) in H3K9M- and H3K36M-induced HSPCs, respectively. Multidimensional scaling analysis indicated that biological replicates were highly consistent and expression profiles for H3K9M, H3K36M and control were distinct (Fig. 5a). Importantly, genes associated with specific hematopoietic lineages were dysregulated between mutant histone samples and control, correlating with the hematopoietic abnormalities observed in vivo (Fig. 5b). For example, we detected a striking increase in the expression of erythroid lineage regulators such as Gata1, Zfpm1, Klf1 and Gfi1b in HSPCs following H3K36M induction, consistent with increased early erythropoiesis in these mice. Similarly, downregulation of important HSC and granulocyte regulators such as Gfi129,30 and Cebpa31,32 correlated with the HSC defect and impaired granulocyte maturation in H3K36M mice. By contrast, regulators of the B-cell lineage, including Pax5 and Ebf1 were noticeably upregulated in HSPCs following H3K9M induction in accordance with the increase in LMPPs and ProB cells. Gene set enrichment analysis (GSEA) for hematopoietic gene expression signatures33,34 revealed that HSPCs from H3K9M mice were enriched for transcriptional programs characteristic of early hematopoietic progenitors (e.g., MPP) and lymphoid progenitors (e.g., CLP), corroborating the differentiation defects in these mice (Fig. 5c and Extended Data 5a). Conversely, H3K36M HSPCs exhibited a marked loss of gene expression patterns associated with HSCs but showed a strong enrichment for expression patterns related to early erythroid development (e.g., Pre-CFUE) (Fig. 5c and Extended Data 5a). Finally, H3K36M, H3K9M and control HSPCs showed similar transcriptional differences and lineage bias after as little as one week of dox induction, suggesting that these programs are directly impacted by loss of H3K36 and H3K9 methylation (Extended Data 5b–c). Together, induction of H3K9M and H3K36M in early progenitors leads to expression changes of important hematopoietic genes, which likely contribute to the observed abnormalities in vivo.
Fig. 5. H3K36M and H3K9M induction in hematopoietic stem and progenitor cells leads to widespread changes in the chromatin landscape and altered gene expression.
(a) Multidimensional scaling analysis based on the union of differentially expressed genes between H3 control and H3K9M or H3K36M samples. Each circle represents a biological replicate. (b) Expression values (log2 fold-change relative to control) for select hematopoietic genes in biological duplicate from HSPCs sorted at 4 weeks of induction. (c) GSEA analysis based on RNA-seq data for HSPCs sorted from H3K9M and H3K36M mice induced for 4 weeks (H3, n=2; H3K9M, n=2; H3K36M, n=2). Enrichment is shown for transcriptional signatures related to multipotent progenitors (MPP50) and common lymphoid progenitors (CLP34) as well hematopoietic stem cells (HSC33) and early erythroid progenitors (Pre-CFUE34). Statistics were generated in accordance with the published GSEA algorithm51. (d) Replicate heat maps showing H3K9me3 levels at loci that lose methylation following H3K9M expression in HSPCs. (e) A meta-analysis profile for H3K9me3 at loci that lose methylation following H3K9M induction in HSPCs. (f) A correlation plot comparing chromatin accessibility between H3- and H3K9M-expressing HPSCs. Loci that lose H3K9me3 signal with H3K9M expression are highlighted. (g) Representative tracks for a locus with decreased H3K9me3 signal following H3K9M expression in HSPCs. (h) A correlation plot showing the ratio of gene body H3K36me3 levels (top panel). The bottom panel displays the same data but indicates differentially expressed transcripts between H3K36M- and H3-expressing cells. (i) Replicate heat maps showing H3K36me3 methylation at gene bodies for genes that are downregulated following H3K36M induction in HSPCs. (j) Meta-analysis profiles for H3K36me3 and H3K27me3 for genes that are downregulated following H3K36M induction in HSPCs. (k) Representative tracks for Gfi1, a gene that decreased expression in HSPCs following H3K36M induction. (l) Replicate heat maps showing H3K27me3 methylation at gene bodies for genes that are upregulated following H3K36M induction in HSPCs. (m) Meta-analysis profiles for H3K36me3 and H3K27me3 for genes that are upregulated following H3K36M induction in HSPCs. (n) Representative tracks for Klf5, a gene that increased expression following H3K36M expression.
H3K9M expression increases chromatin accessibility via loss of H3K9me3
We next performed chromatin immunoprecipitation-sequencing (ChIP-seq) for H3K9me3 on HSPCs purified from induced H3K9M mice. H3K9me3 deposition was predominantly found in extended heterochromatic areas in control cells, including intergenic regions, LINEs and LTR elements (Extended Data 6a). Comparing H3K9M to control samples, we observed a genome-wide reduction of H3K9 methylation (Extended Data 6b). In total, 323 of 576 domains (56%) with broad H3K9me3 enrichment in control cells had reduced methylation in H3K9M cells. Closer inspection revealed that domains with the highest H3K9me3 signal (40/576 or 7%) appeared to resist H3K9M dependent loss of H3K9 methylation (Extended Data 6b). By contrast, regions that showed intermediate enrichment levels in control cells demonstrated substantial loss of H3K9me3 signal (Fig. 5d,e). Thus, H3K9M induction leads to a global reduction of H3K9 trimethylation across heterochromatic regions while sparing certain domains that normally exhibit high levels of H3K9me3.
