Abstract
αMβ2 integrin (complement receptor 3) is a major receptor for phagocytosis in macrophages. In other contexts, integrins’ activities and functions are mechanically linked to actin dynamics through focal adhesions (FAs). We asked whether mechanical coupling of αMβ2 integrin to the actin cytoskeleton mediates phagocytosis. We found that particle internalization was driven by formation of Arp2/3 and formin-dependent actin protrusions that wrapped around the particle. Focal complex-like adhesions formed in the phagocytic cup that contained β2 integrins, FA proteins and tyrosine kinases. Perturbation of talin and Syk demonstrated that a talin-dependent link between integrin and actin and Syk-mediated recruitment of vinculin enable force transmission to target particles and promote phagocytosis. Altering target mechanical properties demonstrated more efficient phagocytosis of stiffer targets. Thus, macrophages use tyrosine kinase signaling to build a mechanosensitive, talin- and vinculin-mediated, FA-like molecular clutch, which couples integrins to cytoskeletal forces to drive particle engulfment.
Introduction
Phagocytosis is an essential function of macrophages, neutrophils and dendritic cells that enables removal, killing and breakdown of particulate material including microbes, dead cells and debris1,2. Phagocytosis is triggered by particle binding to cell surface receptors, which signal to activate morphological changes that lead to particle internalization3,4. αMβ2 integrin, also known as complement receptor (CR) 3, is highly expressed in macrophages, is the main receptor for many pathogens, and mediates clearance of apoptotic and certain cancer cells5,6. CR-mediated phagocytosis involves deposition of serum complement C3b on the particle surface, also called opsonization, and its cleavage into iC3b, which is a specific ligand for αMβ2 integrins7,8.
CR-mediated phagocytosis is thought to require active reorganization of the actin cytoskeleton. It is inhibited by pharmacological disruption of the cytoskeleton9, requires Arp2/310,11, the formin mDia112, and the GTPase RhoA13. However, how actin dynamics are spatially and temporally orchestrated to mediate particle internalization is not known. Early electron microscopy studies suggested that while Fc receptor-mediated phagocytosis involved ruffle-like pseudopods that reach out from the cell surface and wrap around target particles, complement-opsonized targets appeared to sink into the macrophage cytoplasm without involvement of membrane extensions14,15. How actin regulatory proteins control cytoskeletal dynamics to facilitate CR-mediated “sinking phagocytosis” is unclear.
In addition to phagocytosis, integrins also mediate mesenchymal cell migration, where integrin activation, adhesion and traction force are driven by actin dynamics16,17. Whether actin plays a similar role in CR-mediated phagocytosis is unknown. During migration, Arp2/3-mediated actin polymerization pushes against the leading edge plasma membrane, generating forces that drive actin “flow” towards the cell interior18,19. Formation of a linkage between integrins and actin filaments activates integrin to bind ligand and anchors the flowing actin, slowing its rearward motion. Continued assembly of anchored filaments drives forward protrusion of the cell edge and force transmission to integrins, generating traction forces on the ECM20-22. As integrin cytoplasmic tails cannot bind directly to actin filaments, coupling integrin to actin is mediated indirectly by integrin- and actin-binding proteins in FAs that form a so-called “molecular clutch”16. Talin, which binds and activates integrins by its head piece23 and has actin binding sites in its rod domain24, forms a force-transmitting link between integrins and actin during cell migration25, and is required for CR-mediated phagocytosis26. In addition, force across talin unveils vinculin binding sites27, which, in conjunction with FAK-mediated tyrosine phosphorylation of paxillin, promotes vinculin binding to talin and actin, reinforcing the link between integrins, talin and actin at FAs28-30. Paxillin and vinculin association with phagosomes has been observed during Fc- and CR-mediated phagocytosis14, and Syk and the FAK-related tyrosine kinase Pyk2 may function in CR-mediated phagocytosis31,32, suggesting that mechanical coupling of CR to actin by a similar molecular clutch might mediate phagocytosis.
In mesenchymal cells, the FA molecular clutch is sensitive to ECM stiffness33-35, which regulates decisions driving growth, death, differentiation, and migration36-38. In the context of phagocytosis, target stiffness varies enormously ranging from low kPa for mammalian cells39 to hundreds of MPa for bacteria and fungi40,41. In addition, apoptosis42,43 and transformation to malignancy44 are associated with changes in cell stiffness. Thus, CR could render phagocytosis sensitive to mechanical properties of the targets, which would have important implications for discrimination between microbes and living, dead, and transformed cells.
Here, we tested the hypothesis that αMβ2 integrin-mediated phagocytosis by macrophages involves an indirect linkage between actin and integrins, analogous to the molecular clutch characterized in migrating mesenchymal cells. We present evidence that a talin/vinculin-based molecular clutch mechanically couples the forces generated by Arp2/3- and Dia1-mediated actin polymerization to complement-engaged integrins during phagosome formation, promoting fast membrane protrusion around stiff targets.
Results
CR-mediated phagocytosis involves an actin-based reaching mechanism to engulf particle targets
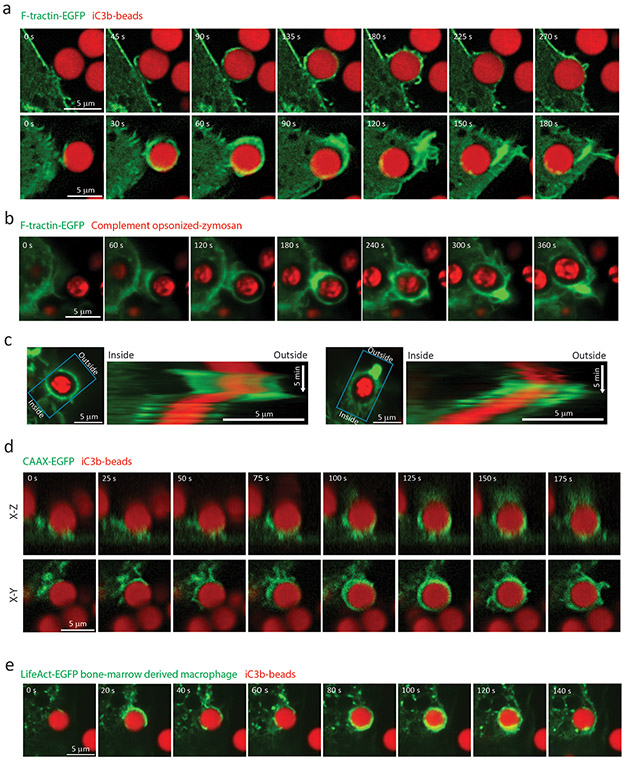
To gain insight into the role of the actin cytoskeleton in CR-mediated phagocytosis, we imaged actin dynamics during particle uptake. β2 integrins were activated with PMA in RAW 264.7 macrophages expressing F-tractin-EGFP, a fluorescent probe for actin filaments45. Macrophages were exposed to iC3b-opsonized microspheres and their internalization followed by time-lapse confocal microscopy. Movies revealed that contact between the cell and particle induced formation of actin-filled protrusions that rapidly advanced along the particle surface (Figure 1a and Movie 1). After full surrounding of the particle by actin protrusions, the particle was propelled inwards by the curved actin cortex flattening (Figure 1a, upper panel), or a burst of actin polymerization between the internalized phagosome and the plasma membrane (Figure 1a lower panel). Once internalized, intermittent bursts of actin assembly occurred on the phagosome, as previously observed46.
Figure 1. CR-mediated phagocytosis involves an actin-based reaching mechanism to engulf target particles.
Time-lapse spinning disc confocal microscopy images of complement mediated particle uptake by RAW 264.7 (a-d) or bone-marrow-derived (e) murine macrophages. a, two examples (upper row and lower row) of RAW macrophages expressing EGFP-F-tractin during phagocytosis of iC3b-opsonized 4.19 μm polystyrene Flash Red microspheres, from more than 10 independent experiments. b, Phagocytosis of complement-opsonized Zymosan A labelled with Texas Red by RAW macrophages expressing EGFP-F-tractin, from 3 independent experiments. c, Images (left) and kymographs (right) from time-lapse movies of phagocytosis of complement-opsonized Zymosan A labelled with Texas Red by RAW EGFP-F-tractin-expressing macrophages. Kymographs were generated by scanning mean intensities within a 6 μm wide line (blue box) along the direction of particle internalization into the cell. d, Three-dimensional time-lapse confocal of a RAW macrophage expressing EGFP-CAAX during phagocytosis of iC3b-opsonized 4.19 μm polystyrene Flash Red microspheres, from 4 independent experiments. Planar view (X-Y, bottom) and axial view (X-Z, top) corresponding to the center of particle are displayed. e, Bone marrow-derived macrophage expressing EGFP-Lifeact, during phagocytosis of an iC3b-opsonized 4.19 μm polystyrene Flash Red microsphere, from 3 independent experiments. Elapsed time shown in seconds. Scale bars are 5 μm, elapsed times are in seconds.
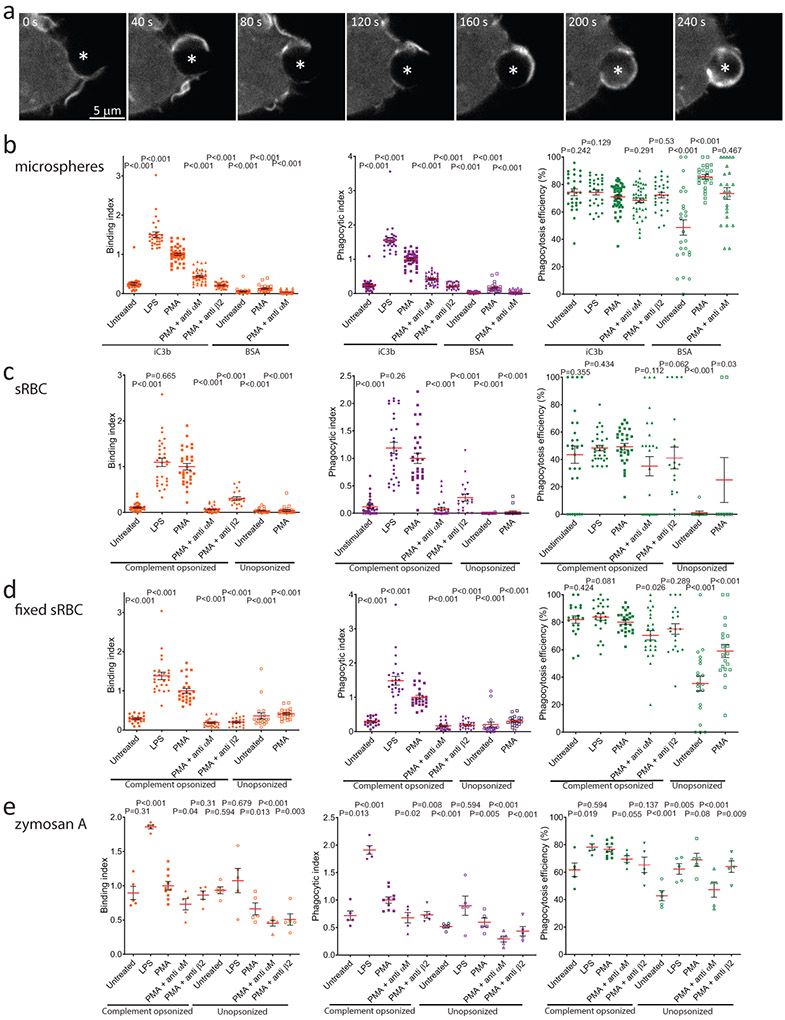
Similar actin protrusions around particles were seen in F-tractin-EGFP-expressing RAW macrophages phagocytosing serum-opsonized zymosan A (a yeast wall particulate preparation that engages αMβ247,48, Figure 1b, Movie 1) or complement-opsonized sheep red blood cells (sRBCs) (Movie 1), as well as in Lifeact-EGFP-expressing mouse bone marrow-derived macrophages or F-tractin-EGFP-expressing THP-1 macrophages phagocytosing iC3b-opsonized microspheres (Figure 1e, Supplementary Figure 1a). The integrin-dependence of this process was verified by incubating PMA-activated macrophages with αM or β2 integrin blocking antibodies, which reduced binding and internalization of iC3b-opsonized microspheres, complement-opsonized native or glutaraldehyde-fixed sRBCs, or zymosan (Supplementary Figure 1b-e). Lipopolysaccharide (LPS), which activates αMβ249, also promoted internalization of iC3b-opsonized microspheres or complement-opsonized-sRBCs via similar actin protrusion dynamics (Supplementary Figure 1b-e, Movie 1-2). Thus, αMβ2 engagement induces actin-based protrusions that reach out and surround target particles.
