Abstract
Venous thromboembolism (VTE) is a significant cause of mortality1, yet its genetic determinants remain incompletely defined. We performed a discovery genome-wide association study in the Million Veteran Program and UK Biobank testing ~13 million DNA sequence variants for association with VTE (26,066 cases; 624,053 controls) and meta-analyzed both studies, followed by independent replication with up to 17,672 VTE cases and 167,295 controls. We identified 22 novel loci, bringing the total number of VTE-associated loci to 33 and subsequently fine-mapped these associations. We developed a genome-wide polygenic risk score for VTE that identifies 5% of the population at equivalent incident VTE risk to carriers of the established F5 Leiden (p.R506Q) and prothrombin G20210A mutations. Our data provide new mechanistic insights into the genetic epidemiology of VTE and suggest a greater overlap among venous and arterial cardiovascular disease than previously suggested.
Keywords: Venous Thromboembolism, population genetics, genome-wide association studies, cardiovascular disease
Editorial summary
Genome-wide analysis of venous thromboembolism identifies 22 new risk loci and facilitates construction of a polygenic risk score. Comparison to arterial vascular disease highlights shared pathophysiology and potential therapeutic strategies.
Venous thromboembolism (VTE) is a complex disease impacted by both environmental1 and genetic determinants2,3, and the narrow-sense heritability of VTE has been estimated to be approximately 30%4. At the time of analysis, genome-wide association studies (GWAS) revealed only 11 loci reaching genome-wide significance4–10, leaving a significant portion of VTE heritability unknown.
Large-scale biobanks linking genetic and diverse phenotypic data in the electronic health record (EHR) are being developed throughout the world11,12. Leveraging two large-scale biobanks – UK Biobank and the Million Veteran Program (MVP) – we sought to: 1) perform a genetic discovery analysis for VTE, 2) evaluate the causal role of blood lipids in VTE, 3) further characterize the role of plasminogen activator inhibitor-1 (PAI-1) in VTE, and 4) develop and evaluate a genome-wide polygenic risk score (PRS) for VTE.
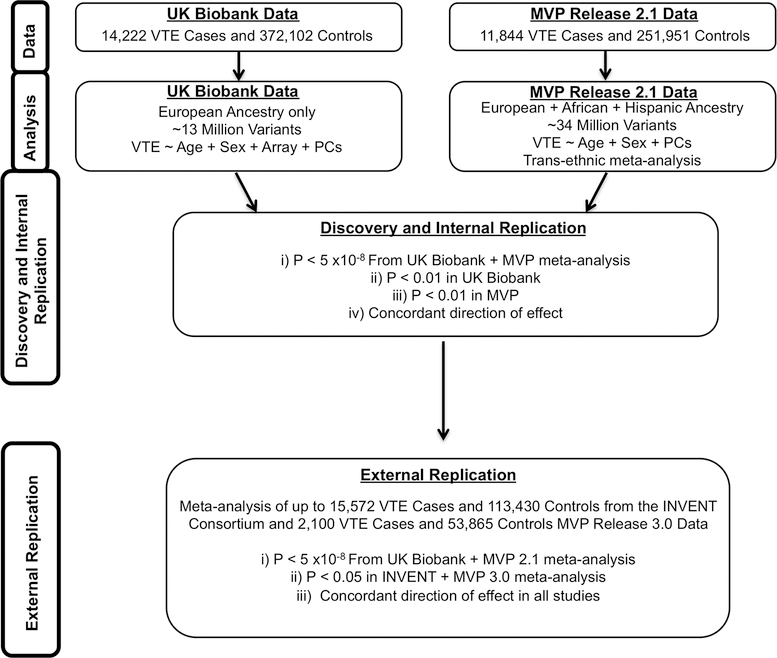
We designed a two-phased VTE discovery GWAS (Fig. 1, Supplementary Fig. 1). In Phase 1, we used MVP release 2.1 data and performed testing for association separately among individuals of European (whites), African (blacks), and Hispanic ancestry and meta-analyzed results across ancestral groups. In UK Biobank, association testing was performed in individuals of European ancestry. We combined statistical evidence across MVP and UK Biobank and set a significance threshold of P < 5 ×10−8 (genome-wide significance), and also required an internal replication P < 0.01 in each of the individual MVP and UK Biobank analyses, with concordant directions of effect, to minimize false positive findings. In Phase 2, an additional round of external replication was performed for lead variants using summary data of up to 15,572 VTE cases and 113,430 disease-free controls from the INVENT consortium13 combined with 2,100 VTE cases and 53,865 controls from MVP 3.0 data, requiring P < 0.05 with consistent direction of effect for successful replication.
Figure 1. Venous thromboembolic disease genetic discovery and replication study design.
In UK Biobank, we performed an association analysis for DNA sequence variants in 14,222 VTE cases and 372,102 controls of European ancestry using logistic regression. These results were combined with association statistics from DNA sequence variants across 3 mutually exclusive ancestry groups in the Million Veteran Program release 2.1 data representing 11,844 VTE cases and 251,951 controls. Data from UK Biobank and MVP were meta-analyzed using an inverse-variance weighted fixed effects method. We set a significance threshold of two-sided P < 5 ×10−8 (genome-wide significance), and also required an internal replication two-sided P < 0.01 in each of the MVP and UK Biobank analyses, with concordant direction of effect, to minimize false positive findings. We subsequently performed external replication using summary data from the INVENT consortium (up to 15,572 VTE cases and 113,430 controls) meta-analyzed with data from MVP 3.0 (2,100 VTE cases and 53,865 controls), requiring an external replication P < 0.05 with a consistent direction of effect.