We failed to detect a direct correlation between altered H3K9me3 levels and gene expression in H3K9M HSPCs compared to control. This is likely due to the broad distribution of H3K9me3 over repetitive and gene-poor areas. We note that although H3K9me3 differences were smaller in magnitude, they tended to span larger areas (Fig. 5e). We therefore hypothesized that H3K9M may exert its regulatory effect indirectly, by changing chromatin organization rather than influencing specific genes or loci. In support of this notion, we discovered a strong association between loss of H3K9me3 signal and increased chromatin accessibility when comparing our ChIP-Seq data with ATAC-Seq data from the same cells (Fig. 5f,g and Extended Data 6c). These results imply that H3K9 methylation predominantly impacts chromatin accessibility in HSPCs, which secondarily influences gene expression of critical hematopoietic genes. The increased expression of repetitive elements including LINEs, SINEs and LTRs we observed after H3K9M induction (Extended Data 6d) are likely due to more subtle changes in H3K9me3 levels and hence escaped detection.
H3K36M expression globally alters the chromatin landscape
We performed ChIP-seq for H3K36me3 and its antagonizing mark, H3K27me3, in HSPCs expressing H3K36M and observed a genome-wide reduction of H3K36me3 (Extended Data 6e). In total, 49.2% of H3K36me3 domains were reduced 1.5-fold or greater in H3K36M cells. Consistent with its connection to active transcription, H3K36me3 in control cells was primarily associated with gene bodies (i.e. exon/intron regions; Extended Data 6f). Overall, H3K36me3 was depleted within genes that were downregulated following induction of H3K36M (Fig. 5h–j and Extended Data 6g), suggesting a connection between loss of the histone mark and transcriptional silencing. Notably, a subset of loci that lost H3K36me3 showed increased H3K27me3 signal at the promoter and in regions adjacent to the gene body (Fig. 5j and Extended Data 6h,i), which is in agreement with the antagonistic relationship between these two histone marks35. Increased H3K27me3 signal was observed over large domains (+/−100 kb from the TSS), suggesting a widespread effect of H3K36M on this mark (Extended Data 6j; upper panel). However, H3K27me3 signal was not simply increased genome-wide, because H3K27me3 levels were unaltered at control loci (Extended Data 6j; lower panel). Several key hematopoietic regulators such as Gfi1 and its co-factor Prdm5, Klf4, Cebpa, and Fli1 demonstrated this pattern, suggesting that they may be directly silenced by H3K36M induced loss of H3K36 methylation and gain of H3K27 methylation (Fig. 5k). Transcriptional silencing of these genes was accompanied by reduced chromatin accessibility of associated regulatory elements, as judged by ATAC-seq profiles (Fig. 5k and Extended Data 6k–m). Moreover, we observed a global correlation between loss of H3K36 methylation and decreased chromatin accessibility when considering all genomic regions (Extended Data 6k). Previous work suggested a link between genic H3K36 methylation and DNA methylation36. We thus performed reduced representation bisulfite sequencing to compare DNA methylation between H3K36M-expressing and control HSPCs. We observed only modest decreases in DNA methylation over gene bodies (Extended Data 6n). Nevertheless, several hematopoietic genes were affected (Extended Data 6n), including Gfi1, which exhibited a decrease in DNA methylation at the gene body (Extended Data 6o), paralleling the observed loss of H3K36me3 (Fig. 5k).
A minority of H3K36me3 domains (3.3%) showed increased signal within gene bodies after H3K36M induction. While expression of the corresponding transcripts increased (Extended Data 6g), they represented only a fraction of the upregulated genes following H3K36M induction (Extended Data 6p), suggesting a secondary, indirect mechanism of regulation. Indeed, when we examined chromatin patterns for upregulated genes, we discovered that H3K36me3 levels were largely unaffected. However, the TSSs of these genes exhibited decreased H3K27me3 signal in H3K36M cells compared to control (Fig. 5l,m). In line with the phenotypes we observed in H3K36M-expressing mice, this subset of genes included ectopically expressed hematopoietic regulators such as Klf5, Zfpm1, and Lmna (Fig. 5n and Extended Data 6q,r). This finding is consistent with previous observations that H3K36M induction leads to a genomic change in H3K27me3 signal, correlating with the transcriptional reactivation of genes that are normally silenced by the PRC complex10. Collectively, our integrative genomic analyses suggest that H3K36M induction impacts cell fate through both direct and indirect mechanisms.