A further prediction of a reaching mechanism is that protrusions should emanate before the particle is pulled toward the cell interior, while in a sinking mechanism, movement of the particle toward the cell interior should precede internalization. The timing of internalization relative to inward movement was determined from kymographs along the axis of particle displacement in movies of F-tractin-GFP-expressing RAW macrophages during phagocytosis of serum-opsonized zymosan. This revealed that actin-filled protrusions extended to the distal side of the particle before the particle moved toward the cell interior (Figure 1c). In addition, three-dimensional confocal imaging of the plasma membrane marker CAAX-EGFP showed that membrane protrusions emanated from the cell surface after particle binding and wrapped around and fully internalized the target particle prior to its inward movement (Figure 1d and Movie 3). These protrusions were distinct form the previously described ruffles that grab particles before uptake50. Together, these results show that CR-mediated phagocytosis involves a reaching mechanism in which actin-based protrusions extend along a target particle surface to mediate phagosome formation, and that particles are actively driven to the cell interior by cortical actin dynamics.
Arp2/3 and mDia1 contribute to specific aspects of actin dynamics during CR-mediated phagocytosis.
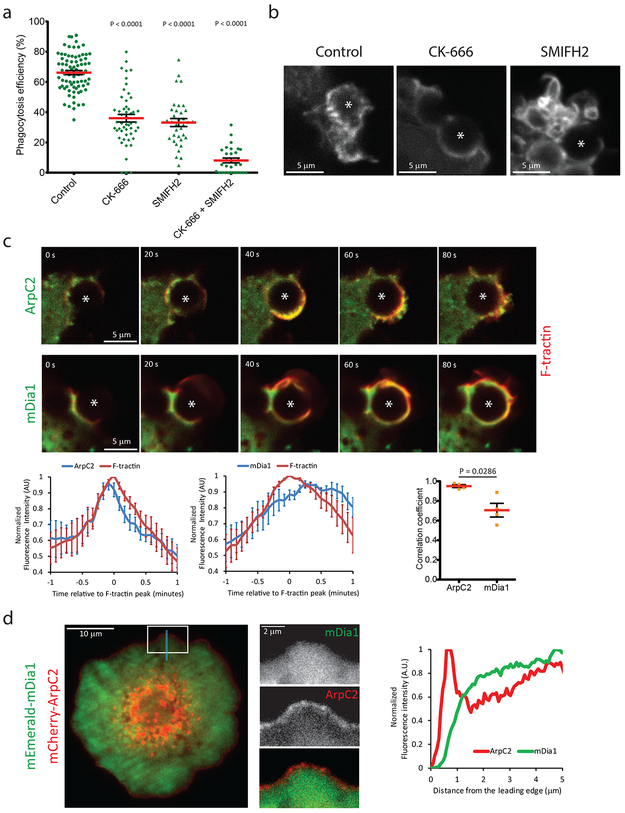
We next sought to delineate the mechanism of actin assembly in phagocytosis. Pharmacological inhibition of Arp2/3 with CK-66651 or formins with SMIFH252 significantly reduced phagocytosis efficiency of iC3b-opsonized microspheres (Figure 2a). Treatment with CK-666 together with SMIFH2 further reduced phagocytosis efficiency, suggesting that Arp2/3 and formins play distinct roles in phagosome formation. Confocal imaging of F-tractin-mCherry revealed for the small fraction of successful phagocytic events that CK-666 treatment blocked cell surface ruffling, with most particles making contact with non-ruffled regions of the cell. Once contacted, particles formed a hemispherical divot in the cell surface, followed by formation of a thin actin-filled protrusion that advanced along the particle surface to internalize it (Figure 2b, Movie 3), and bursts of actin driving phagosome movement were absent. In contrast, SMIFH2 treatment caused excessive ruffling over the entire cell surface, which mediated contact with particles. Although cell surface ruffles were not tightly coupled to the particle surface, they eventually did envelope and internalize particles (Figure 2b Movie 4). Thus, Arp2/3 drives particle capture by ruffles, initiation of the phagocytic cup, and phagosome movement, while formins couple actin protrusions to the particle surface.
Figure 2. Arp2/3 and mDia1 contribute to specific aspects of actin dynamics during CR-mediated phagocytosis.
a, Percent of bound particles that were internalized after perturbation of Arp2/3, formins, myosin II, and ROCK. RAW 264.7 macrophages were incubated in cytoskeletal drugs (Control 0.1%, DMSO (n= 79 fields from 9 independent experiments), CK-666 100 μM (n= 51 fields from 8 independent experiments), SMIFH2 20 μM (n= 35 fields from 7 independent experiments), CK-666 100 μM + SMIFH2 20 μM (n= 30 fields from 3 independent experiments) for 15 minutes prior to addition of iC3b-opsonized 5.15 μm polystyrene beads and internalization assayed one hour later. b, Confocal images from time-lapse movies of RAW macrophages expressing EGFP-F-tractin during the intermediate stage of iC3b-opsonized polystyrene bead engulfment, under the following treatment conditions: control 0.1% DMSO, CK-666 100 μM, SMIFH2 20 μM, from 3 independent experiments. c, Time-lapse confocal microscopy (elapsed time in seconds) of RAW 264.7 macrophage co-expressing mEmerald-ArpC2 (top row, green) or mEmerald-mDia1 (middle row, green) and mCherry-F-tractin (red) during phagocytosis of iC3b-opsonized 5.15 μm polystyrene beads. Fluorescence intensities were measured in a 6 μm diameter circular ROI, centered on the phagocytosed bead, and normalized to the maximal intensity and plotted over time relative to the peak of F-tractin recruitment (bottom row, left, center). Correlation between actin nucleator and F-tractin fluorescence intensities were calculated individually for each experiment, n=4 experiments. d, TIRF microscopy of a RAW macrophage co-expressing mEmerald-mDia1 (top and green) and mCherry-ArpC2 (middle and red) during formation of a frustrated phagocytic cup on an anti-αMβ2-coated coverslip, from 3 independent experiments. White box shows location of the inset. Blue bar shows location of the line scan of fluorescence intensities presented in the right panel. Scale bars are 5 μm, except d left panel is 2 μm. Error bars represent SEM, p values are from two tailed Mann-Whitney test. Numerical source data are provided in Statistical Source Data Figure 2.
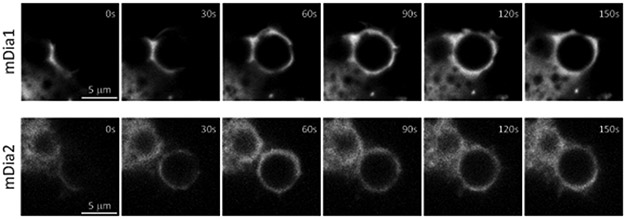
We next examined how Arp2/3 and formins coordinate actin dynamics during phagocytosis. We co-expressed mCherry-F-tractin together with either mEmerald-ArpC2 as a marker of the Arp2/3 complex, or mEmerald-tagged mDia1 or mDia2 as markers of the formins that have been previously implicated in CR-mediated phagocytosis12. Time-lapse confocal movies showed that ArpC2 and mDia1 were enriched at the cell-particle contact and during phagosome formation, whereas mDia2 was not (Figure 2c, Supplementary Figure 2, Movie 5). ArpC2 filled protrusions as they advanced along the particle surface and concentrated in actin bursts that propelled particles inwards. In contrast, mDia1 only extended partway around particles from the contact site during the initial phase of protrusion advance, but filled the cup during later phases of protrusion and phagosome closure. To determine their respective recruitment kinetics , we measured ArpC2 or mDia1 and F-tractin fluorescence intensities in a region surrounding the particle over time (Figure 2c bottom left and center). This showed that ArpC2 was recruited concomitantly with F-tractin and reached its maximum ≈5 seconds prior to the peak in F-tractin intensity. In contrast, mDia1 intensity increased more slowly than that of F-tractin, reaching its maximum and remaining stable 15 seconds after the F-tractin peak. This was supported by temporal cross-correlation analysis, which showed a higher correlation coefficient for ArpC2 and F-tractin intensities than for mDia1 and F-tractin intensities (Figure 2c bottom right), suggesting that Arp2/3 may be the primary driver of actin assembly in the phagocytic cup.
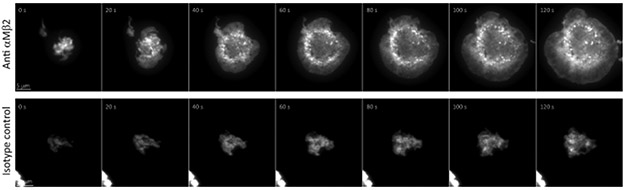
To better visualize the precise position of Arp2/3 and mDia1 relative to the leading edge of the phagocytic cup, we employed the frustrated phagocytosis model53. Coverslips were coated with an antibody specific to the iC3b binding site of αMβ2 integrin54, which promotes formation of abortive phagocytic cups on the coverslip surface that can be imaged by total internal reflection fluorescence microscopy (TIRFM). Of note, plating on isotype control antibody-coated coverslips (Supplementary Figure 3, Movie 6) abolished cell spreading and frustrated cup formation. Time-lapse TIRFM of RAW macrophages co-expressing ArpC2-mCherry and mDia1-mEmerald showed that Arp2/3 concentrated at vesicles in the cell center as well as along the leading edge of the phagocytic cup during the expansion phase, whereas mDia1 was recruited behind ArpC2 (Figure 2d and Movie 7). Super-resolution structured illumination TIRFM (TIRF-SIM) of fixed frustrated cups stained with fluorescent phalloidin revealed a dense actin network within a few microns from the leading edge, similar to the network formed by Arp2/3 in lamellipodia18 (Figure 4b and Movie 8). This was followed by a sparser network of circumferential bundles, reminiscent of actin in a lamellum55, albeit lacking thick stress fibers. Together, these results suggest that Arp2/3-mediated polymerization drives the leading edge of protrusions to mediate target capture and phagocytic cup formation and also propels phagosomes into the cell interior after internalization, whereas mDia1 follows Arp2/3 in the phagocytic cup to mediate actin assembly that couples advancing protrusions to the particle surface.
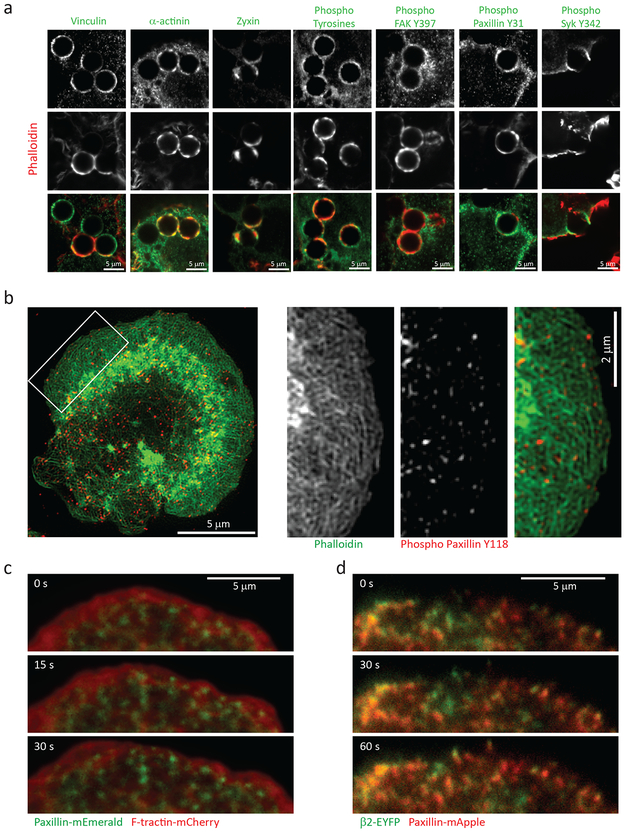
Figure 4. β2 integrins mediate the formation of focal complex-like signaling platforms at the phagosome.
a, Immunofluorescence localization of vinculin, a-actinin, zyxin, phospho-tyrosine, FAK phosphorylated on tyrosine 397 (Y397), paxillin phosphorylated on tyrosine 31 (Y31), syk phosphorylated on tyrosine 342 (Y342) (top panels, and green) in RAW macrophages phagocytosing iC3b-opsonized 5.15 μm polystyrene beads, imaged by confocal microscopy. Actin filaments were labeled with fluorescent phalloidin (middle panels, and red). b, Immunofluorescence of paxillin phosphorylated on tyrosine 118 (Y118, middle right and red) and labeling of actin filaments with fluorescent phalloidin (middle left and green), at the frustrated phagocytic cup of a RAW macrophage on an anti-αMβ2 coated coverslips, imaged by TIRF-SIM. c, Time-lapse TIRF microscopy of the leading edge of a RAW macrophage expressing mEmerald-Paxillin (green) and mCherry-F-tractin (red) during formation of a frustrated phagocytic cup on an anti-αMβ2 coated coverslip. d, Time lapse TIRF microscopy of a RAW macrophage co-expressing αM, β2-EYFP (green) and mApple-Paxillin (red) during formation of a frustrated phagocytic cup on an anti-αMβ2 coated coverslip. Images are representative examples from 3 independent experiments. Scale bars are 5 μm, except b left panel is 2 μm. Elapsed times are in seconds.