Abbreviations: MVP, Million Veteran Program; VTE, Venous thromboembolism; PCs, Principal Components
In MVP, the discovery analysis was composed of 11,844 VTE cases (8,929 white, 2,261 black, 654 Hispanic) and 211,753 controls from the MVP release 2.1 data. In UK Biobank we identified 14,222 VTE cases and 372,102 controls. The baseline characteristics for both cohorts are presented in Supplementary Tables 1–2. VTE cases were more likely to be older, have a history of smoking, a higher body-mass index, and have type 2 diabetes. Following trans-ethnic meta-analysis across MVP and UK Biobank, a total of 2,706 variants at 39 loci met a genome-wide significance threshold, with P < 0.01 and concordant effect directions in both datasets (Supplementary Fig. 2–5). The F5 Leiden variant, rs6025 (p.R506Q, NC_000001.10:g.169519049T>C), was the top association result (2.5% frequency for the T allele; OR =2.53; 95%CI: 2.43–2.64; P < 1.0×10−300). We replicated all 11 previously described genome-wide VTE loci, and identified 28 candidate novel VTE loci brought forward for external replication (Supplementary Tables 3–4). Of the 28 candidate novel loci, 22 successfully replicated in an independent set of up to 17,672 VTE cases and 167,295 controls (Supplementary Tables 5–6).
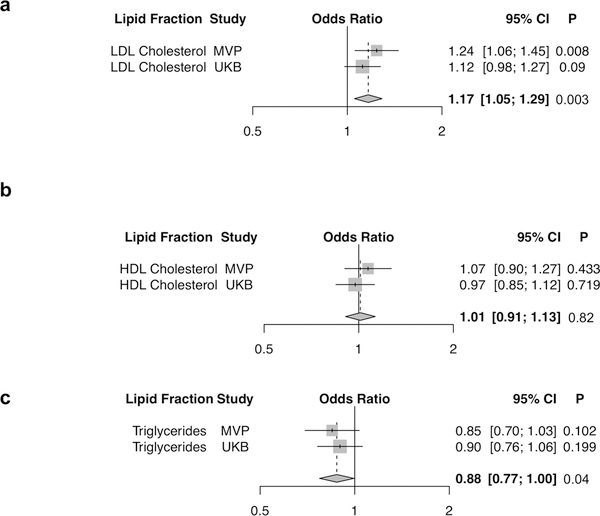
One large randomized controlled trial showed that LDL cholesterol-lowering with a statin versus placebo led to a reduced risk of venous thromboembolic events14. We sought to explore potential causal relationships of blood lipids with VTE development by performing a multivariate Mendelian randomization analysis using a weighted polygenic score of 222 lipid-associated variants from the Global Lipids Genetics Consortium and summary data from the MVP release 2.1 and UK Biobank VTE GWAS restricted to Europeans (Supplementary Table 7)15. We observed that a 1-standard deviation of genetically-elevated LDL cholesterol was associated with an increased risk of VTE (ORLDL = 1.17, 95% CI =1.05–1.29, PLDL = 0.003). In contrast, both a 1-standard deviation of genetically-elevated HDL cholesterol and a 1-standard deviation of genetically-elevated triglycerides were not associated with risk of VTE [ORHDL = 1.01, 95% CI =0.91–1.13, PHDL =0.82; ORTriglycerides = 0.88, 95% CI =0.77–1.00, PTriglycerides = 0.04] after Bonferroni correction (P < 0.016 = [0.05/3 lipid fractions]). An MR-Egger analysis16 indicated no pleiotropic biases of our lipid genetic instruments [MR-Egger intercept P > 0.05 for all 3 lipid fractions (Supplementary Table 8, Fig. 2)]. A PheWAS (1), an analysis of how DNA sequence variants differ in their contribution to vascular disease risk in the arterial and venous territories (2), an examination of VTE risk variant-pQTL associations (3), and results of a VTE fine-mapping analysis including a 99% credible set of 4 variants at the ZFPM2 locus which were genome-wide trans-pQTL associations with plasma PAI-1 concentration (4, Supplementary Table 9) are provided in the Supplementary Results.
Figure 2. Blood lipids and VTE risk.
Association of the 222 variant lipid genetic risk score with VTE in a multivariable Mendelian randomization analysis. Logistic regression odds ratios are displayed per 1-standard deviation genetically increased a) LDL cholesterol, b) HDL cholesterol, and c) triglycerides. Wald statistic two-sided values of P are displayed. Summary-level lipids data from up to 319,677 participants of the Global Lipids Genetics Consortium15, and VTE association data from MVP (N = 8,929 cases; 181,337 controls) and UK Biobank (N = 14,222 cases; 372,102 controls) were used for this analysis. Gray boxes reflect the inverse-variance weight for each study.
Abbreviations: HDL, High-Density Lipoprotein; LDL, Low-Density Lipoprotein; MVP, Million Veteran Program; UKB, UK Biobank
Given the known role of PAI-1 in venous thrombosis and fibrinolysis in model systems17, we hypothesized that the ZFPM2 VTE GWAS and the PAI-1 trans-pQTL associations may represent colocalizing signals at the ZFPM2 locus. We used a recently described colocalization analysis pipeline18 to compute the colocalization posterior probability (CLPP) for the ZFPM2 locus. Using European MVP release 2.1 and UK Biobank European VTE meta-analyzed summary statistics, PAI-1 pQTL results in human plasma from the INTERVAL study19, and reference LD information of 503 European participants from 1000 Genomes20 phase 3 whole genome sequencing data, we calculated a CLPP of 0.203 at this locus. Previous work suggests that a CLPP > 0.01 is indicative of a “reasonably high” probability of colocalization18,21, and the LocusCompare plot at this site further indicates that the ZFPM2 VTE GWAS and PAI-1 pQTL associations likely represent a true colocalization event (Supplementary Fig. 6).