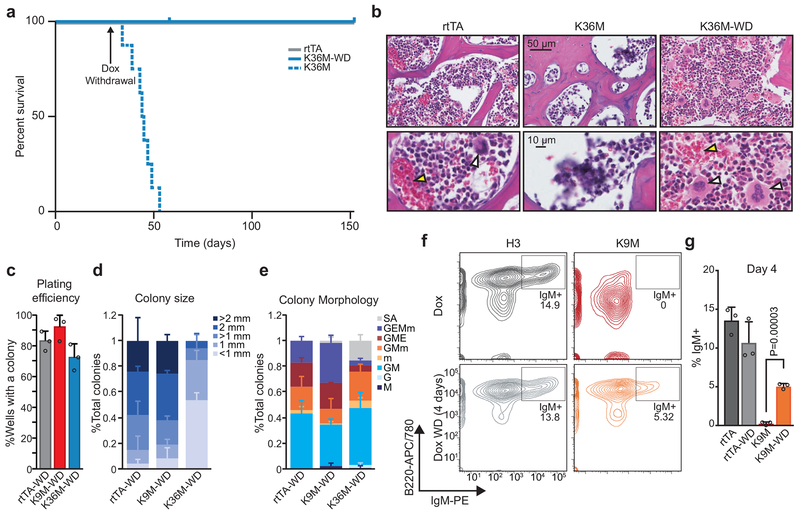
H3K9M and H3K36M dependent differentiation defects are largely reversible
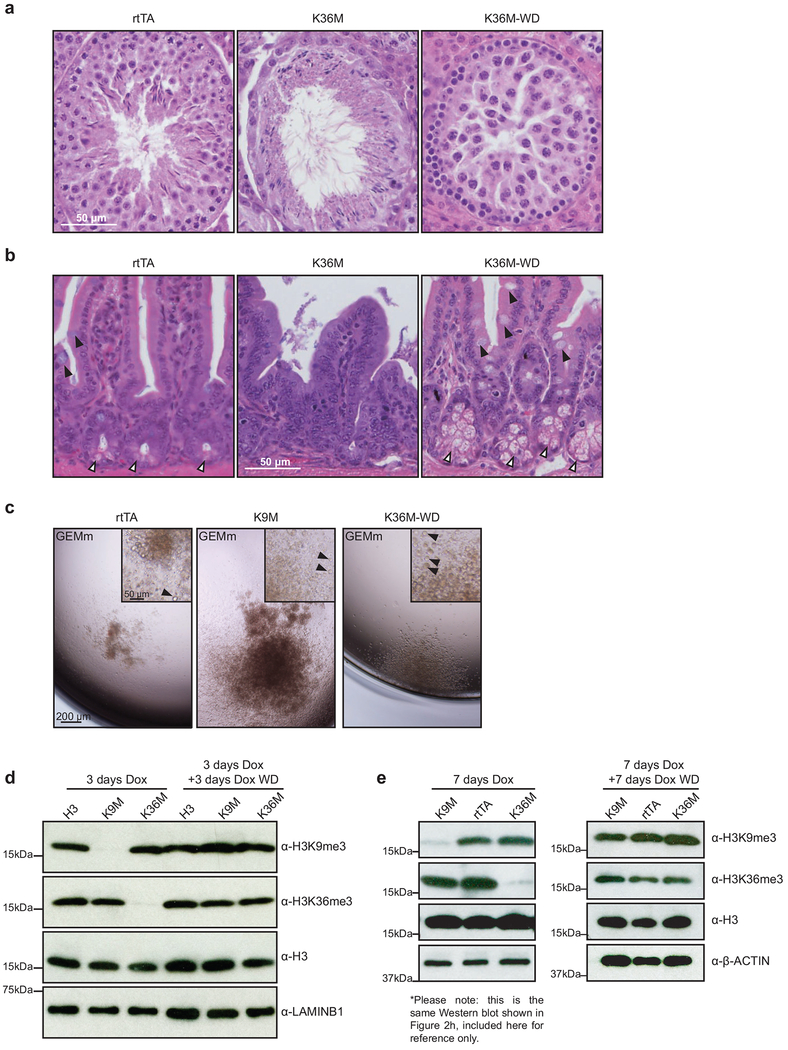
To test whether the HSC defects associated with H3K36M expression were reversible, we induced H3K36M mice and then withdrew dox (referred to hereafter as H3K36M-WD). Strikingly, H3K36M-WD mice survived long-term (Fig. 6a). Histological analysis of mice 4 weeks after dox removal demonstrated robust recovery of bone marrow cellularity (Fig. 6b) and large numbers of megakaryocytes (Fig. 6b). Moreover, histological examination of the testes revealed robust expansion of spermatocytes, indicating recovery of testicular atrophy (Extended Data 7a). Likewise, Paneth and goblet cells were again detectable in H3K36M-WD mice, suggesting rescue of the secretory lineage defect (Extended Data 7b).
Fig. 6. The physiological consequences of suppressing H3K9 and H3K36 methylation are reversible both in vivo and in vitro.
(a) A survival curve for transgenic mice following dox administration and removal at 4 weeks (rtTA, 6 mice; H3K36M, 8 mice; H3K36M-WD, 8 mice). (b) Histological analysis of bone from mice after 4 weeks of dox induction and 4 weeks of dox withdrawal. Scale bar=50 μm (upper panels) and 10 μm (lower panels). White arrowheads indicate megakaryocytes, yellow arrowheads indicate mature erythroid cells. (c) Quantification of plating efficiency for methylcellulose assays with dox removal. Columns represent the mean and error bars represent standard deviation of the mean for biological replicates (rtTA, n=3; H3K9M, n=3; H3K36M, n=3). (d) Quantification of colony size from methycellulose assays with dox removal. Columns represent the mean and error bars represent standard deviation of the mean for biological replicates (rtTA, n=3; H3K9M, n=3; H3K36M, n=3). (e) Quantification of colony morphology from methycellulose assays with dox removal. SA=sparse, atypical; G=granulocyte; M=monocyte; E=erythrocyte; m=megakaryocyte. Columns represent the mean and error bars represent standard deviation of the mean for biological replicates (rtTA, n=3; H3K9M, n=3; H3K36M, n=3). (f) Flow cytometry analysis for IgM+ B cells. Frequencies for each cell type are indicated as a percentage of the parent gate. (g) Quantification of IgM+ B cells. Columns represent the mean and error bars represent standard deviation of the mean for biological replicates (H3, n=3; H3K9M, n=3; H3K36M, n=3). Statistical significance was determined using a two-tailed unpaired Student’s t-test (**** p<0.0001). Data in b,f are representative of 3 biological replicates.