Two distinct actin networks are differentially coupled to the target particle to drive protrusion of the phagocytic cup.
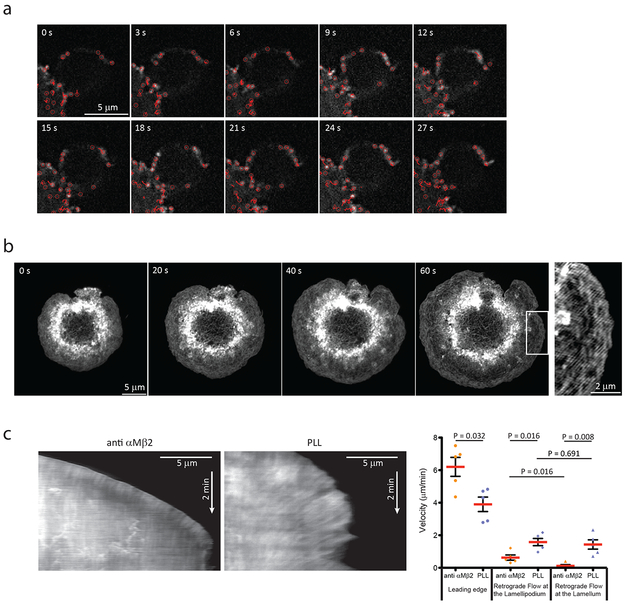
Actin polymerization at the leading edge of a migrating cell can drive either retrograde flow of the cytoskeleton within the cell, or, if coupled through an integrin-mediated molecular clutch to the extracellular substrate, can drive leading edge protrusion16. To determine if similar actin dynamics mediate phagocytosis, we employed confocal fluorescent speckle microscopy (FSM)56 of actin dynamics in RAW macrophages expressing actin-mEos3.2 by photo-switching at a low rate during phagocytosis of iC3b-opsonized microspheres. Automated detection and tracking57 revealed that fluorescent actin speckles appeared sequentially at the advancing tip of the phagocytic cup at about the same rate as leading edge protrusion, and after appearance, remained nearly immobile relative to the particle and did not undergo retrograde flow along the particle surface (Figure 3a and Movie 9). This suggests that actin assembly at the leading edge of the phagocytic cup drives protrusion without retrograde flow. To rule out the possibility of flow off-axis to the confocal imaging plane, we analyzed actin dynamics in flat phagocytic cups during frustrated phagocytosis. Time-lapse TIRF-SIM imaging of F-tractin-EGFP showed that shortly after coverslip attachment, the peripheral lamellipodium-like zone, and more proximal, lamella-like zone expanded outward until the cell reached a maximal spread area (Figure 3b and Movie 8). Kymographs perpendicular to the cell edge revealed cup protrusion at 6 ± 0.7 μm.min−1, with slow actin retrograde flow (0.6 ± 0.1 μm.min−1) occurring in the lamellipodia-like zone, together indicating an actin network assembly rate of ≈6.6 μm.min−1 (sum of protrusion and retrograde flow) (Figure 3c). In contrast to the flow in the periphery, retrograde flow was virtually absent in the lamella-like zone (Figure 3c). To test whether αMβ2 integrin engagement was required to couple actin to the substrate, we analyzed actin dynamics in macrophages spread on poly-L-lysine to allow adhesion without integrin engagement. Movies and kymograph analysis of F-tractin-EGFP showed that compared to plating on anti-αM, in cells plated on poly-L-lysine, protrusion (3.9 ± 0.4 μm.min−1) and actin network assembly (5.5 μm.min−1) were slower and retrograde flow was nearly 3x faster (1.6 ± 0.2 μm.min−1, Figure 3c and Movie 9). Together, these results indicate that polymerization of two distinct actin networks are differentially coupled by αMβ2-mediated adhesion to complement-opsonized target particles to drive protrusion of the phagocytic cup: A peripheral, Arp2/3-mediated network that is partially coupled to the particle surface, and a more proximal, mDia1-generated network that is fully particle-coupled.
Figure 3. Coupling of the actin cytoskeleton to the target enables fast protrusion at the forming phagosome.
a, Fluorescent speckle microscopy during phagocytosis of iC3b-opsonized 5.15 μm polystyrene beads by RAW macrophages expressing Actin-mEos3.2, photoswitched with 405 nm light, and imaged with 561 nm light by time-lapse spinning disk confocal microscopy, from 4 independent experiments. Images were aligned with respect to the ingested particle. Red circles represent detected speckles in the corresponding frame, red lines represent tracks of the speckles from the first time point of their detection to the current frame, as analyzed using automated speckle-tracking software. b, Time lapse TIRF-SIM images of a RAW macrophage expressing F-tractin-mNeonGreen during formation of a frustrated phagocytic cup on anti-αMβ2 coated coverslip, from 3 independent experiments. c, Examples of kymographs generated by line scans perpendicular to the leading edge taken from time-lapse movies during frustrated phagocytosis by F-tractin-EGFP-expressing RAW macrophages spreading on anti-αMβ2-coated coverslips (left) or poly-L-lysine (PLL)-coated coverslips in the presence of 2 mM EDTA to inhibit integrin-ligand engagement (center). Initial leading edge protrusion and actin retrograde flow velocities (lamellipodium= 0-3μm from the leading edge, lamellum= 3-8μm from the leading edge) were quantified from such kymographs (n=5 experiments). Scale bars are 5 μm, except b left panel is 2 μm. Error bars represent SEM, p values are from two tailed Mann-Whitney test. Numerical source data are provided in Statistical Source Data Figure 3.
β2 integrins mediate the formation of focal complex-like signaling platforms at the phagosome
We next sought to determine if complement-engaged integrins were coupled to actin by a FA-like molecular clutch. We first immunolocalized FA structural and signaling proteins in RAW macrophages that were fixed after five-minute incubation with iC3b-opsonized microspheres. Confocal imaging revealed patches on the phagosome surface of vinculin, α-actinin, and zyxin, actin-associated proteins typically found in FAs and podosomes58 (Figure 4a). Moreover, phospho-tyrosine, phospho-FAK (tyrosine 397), phospho-paxillin (tyrosines 31 and 118) and phospho-Syk (tyrosine 342) were all enriched in patches on phagosomes, indicative of active integrin signaling (Figure 4a-b). To better characterize the structure and dynamics of adhesions in the phagocytic cup, we imaged frustrated phagocytic cups by TIRF-SIM. Localization of actin and phospho-paxillin in fixed cells showed near-diffraction limited puncta lacking actin enrichment that were sparsely distributed throughout the cup (Figure 4b), similar to focal complexes formed by macrophages adhered to ECM59. Lack of localization of expressed ArpC2-mCherry to such puncta (Figure 2d and Movie 6) confirmed they were not podosomes58,60. Analysis of paxillin-mEmerald dynamics by time-lapse TIRF microscopy showed that puncta formed a micron or so behind the leading edge and remained stable throughout spreading (Figure 4c, Movie 11). Co-expression of un-tagged αM integrin, EYFP-β2 integrin, and mApple-paxillin showed that paxillin puncta were enriched in β2 integrin, confirming they were sites of integrin engagement (Figure 4d and Movie 12). This demonstrates that phagocytosis is mediated by discrete integrin adhesions that bear the composition, signaling, morphological and dynamic characteristics of focal complexes.
Mechanical coupling of integrins to the actin cytoskeleton by talin enhances particle engulfment
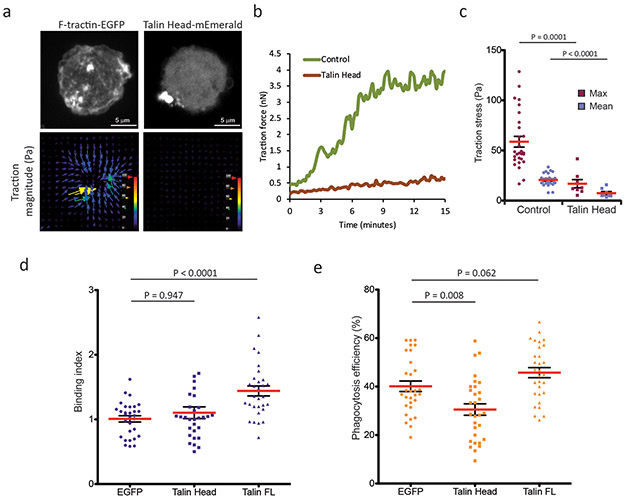
We next sought to examine the transmission of actin-generated forces to complement-engaged integrins during phagocytosis. We characterized the stresses exerted by RAW macrophages on a phagocytic target by performing time-lapse traction force microscopy (TFM)61 on fiducial-marked polyacrylamide gels functionalized with anti-αM integrin antibody during frustrated phagocytosis (Figure 5a). This showed that force increased during the expansion of the phagocytic cup and peaked as the cell reached its maximal spread area (Figure 5b). After spreading, stresses fluctuated throughout the cup with an average peak of 58.8 ± 5.3 Pa (Figure 5c, Movie 13). The highest stresses localized to the periphery of the cup and were directed toward the center where stresses were below the detection limit (Figure 5a). Together with our observations of actin dynamics above, this shows that the direction of traction stresses correlate with the direction of actin flow within the phagocytic cup.
Figure 5. Mechanical coupling of integrins to the actin cytoskeleton by talin enhances particle engulfment.
a, Examples from time-lapse traction force microscopy of RAW macrophages expressing F-tractin-EGFP (left) or talin head domain-mEmerald (right), during frustrated phagocytosis on anti-αMβ2 coated 4 kPa polyacrylamide gels. Top: spinning disk confocal images, bottom: stress vectors calculated from the displacement of fluorescent 40nm particles incorporated in the gel, pseudocolor scale and vector lengths represent traction stress magnitude. b, Quantification of the total traction force generated within the phagocytic cup over time by RAW macrophages (green: mock-transfected control, brown: expressing talin head domain-mEmerald) from a representative experiment. c, Quantification of traction stress within the phagocytic cup during spreading of control RAW macrophages (n= 29 cells) or macrophages expressing Talin Head-mEmerald (n=8 cells). Maximal (red) and mean (blue) stress within the phagocytic cup were measured at the time point when the cup reached its full size. d, Binding index (relative to EGFP-expressing control) and (e) phagocytosis efficiency (percentage of internalized relative to contacted beads) after 1 hour incubation of iC3b-opsonized 5.15 μm polystyrene beads with RAW macrophages expressing EGFP, talin head domain-mEmerald, or full-length (FL) talin-mEmerald (n= 30 fields). Experiments were repeated 3 times independently. Scale bars are 5 μm, error bars represent SEM, p values are from two tailed Mann-Whitney test. Numerical source data are provided in Statistical Source Data Figure 5.
We then probed the molecular mechanism of force transmission to the phagocytic target. Talin forms a force-bearing link between integrin and actin in migrating cells, and over-expression of its head domain disrupts this link25. Analysis of macrophages over-expressing talin head-mEmerald in the frustrated phagocytosis TFM assay showed a 71% reduction of traction stresses compared to contol (Figure 5a-c, Movie 13). In a phagocytosis assay, over-expression of full-length talin significantly increased particle binding and internalization, in agreement with its known effects on integrin activation23. In contrast, talin head over-expression had no effect on binding, but significantly reduced particle internalization (Figure 5d-e, Movie 14). Thus, a talin-mediated linkage between integrin and actin is required for force generation on target particles and efficient internalization.