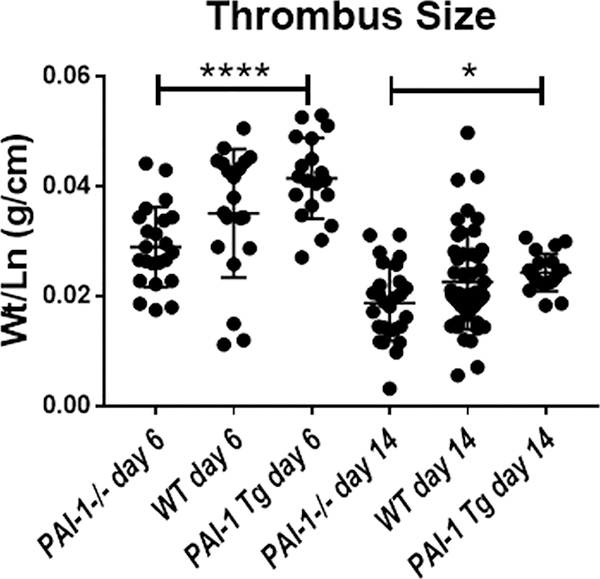
PAI-1 influences thrombosis by directly inhibiting conversion of plasminogen to plasmin and indirectly via disrupting the interaction of circulating monocytes with glycoprotein vitronectin within the thrombus and adjacent vein wall22. Monocytes are a key source of factor III (tissue factor) as well as matrix metalloproteinases during thrombus clearance23,24. Given the colocalization between PAI-1 concentration and human VTE, we sought experimentally to determine the impact of PAI-1 levels on venous thrombus size in an experimental DVT model utilizing transgenic mice. PAI-1−/− mice have no circulating active PAI-1, whereas those overexpressing PAI-1 (PAI-1 Tg), have levels approximately 137-fold greater than wild-type C57B/L6 (WT) mice25. At 6 days following IVC occlusion with generation of thrombus, the PAI-1 overexpressing mice had 1.5-fold larger thrombus size compared to PAI-1−/− mice, with the WT mice demonstrating an intermediate phenotype. This difference persists during late thrombus resolution, at day 14 (Fig. 3), demonstrating progressive impairment in thrombus clearance in the setting of increasing PAI-1 protein levels.
Figure 3. Functional assessment of PAI-1 in murine models.
Inferior vena cava venous thrombus size was measured at day 6 and day 14 after inferior vena cava ligation in PAI-1 Tg (day 6 N = 19; day 14 N = 20), wild type (day 6 N = 20; day 14 N = 49), and PAI-1 −/− mice (day 6 N =23; day 14 N =27). Thrombus size was observed to be larger in the PAI-1 Tg mice compared to PAI-1−/− mice (one-way analysis of variance followed by Tukey’s multiple comparisons post hoc test, *p=0.02, ****p<0.0001). A scatter dot plot depicting mean thrombus size ± standard deviation is shown.
Abbreviations: PAI-1, Plasminogen Activator Inhibitor-1; Tg, Transgenic; WT, Wild Type
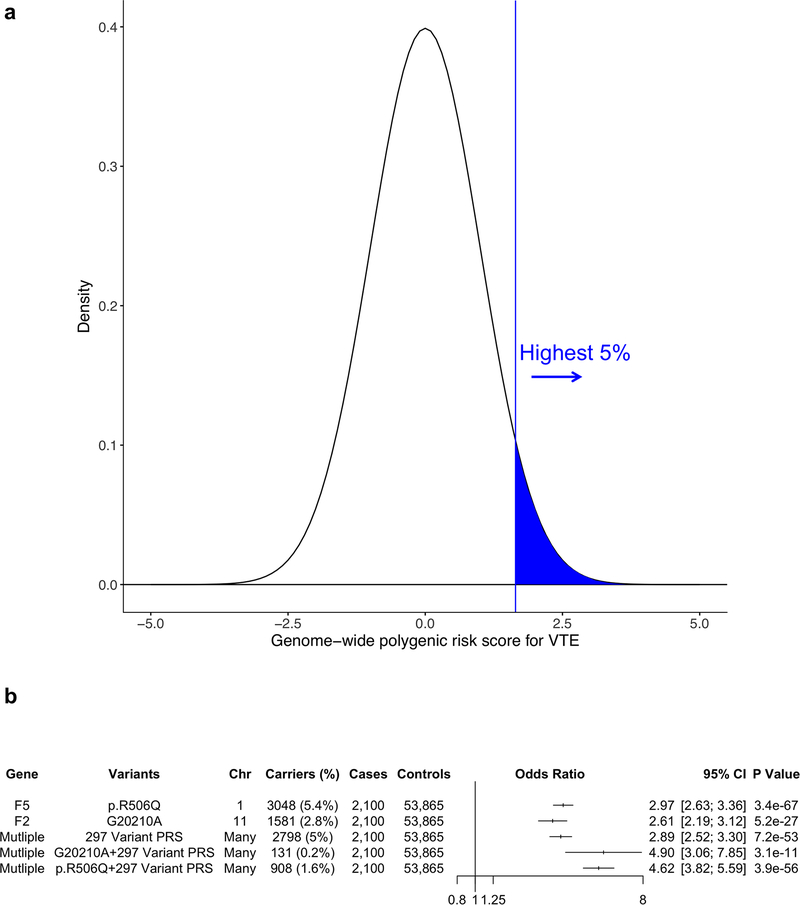
Finally, we sought to examine the contribution of polygenic inheritance on VTE risk. Currently, the F5 Leiden (p.R506Q) and F2 (prothrombin) G20210A mutations, low-frequency variants which confer a 2–3-fold risk of VTE, are frequently tested in clinical settings to evaluate the role of inherited thrombophilia predisposing to acute thrombotic syndromes. Given the individual associations of common genetic variants with VTE, heritable VTE risk may also be explained by an aggregate of common variant VTE risk alleles26. We hypothesized that those at the right tail of the normally distributed VTE PRS (highest 5%) would be at significantly increased VTE risk (Fig. 4a).
Figure 4. Genome-wide polygenic risk score for VTE.
a) Distribution of the PRSVTE in the MVP release 3.0 dataset (n = 55,965). The x-axis represents the PRS with values transformed to have a mean of 0 and standard deviation of 1. The region shaded in blue represents those with the highest 5% of PRSVTE values.
b) VTE odds ratios in MVP release 3.0 data for carriers of the F5 p.R506Q and F2 G20210A mutations. In addition, the odds ratio for individuals with the highest 5% PRSVTE compared to individuals among the lower 95% of PRSVTE, as well as for carriers of the F5 p.R506Q and F2 G20210A mutations within the highest 5% PRSVTE are depicted. Wald statistic two-sided values of P are displayed.