To confirm these results in a controlled in vitro system, we performed single-cell methylcellulose assays using LT-HSCs from H3K9M and H3K36M mice that had been induced for 4 weeks, followed by culture in dox-free media. Notably, plating efficiency for H3K36M HSCs recovered to levels commensurate with control and H3K9M mice upon dox withdrawal (Fig. 6c), although colony size remained smaller for H3K36M cells following the 12-day differentiation time course (Fig. 6d). This observation suggested that the self-renewal capacity of H3K36M cells recovered but the proliferative potential was not fully restored during the examined time period following transgene removal. Importantly, however, these colonies contained all expected differentiated cell types including megakaryocytes and mature erythroid cells, consistent with reversal of the differentiation defects (Fig. 6e and Extended Data 7c).
To determine whether the consequences of H3K9M expression were reversible, we examined B cell maturation37 using a well-defined in vitro system. We again induced mutant and control mice with dox for 4 weeks and then sorted ProB cells (Hardy fraction C37) onto ST2 stromal cells with or without dox. H3K9M cells maintained in dox were unable to differentiate into IgM+ B cells, which was consistent with the in vivo phenotype and confirmed the lineage-intrinsic nature of the B-cell differentiation defect (Fig. 6f,g). However, H3K9MWD samples demonstrated a significant recovery of differentiation capacity after only 4 days of culture without dox (Fig. 6f,g). Importantly, Western blot analysis revealed that histone methylation levels recovered following dox withdrawal both in vitro (EB assays) and in vivo (HSPCs) (Extended Data 7d,e). Thus, the methylation changes and differentiation blocks imposed by H3K36M and H3K9M induction are largely reversible.
Long-term induction of H3K9M or H3K9I leads to aggressive T cell leukemia
Previous studies showed that replacement of histone lysine residues with isoleucine (e.g., H3K9I) blocks methylation in a manner similar to methionine substitution9,38. Analogous to our K-to-M mutants, we generated dox-inducible H3K9I ES cells and mice. We then examined hematopoiesis after 4 weeks of dox treatment. Like H3K9M mice, H3K9I mice exhibited expanded MkPs, increased platelet counts (Extended Data 8, a–c) and a block during the ProB cell-to-PreB cell transition (Extended Data 8, d–g) as well as increased MPPs and LMPPs but normal numbers of LT-HSCs (Extended Data 8, h–m). These results demonstrate that H3K9I mice closely phenocopy H3K9M mice.
Chronic loss of H3K9 methylation has been implicated in impaired viability and increased tumorigenesis39. To explore the long-term impact of H3K9M or H3K9I induction in vivo, we followed cohorts of H3K9M, H3K9I, rtTA and H3 wild-type mice continuously treated with dox. Remarkably, most H3K9M and H3K9I mice but none of the control mice died by 12 months of dox treatment (Fig. 7a). Analysis of 4 moribund animals (one H3K9M mouse and 3 H3K9I mice) showed enlarged spleens and lymph nodes and histologic examination revealed infiltration by blast cells, suggestive of leukemia/lymphoma (Fig. 7b). Flow-cytometry analysis of bone marrow from these mice confirmed increased numbers of CD4 or CD8 single-positive cells or aberrant CD4/CD8 doublepositive cells (Fig. 7c), indicative of T-cell leukemia/lymphoma. We unequivocally confirmed leukemia by demonstrating engraftment of cells and lethality (1–2 months) upon transplantation into syngeneic recipients (Fig. 7d). Thus, chronic H3K9I/M expression predisposes mice to aggressive T-cell leukemia/lymphoma.
Fig. 7. Prolonged expression of H3K9M is associated with T cell leukemia/lymphoma.
(a) A survival curve for H3K9I/M transgenic mice with long-term dox administration (rtTA, n=4 mice; H3, n=8; H3K9M/I, n=14). Statistical significance was determined using a two sided Log-rank (Mantel-Cox) test. (b) Representative spleens and lymphnodes from control (rtTA) and K9I mice. Scale bar=1cm (spleens) or 5 mm (lymphnodes; H3, 3 mice; K9I, 4 mice; one representative example of two independent transplant experiments from independent primary leukemia-bearing mice is shown). (c) Flow cytometry analysis for T cells in bone marrow of mice with long-term dox induction (left) and a representative transplant recipient (right). Frequencies for each cell type are indicated as a percentage of the parent gate (H3, 3 mice; K9I, 4 mice; one representative example of two independent transplant experiments from independent primary leukemia-bearing mice is shown). (d) A survival curve for transplant recipients of bone marrow from a mouse that developed acute T cell leukemia/lymphoma or control bone marrow after long-term dox induction (H3, n=3; K9I, n=4; one representative example of two independent transplant experiments from independent primary leukemia-bearing mice is shown). Statistical significance was determined using a two sided Log-rank (Mantel-Cox) test. (e) A summary of the effects of H3K9M and H3K36M induction on hematopoiesis. (f) Potential uses of the K-to-M system to study diverse biological processes.