Syk kinase activity is required for vinculin recruitment and strengthening force transmission for optimal particle uptake
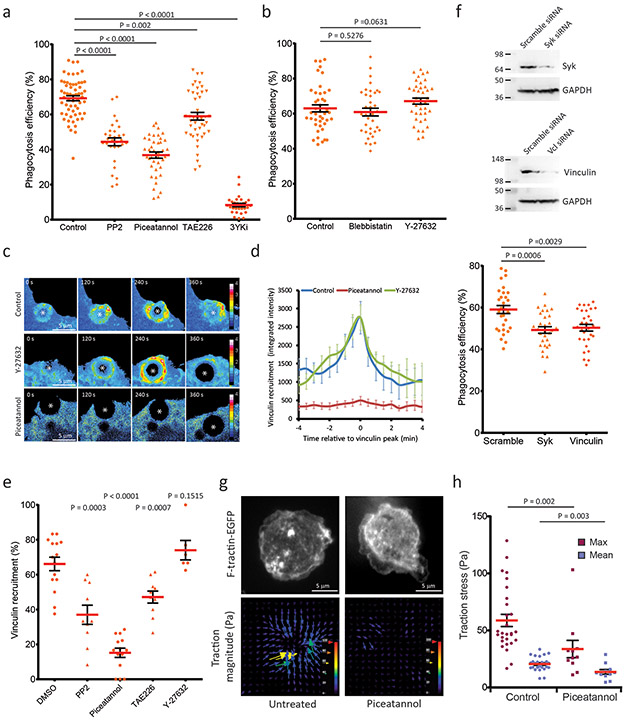
We next sought to test whether tyrosine kinases or Rho-associated kinase (ROCK)-mediated myosin II activity promote molecular clutch reinforcement in phagocytosis, as shown for FAs in cell migration29,62. Pharmacological inhibition showed that phagocytosis efficiency of iC3b-opsonized microspheres was strongly reduced by treatment with either the Syk inhibitor piceatannol63 or the Src-family kinase inhibitor PP264, with weaker albeit significant reduction by the FAK/Pyk2 inhibitor TAE22665 (Figure 6a). Combined treatment with the three inhibitors abolished microsphere internalization almost entirely, suggesting that the tyrosine kinases have non-redundant functions. In contrast, inhibition of myosin II with blebbistatin66 or ROCK with Y-2763267 did not significantly affect phagocytosis efficiency, indicating that myosin II-mediated contractility was not required for particle internalization (Figure 6b).
Figure 6. Syk kinase activity is required for vinculin-mediated clutch reinforcement and optimal particle uptake.
a, b, e, Phagocytosis efficiency in RAW macrophages upon treatment with DMSO (n= 60 fields, from 6 independent experiments) PP2 (20 μM, n= 30 fields, form 3 experiments), piceatannol (50 μM, n= 40 fields, from 4 experiments), TAE226 (5 μM, n= 44 fields, from 5 experiments), or the three inhibitors combined (3YKi) (n= 29 fields, from 3 experiments) (a), upon treatment with DMSO (n= 43 fields), blebbistatin (50 μM, n= 36 fields) or Y-27632 (10 μM, n= 40 fields), from 5 independent experiments (b), or after siRNA knockdown of Syk or vinculin (n= 30 fields from 3 experiments) (f). f upper panels show representative western blots from 3 independent experiments of Syk, vinculin and GAPDH protein levels, . c, Time-lapse of vinculin concentration relative to a soluble marker during phagocytosis of iC3b-opsonized microspheres by macrophages treated with DMSO (control, upper row), Y-27632 (10 μM), or piceatannol (50 μM). d, Quantitation of vinculin-mEmerald relative concentration at the phagosome (control: n= 10 cells; piceatannol: n= 6 cells, Y-27632: n= 12 cells). Time expressed relative to the peak of vinculin-mEmerald recruitment. e, Percentage of phagosome showing a recruitment of vinculin-mEmerald during phagocytosis of iC3b-opsonized microspheres, imaged by confocal microscopy with a single plan 2.5 μm above the the coverslip, every 30 seconds. Macrophages were imaged in the presence of 0.1% DMSO (n= 15 fields), 50 μM piceatannol (n= 14 fields) 20 μM PP2 (n= 10 fields), 10 μM TAE226 (n= 10 fields), or 10 μM Y-27632 (n= 6 fields). g, Time-lapse traction force microscopy of untreated (control) or piceatannol-treated (50 μM) macrophages expressing F-tractin-EGFP during frustrated phagocytosis. Top: confocal images of F-tractin-EGFP, bottom: stress vectors, pseudocolor scale and vector lengths represent traction stress magnitude. Experiments were repeated 3 times independently. h, Quantification of traction stress within the phagocytic cup in control (n= 29 cells) or piceatannol-treated macrophages (n=12 cells). Maximal (red) and mean (blue) stress measured when the cup reached its full size. Scale bars are 5 μm,error bars represent SEM, p values from two-tailed Mann-Whitney test. Numerical source data are provided in Statistical Source Data Figure 6.
We next examined the role of tyrosine kinases and contractility in regulation of vinculin. In migrating cells, Src- and FAK-mediated phosphorylation of paxillin and ROCK-mediated myosin II activity promote vinculin recruitment to FAs29, where vinculin engages actin to enhance force transmission to ECM-bound integrins30. We quantified the effects of perturbations on vinculin recruitment to phagosomes by ratiometric imaging of vinculin-mEmerald and soluble FusionRed protein (enabling correction for volume effects68). This showed that Syk inhibition abolished vinculin recruitment to puncta on the phagosome, Src or FAK/Pyk2 inhibition caused partial reduction, while ROCK inhibition had no significant effect (Figure 6c-e, Movie 15). We confirmed the role of Syk and vinculin in phagocytosis by siRNA silencing, which each significantly decreased phagocytosis efficiency (Figure 6f). We then assessed whether Syk activity was important for force transmission to integrins. TFM analysis of frustrated phagocytosis showed that treatment with piceatannol significantly reduced traction force (Figure 6g-h, Movie 13). Together, these results indicate that multiple families of tyrosine kinases contribute to CR-mediated phagocytosis, but Syk activity specifically promotes vinculin recruitment to the phagosome, enhances force transmission to the target particle, and increases particle uptake.
CR-mediated phagocytosis is mechanosensitive
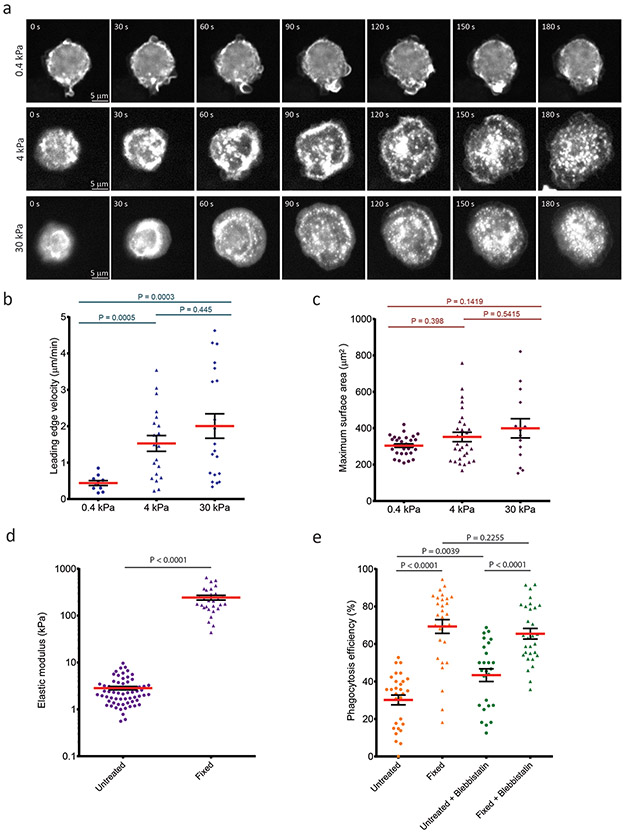
We finally sought to determine whether CR-dependent phagocytosis was mechanosensitive. We first analyzed the response of macrophages to ligand-coated polyacrylamide gels of different stiffness during frustrated phagocytosis. We found that cells formed a phagocytic cup on gel stiffnesses above 0.4 kPa (Figure 7a, Movie 16). Kymograph analysis showed that after plating, the initial velocity of leading edge protrusion increased directly with gel stiffness, whereas the final spread area was unaffected (Figure 7b-c). To determine if differences in cup formation in response to changes in target stiffness during frustrated phagocytosis translated to changes in phagocytosis, we altered sRBC stiffness by treatment with glutaraldehyde. Measurements by atomic force microscopy showed that 50 mM glutaraldehyde treatment for 1 min increased the elastic modulus of sRBCs by nearly 10-fold (from 2.8 ± 0.2 kPa to 242.6 ± 27.6 kPa , Figure 7d). Phagocytosis assays showed that while untreated sRBCs were modestly internalized (30.1 ± 2.6 % efficiency), stiff sRBCs were engulfed with nearly threefold higher efficiency (69.3 ± 3.6 %), and this difference was unaffected by inhibition of myosin II with blebbistatin (Figure 7e). Together, these results show that CR-mediated phagocytosis involves a myosin II-independent mechanosensitive molecular clutch.
Figure 7. Molecular clutch-mediated mechanosensing regulates phagocytosis.
a, Representative spinning disk confocal images of RAW macrophages expressing F-tractin-EGFP, from time-lapse movies of frustrated phagocytosis on anti-αMβ2-coated 0.4 kPa, 4 kPa, and 30 kPa polyacrylamide gel substrates. b, Quantification of the velocity of the edge of the phagocytic cup determined from movies of frustrated phagocytosis of RAW macrophages on anti-αMβ2-coated 0.4 kPa (n= 10 cells), 4 kPa (n= 20 cells), and 30 kPa (n= 20 cells) polyacrylamide gel substrates. c, Quantification of the phagocytic cup maximum surface area determined from movies of frustrated phagocytosis of RAW macrophages on anti-αMβ2-coated 0.4 kPa (n= 30 cells), 4 kPa (n= 30 cells), and 30 kPa (n= 14 cells) polyacrylamide gel substrates. d, Elastic moduli determined by atomic force microscopy of complement-opsonized untreated sheep red blood cells (n = 66 cells) or sheep red blood cells fixed with 0.05% glutaraldehyde for 1 minute (n= 29 cells). e, Percent of bound complement-opsonized untreated sheep red blood cells (circles) or sheep red blood cells fixed with 0.05% glutaraldehyde (triangles) that were internalized by RAW macrophages in absence (orange) or presence of 50 μM blebbistatin (green) (n= 30 fields from 3 independent experiments). Scale bars are 5 μm, error bars represent SEM, p values are from two tailed Mann-Whitney test. Numerical source data are provided in Statistical Source Data Figure 7.
Discussion
This study demonstrates that contrary to the prevailing view2,14,15, CR-mediated phagocytosis does not proceed by particles sinking into the cell, but is driven by actin-based protrusions that extend along the particle to mediate internalization. Given the rapidity of phagocytosis (≈2-5 min), protrusion formation could have been missed by electron microscopy, whereas live cell imaging enables visualization of the entire process. Similar observations were made with mouse and human macrophage cell lines, as well as mouse primary macrophages, suggesting that this mechanism is conserved.
We show that CR-mediated phagocytosis and migration of mesenchymal cells utilize similar mechanisms. In both systems, Arp2/3-mediates polymerization of a dense actin network drives protrusion of the leading edge, while mDia1 follows Arp2/3 to promote integrin-dependent coupling to the particle surface. Phagocytic cups contain clusters of αMβ2 integrins and FA proteins, suggesting that actin is coupled to integrins by FA machinery. Indeed, we demonstrate that talin, vinculin and tyrosine kinases play critical roles in transmission of forces generated by the actin cytoskeleton to ligand-engaged αMβ2 integrins, acting as a molecular clutch that drives phagocytosis. We find that macrophages form phagocytic cups faster on stiffer targets and engulf them more efficiently, indicating that CR-mediated phagocytosis is mechanosensitive. These findings highlight the conserved nature of the molecular clutch to mediate different cell functions in mesenchymal and immune cells.