Abbreviations: VTE, Venous Thromboembolism; PRS, Polygenic Risk Score; Chr, Chromosome; MVP, Million Veteran Program; CI, Confidence Interval
We generated a 297-variant VTE PRS using a pruning and thresholding method (R2 < 0.2, P < 1×10−5) from European MVP release 2.1 and UK Biobank European VTE meta-analyzed summary statistics (Supplementary Table 10). Notably, we excluded the LD blocks (R2 > 0.2) containing the F5 p.R506Q and F2 G20210A variants from the PRS. We first assessed the associated VTE risk for the 5% of individuals with the highest PRSVTE relative to the rest of the population using prevalent data from MVP release 3.0, a set of 2,100 VTE cases and 53,865 VTE controls entirely independent from the individuals in the MVP discovery GWAS. We observed that the 2,798 individuals in MVP release 3.0 with the 5% highest PRSVTE had 2.89-fold increased risk of VTE relative to the rest of the population (ORPRS = 2.89, 95% CI =2.52–3.30, PPRS =7.2×10−53). This effect estimate was similar in magnitude to those observed for F5 p.R506Q (ORF5 = 2.97, 95% CI = 2.63–3.36, PF5 =3.4×10−67) and F2 G20210A (ORF2 = 2.61, 95% CI = 2.19–3.12, PF2 =5.2×10−27) [Fig. 4b]. In addition, we observed that this risk was further compounded for individuals among the top 5% with increased polygenic VTE risk who were also F5 Leiden or F2 G20210A carriers.
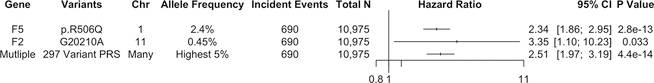
We sought replication of our PRS findings using incident VTE data from the prospective Women’s Health Initiative (WHI) Hormone Trial (HT). In total, among 10,975 European women prospectively followed for up to 25 years in the WHI-HT, 690 incident VTE events were identified among participants with genetic data. Demographic and clinical characteristics for WHI participants in our VTE incident event analysis are shown in Supplementary Table 11. We estimated the risk for carriers of F5 p.R506Q and F2 G20210A mutations as well as those among the 5% highest PRSVTE through Cox proportional hazards models. We observed that F5 p.R506Q carriers were at greater than 2-fold risk of developing VTE [Hazard Ratio (HRF5) = 2.34, 95% CI = 1.86–3.35, PF5 =2.8×10−13], and the F2 G20210A mutation was nominally associated with increased VTE risk [HRF2 = 3.35, 95% CI = 1.10–10.23, PF2 =0.033]. The 549 individuals in WHI with the 5% highest PRSVTE had 2.51-fold risk of incident VTE relative to the rest of the population [HRPRS = 2.51, 95% CI =1.97–3.19, PPRS =4.4×10−14] as depicted in Figure 5. Much like in MVP, the risk among the 5% of the population with the highest PRSVTE in WHI was comparable in effect size to that of large-effect, monogenic mutations in F5 and F2.
Figure 5. Genome-wide polygenic risk score and incident VTE events.
Hazard ratios calculated from the Cox Proportional hazards model for incident VTE events in the Women’s Health Initiative study for carriers of the F5 p.R506Q and F2 G20210A mutations. The hazard ratio for individuals with the highest 5% PRSVTE compared to individuals among the lower 95% of PRSVTE is also depicted. Two-sided values of P are displayed.
Abbreviations: VTE, Venous Thromboembolism; PRS, Polygenic Risk Score; Chr, Chromosome; CI, Confidence Interval
These findings permit several conclusions. First, our results lend human genetic support to LDL cholesterol lowering as a preventive strategy for VTE. In the JUPITER (Justification for the Use of statins in Prevention: an Interventional Trial Evaluating Rosuvastatin) trial, administration of 20mg of rosuvastatin in asymptomatic participants resulted in a reduced occurrence of symptomatic VTE14. This implies that the apparent VTE risk reduction from statins may be due to on-target lowering of lipoproteins, much like the benefits observed for multiple atherosclerotic syndromes27,28. Second, partial antagonism of PAI-1 as a preventive treatment for VTE deserves further consideration. In our analysis, we noted colocalizing ZFPM2 VTE GWAS and PAI-1 pQTL associations and observed PAI-1 overexpressing mice had 1.5-fold larger thrombus size compared to PAI-1−/− mice in an inferior vena cava ligation model. These results suggest that imbalance in the thrombosis-fibrinolysis pendulum in the human condition may lead to development of pathologic VTE, whereas lower active PAI-1 levels may allow for resolution of incidental venous thrombosis prior to becoming clinically relevant. Third, our data provide further evidence for the utility of polygenic risk prediction in the clinical realm. In a recent publication by Khera and colleagues29, the authors generated expanded PRS, and demonstrate that those within the right tail of the distribution have a >3-fold increased risk of developing the disease, akin to carriers of monogenic mutations. We build on these findings by extending polygenic scoring to incident VTE events, where we observed similar magnitudes of effect for our PRSVTE and the F5 p.R506Q/F2 G20210A mutations. Our data suggest that extending current thrombophilia genetic panels to include testing for polygenic VTE risk would significantly increase the yield of current genetic testing and may be warranted.
Our study should be interpreted within the context of its limitations. First, our VTE phenotype is based on EHR data and may result in misclassification of case status. Such misclassification should, however, reduce statistical power for discovery and bias results toward the null. Second, while our colocalization analysis and murine functional data support the role of PAI-1 in VTE, further research is needed to fully understand the causal variant at the ZFPM2 locus and its underlying mechanism. Lastly, while those with the highest PRSVTE are at increased risk for VTE, the PRS’s mechanism of action represents a combination of many causal risk factors, rather than one single pathway that leads to disease. However, assessment of individual risk may help identify a subpopulation more likely to benefit from thromboprophylaxis during periods of increased risk - for instance perioperatively30 or during hospitalizations for acute, medical illness31.
In conclusion, our data provide new mechanistic insights into the genetic epidemiology of VTE and suggest a greater intersection between blood lipids, VTE, and arterial vascular disease than previously understood.
Online Methods
Study Populations
We conducted genetic association analyses using DNA samples and phenotypic data from two cohorts: the Million Veteran Program (MVP) and UK Biobank. In MVP, individuals aged 19 to over 100 years were recruited from 63 VA Medical Centers across the United States. In our initial MVP analysis, we evaluated 11,844 VTE cases (8,929 white, 2,261 black, 654 Hispanic) and 211,753 VTE-free controls.