DISCUSSION
Using histone mutants as tools, we probed the impact of two crucial histone modifications on cell identity and differentiation in mammals. Our study identified unexpectedly specific and context dependent roles for H3K36 and H3K9 methylation in cell fate regulation, including pluripotent stem cell differentiation, testis and gut regeneration as well as blood cell homeostasis (Fig. 7e). We anticipate that our mouse models will be equally useful in studying the role of histone methylation in other biological contexts. Indeed, during the preparation of our manuscript a study was published that characterized the effects of ectopic H3K36M expression on adipogenesis in mice40. In agreement with our observations in blood, the authors found that expression of H3K36M impairs differentiation of progenitors by altering the distribution of H3K36me3 and H3K27me3.
Our system holds key advantages over knockout approaches for histone modifying enzymes. First, our K-to-M and K-to-I mutations elicit global suppression of methylation at their respective sites, without the need for genetic manipulation of multiple factors. Second, expression of these mutations results in decreased levels, but not complete absence of histone marks, which is advantageous in cases where a complete loss of histone methylation is toxic. Finally, our inducible systems can probe the reversibility of observed cellular phenotypes and underlying chromatin changes. Indeed, we demonstrate that the effect of histone K-to-M mutations is largely reversible as differentiation defects recovered just days after dox withdrawal. We recognize that transgenic and heterogeneous expression of histone mutants in certain cellular contexts19,20 is a potential caveat of our approach compared to KMT knockouts. Moreover, the mechanisms by which histone mutants inhibit methylation may be complex and involve sequestering of KMTs/KDMs on chromatin16,41–44, aberrant histone acetylation and exclusion of KMTs17,45 and/or global redistribution of histone marks46. Regardless of the precise mechanisms, our inducible mutant mice offer a fresh perspective at understanding the physiological role of histone methylation, thus complementing conventional knockout strategies for KMTs.
Previous studies suggested that K-to-M mutations drive tumorigenesis by locking cells into a more primitive developmental state10,14,47. Our observations that H3K36M and H3K9M/H3K9I mice exhibit differentiation blocks in multiple lineages and that a subset of H3K9M/H3K9I mice succumb to T-cell leukemia/lymphoma support this concept10,12,14. We speculate that an increase in the fraction of progenitor cell populations provides an expanded pool of cells that select for secondary oncogenic mutations, such as LMPPs in the case of T cell leukemia in H3K9M/H3K9I mice. Further studies are needed to test this hypothesis and to establish if the oncogenic effects of K-to-M mutants are reversible. Our tools may also prove useful to study histone modifications and physiological processes that are associated with changes in chromatin such as tissue regeneration following injury48 and aging49 (Fig. 7f). Finally, our system will allow to study tissue-specific effects of K-to-M mutants in combination with models that express rtTA under cell type-specific promoters. In summary, our study has uncovered unexpected, direct effects for H3K36 and H3K9 methylation in cell fate and provides a powerful resource to study the functional role of chromatin signaling in both normal and pathological contexts.
Extended Data
Extended Data 1. Expression of H3K9M and H3K36M suppresses methylation and blocks differentiation of pluripotent stem cells.
(a) Western blot analysis for tri-methyl H3K9 and H3K36 with and without mutant histone expression in mouse ES cells. (b) Pie charts showing the distribution of ATAC-seq peaks in EBs expression H3, H3K9M, and H3K36M. (c) Images of teratomas derived from ES cells expressing H3, H3K9M, or H3K36M. Scale bar=200 μm; endo=endoderm, meso=mesoderm, ecto=ectoderm. (d) qRT-PCR analysis for knockdown of H3K9 and H3K36 KMTs. One sample is shown. (e) Flow cytometry for Rex1-GFP at days 2 and 3 of embryoid body formation. (f) Western blot analysis showing H3K36me3 levels following knockdown of H3K36 KMTs. (g) Western blot analysis showing H3K9me3 levels following knockdown of H3K9 KMTs. See source data for full membrane Western blot images. Data in a,c,e,f,g are representative of 3 independent experiments.
Extended Data 2. In vivo expression of H3K9M and H3K36M disrupts tissue homeostasis.