Despite many commonalities, subtle differences exist that may be critical to the different cellular processes. In migrating cells, most leading edge adhesions disassemble in the lamellum, where formin-dependent polymerization and myosin II-mediated bundling of actin stress fibers drive elongation of remaining adhesions into mature FAs69-72. In CR-mediated phagocytosis, adhesions formed concomitantly with cup protrusion, however they neither matured, turned over, nor formed podosome-like ring structures, suggesting that they are similar to focal complexes72. In migrating cells, lamellipodial actin polymerization drives retrograde flow, which slows down when engaged to ECM-bound integrins via a talin-mediated molecular clutch55,73 that transmits traction forces to the ECM, and drives leading edge protrusion20,21. Actin flow slows further in the lamella as the clutch is reinforced by the recruitment of vinculin, mediated by myosin II activity and paxillin phosphorylation by FAK29,74,75. Variability in the degree of actin-integrin engagement in the lamella is thought to regulate the speed of migration76. In the phagocytic cup, actin retrograde flow occurred in the lamellipodium-like network at the cup edge, but completely ceased in the lamellum-like zone, indicating full engagement of actin to complement-bound integrins, consistent with the observed fast cup protrusion. Recruitment of vinculin was independent of myosin II contractility, and the low traction force and lack of stress fibers support the notion that contractility is low in the cup. Since Vinculin can be recruited to FAs in a myosin-independent manner by interaction with phosphorylated paxillin29, or when vinculin conformation is open74, Syk could recruit vinculin to αMβ2 adhesions through phosphorylation of paxillin or vinculin. Thus, comparing the cell migration and phagocytic machinery, the low contractility and tight coupling between actin and integrins induced by Syk-mediated recruitment of vinculin during phagocytosis may shift the balance of actin-driven forces toward less retrograde flow and faster protrusion of the leading edge to promote rapid and efficient target internalization.
What might phagocytic preference for stiff targets be good for? Such mechanosensitivity may influence target discrimination, as cells stiffen during apoptosis, and microbes are much stiffer than mammalian cells40-43. Such preference could affect antigen uptake and subsequent presentation, thus affecting the ability to mount an adaptive immune response. Mechanosensitivity could also bear on particle-based drug delivery, where phagocytosis reduces particles reaching target tissues77. Although FcR-mediated phagocytosis has also been reported to be mechanosensitive78, how these receptors are linked to the cytoskeleton is not known. Elucidating the mechanisms of phagocytic mechanosensitivity could enable engineering of dendritic cells or macrophages with improved anti-tumor response79 or drug-bearing particles that evade phagocytosis.
Methods
Cell culture
RAW 264.7 murine macrophage cell line and THP-1 human monocytic cell line were obtained from the American Type Culture Collection and cultured in RPMI 1640 ATCC modification (Gibco) with 10% heat inactivated FBS (Atlanta), at 37°C in a 5% CO2 incubator. THP-1 cells were differentiated into macrophages with 100 ng/ml of PMA (Sigma) for 72 hours. Transfections were performed using Amaxa nucleofactor (Lonza), according to the manufacturer’s instructions.
To prepare bone marrow derived macrophages, femurs and tibias were dissected from euthanized C57B/6 LifeAct-GFP mice. The bone marrow was extracted by perfusion of RMPI 1640 (Gibco) with a 21-gauge needle to produce a single-cell suspension, which was plated in RPMI 1640 with 10% heat inactivated FBS, 10 ng/mL murine M-CSF (Peprotech) and 100 U/mL penicillin, 100 μg/mL streptomycin and 25 μg/mL amphotericin B (Gibco). After removal of non-adherent cells, macrophages were cultured for 7 days at 37°C in a 5% CO2 incubator to allow complete maturation. The animal protocol was approved by the NHLBI Animal Care and Use Committee, and all protocols are compliant with ethical regulations stipulated by the Animal Care and Use Committee.
Reagents
CK-666 and SMIFH2 were purchased from EMD Millipore. Blebbistatin was obtained from TRC Canada. Y-27632, PP2 and Piceatannol were from Calbiochem. TAE226 was obtained from Selleckchem.
Monoclonal anti-CD11b antibodies clone M1/70, anti-CD18 clone GAME-46, the rat IgGb2 isotype controls and anti Syk Y348 (Y342 in mouse) were purchased from BD pharmingen. The ant Syk, clone D3Z1E, and anti GAPDH, clone D16H11, were from Cell Signaling. The anti iC3b antibody was from GeneTex. Anti-BSA and anti-1.3 β glucan antibodies were from Abcam. The anti-sRBC IgM was from MyBioSource. The anti-sRBC IgG was from MP Bio. Anti-vinculin and -α-actinin antibodies were from Sigma Aldrich. The anti-phospho tyrosine antibody clone 4G10 was from EMD Millipore. Anti-phospho FAK Y397, paxillin Y31 and paxillin Y118 were from Invitrogen. The anti-zyxin antibody was a generous gift from Mary Beckerle. Secondary antibodies were obtained from Jackson Immunoresearch. Alexa Fluor 488 phalloidin was purchased from Fischer scientific.
Plasmids for the expression of F-tractin-EGFP, F-tractin-mCherry, F-tractin-mNeonGreen, CAAX-mEGFP, ArpC2-mEmerald, ArpC2-mCherry, mDia1-mEmerald, Actin-mEos3.2, Paxillin-mEmerald, Paxillin-mApple, Talin Head-mEmerald, Talin-mEmerald, Vinculin-mEmerald, mEGFP, and FusionRed were obtained from Michael Davidson. Plasmids for the expression of Mac-1 and integrin β2-EYFP were purchased from Addgene.
siRNAs were purchased from Ambion. Specific sequences for silencing of Syk were 5’-GCAAUUACCUGGGUGGUUUtt-3’and 5’-CCAUCGAGAGGGAACUUAAtt-3’. Specific sequences for silencing of Vcl were 5’-GCUCGGAAAUGGUCUAGCAtt-3’ and 5’-CAGAGAUGCUGGUUCAUAAtt-3’.
Phagocytosis assays
Polystyrene microspheres (Bangs laboratories) were opsonized with purified iC3b (Complement Technology) at 100 μg/ml in PBS, or BSA at 1 mg/ml, overnight at 4˚C. Sheep red blood cells (MP Bio) were labelled with Alexa Fluor 647 succinimidyl ester (Invitrogen) for 1 hour at room temperature, then washed in PBS and incubated with an anti-sheep RBC rabbit IgM (MyBioSource MBS524107, 1:200 dilution) for 1 hour at room temperature. After washes, RBCs were incubated in 20% C5 deficient serum (Sigma-Aldrich) at 37˚C for 20 minutes. Texas Red-labelled zymosan A (Invitrogen) was opsonized with 20% C5 deficient serum (Sigma-Aldrich) at 37˚C for 1 hour. Macrophages were plated overnight on # 1.5 glass coverslips (Corning), then incubated with 150 ng/ml of PMA in serum free RPMI-1640, and the indicated blocking antibodies for CD11b (BD Pharmingen 553307), CD18 (BD Pharmingen 553341) or rat IgG2b isotype control (BD Pharmingen 553985) at 10 μg/mL, 15 minutes before adding particles. For live cell microscopy, imaging was started immediately after particle addition. For quantification of phagocytosis, cells were incubated with opsonized particles for 1 hour at 37˚C, then rinsed twice and fixed with 4% paraformaldehyde (Electron Microscopy Science) in PBS for 20 minutes. Outside opsonized particles were labeled by immunofluorescence with an anti-iC3b antibody (GeneTex GTX40522, 1:2000 dilution), and nuclei were stained with Hoechst 33342 (Thermo scientific). Outside BSA-coated microspheres, unopsonized sRBC and zymosan A were labelled with an anti-BSA (Abcam ab186531, 1:100 dilution), anti-sRBC (MP Bio 55806, 1:200 dilution) or anti-1,3 β glucan antibody (Abcam ab233743, 1:200 dilution), respectively. Image fields of view containing cells were selected randomly and acquired with a 20× 0.45 NA objective. At least 1000 cells were quantified per condition for each independent experiment. Binding index (total cell-associated particles normalized to the number of cell-associated particles in the reference condition, in this case PMA-activated cells), phagocytic index (total internalized particles per cell normalized to internalized particles in the PMA reference condition). We include both surface associated particles and internalized particles when counting “binding” because it is assumed that a particle had to have bound to the cell to in order to be internalized. Phagocytosis efficiency was defined as the percentage of cell-associated particles that were internalized. Silencing of Syk or Vinculin expression was verified by western blotting of cell lysates from the same sample of transfected cells, using anti Syk (Cell Signaling 13198S, 1:1000 dilution) and anti Vinculin (Sigma-Aldrich V9131, 1:2000 dilution) antibodies, respectively. To control for equal loading, the lower parts of the membranes were cut after protein transfer and processed for anti GAPDH western blotting (Cell Signaling 5174, 1:5000 dilution).
Frustrated phagocytosis
Glass coverslips #1.5 were coated with neutravidin (Thermo scientific) at 0.5 mg/mL in PBS, at 4°C for 4 hours. After washes, coverslips were incubated with 10 μg/mL biotin-conjugated M1/70 (BD Pharmingen 553309) or rat IgG2b isotype control (BD Pharmingen 553987) antibodies at 4°C overnight. Coverslips were then blocked with PLL-PEG (Susos) at 0.1 mg/mL, at 4°C for at least 2 hours. Macrophages were suspended in ice cold PBS with 2 mM EDTA by scraping, then washed in serum free RMPI 1640, supplemented with 25 mM HEPES pH 7.3 and 150 ng/ml PMA 15 minutes before addition onto the coverslips. Cells were imaged immediately for live cell microscopy, or fixed with 4% PFA after 5 minutes and processed for immunofluorescence.
For frustrated phagocytosis on polyacrylamide gels and traction force microscopy, 40 μm thick gels with 40 nm TransFluoSpheres (633/720) (Thermo scientific) were prepared on glass coverslips as described before35. Neutravidin (0.5 mg/mL) was crosslinked to the surface of the gels using Sulfo-SANPAH at 1mg/ml (Thermo scientific). After washes, gels were incubated with biotin-conjugated M1/70 (BD Pharmingen 553309) or rat IgG2b isotype control (BD Pharmingen 553987) antibodies at 10 μg/mL, at 4°C overnight.
Microscopy
All live cell imaging was performed at 37˚C in RPMI 1640, supplemented with 25 mM HEPES pH 7.3 and 150 ng/ml PMA. Microscopy was performed on Nikon Eclipse Ti or Ti2 microscopes equipped with Perfect Focus™, a TIRF illuminator, a Yokogawa CSU-X1 or CSU-W1 spinning disc and either a Photometrics Coolsnap Myo or a Hamamatsu Orca-flash 4.0 v3 camera. Illumination was provided by fiber-optically (Oz Optics) coupled solid state lasers (45 mW 405 nm (for photoconversion), 100 mW 488 nm (for EGFP, EYFP and mEmerald excitation) and 200 mW mW 561 nm (for mCherry, mEos3.2 and Texas red excitation) and 100mW 655 nm (for fluorescent beads ) from Spectral Applied Research, and emission was collected via a multibandpass dichroic and single bandpass emission filters (525/50 nm for green, 605/52 nm for red) and a long pass LP647nm for far red (Chroma) in an electronic filter wheel (Sutter instruments). Pairs of images in each channel were captured in rapid succession at the noted intervals over time for multi-color time-lapse imaging. Microscope functions were controlled by Elements (Nikon) or Metamorph (GE) software. Stage temperature was held constant with an airstream incubator (Nevtek). Phagocytosis of particles was imaged by spinning disk confocal microscopy with a 100× 1.49 NA oil objective for 2D imaging, and a 60× 1.27 NA water objective for 3D imaging. Frustrated phagocytosis on polyacrylamide gels was imaged by spinning disk confocal microscopy with a 60× 1.2 NA water objective. Frustrated phagocytosis on glass coverslips was imaged by TIRF microscopy with a 60× 1.49 NA oil objective. TIRF-SIM was performed on a DeltaVision OMX SR microscope, equipped with a 60× 1.42 NA oil objective. SIM images were reconstructed using 0.02 for the wiener filter, and aligned using softWoRx (Applied Precision). Images were analyzed and quantified using ImageJ 1.52 (NIH) and MetaMorph 7.7 (Meta imaging). For three-dimensional imaging, confocal stacks were deconvolved with NIS Elements (Nikon).
Quantification of protein recruitment during phagosome formation
For quantification of ArpC2, mDia1 and F-tractin recruitment to phagosomes, confocal images were acquired every 5 seconds. Integrated fluorescence intensities were measured in a 6 μm diameter circular ROI, centered on the phagocytosed bead. After background subtraction, measurements were normalized to the maximal intensity and plotted over time relative to the peak of F-tractin recruitment. For each phagosome, recruitment of the fluorescent actin nucleator and fluorescent F-tractin were compared using the cross-correlation function in Matlab.