In UK Biobank, individuals aged 45 to 69 years old were recruited from across the United Kingdom for participation. In this study, we identified 14,222 VTE cases and 372,102 controls of European ancestry. Further details of cohort descriptions and disease definitions are described in the Supplementary Note. All studies received ethical and study protocol approval by their appropriate Institutional Review Boards and informed consent was obtained from all participants. Additional information regarding experimental design and participants are provided in the Life Sciences Reporting Summary.
In addition, we examined incident VTE data from the WHI randomized clinical trial of Hormone Therapy (HT) for our PRS analysis. The overall design of the WHI study has been described previously32. In brief, at the inception of the WHI study (1993–1998), 161,808 postmenopausal women between the ages of 50 and 79 years were eligible for inclusion in multiple clinical trials. Exclusion criteria related to the presence of medical conditions predisposing to shortened survival or safety concerns. The protocol and consent forms were approved by institutional review committees and all participants provided written informed consent. The WHI-HT initially comprised 27,347 postmenopausal women who were randomized to receive either estrogen plus progestin or estrogen alone versus placebo until the trials were stopped early in July 2002 and March 2004, respectively. All WHI-HT participants subsequently continued to be followed without intervention until close-out. Of the various components of WHI, VTE was adjudicated by physician adjudicators for participants who were enrolled in the HT trials.
Genetic Data and Quality Control for Association Analysis
DNA extracted from whole blood was genotyped in MVP using a customized Affymetrix Axiom biobank array, the MVP 1.0 Genotyping Array. Veterans (U.S. military personnel) of three mutually exclusive ethnic groups were identified for analysis: 1) non-Hispanic whites (European ancestry), 2) non-Hispanic blacks (African ancestry), and 3) self-identified Hispanics. After pre-phasing using EAGLE33 v2, genotypes from the 1000 Genomes Project20 phase 3, version 5 reference panel were imputed into MVP participants via Minimac3 software34. Ethnicity-specific principal component analysis was performed using the EIGENSOFT software35. Additional details of quality control procedures used to assign ancestry and perform genotype imputation are described in the Supplementary Note.
In MVP, sample and variant quality control was performed as previously described36. In brief, duplicate samples, samples with more heterozygosity than expected, an excess (>2.5%) of missing genotype calls, or discordance between genetically inferred sex and phenotypic gender were excluded. In addition, one individual from each pair of related individuals (kinship > 0.0884 as measured by the KING37 software) were removed. In total, we identified 312,571 multi-ethnic participants passing quality control from the MVP release 2.1 data (used in association analysis), and another 69,578 from the MVP release 3.0 data used for the PRS analysis.
Following imputation, variant-level quality control was performed using the EasyQC R package38 and exclusion metrics included: ancestry-specific Hardy-Weinberg equilibrium P <1×10−20, posterior call probability < 0.9, imputation quality <0.3, minor allele frequency (MAF) < 0.0003, call rate < 97.5% for common variants (MAF > 1%), and call rate < 99% for rare variants (MAF < 1%). Variants were also excluded if they deviated > 10% from their expected allele frequency based on reference data from the 1000 Genomes Project20. Following variant-level quality control, we obtained 19.9 million, 31.9 million, and 28.1 million DNA sequence variants for analysis in white, black, and Hispanic participants, respectively.
In UK Biobank, analysis was performed separately in white individuals after genotyping using either the UK BiLEVE or UK Biobank Axiom Arrays. Approximately 500,000 individuals were genotyped and subsequently imputed to the haplotype reference consortium (HRC) and UK10K reference panels (UK Biobank v3 release). Details of these procedures are described elsewhere39. We performed genome-wide association testing for VTE in the UK Biobank using all variants in the v3 release with MAF > 0.3% and imputation quality INFO > 0.4. To avoid potential population stratification, only European-ancestry samples were included in the analysis. This subset was selected based on self-reported white ethnicity that was subsequently confirmed using genetic principal components analysis. Outliers within the self-reported white samples in the first 6 principal components of ancestry were detected and subsequently removed using the R package aberrant40. In addition, individuals with sex chromosome aneuploidy (neither XX or XY), discordant self-reported and genetic sex, or excessive heterozygosity or missingness, as defined centrally by the UK Biobank were removed. Finally, one individual from each pair of second-degree or closer relatives (kinship > 0.0884) was removed, selectively retaining VTE cases when possible.
VTE Discovery Association Analysis
In MVP, genotyped and imputed DNA sequence variants were tested for association with VTE through logistic regression adjusting for age, sex, and 5 principal components of ancestry assuming an additive model using the SNPTEST (mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html) statistical software program. In our discovery analysis, we performed association analyses using MVP release 2.1 data separately for each ancestral group (whites, blacks, and Hispanics) and then meta-analyzed using an inverse-variance weighted fixed effects method implemented in the METAL software program41. We excluded variants with a high amount of heterogeneity (I2 statistic > 75%) across the three ancestries. In UK Biobank, association testing was performed using a logistic regression model adjusted for age at baseline, sex, genotyping array, and the first 5 principal components of ancestry. All testing was performed in PLINK2 (https://www.cog-genomics.org/plink/2.0/).
We combined results across MVP release 2.1 and UK Biobank cohorts using inverse-variance weighted fixed effects meta-analysis and set a significance threshold of P < 5 ×10−8 (genome-wide significance). In addition, we also required an internal replication P < 0.01 in each of the MVP and UK Biobank analyses (e.g. MVP discovery and subsequent UK Biobank replication, and vice versa), with concordant direction of effect, to minimize false positive findings. Novel loci were defined as being greater than 500,000 base-pairs away from a known VTE genome-wide associated lead variant. Additionally, European linkage disequilibrium information from the 1000 Genomes Project20 was used to determine independent variants where a locus extended beyond 500,000 base-pairs. All logistic regression P values were two-sided. For X chromosome analyses, male genotypes were coded as if they were homozygous diploid for the observed allele.
Replication
In Phase 2, an additional round of external replication was performed for lead variants using summary data of up to 15,572 VTE cases and 113,430 disease-free controls from the INVENT consortium’s current VTE meta-analysis13 combined with 2,100 VTE cases and 53,865 controls from MVP 3.0 data. Of note, UK Biobank data was excluded from the summary statistics provided by INVENT. We defined significant novel associations as those that were at least nominally significant in replication (P<0.05) with consistent direction of effect and had an overall P < 5×10−8 (genome-wide significance) in the discovery and replication cohorts combined.