(a) Histological analysis of testis in mice induced for 4 weeks. Scale bar=20 μm. (b) GSEA based on RNA-seq data from testes isolated from H3K9M and H3K36M mice induced for 4 weeks (H3, n=2; H3K9M, n=2; H3K36M, n=2). Enrichment is shown for transcriptional signatures52 related to spermatogenesis. Statistics were generated in accordance with the published GSEA algorithm51 (c) Gene tracks showing expression for genes characteristic of spermatogonia (left panel) or spermatocytes (right panel). (d) Histological analysis of intestine in mice induced for 4 weeks. Black and white arrowheads indicate goblet cells and paneth cells, respectively. Scale bar=20 μm. (e) Staining for markers of Paneth cells (Lysozyme; left panel) and goblet cells (periodic acid Schiff; right panel) (f) GSEA based on RNA-seq data from intestine isolated from H3K9M and H3K36M mice induced for 4 weeks (H3, n=2; H3K9M, n=2; H3K36M, n=2). Enrichment is shown for transcriptional signatures related to secretory lineages53 and Goblet cells54. Statistics were generated in accordance with the published GSEA algorithm51. (g) Gene tracks showing expression for genes characteristic of secretory lineages (left panel) or goblet cells (right panel). (h) Tibia from mice expressing mutant histones for 4 weeks. Scale bar=5 mm. Data in a,d,e,h are representative of 3 independent experiments.
Extended Data 3. Loss of methylation on H3K9M and H3K36M induction cause abnormalities in the erythroid, megakaryocyte, granulocyte and T cell lineages.
(a) cKit expression in early erythroid progenitors. Data are representative of three independent experiments. (b) Quantification of cKit positive erythroid progenitors (right panel). Columns represent the mean and error bars represent standard deviation of the mean for biological replicates (rtTA, n=6; H3K9M, n=3; H3K36M, n=3). Statistical significance was determined using a two-tailed unpaired Student’s t-test. (c) Time course analysis for hemoglobin assessed by CBC. Columns represent the mean and error bars represent standard deviation of the mean for biological replicates (rtTA, n=5; H3K9M, n=4; H3K36M, n=3). Statistical significance was determined using the Holm-Sidak method. (d) An infarcted spleen from a H3K9M mouse induced for 4 weeks. Scale bar=5 mm. Infarctions were present in 2 out of 3 mice inspected.(e) Flow cytometry analysis for granulocyte cells. Gr1+ cells are shown in green.(f) Granularity of transgenic cells assessed by side scatter. (g) Cytospin images of Gr1+ granulocytes sorted from H3K36M and rtTA mice. Scale bar=5 μm. (h) Flow cytometry analysis for thymocytes. Frequencies for each cell type are indicated as a percentage of the parent gate. (i) Quantification of CD4/CD8 double negative thymocytes (left panel) and double positive CD4/CD8 thymocytes (right panel). Columns represent the mean and error bars represent standard deviation of the mean for biological replicates (rtTA, n=6; H3K9M, n=3; H3K36M, n=3). Statistical significance was determined using a two-tailed unpaired Student’s t-test. Data in e,f,g,h are representative of 3 independent experiments.
Extended Data 4. Mutant histone expression impacts HSC function in competitive bone marrow transplantation assays.
(a) Frequency of granulocytes derived from donor bone marrow (CD45.2+) harboring control or mutant histones before and after 12 weeks of induction with doxycyline. Columns represent the mean and error bars represent standard deviation of the mean for biological replicates (n=4). Statistical significance was determined using a twotailed unpaired Student’s t-test. (b) Flow cytometry analysis for donor-derived, CD45.2+ LKS cells, hematopoietic progenitor cells, and stem cells at the conclusion of competitive bone marrow transplant assays. Frequencies for each cell type are indicated as a percentage of the parent gate. (c) Flow cytometry showing granularity of both wild-type competitor marrow (CD45.1) and marrow expressing histone transgenes (CD45.2) as assessed by side scatter. (d) Brightfield images of colonies in methylcellulose. Black arrowheads indicate megakaryocytes. Scale bar=200 μm or 50. (e) May Grünwald Giemsa stains of cells grown in methylcellulose. Representative images are shown for rtTA, H3K9M, and H3K36M. Scale bar=20 μm. Black arrowheads indicate megakaryocytes, white arrowheads indicate granulocytes, and red arrowheads indicate erythroid cells. Data in b,c,d,e are representative of 3 independent experiments.
Extended Data 5. Mutant histone expression impacts HSC gene expression.
(a) GSEA analysis based on RNA-seq data for hematopoietic stem and progenitor cells sorted from H3K9M and H3K36M mice induced for 4 weeks (H3, n=2; H3K9M, n=2; H3K36M, n=2). Enrichment is shown for transcriptional signatures related to Lymphoid/Myeloid progenitors (CMP, LMPP, and proB)33 and B cells50 (K9M) as well as erythroid cells33 and B cells50 (K36M). Statistics were generated in accordance with the published GSEA algorithm51. (b) Principal component analysis based on the union of differentially expressed genes between rtTA control and H3K9M or H3K36M samples. Each circle represents a biological replicate (rtTA, n=3; H3K9M, n=3; H3K36M, n=3) while squares represent the mean. (c) Expression (RPKM) values for select genes relative to average expression in rtTA replicates (shown in log2 scale) for biological triplicates from LKS cells sorted at day 7 of induction.
Extended Data 6. H3K36M/H3K9M induction change the chromatin landscape in HSPCs.