For quantitative measurement of vinculin recruitment to phagosomes, confocal images were acquired every 30 seconds. Correction for variations in cell thickness within the confocal slice was performed as described previously68. Briefly, expression of freely diffusing FusionRed was employed as a volume reference and each cell was treated separately. After background subtraction, each cell was isolated using a mask created with the mEmerald-vinculin channel. Images were normalized by the mean intensity of the entire cytoplasm. Subtraction of the volume (FusionRed) image from the mEmerald-vinculin image created an image Re, where the intensity of each pixel represents the amount of protein recruited (positive values) or excluded (negative values), relative to the average concentration of protein within the cytoplasm. For each phagosome, vinculin relative recruitment was quantified from the Re image, in a 6 μm diameter circular ROI centered on the phagocytosed bead.
Kymograph analysis of actin retrograde flow and cell protrusion
Kymographs for cell protrusion and actin flow measurements were obtained by plotting the fluorescence intensities of the GFP channel along a line perpendicular to the leading edge of the cell (X axis), over the time course of the experiment (Y axis), using ImageJ. Slope angles were measured for the initial protrusion of the leading edge, the actin retrograde flow at the leading edge (lamellipodium) and in the lamellum, and converted into velocity.
Fluorescent speckle microscopy
RAW macrophages expressing Actin-mEos3.2 were imaged at by spinning disc confocal microscopy. A low level of 405 nm laser light was employed for the photoconversion of a small number of mEos3.2-actin molecules from green to red for every time point, and images of red fluorescence were acquired every 3 seconds. To analyze the flow relative to the particle, image sequences for each phagocytosis event were cropped and realigned in reference to the particle position using ImageJ. Fluorescent speckles were then detected and tracked with the QFSM software from the Danuser lab in Matlab R2016a (Mathworks), as described previously57.
Immunofluorescence
For immunofluorescence of endogenous adhesion proteins, cells were incubated with opsonized particles, fixed for 10 minutes at 37˚C, then rinsed twice and fixed with 4% paraformaldehyde (Electron microscopy science) in cytoskeleton buffer (10 mM MES pH 6.1, 138 mM KCl, 3 mM MgCl2, 2 mM EGTA) for 20 minutes. Free aldehydes were quenched 100 mM glycine for 10 minutes, and cells were permeabilized with 0.5% Triton X-100 for 5 minutes. Endogenous proteins were immuno-labelled using the following antibodies: anti α-Actinin (Sigma-Aldrich A5044, 1:500 dilution), anti Vinculin (Sigma-Aldrich V9131, 1:500 dilution), anti Zyxin (B71, 1:600 dilution), anti phosphotyrosine (EMD Millipore 05-321, 1:300 dilution), anti phospho-FAK Y397 (Invitrogen 44722G, 1:100 dilution), anti phospho-Paxillin Y31 (Invitrogen 44720G, 1:100 dilution), anti phospho-Paxillin Y118 (Invitrogen 44722G, 1:100 dilution), anti Syk Y348 (BD Pharmingen 558167, 1:200 dilution). Cy3-conjugated secondary antibodies (Jackson) and Alexa Fluor 488 phalloidin (Fischer scientific) were used at 1:400 dilution. Samples were mounted on glass slides with Dako (Agilent).
Traction force microscopy
Polyacrylamide gels (4.07 kPa shear modulus, 40 um thickness) were prepared on glass coverslips with embedded 40 nm fluorescent beads (TransFluoSpheres (633/720), Thermo scientific) as described before35. An average of 10 images of the beads in the gels was acquired before addition of macrophages as a no-stress reference. Macrophages were added and beads (as markers of gel deformation deformation) and expressed fluorescent proteins imaged every 10 seconds during frustrated phagocytosis. Bead displacements relative to the reference position were determined using particle image velocimetry, and the corresponding contractile energy was estimated with the Fourier transform traction cytometry, using the ImageJ plugins previously described61. The phagocytic cup area was determined by manually drawing a region of interest following the leading edge of the cup in the fluorescent protein channel. Maximal and mean stress within the phagocytic cup were measured at the time point when the cup reached its maximal surface area. For measurement of traction force over time, mean magnitude of traction stress within the cup was measured and reported to the surface area of the cup.
Atomic force microscopy
Sheep RBCs were plated on a glass-bottom dish (Willco Wells) pre-coated with 0.01% poly-L-lysine (Life Technologies) and immersed in phosphate-buffered saline solution (Life Technologies). Force mapping atomic force microscopy experiments were performed using a Bruker Bioscope Catalyst AFM system (Bruker) mounted on an inverted Axiovert 200M microscope (Zeiss) equipped with a confocal laser scanning microscope 510 Meta (Zeiss) and a 40x objective lens (0.95 NA, Plan-Apochromat, Zeiss) with temperature at 37°C using a heated stage (Bruker). A soft silicon nitride AFM probe (MLCT, Bruker) was used with tip dimensions: height of 2.5-8 μm, nominal radius of 20 nm, and nominal half-opening angle of 18°. The AFM microcantilevers were pre-calibrated using the standard thermal noise fluctuation method. The estimated spring constants for microcantilevers used were 0.02 – 0.06 N/m. After calibration, the AFM probe was moved on top of a cell. Five successive curves were performed on each cell. The deflection set-point was set to 20 nm yielding applied forces between 400 pN to 1 nN. The force mapping scan speed used was 0.5 Hz. Young’s modulus was determined from the force distance curves using NanoScope Analysis software (Bruker). For fitting, indentation depths between 0-100 nm were consistent in yielding good fits (R2>0.9). Curves with poor fits R2<0.9 were discarded from the analysis. The cellular elastic Young’s modulus (E; Pa) was computed by fitting each recorded force-distance curve with the Sneddon’s contact mechanics model for indenting an infinite isotropic elastic half-space with a conical indenter80:
, where F is the applied force, α is the tip half-opening angle and δ is the sample mean indentation.
Statistics and Reproducibility
Statistical analysis was performed using Prism 7 (GraphPad). Error bars represented standard errors of the mean, n values were the number of independent experiments. Statistical comparison of the data was performed by two-tailed Mann-Whitney test, which is a nonparametric test that does not assume Gaussian distributions.
Reporting Summery
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. CR-mediated phagocytosis is blocked by anti-integrin antibodies and divalent cation chelation.
a, time-lapse spinning disc confocal microscopy images of a human THP-1 macrophage expressing F-tractin-mCherry during phagocytosis of iC3b-opsonized 5.15 μm polystyrene microspheres, representative of three independent experiments.. Elapsed time shown in seconds. b-e, binding index (fraction of cell-associated particles relative to PMA control), phagocytosis index (fraction of internalized particles relative to PMA control) and phagocytosis efficiency (percentage of internalized particles relative to all cell-associated particles) after 1 hour incubation of iC3b-opsonized or BSA-coated 5.15 μm polystyrene beads (b), complement-opsonized or unopsonized sheep red blood cells (c), complement-opsonized or unopsonized sheep red blood cells previously fixed with glutaraldehyde (d) and complement-opsonized or unopsonized zymosan A (e). RAW 264.7 macrophages were preincubated with 10 μg/mL LPS, or 150 ng/mL PMA, without or with blocking antibodies to αM or β2 integrins. Error bars represent SEM. P values were calculated for each individual condition compared the PMA control using two tailed Mann-Whitney test. b, iC3b-beads untreated n= 28 fields, LPS n= 30 fields, PMA n= 47 fields, PMA + anti αM n= 44 fields, PMA + anti β2 n= 30 fields; BSA-beads n= 25 fields, from 3 independent experiments. c, opsonized sRBCs n= 30 fields except, PMA + anti β2 n= 20 fields; unopsonized sRBCs n= 20 fields, from 3 independent experiments. d, opsonized fixed-sRBCs with LPS, PMA, or PMA + anti αM: n= 25 fields, others n= 20 fields, form 3 independent experiments. e, opsonized zymosan with PMA: n= 10 fields, others n= 5 fields. Scale bars are 5 μm, elapsed times are in seconds. Numerical source data are provided in Statistical Source Extended Data Figure 1.
Extended Data Fig. 2. The formin mDia1, but not mDia2, is transiently recruited to the forming phagosome.
Time-lapse spinning disc confocal microscopy images of RAW 264.7 macrophages expressing mDia1-mEmerald (top) or mDia2-mEmerald (bottom), during phagocytosis of iC3b-opsonized 5.15 μm polystyrene microspheres. Representative examples from three independent experiments. Scale bars are 5 μm, elapsed time are in seconds.
Extended Data Fig. 3. Formation of a frustrated phagocytic cup requires integrin engagement.
Time-lapse TIRF microscopy of a RAW 264.7 macrophage expressing EGFP-F-tractin during formation of a frustrated phagocytic cup on an anti-αMβ2-coated coverslip (top) or on an isotype control antibody (bottom). Representative examples from three independent experiments. Scale bars are 5 μm, elapsed time are in seconds.
Supplementary Material
Supplementary Video 1 Actin dynamics during CR-mediated phagocytosis of diverse target particles. Time-lapse spinning disc confocal microscopy of RAW 264.7 macrophages expressing EGFP-F-tractin to mark actin filaments during phagocytosis of iC3b-opsonized 4.19 μm polystyrene Flash Red microspheres (left, iC3b-bead), complement-opsonized Zymosan A labelled with Texas Red (middle, Comp-zymosan), complement-opsonized sheep red blood cells labelled with Alexa Fluor 647 (right, Comp-sRBC). A single confocal image plane corresponding to the center of particle is shown over time (minutes: seconds) in each panel. Representative examples from more than 10 phagocytosis events over at least 3 independent experiments.
Supplementary Video 2 Comparison of actin dynamics during CR-mediated phagocytosis upon CR activation by PMA or LPS. Time-lapse spinning disc confocal microscopy of RAW 264.7 macrophages expressing EGFP-F-tractin to mark actin filaments during phagocytosis of iC3b-opsonized 4.19 μm polystyrene Flash Red microspheres (iC3b-bead), upon activation by PMA 150 ng/mL or LPS 10 μg/ml. A single confocal image plane corresponding to the center of particle is shown over time (minutes: seconds) in each panel. Representative examples from 3 independent experiments.
Supplementary Video 3 Membrane dynamics in 3D during CR-mediated phagocytosis. Three-dimensional time-lapse spinning disc confocal microscopy of RAW 264.7 macrophages expressing EGFP-CAAX to mark the plasma membrane during phagocytosis of iC3b-opsonized 4.19 μm polystyrene Flash Red microspheres. A single confocal image plane (X-Y, left) and a reconstructed axial image plane (X-Z, right) corresponding to the center of particle is shown over time (minutes: seconds) in each panel. Representative examples from 4 independent experiments.
Supplementary Video 4 Actin dynamics during CR-mediated phagocytosis with perturbation of actin nucleators. Time-lapse spinning disc confocal microscopy of RAW 264.7 macrophages expressing EGFP-F-tractin to mark actin filaments during phagocytosis of iC3b-opsonized 5.15 μm polystyrene microspheres: untreated (control, left), 100 μM CK-666 (CK-666, middle), 20 μM SMIFH2 (SMIFH2, right). A single confocal image plane corresponding to the center of particle is shown over time (minutes: seconds) in each panel. Representative examples from 3 independent experiments.
Supplementary Video 5 Recruitment dynamics of ArpC2 and mDia1 during CR-mediated phagocytosis. Time-lapse spinning disc confocal microscopy of RAW 264.7 macrophages co-expressing mEmerald-ArpC2 (top left and green) or mEmerald-mDia1 (bottom left and green) and mCherry-F-tractin (middle panels and red) during phagocytosis of iC3b-opsonized 5.15 μm polystyrene microspheres. A single confocal image plane corresponding to the center of particle is shown over time (minutes: seconds) in each panel. Representative examples from 4 independent experiments.
Supplementary Video 6 Formation of a frustrated phagocytic cup requires integrin engagement. Time-lapse TIRF microscopy of a RAW 264.7 macrophage expressing EGFP-F-tractin during formation of a frustrated phagocytic cup on an anti-αMβ2-coated coverslip (Anti-Mac-1, left) or on an isotype control antibody (right). Elapsed time shown in minutes: seconds. Representative example from 3 independent experiments.
Supplementary Video 7 Arp2/3 dynamics during CR-mediated frustrated phagocytosis. Time-lapse TIRF microscopy of a RAW 264.7 macrophage co-expressing and mCherry-ArpC2 (left and red) mEmerald-mDia1 (middle and green) during formation of a “frustrated phagocytic cup” on anti-αMβ2 antibody-coated coverslip. Elapsed time shown in minutes: seconds. Representative example from 3 independent experiments.