Venous Thromboembolism Disease Definitions
From the 312,571 multi-ethnic participants in MVP release 2.1, and 69,578 European participants in MVP release 3.0, individuals were defined as having VTE based on possessing at least two of the ICD-9/10 codes outlined in Supplementary Table 12 in their EHR. Individuals were defined as controls if did not meet the definition of a VTE case and their EHR reflected 2 or more separate encounters in the Veterans Affairs Healthcare System in each of the two years prior to enrollment in MVP. In UK Biobank, individuals were defined as having VTE based on the definition by Klarin and colleagues as previously described4. All other individuals were defined as controls.
Lipids and VTE Mendelian Randomization Analysis
Summary-level data for 222 genome-wide lipids-associated variants were obtained from the publicly available data from the Global Lipids Genetics Consortium15 using a previously described genetic risk score instrument36. As previously described, cohorts either excluded participants on statins or adjusted total cholesterol and LDL cholesterol (by dividing by 0.8 or 0.7, respectively) if a statin was prescribed. One variant, rs77375493, was excluded from the current analysis after not passing quality control. We then utilized results from the MVP and UK Biobank GWAS meta-analysis restricted to Europeans. The effect alleles were matched with all lipid and VTE summary data and 3 different Mendelian randomization analyses were performed: 1) inverse-variance weighted; 2) multivariable; 3) MR-Egger to account for pleiotropic bias. First, we performed inverse-variance weighted Mendelian randomization using each set of variants for each lipid trait as instrumental variables. This method, however, does not account for possible pleiotropic bias. Therefore, we next performed inverse-variance weighted multivariable Mendelian randomization. This method adjusts for possible pleiotropic effects across the included lipid traits in our analyses using effect estimates from the variant-VTE outcome and effect estimates from variant-LDL cholesterol, variant-HDL cholesterol, and variant-triglycerides as predictors in 1 multivariable model. We additionally performed MR-Egger as previously described16. This technique can be used to detect bias secondary to unbalanced pleiotropy in Mendelian randomization studies. In contrast to inverse-variance weighted analysis, the regression line is unconstrained, and the intercept represents the average pleiotropic effects across all variants. Bonferroni-corrected 2-sided P values (P=0.016; 0.05/3) for 3 tests were used to declare statistical significance. Analysis was performed using the R software program (version 3.2.1; Vienna, Austria).
Colocalization of ZFPM2 VTE GWAS and PAI-1 plasma pQTL Signals
To evaluate whether there was evidence of colocalization across the VTE GWAS and PAI-1 pQTL studies, we used European MVP release 2.1 and UK Biobank European VTE meta-analyzed summary statistics and PAI-1 pQTL results from the INTERVAL study19. For the 2,178 variants within the 1-megabase region surrounding the lead ZFPM2 lead VTE GWAS variant, we performed a locus-wide colocalization analysis using FINEMAP42 to generate posterior causal probabilities for each of these variants in the GWAS and the pQTL analyses. We used the European superpopulation subset of the 1000 Genomes20 phase 3 whole genome sequence data as a reference for the linkage disequilibrium statistics, and assumed only 1 causal variant at the locus. We then analyzed these posterior probabilities with a publicly available pipeline18 to compute the CLPP for the entire locus as previously described21. The R package LocusCompareR was used to visualize the colocalizing signals.
Functional Assessment of PAI-1 in Murine Models
Male C57BL/6 (WT) mice (Jackson Laboratory, Farmington, CT), PAI-1−/− (backcrossed 5–10 generations on C57BL/6 mice) and PAI-1 over-expressing mice (PAI-1 Tg, backcrossed 5–10 generations on C57BL/6 background) were utilized in this study25,43,44. Previous data comparing homozygous littermates to wild type C57BL/6 controls demonstrated identical phenotype with regards to venous thrombosis with regards to size and cellular composition25,44. Therefore, in the interest of humane and responsible animal use, wild type C57BL/6 mice (WT) were utilized as controls. Animals underwent a well-characterized DVT model, stasis inferior vena cava (IVC) thrombosis, at 8–10 weeks of age and 20–25 grams body weight24,25,45–47. Isoflurane 2% was administered as inhaled anesthetic. A midline laparotomy was performed, the retroperitoneum exposed, and dorsal IVC branches were interrupted with electrocautery. The infrarenal IVC and any accompanying side branches caudal to the left renal vein were ligated with 7–0 prolene (Ethicon, Inc., Somerville, NJ) to generate blood stasis. A running continuous 5–0 Vicryl suture was used to close the fascia and Vetbond tissue adhesive was applied for skin closure (3M Animal Care Products, St. Paul, MN). Mice were euthanized at 6 and 14 days post-thrombosis. The IVC and its associated thrombus were weighed (grams) and measured (centimeters) for weight to length analysis of thrombus size24,48,49. GraphPad Prism software version 6.0 was used to analyze the thrombus size. Data is presented as the mean +/− the standard deviation. Statistical significance amongst multiple groups was determined using one-way analysis of variance followed by Tukey’s multiple comparisons post hoc test. A value of P < 0.05 was considered significant. All work was approved by the University of Michigan, University Committee on Use and Care of Animals and was performed in compliance with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health.
VTE Polygenic Risk Score Generation
Polygenic risk scores (PRS) represent an individual’s risk of a given disease conferred by the cumulative impact of many common DNA sequence variants. A weight is assigned to each genetic variant based on its strength of association with disease risk (β). Individuals are then additively scored in a weighted fashion based on the number of risk alleles they carry for each variant in the PRS.
To generate our score, we used summary statistics from the combined MVP release 2.1 and UK Biobank VTE summary statistics restricted to Europeans (23,151 VTE cases, 553,439 controls) and a linkage disequilibrium panel of 20,000 randomly selected European samples from UK Biobank. We restricted variants to those present in both MVP release 2.1 and UK Biobank VTE summary statistics with a consistent direction of effect. To increase the number of independent variants included in our score, we performed a pruning and thresholding analysis using the linkage disequilibrium-driven clumping procedure in PLINK version 1.90b (--clump). In brief, this algorithm formed “clumps” around variants with VTE association P < 1×10−5 and with an R2 > 0.2 based on the linkage disequilibrium reference. From our initial set of summary statistics, the algorithm selects only 1 associated variant from each clump below our pre-specified P value threshold. The final output from this procedure generated a score of 299 independent (R2 <0.2), VTE associated (P < 1×10−5) variants, representing the strongest disease-associated variant for each linkage disequilibrium-based clump across the genome. From this 299 variant PRS, we then removed the clumps containing the F5 p.R506Q and F2 G20210A variants, resulting in a 297 variant PRSVTE for downstream analysis.