(a) Distribution of H3K9me3 across genomic elements. (b) Heat-maps showing H3K9me3 methylation levels genome-wide following four weeks of H3 or H3K9M induction (averages, two replicates). (c) Representative tracks for loci with decreased H3K9me3 signal following H3K9M expression. (d) RNA-seq scatter plot based on RPKM. Red lines marks 1.5-fold difference between WT and K9M, gray line marks RPKM=1 (cutoff for expressed repeats). Differentially expressed repeats (1.5-fold, FDR<0.1; RNA-Seq data) are marked in orange and green dots. Red circles mark loci where H3K9me3 is differentially bound. Repeats overlapping protein-coding exons were excluded. (e) Heat-maps showing H3K36me3 methylation levels genome-wide following four weeks of H3 or H3K36M induction (averages, two replicates). (f) Distribution of H3K36me3 across genomic elements. (g) Levels of H3K36me3 in genes with increased expression, decreased expression, and all genes in H3K36M vs control HSPCs. The center bar represents the mean and the whiskers represent the standard deviation of the mean. (h) Replicate heat-maps showing H3K27me3 methylation for genes that are downregulated following H3K36M induction. (i) Correlation plot comparing H3K27me3 levels in H3K36M vs. H3 HSPCs. Levels of H3K36me3 overlaid on data points. (j) Meta-analysis showing H3K27me3 levels for a 200 kb region surrounding genes that are downregulated with H3K36M expression (top) and at random regions (bottom). (k) ATAC-seq scatter plot. Loci with reduced H3K36me3 following H3K36M expression are in blue. (l) Replicate tracks for Gfi1 in Fig. 5g. (m) Representative tracks for Prdm5, which is downregulated following H3K36M expression. (n) Correlation plots comparing gene body methylation between H3 or H3K36M HSPCs. Significant differences in green (left), hematopoietic genes in blue (right). (o) Gene tracks for Gfi1 showing DNA methylation in H3 or H3K36M HSPCs. (p) Replicate heat-maps showing H3K36me3 methylation at gene bodies for genes upregulated in H3K36M HSPCs. (q) Replicate tracks for Klf5 in Fig. 5h. (r) Representative tracks for Zfpm1, which is upregulated following H3K36M expression. N numbers in d represent repetitive elements. N numbers in g,n represent genes. All n numbers are stated in the figure.
Extended Data 7. Reversibility of K-to-M dependent phenotypes.
(a) Histological analysis of testis in mice induced for 4 weeks with an additional 4 weeks of dox withdrawal. Scale bar=50 μm. (b) Histological analysis of intestine in mice induced for 4 weeks with an additional 4 weeks of dox withdrawal. Black and white arrowheads indicate goblet cells and paneth cells, respectively. Scale bar=50 μm. (c) Brightfield images of colonies in methylcellulose with dox removal. Black arrowheads indicate megakaryocytes. Scale bar=200 μm or 50 μm. (d) Western blot analysis for tri-methyl H3K9 and H3K36 from EBs induced for 3 days with dox and EBs induced for 3 days with dox followed by 3 days of dox withdrawal. (e) Western blot analysis for H3K9me3 and H3K36me3 from HSPCs in mice induced for 7 days with dox and mice induced for 7 days with dox followed by 7 days of dox withdrawal. See source data for full membrane Western blot images. Data in a-e are representative of 3 independent experiments.
Extended Data 8. K9I mice phenocopy K9M mice.
(a) Flow cytometry analysis of megakaryocyte progenitors (MkPs) cells, 4 weeks of dox. Frequencies as percentages of the parent gate. (b) Quantification of MkPs in bone marrow. Columns represent the mean +/− standard deviation (rtTA, n=1; H3K9I, n=3, biological replicates). (c) Platelet counts after 4 weeks of dox. Columns represent the mean +/− standard deviation (rtTA, n=1; H3K9I, n=3, biological replicates). (d) Flow cytometry of B cells in the bone marrow, 4 weeks of dox. Frequencies as percentages of the parent gates. (e) Quantification of mature B cells and IGM+ B cells. Columns represent the mean +/− standard deviation (rtTA, n=1; H3K9I, n=2, biological replicates). (f) Flow cytometry analysis of B cell progenitors in bone marrow, 4 weeks of dox. Frequencies as percentages of parent gates. (g) Quantification of ProB cells and PreB cells. Columns represent the mean +/− standard deviation (rtTA, n=1; H3K9I, n=2, biological replicates). (h) Flow cytometry analysis of LKS cells. Frequencies as percentages of parent gates. (i) Quantification of LKS cells in bone marrow. Columns represent the mean +/− standard deviation (rtTA, n=1; H3K9I, n=3, biological replicates). (j) Flow cytometry of stem cell populations gated on LKS cells. Frequencies indicated as percentages of parent gates. (k) Quantification of LMPPs, defined as Flt3high cells (top 25% of Flt3+ cells set on rtTA control). Columns represent means and error bars standard deviation (rtTA, n=1; H3K9I, n=3, biological replicates). (l) Flow cytometry of SLAM markers gated on LKS cells. Frequencies as percentages of parent gates. (m) Quantification of MPPs (left panel) and long-term HSCs (right panel) in bone marrow. Columns represent the mean +/− standard deviation (rtTA, n=1; H3K9I, n=3 biological replicates). Data in a,d,f,h,j,l are representative of 3 independent experiments.