Supplementary Video 8 Super-resolution microscopy of actin dynamics during CR-mediated frustrated phagocytosis. Time-lapse TIRF-SIM of a RAW 264.7 macrophage expressing mNeonGreen-F-tractin during formation of a “frustrated phagocytic cup” on anti-αMβ2 antibody-coated coverslip. Elapsed time shown in minutes: seconds. Representative example from 3 independent experiments.
Supplementary Video 9 Fluorescent speckle microscopy of actin dynamics during CR-mediated phagocytosis. A RAW 264.7 macrophage expressing green-to-red photoconvertible Actin-mEos3.2 during phagocytosis of iC3b-opsonized 5.15 μm polystyrene microspheres was exposed to a low level of 405 nm light and photo-converted red fluorescent actin recorded by time-lapse spinning disc confocal microscopy. A single confocal image plane corresponding to the center of particle is shown over time (minutes: seconds) in each panel. Images in the time-lapse were aligned relative to the negative image of the bead and individual fluorescent speckles were detected (red circles) and tracked (red lines) with qFSM automated image analysis software. Elapsed time shown in minutes: seconds. Representative example from 4 independent experiments.
Supplementary Video 10 Actin dynamics during frustrated phagocytosis in the absence of integrin engagement. Time-lapse TIRF microscopy of a RAW 264.7 macrophage expressing mCherry-F-tractin during formation of a “frustrated phagocytic cup” on a poly-L-lysine-coated coverslip. Elapsed time shown in minutes: seconds. Representative example from 3 independent experiments.
Supplementary Video 11 Adhesion dynamics during CR-mediated frustrated phagocytosis. Time-lapse TIRF microscopy of a RAW 264.7 macrophage expressing mEmerald-Paxillin (green) and mCherry-F-tractin (red) during formation of a “frustrated phagocytic cup” on anti-αMβ2 antibody-coated coverslip. Elapsed time shown in minutes: seconds. Representative example from 3 independent experiments.
Supplementary Video 12 β2 integrin dynamics during CR-mediated frustrated phagocytosis. Time-lapse TIRF microscopy of a RAW 264.7 macrophage expressing co-expressing αM, β2-YFP (green) and mApple-paxillin (red) during formation of a “frustrated phagocytic cup” on anti-αMβ2 antibody-coated coverslip. Elapsed time shown in minutes: seconds. Representative example from 3 independent experiments.
Supplementary Video 13 Traction stress dynamics during CR-mediated frustrated phagocytosis with inhibition of Syk kinase or overexpression of talin head domain. Top panels: Time lapse spinning disc confocal microscopy of RAW 264.7 macrophages expressing EGFP-F-tractin with (middle) or without (left) treatment with 50 μM piceatannol, or expressing mEmerald-Talin Head (right), during frustrated phagocytosis on anti-αMβ2 antibody-coated 4 kPa polyacrylamide gels. A single confocal image plane at the gel surface is shown over time (minutes: seconds) in each panel. Bottom panels: Time-lapse of stress vectors computed from images of fluorescent beads embedded in the polyacrylamide gel substrate, pseudocolor scale and vector lengths represent the magnitude of traction stresses. Elapsed time shown in minutes: seconds. Representative examples from 3 independent experiments.
Supplementary Video 14 Effect of of talin head domain over-expression on CR-mediated phagocytosis. Time-lapse spinning disc confocal microscopy of RAW 264.7 macrophages co-expressing mEmerald-talin head domain (green) during phagocytosis of iC3b-opsonized 4.19 μm Flash Red polystyrene microspheres (red). A single confocal image plane corresponding to the center of particle is shown over time (minutes: seconds) in each panel. Elapsed time shown in minutes: seconds. Representative example from 3 independent experiments.
Supplementary Video 15 Vinculin localization to the phagosome is Syk kinase dependent. Time-lapse of vinculin concentration relative to a soluble marker from spinning-disk microscopy during phagocytosis of iC3b-opsonized 5.15 μm polystyrene beads by RAW 264.7 macrophages expressing vinculin-mEmerald and FusionRed, treated with DMSO (control), ROCK inhibitor (Y-27632, 10 μM), or Syk inhibitor (piceatannol, 50 μM). Heat maps were generated by subtracting mean-normalized fusion red intensity from mean-normalized vinculin-mEmerald intensity. Elapsed time in minutes:seconds. Representative examples from 3 independent experiments.
Supplementary Video 16 Formation of frustrated phagocytic cups is enhanced by target stiffness. Time-lapse spinning disk confocal microscopy of RAW 264.7 macrophages expressing F-tractin-EGFP during spreading on anti-αMβ2-coated 0.4 kPa, 4 kPa, and 30 kPa polyacrylamide gel substrates. Elapsed time shown in hours:minutes. Representative examples from 3 independent experiments.
Footnotes
Data availability
Statistical source data supporting figures 2-7 and supplementary figure 1 are provided in supplementary table 1. All data supporting the findings of this study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
References
- 1.Lim JJ, Grinstein S & Roth Z Diversity and Versatility of Phagocytosis: Roles in Innate Immunity, Tissue Remodeling, and Homeostasis. Front. Cell. Infect. Microbiol 7, 191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Underhill DM & Goodridge HS Information processing during phagocytosis. Nat. Rev. Immunol 12, 492–502 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaumouillé V & Grinstein S Molecular Mechanisms of Phagosome Formation. Microbiol. Spectr 4, (2016). [DOI] [PubMed] [Google Scholar]
- 4.Swanson JA Shaping cups into phagosomes and macropinosomes. Nat Rev Mol Cell Biol 9, 639–49 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown EJ Complement receptors and phagocytosis. Curr. Opin. Immunol 3, 76–82 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Chen J et al. SLAMF7 is critical for phagocytosis of haematopoietic tumour cells via Mac-1 integrin. Nature 544, 493–497 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman SL, Devery-Pocius JE, Ross GD & Henson PM Phagocytosis by human monocyte-derived macrophages. Independent function of receptors for C3b (CR1) and iC3b (CR3). Complement Basel Switz. 1, 213–227 (1984). [PubMed] [Google Scholar]
- 8.Xu S, Wang J, Wang J-H & Springer TA Distinct recognition of complement iC3b by integrins αXβ2 and αMβ2. Proc. Natl. Acad. Sci. U. S. A 114, 3403–3408 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newman SL, Mikus LK & Tucci MA Differential requirements for cellular cytoskeleton in human macrophage complement receptor- and Fc receptor-mediated phagocytosis. J. Immunol. Baltim. Md 1950 146, 967–974 (1991). [PubMed] [Google Scholar]
- 10.May RC, Caron E, Hall A & Machesky LM Involvement of the Arp2/3 complex in phagocytosis mediated by FcgammaR or CR3. Nat. Cell Biol 2, 246–248 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Rotty JD et al. Arp2/3 Complex Is Required for Macrophage Integrin Functions but Is Dispensable for FcR Phagocytosis and In Vivo Motility. Dev. Cell 42, 498–513.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colucci-Guyon E et al. A role for mammalian diaphanous-related formins in complement receptor (CR3)-mediated phagocytosis in macrophages. Curr. Biol. CB 15, 2007–2012 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Caron E & Hall A Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science 282, 1717–1721 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Allen LA & Aderem A Molecular definition of distinct cytoskeletal structures involved in complement- and Fc receptor-mediated phagocytosis in macrophages. J. Exp. Med 184, 627–637 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan G Differences in the mode of phagocytosis with Fc and C3 receptors in macrophages. Scand. J. Immunol 6, 797–807 (1977). [DOI] [PubMed] [Google Scholar]
- 16.Case LB & Waterman CM Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol 17, 955–963 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gauthier NC & Roca-Cusachs P Mechanosensing at integrin-mediated cell-matrix adhesions: from molecular to integrated mechanisms. Curr. Opin. Cell Biol 50, 20–26 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Svitkina TM & Borisy GG Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol 145, 1009–1026 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Theriot JA & Mitchison TJ Comparison of actin and cell surface dynamics in motile fibroblasts. J. Cell Biol 119, 367–377 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardel ML et al. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J. Cell Biol 183, 999–1005 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu K, Ji L, Applegate KT, Danuser G & Waterman-Storer CM Differential transmission of actin motion within focal adhesions. Science 315, 111–115 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Zhu J et al. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell 32, 849–861 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calderwood DA et al. The Talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem. 274, 28071–28074 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Hemmings L et al. Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J. Cell Sci 109 ( Pt 11), 2715–2726 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Giannone G, Jiang G, Sutton DH, Critchley DR & Sheetz MP Talin1 is critical for force-dependent reinforcement of initial integrin-cytoskeleton bonds but not tyrosine kinase activation. J. Cell Biol 163, 409–419 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim J et al. An essential role for talin during alpha(M)beta(2)-mediated phagocytosis. Mol. Biol. Cell 18, 976–985 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.del Rio A et al. Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphries JD et al. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol 179, 1043–1057 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD & Waterman CM Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol 188, 877–890 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thievessen I et al. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol 202, 163–177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paone C et al. The Tyrosine Kinase Pyk2 Contributes to Complement-Mediated Phagocytosis in Murine Macrophages. J. Innate Immun 8, 437–451 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Y et al. Protein-tyrosine kinase Syk is required for pathogen engulfment in complement-mediated phagocytosis. Blood 107, 4554–4562 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Chan CE & Odde DJ Traction dynamics of filopodia on compliant substrates. Science 322, 1687–1691 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Elosegui-Artola A et al. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat. Cell Biol. 18, 540–548 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Plotnikov SV, Pasapera AM, Sabass B & Waterman CM Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell 151, 1513–1527 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engler AJ, Sen S, Sweeney HL & Discher DE Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Levental KR et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139, 891–906 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pelham RJ & Wang Y l. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. U. S. A 94, 13661–13665 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuznetsova TG, Starodubtseva MN, Yegorenkov NI, Chizhik SA & Zhdanov RI Atomic force microscopy probing of cell elasticity. Micron 38, 824–833 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Amir A, Babaeipour F, McIntosh DB, Nelson DR & Jun S Bending forces plastically deform growing bacterial cell walls. Proc. Natl. Acad. Sci. U. S. A 111, 5778–5783 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng Y, Sun M & Shaevitz JW Direct measurement of cell wall stress stiffening and turgor pressure in live bacterial cells. Phys. Rev. Lett 107, 158101 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Lam WA, Rosenbluth MJ & Fletcher DA Chemotherapy exposure increases leukemia cell stiffness. Blood 109, 3505–3508 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nikolaev NI, Müller T, Williams DJ & Liu Y Changes in the stiffness of human mesenchymal stem cells with the progress of cell death as measured by atomic force microscopy. J. Biomech 47, 625–630 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Swaminathan V et al. Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer Res. 71, 5075–5080 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belin BJ, Goins LM & Mullins RD Comparative analysis of tools for live cell imaging of actin network architecture. Bioarchitecture 4, 189–202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bohdanowicz M, Cosío G, Backer JM & Grinstein S Class I and class III phosphoinositide 3-kinases are required for actin polymerization that propels phagosomes. J. Cell Biol 191, 999–1012 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia Y et al. The beta-glucan-binding lectin site of mouse CR3 (CD11b/CD18) and its function in generating a primed state of the receptor that mediates cytotoxic activation in response to iC3b-opsonized target cells. J. Immunol. Baltim. Md 1950 162, 2281–2290 (1999). [PubMed] [Google Scholar]
- 48.Ross GD, Cain JA & Lachmann PJ Membrane complement receptor type three (CR3) has lectin-like properties analogous to bovine conglutinin as functions as a receptor for zymosan and rabbit erythrocytes as well as a receptor for iC3b. J. Immunol. Baltim. Md 1950 134, 3307–3315 (1985). [PubMed] [Google Scholar]