VTE Polygenic Risk Score Analysis
From the 69,578 MVP release 3.0 participants (none of whom were included in the VTE discovery analysis), we identified 2,100 prevalent VTE cases and 53,865 controls. We first assessed the associated VTE risk for the 5% of individuals with the highest PRSVTE relative to the rest of the population using logistic regression adjusting for age, sex, and 5 principal components of ancestry. We then tested the association of the F5 p.R506Q and F2 G20210A variants among the 5% of individuals with the highest PRSVTE relative to the rest of the population in the MVP release 3.0 data using an identical logistic regression model.
We replicated our findings using incident VTE data from the WHI. Data used in this analysis included genetic data from WHI-HT participants derived from three separate GWAS sub-studies: 1) the WHI Genomics and Randomized Trials Network study (WHI-GARNET, 457 incident VTE events among 4,233 participants), (2) the WHI Memory Study (WHIMS, 180 incident VTE events among 5,637 participants), and (3) the WHI Long Life Study (WHI-LLS, 53 incident VTE events among 1,105 participants). All individuals included in the incident event analysis were of European ancestry. Specific details of each WHI sub-study including genotyping, study design, and imputation are included in the Supplementary Note. Cox proportional hazards models were used to estimate hazard ratios (HR) and 95% confidence intervals for the associations of the F5 p.R506Q and F2 G20210A mutations with VTE adjusting for age, 10 principal components of ancestry, and hormone therapy intervention status during the active phase of the WHI-HT. We then tested the associated VTE risk for the 5% of individuals with the highest PRSVTE relative to the rest of the population using Cox proportional hazards models adjusting for age, 10 principal components of ancestry, and hormone therapy intervention status during the active phase of the WHI-HT. Results from WHIMS, WHI-LLS, and WHI-GARNET were combined using an inverse-variance weighted fixed effects meta-analysis. Bonferroni-corrected 2-sided P values (P=0.016; 0.05/[2 variants + 1 PRSVTE) for 3 tests were used to declare statistical significance. Analyses were performed using the R software program (version 3.2.1).
Data availability
The full summary level association data from the MVP trans-ancestry VTE meta-analysis from this report are available upon request through dbGAP, accession code phs001672.v2.p1. Data contributed by CARDIoGRAMplusC4D investigators are available online (http://www.CARDIOGRAMPLUSC4D.org/). Data on large artery stroke have been contributed by the MEGASTROKE investigators and are available online (http://www.megastroke.org/). The genetic and phenotypic UK Biobank data are available upon application to the UK Biobank.
Supplementary Material
Acknowledgments
Funding from the Department of Veterans Affairs Office of Research and Development, Million Veteran Program Grant #MVP000. This publication does not represent the views of the Department of Veterans Affairs or the United States Government. This research was also supported by three additional Department of Veterans Affairs awards (I01-01BX03340 [K Cho/P Wilson], I01-BX003362 [P Tsao/KM Chang], and I01-CX001025 [P Wilson]) and used resources and facilities at the VA Informatics and Computing Infrastructure (VINCI), VA HSR RES 13-457. S Damrauer is supported by the Veterans Administration [IK2-CX001780]. S Kathiresan is supported by a Research Scholar award from the Massachusetts General Hospital (MGH), the Donovan Family Foundation, and the National Institutes of Health [R01HL127564]. P Natarajan is supported by the NIH/NHLBI [K08HL140203 and R01HL142711]. J Concato is now with the U.S. Food and Drug Administration. D Trégouët was financially supported by the “EPIDEMIOM-VTE” Senior Chair from the Initiative of Excellence of the University of Bordeaux. C Kabrhel is supported by the NIH [HL116854]. Data on coronary artery disease have been contributed by the CARDIoGRAMplusC4D investigators. Data on large artery stroke have been contributed by the MEGASTROKE investigators. The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgements.html. The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. For a list of all the investigators who have contributed to WHI science, see: https://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Long%20List.pdf. This research has been conducted using the UK Biobank resource, application 7089.
Competing Interests: Dr. Natarajan reports grant support from Amgen, Apple, and Boston Scientific, and consulting income from Apple, all unrelated to the submitted work. Dr. Kathiresan reports grant support from Regeneron and Bayer, grant support and personal fees from Aegerion, personal fees from Regeneron Genetics Center, Merck, Celera, Novartis, Bristol-Myers Squibb, Sanofi, AstraZeneca, Alnylam, Eli Lilly, and Leerink Partners, personal fees and other support from Catabasis, and other support from San Therapeutics outside the submitted work. He is also the chair of the scientific advisory board at Genomics plc and the CEO of Verve Therapeutics. Dr. DuVall reports grants to his institution in the last three years outside the submitted work: AbbVie Inc., Anolinx LLC, Astellas Pharma Inc., AstraZeneca Pharmaceuticals LP, Boehringer Ingelheim International GmbH, Celgene Corporation, Eli Lilly and Company, Genentech Inc., Genomic Health, Inc., Gilead Sciences Inc., GlaxoSmithKline PLC, Innocrin Pharmaceuticals Inc., Janssen Pharmaceuticals, Inc., Kantar Health, Myriad Genetic Laboratories, Inc., Novartis International AG, and PAREXEL International Corporation. Dr. Kabrhel reports grants to his institution from Janssen, Diagnostica Stago and Siemens Healthcare Diagnostics, for research related to VTE, but not related to the current work.