Acknowledgements
We thank Maris Handley, Amy Galvin, Marianne Gesner, and Eric Surette of the MGH/HSCI Rodent Histopathology Core for technical assistance. We also thank members of the Hochedlinger lab for helpful discussions. A.J.H. is supported by an American Cancer Society - New England Division - Ellison Foundation Postdoctoral Fellowship (PF-15-130-01-DDC). B.D.S. was supported by an EMBO long-term Fellowship (#ALTF 1143-20l5) and an MGH Tosteson and FMD postdoctoral fellowship. K.H. was supported by funds from the MGH, NIH (R01 HD058013-06) and the Gerald and Darlene Jordan Chair in Regenerative Medicine. H.H. was supported by a Hyundai Hope on Wheels Scholar Grant. J.B. is grateful for support from the NIH (1F32HD078029-01A1). R.I.S. was supported by funds from the NIH (P30-DK40561). The authors declare no competing interests.
REFERENCES
- 1.Faivre G, Lamarche M, Gilgenkrantz JM, Royer R & Weiller M [Digitalis-quinidine combination. Clinical and experimental aspects]. Actual Cardiol Angeiol Int (Paris) 16, 209–23 (1967). [PubMed] [Google Scholar]
- 2.Greer EL & Shi Y Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 13, 343–57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pengelly AR, Copur O, Jackle H, Herzig A & Muller J A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339, 698–9 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Maze I, Noh KM, Soshnev AA & Allis CD Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nat Rev Genet 15, 259–71 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller SA, Mohn SE & Weinmann AS Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell 40, 594–605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shpargel KB, Sengoku T, Yokoyama S & Magnuson T UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet 8, e1002964 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim E et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 23, 839–52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu K et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 338, 1465–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis PW et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu C et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352, 844–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behjati S et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet 45, 1479–82 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang D et al. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 352, 1344–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartzentruber J et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–31 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Papillon-Cavanagh S et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat Genet 49, 180–185 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herz HM et al. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science 345, 1065–70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayaram H et al. S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3. Proc Natl Acad Sci U S A 113, 6182–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohammad F et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med 23, 483–492 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Mohammad F & Helin K Oncohistones: drivers of pediatric cancers. Genes Dev 31, 2313–2324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beard C, Hochedlinger K, Plath K, Wutz A & Jaenisch R Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44, 23–8 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Hochedlinger K, Yamada Y, Beard C & Jaenisch R Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121, 465–77 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Wray J et al. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol 13, 838–45 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ceol CJ et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 471, 513–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inagawa M et al. Histone H3 lysine 9 methyltransferases, G9a and GLP are essential for cardiac morphogenesis. Mech Dev 130, 519–31 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Bilodeau S, Kagey MH, Frampton GM, Rahl PB & Young RA SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev 23, 2484–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo X et al. The histone methyltransferase Setd2 is required for expression of acrosin-binding protein 1and protamines and essential for spermiogenesis in mice. J Biol Chem (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sitnicka E et al. Key role of flt3 ligand in regulation of the common lymphoid progenitor but not in maintenance of the hematopoietic stem cell pool. Immunity 17, 463–72 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Adolfsson J et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 121, 295–306 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Zhang YL et al. Setd2 deficiency impairs hematopoietic stem cell self-renewal and causes malignant transformation. Cell Res 28, 476–490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hock H et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 431, 1002–7 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Hock H et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity 18, 109–20 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Ye M et al. C/EBPa controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat Cell Biol 15, 385–94 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang DE et al. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci U S A 94, 569–74 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng SY, Yoshida T, Zhang J & Georgopoulos K Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity 30, 493–507 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pronk CJ et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 1, 428–42 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Yuan W et al. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 286, 7983–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baubec T et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520, 243–7 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Hardy RR, Carmack CE, Shinton SA, Kemp JD & Hayakawa K Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med 173, 1213–25. (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehnertz B et al. H3(K27M/I) mutations promote context-dependent transformation in acute myeloid leukemia with RUNX1 alterations. Blood 130, 2204–2214 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Peters AH et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–37 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Zhuang L et al. Depletion of Nsd2-mediated histone H3K36 methylation impairs adipose tissue development and function. Nat Commun 9, 1796 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shan CM et al. A histone H3K9M mutation traps histone methyltransferase Clr4 to prevent heterochromatin spreading. Elife 5(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang S et al. Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase. Genes Dev 30, 1611–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Justin N et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun 7, 11316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiao L & Liu X Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 350, aac4383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piunti A et al. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med 23, 493–500 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang D et al. H3.3K27M mutant proteins reprogram epigenome by sequestering the PRC2 complex to poised enhancers. Elife 7(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Funato K, Major T, Lewis PW, Allis CD & Tabar V Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science 346, 1529–33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blanpain C & Fuchs E Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science 344, 1242281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Booth LN & Brunet A The Aging Epigenome. Mol Cell 62, 728–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Graaf CA et al. Haemopedia: An Expression Atlas of Murine Hematopoietic Cells. Stem Cell Reports 7, 571–582 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–50 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Green CD et al. A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq. Dev Cell 46, 651–667 e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lo YH et al. Transcriptional Regulation by ATOH1 and its Target SPDEF in the Intestine. Cell Mol Gastroenterol Hepatol 3, 51–71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haber AL et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagy A, Rossant J, Nagy R, Abramow-Newerly W & Roder JC Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A 90, 8424–8. (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eggan K et al. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci U S A 98, 6209–14. (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]