- 49.Caron E, Self AJ & Hall A The GTPase Rap1 controls functional activation of macrophage integrin alphaMbeta2 by LPS and other inflammatory mediators. Curr. Biol. CB 10, 974–978 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Patel PC & Harrison RE Membrane ruffles capture C3bi-opsonized particles in activated macrophages. Mol Biol Cell 19, 4628–39 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nolen BJ et al. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460, 1031–1034 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rizvi SA et al. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol 16, 1158–1168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wright SD & Silverstein SC Phagocytosing macrophages exclude proteins from the zones of contact with opsonized targets. Nature 309, 359–361 (1984). [DOI] [PubMed] [Google Scholar]
- 54.Sanchez-Madrid F, Simon P, Thompson S & Springer TA Mapping of antigenic and functional epitopes on the alpha- and beta-subunits of two related mouse glycoproteins involved in cell interactions, LFA-1 and Mac-1. J. Exp. Med 158, 586–602 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ponti A, Machacek M, Gupton SL, Waterman-Storer CM & Danuser G Two distinct actin networks drive the protrusion of migrating cells. Science 305, 1782–1786 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Danuser G & Waterman-Storer CM Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annu. Rev. Biophys. Biomol. Struct 35, 361–387 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Mendoza MC, Besson S & Danuser G Quantitative fluorescent speckle microscopy (QFSM) to measure actin dynamics. Curr. Protoc. Cytom Chapter 2, Unit2.18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Albiges-Rizo C, Destaing O, Fourcade B, Planus E & Block MR Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J. Cell Sci 122, 3037–3049 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pixley FJ Macrophage Migration and Its Regulation by CSF-1. Int. J. Cell Biol 2012, 501962 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Linder S et al. The polarization defect of Wiskott-Aldrich syndrome macrophages is linked to dislocalization of the Arp2/3 complex. J. Immunol. Baltim. Md 1950 165, 221–225 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Martiel J-L et al. Measurement of cell traction forces with ImageJ. Methods Cell Biol. 125, 269–287 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Choquet D, Felsenfeld DP & Sheetz MP Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell 88, 39–48 (1997). [DOI] [PubMed] [Google Scholar]
- 63.Oliver JM, Burg DL, Wilson BS, McLaughlin JL & Geahlen RL Inhibition of mast cell Fc epsilon R1-mediated signaling and effector function by the Syk-selective inhibitor, piceatannol. J. Biol. Chem 269, 29697–29703 (1994). [PubMed] [Google Scholar]
- 64.Hanke JH et al. Discovery of a Novel, Potent, and Src Family-selective Tyrosine Kinase Inhibitor STUDY OF Lck- AND FynT-DEPENDENT T CELL ACTIVATION. J. Biol. Chem 271, 695–701 (1996). [DOI] [PubMed] [Google Scholar]
- 65.Liu T-J et al. Inhibition of both focal adhesion kinase and insulin-like growth factor-I receptor kinase suppresses glioma proliferation in vitro and in vivo. Mol. Cancer Ther 6, 1357–1367 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Straight AF et al. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science 299, 1743–1747 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Uehata M et al. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature 389, 990–994 (1997). [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y, Hoppe AD & Swanson JA Coordination of Fc receptor signaling regulates cellular commitment to phagocytosis. Proc. Natl. Acad. Sci. U. S. A 107, 19332–19337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gupton SL, Eisenmann K, Alberts AS & Waterman-Storer CM mDia2 regulates actin and focal adhesion dynamics and organization in the lamella for efficient epithelial cell migration. J. Cell Sci 120, 3475–3487 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Hotulainen P & Lappalainen P Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol 173, 383–394 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riveline D et al. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J. Cell Biol 153, 1175–1186 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choi CK et al. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol 10, 1039–1050 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang G, Giannone G, Critchley DR, Fukumoto E & Sheetz MP Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature 424, 334–337 (2003). [DOI] [PubMed] [Google Scholar]
- 74.Carisey A et al. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr. Biol. CB 23, 271–281 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Galbraith CG, Yamada KM & Sheetz MP The relationship between force and focal complex development. J. Cell Biol 159, 695–705 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jurado C, Haserick JR & Lee J Slipping or gripping? Fluorescent speckle microscopy in fish keratocytes reveals two different mechanisms for generating a retrograde flow of actin. Mol. Biol. Cell 16, 507–518 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Allen TM & Cullis PR Drug Delivery Systems: Entering the Mainstream. Science 303, 1818–1822 (2004). [DOI] [PubMed] [Google Scholar]
- 78.Beningo KA & Wang Y Fc-receptor-mediated phagocytosis is regulated by mechanical properties of the target. J. Cell Sci 115, 849–856 (2002). [DOI] [PubMed] [Google Scholar]
- 79.Cornel AM, van Til NP, Boelens JJ & Nierkens S Strategies to Genetically Modulate Dendritic Cells to Potentiate Anti-Tumor Responses in Hematologic Malignancies. Front. Immunol 9, 982 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sneddon IN Fourier Transforms Ch. 10 (1951).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1 Actin dynamics during CR-mediated phagocytosis of diverse target particles. Time-lapse spinning disc confocal microscopy of RAW 264.7 macrophages expressing EGFP-F-tractin to mark actin filaments during phagocytosis of iC3b-opsonized 4.19 μm polystyrene Flash Red microspheres (left, iC3b-bead), complement-opsonized Zymosan A labelled with Texas Red (middle, Comp-zymosan), complement-opsonized sheep red blood cells labelled with Alexa Fluor 647 (right, Comp-sRBC). A single confocal image plane corresponding to the center of particle is shown over time (minutes: seconds) in each panel. Representative examples from more than 10 phagocytosis events over at least 3 independent experiments.
Supplementary Video 2 Comparison of actin dynamics during CR-mediated phagocytosis upon CR activation by PMA or LPS. Time-lapse spinning disc confocal microscopy of RAW 264.7 macrophages expressing EGFP-F-tractin to mark actin filaments during phagocytosis of iC3b-opsonized 4.19 μm polystyrene Flash Red microspheres (iC3b-bead), upon activation by PMA 150 ng/mL or LPS 10 μg/ml. A single confocal image plane corresponding to the center of particle is shown over time (minutes: seconds) in each panel. Representative examples from 3 independent experiments.
Supplementary Video 3 Membrane dynamics in 3D during CR-mediated phagocytosis. Three-dimensional time-lapse spinning disc confocal microscopy of RAW 264.7 macrophages expressing EGFP-CAAX to mark the plasma membrane during phagocytosis of iC3b-opsonized 4.19 μm polystyrene Flash Red microspheres. A single confocal image plane (X-Y, left) and a reconstructed axial image plane (X-Z, right) corresponding to the center of particle is shown over time (minutes: seconds) in each panel. Representative examples from 4 independent experiments.
Supplementary Video 4 Actin dynamics during CR-mediated phagocytosis with perturbation of actin nucleators. Time-lapse spinning disc confocal microscopy of RAW 264.7 macrophages expressing EGFP-F-tractin to mark actin filaments during phagocytosis of iC3b-opsonized 5.15 μm polystyrene microspheres: untreated (control, left), 100 μM CK-666 (CK-666, middle), 20 μM SMIFH2 (SMIFH2, right). A single confocal image plane corresponding to the center of particle is shown over time (minutes: seconds) in each panel. Representative examples from 3 independent experiments.
Supplementary Video 5 Recruitment dynamics of ArpC2 and mDia1 during CR-mediated phagocytosis. Time-lapse spinning disc confocal microscopy of RAW 264.7 macrophages co-expressing mEmerald-ArpC2 (top left and green) or mEmerald-mDia1 (bottom left and green) and mCherry-F-tractin (middle panels and red) during phagocytosis of iC3b-opsonized 5.15 μm polystyrene microspheres. A single confocal image plane corresponding to the center of particle is shown over time (minutes: seconds) in each panel. Representative examples from 4 independent experiments.
Supplementary Video 6 Formation of a frustrated phagocytic cup requires integrin engagement. Time-lapse TIRF microscopy of a RAW 264.7 macrophage expressing EGFP-F-tractin during formation of a frustrated phagocytic cup on an anti-αMβ2-coated coverslip (Anti-Mac-1, left) or on an isotype control antibody (right). Elapsed time shown in minutes: seconds. Representative example from 3 independent experiments.
Supplementary Video 7 Arp2/3 dynamics during CR-mediated frustrated phagocytosis. Time-lapse TIRF microscopy of a RAW 264.7 macrophage co-expressing and mCherry-ArpC2 (left and red) mEmerald-mDia1 (middle and green) during formation of a “frustrated phagocytic cup” on anti-αMβ2 antibody-coated coverslip. Elapsed time shown in minutes: seconds. Representative example from 3 independent experiments.
Supplementary Video 8 Super-resolution microscopy of actin dynamics during CR-mediated frustrated phagocytosis. Time-lapse TIRF-SIM of a RAW 264.7 macrophage expressing mNeonGreen-F-tractin during formation of a “frustrated phagocytic cup” on anti-αMβ2 antibody-coated coverslip. Elapsed time shown in minutes: seconds. Representative example from 3 independent experiments.
Supplementary Video 9 Fluorescent speckle microscopy of actin dynamics during CR-mediated phagocytosis. A RAW 264.7 macrophage expressing green-to-red photoconvertible Actin-mEos3.2 during phagocytosis of iC3b-opsonized 5.15 μm polystyrene microspheres was exposed to a low level of 405 nm light and photo-converted red fluorescent actin recorded by time-lapse spinning disc confocal microscopy. A single confocal image plane corresponding to the center of particle is shown over time (minutes: seconds) in each panel. Images in the time-lapse were aligned relative to the negative image of the bead and individual fluorescent speckles were detected (red circles) and tracked (red lines) with qFSM automated image analysis software. Elapsed time shown in minutes: seconds. Representative example from 4 independent experiments.
Supplementary Video 10 Actin dynamics during frustrated phagocytosis in the absence of integrin engagement. Time-lapse TIRF microscopy of a RAW 264.7 macrophage expressing mCherry-F-tractin during formation of a “frustrated phagocytic cup” on a poly-L-lysine-coated coverslip. Elapsed time shown in minutes: seconds. Representative example from 3 independent experiments.
Supplementary Video 11 Adhesion dynamics during CR-mediated frustrated phagocytosis. Time-lapse TIRF microscopy of a RAW 264.7 macrophage expressing mEmerald-Paxillin (green) and mCherry-F-tractin (red) during formation of a “frustrated phagocytic cup” on anti-αMβ2 antibody-coated coverslip. Elapsed time shown in minutes: seconds. Representative example from 3 independent experiments.
Supplementary Video 12 β2 integrin dynamics during CR-mediated frustrated phagocytosis. Time-lapse TIRF microscopy of a RAW 264.7 macrophage expressing co-expressing αM, β2-YFP (green) and mApple-paxillin (red) during formation of a “frustrated phagocytic cup” on anti-αMβ2 antibody-coated coverslip. Elapsed time shown in minutes: seconds. Representative example from 3 independent experiments.
Supplementary Video 13 Traction stress dynamics during CR-mediated frustrated phagocytosis with inhibition of Syk kinase or overexpression of talin head domain. Top panels: Time lapse spinning disc confocal microscopy of RAW 264.7 macrophages expressing EGFP-F-tractin with (middle) or without (left) treatment with 50 μM piceatannol, or expressing mEmerald-Talin Head (right), during frustrated phagocytosis on anti-αMβ2 antibody-coated 4 kPa polyacrylamide gels. A single confocal image plane at the gel surface is shown over time (minutes: seconds) in each panel. Bottom panels: Time-lapse of stress vectors computed from images of fluorescent beads embedded in the polyacrylamide gel substrate, pseudocolor scale and vector lengths represent the magnitude of traction stresses. Elapsed time shown in minutes: seconds. Representative examples from 3 independent experiments.
Supplementary Video 14 Effect of of talin head domain over-expression on CR-mediated phagocytosis. Time-lapse spinning disc confocal microscopy of RAW 264.7 macrophages co-expressing mEmerald-talin head domain (green) during phagocytosis of iC3b-opsonized 4.19 μm Flash Red polystyrene microspheres (red). A single confocal image plane corresponding to the center of particle is shown over time (minutes: seconds) in each panel. Elapsed time shown in minutes: seconds. Representative example from 3 independent experiments.
Supplementary Video 15 Vinculin localization to the phagosome is Syk kinase dependent. Time-lapse of vinculin concentration relative to a soluble marker from spinning-disk microscopy during phagocytosis of iC3b-opsonized 5.15 μm polystyrene beads by RAW 264.7 macrophages expressing vinculin-mEmerald and FusionRed, treated with DMSO (control), ROCK inhibitor (Y-27632, 10 μM), or Syk inhibitor (piceatannol, 50 μM). Heat maps were generated by subtracting mean-normalized fusion red intensity from mean-normalized vinculin-mEmerald intensity. Elapsed time in minutes:seconds. Representative examples from 3 independent experiments.
Supplementary Video 16 Formation of frustrated phagocytic cups is enhanced by target stiffness. Time-lapse spinning disk confocal microscopy of RAW 264.7 macrophages expressing F-tractin-EGFP during spreading on anti-αMβ2-coated 0.4 kPa, 4 kPa, and 30 kPa polyacrylamide gel substrates. Elapsed time shown in hours:minutes. Representative examples from 3 independent experiments.