References
- 1.Heit JA Epidemiology of venous thromboembolism. Nat Rev Cardiol 12, 464–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertina RM et al. Mutation in blood coagulation factor V associated with resistance to activated protein C. Nature 369, 64–7 (1994). [DOI] [PubMed] [Google Scholar]
- 3.Poort SR, Rosendaal FR, Reitsma PH & Bertina RM A common genetic variation in the 3’-untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increase in venous thrombosis. Blood 88, 3698–703 (1996). [PubMed] [Google Scholar]
- 4.Klarin D, Emdin CA, Natarajan P, Conrad MF & Kathiresan S Genetic Analysis of Venous Thromboembolism in UK Biobank Identifies the ZFPM2 Locus and Implicates Obesity as a Causal Risk Factor. Circ Cardiovasc Genet 10(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinds DA et al. Genome-wide association analysis of self-reported events in 6135 individuals and 252 827 controls identifies 8 loci associated with thrombosis. Hum Mol Genet (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heit JA et al. A genome-wide association study of venous thromboembolism identifies risk variants in chromosomes 1q24.2 and 9q. J Thromb Haemost 10, 1521–31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Germain M et al. Meta-analysis of 65,734 individuals identifies TSPAN15 and SLC44A2 as two susceptibility loci for venous thromboembolism. Am J Hum Genet 96, 532–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernandez W et al. Novel genetic predictors of venous thromboembolism risk in African Americans. Blood 127, 1923–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang W et al. A genome-wide association study for venous thromboembolism: the extended cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium. Genet Epidemiol 37, 512–521 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tregouet DA et al. Common susceptibility alleles are unlikely to contribute as strongly as the FV and ABO loci to VTE risk: results from a GWAS approach. Blood 113, 5298–303 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Collins R What makes UK Biobank special? The Lancet 379, 1173–1174 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Gaziano JM et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol 70, 214–23 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Lindstrom S et al. Genomic and Transcriptomic Association Studies Identify 16 Novel Susceptibility Loci for Venous Thromboembolism. Blood (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glynn RJ et al. A randomized trial of rosuvastatin in the prevention of venous thromboembolism. N Engl J Med 360, 1851–61 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu DJ et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat Genet (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowden J, Davey Smith G & Burgess S Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44, 512–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eitzman DT, Westrick RJ, Nabel EG & Ginsburg D Plasminogen activator inhibitor-1 and vitronectin promote vascular thrombosis in mice. Blood 95, 577–80 (2000). [PubMed] [Google Scholar]
- 18.Liu B, Gloudemans MJ, Rao AS, Ingelsson E & Montgomery SB Abundant associations with gene expression complicate GWAS follow-up. Nature Genetics 51, 768–769 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun BB et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hormozdiari F et al. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am J Hum Genet 99, 1245–1260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fogo AB Renal fibrosis: not just PAI-1 in the sky. J Clin Invest 112, 326–8 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pawlinski R & Mackman N Cellular sources of tissue factor in endotoxemia and sepsis. Thromb Res 125 Suppl 1, S70–3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henke PK et al. Deep vein thrombosis resolution is modulated by monocyte CXCR2-mediated activity in a mouse model. Arterioscler Thromb Vasc Biol 24, 1130–7 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Obi AT et al. Plasminogen activator-1 overexpression decreases experimental postthrombotic vein wall fibrosis by a non-vitronectin-dependent mechanism. J Thromb Haemost 12, 1353–63 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wassel CL et al. A genetic risk score comprising known venous thromboembolism loci is associated with chronic venous disease in a multi-ethnic cohort. Thromb Res 136, 966–73 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ridker PM et al. Rosuvastatin to prevent vascular events in men and women with elevated C-reactive protein. N Engl J Med 359, 2195–207 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Mihaylova B et al. The effects of lowering LDL cholesterol with statin therapy in people at low risk of vascular disease: meta-analysis of individual data from 27 randomised trials. Lancet 380, 581–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khera AV et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 50, 1219–1224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bahl V et al. A validation study of a retrospective venous thromboembolism risk scoring method. Ann Surg 251, 344–50 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Anderson FA Jr. & Spencer FA Risk factors for venous thromboembolism. Circulation 107, I9–16 (2003). [DOI] [PubMed] [Google Scholar]
Online Methods References
- 32.The Women’s Health Initiative Study Group. Design of the Women’s Health Initiative clinical trial and observational study. Control Clin Trials 19, 61–109 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Loh PR, Palamara PF & Price AL Fast and accurate long-range phasing in a UK Biobank cohort. Nat Genet 48, 811–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howie B, Fuchsberger C, Stephens M, Marchini J & Abecasis GR Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44, 955–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38, 904–9 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Klarin D et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nature Genetics (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manichaikul A et al. Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winkler TW et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc 9, 1192–212 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bycroft C et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bellenguez C, Strange A, Freeman C, Donnelly P & Spencer CC A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics 28, 134–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willer CJ, Li Y & Abecasis GR METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benner C et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eitzman DT et al. Bleomycin-induced pulmonary fibrosis in transgenic mice that either lack or overexpress the murine plasminogen activator inhibitor-1 gene. J Clin Invest 97, 232–7 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baldwin JF et al. The role of urokinase plasminogen activator and plasmin activator inhibitor-1 on vein wall remodeling in experimental deep vein thrombosis. J Vasc Surg 56, 1089–97 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wojcik BM et al. Interleukin-6: a potential target for post-thrombotic syndrome. Ann Vasc Surg 25, 229–39 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Diaz JA et al. Critical review of mouse models of venous thrombosis. Arterioscler Thromb Vasc Biol 32, 556–62 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Obi AT et al. Endotoxaemia-augmented murine venous thrombosis is dependent on TLR-4 and ICAM-1, and potentiated by neutropenia. Thromb Haemost 117, 339–348 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henke PK et al. Targeted deletion of CCR2 impairs deep vein thombosis resolution in a mouse model. J Immunol 177, 3388–97 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Laser A et al. Deletion of cysteine-cysteine receptor 7 promotes fibrotic injury in experimental post-thrombotic vein wall remodeling. Arterioscler Thromb Vasc Biol 34, 377–85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full summary level association data from the MVP trans-ancestry VTE meta-analysis from this report are available upon request through dbGAP, accession code phs001672.v2.p1. Data contributed by CARDIoGRAMplusC4D investigators are available online (http://www.CARDIOGRAMPLUSC4D.org/). Data on large artery stroke have been contributed by the MEGASTROKE investigators and are available online (http://www.megastroke.org/). The genetic and phenotypic UK Biobank data are available upon application to the UK Biobank.