Abstract
Transcriptional control can be used to program cells to label proteins with non-canonical amino acids by regulating the expression of orthogonal aminoacyl tRNA synthetases (aaRSs). However, we cannot yet program cells to control labeling in response to aaRS and ligand binding. To identify aaRSs whose activities can be regulated by interactions with ligands, we used a combinatorial approach to discover fragmented variants of Escherichia coli methionyl tRNA synthetase (MetRS) that require fusion to associating proteins for maximal activity. We found that these split proteins could be leveraged to create ligand-dependent MetRS using two approaches. When a pair of MetRS fragments was fused to FKBP12 and the FKBP-rapamycin binding domain of mTOR, and mutations were introduced that direct substrate specificity towards azidonorleucine (Anl), Anl metabolic labeling was significantly enhanced in growth medium containing rapamycin, which stabilizes the FKBP12-mTOR complex. In addition, fusion of MetRS fragments to the termini of the ligand-binding domain of the estrogen receptor yielded proteins whose Anl metabolic labeling was significantly enhanced when 4-hydroxytamoxifen was added to the growth medium. These findings suggest that split MetRS can be fused to a range of ligand-binding proteins to create aaRS whose metabolic labeling activities depend upon post-translational interactions with ligands.
Keywords: amino acyl tRNA synthetase, domain insertion, metabolic labeling, non-canonical amino acid, protein engineering, protein fragment complementation, protein-protein interaction
Introduction
In synthetic biology, cells are programmed to dynamically adjust their metabolic activity as environmental conditions change by altering transcription, translation, and protein degradation. To understand the proteome-wide consequences of these programs, variation in protein content can be measured using mass spectrometry.1,2 When performing proteomic measurements, it can be challenging to obtain spatial and temporal resolution within complex cellular populations, especially in multicellular organisms that are made up of many cell types. One strategy that has emerged to improve the spatial and temporal control over mass spectrometry used for proteomics is bioorthogonal non-canonical amino acid tagging (BONCAT).3 In BONCAT, a complex cell population is provided a non-canonical amino acid bearing a bioorthogonal functional group, and a natural or engineered aminoacyl tRNA synthetase (aaRS) is used to charge cellular tRNA with the non-canonical amino acid.4–7 Proteome labeling then arises as the charged tRNA is used for protein translation. Proteins bearing bioorthogonal functional groups can be selectively reacted to add affinity tags7,8 for enrichment and purification or reacted to add fluorescent dyes9,10 for visualization of labeled proteins and cells.
Orthogonal aaRS have been developed that allow for transcriptional control over BONCAT by introducing mutations into natural aaRS that direct substrate specificity towards non-canonical amino acids.11–14 For example, directed evolution was used to identify mutations (L13N/Y260L/H301L) within E. coli MetRS that allow this protein to efficiently utilize azidonorleucine (Anl) as a substrate.15 By coupling the transcription and translation of this mutant protein (NLL-MetRS) to specific cells in a community, cell selective Anl metabolic labeling can be achieved.16,17 Because the activity of a single promoter only affords limited spatial control in communities of cells experiencing a range of environmental conditions, a two-fragment NLL-MetRS was developed that functions as a bipartite transcriptional AND gate,18 similar to that realized with other proteins.19 With this split NLL-MetRS, improved spatial control over BONCAT could be achieved by linking NLL-MetRS fragment expression to a pair of conditional promoters that were activated by two different environmental cues, such that metabolic labeling only occurred in the subset of cells where the first and second promoters were activated.18 Thus, this split NLL-MetRS only charged tRNA with Anl efficiently when the first AND the second fragments were transcribed. While this split protein AND gate allows for increased spatial and temporal control over BONCAT using transcriptional programs, whose design can at times be automated,20 the activity of these split proteins can only be switched on by environmental conditions that alter the activity of promoters. This limitation in regulation could be overcome by developing new components for transcriptional regulation21 or by using protein engineering to generate aaRS whose activities are dependent upon post-translational regulation such as ligand binding.
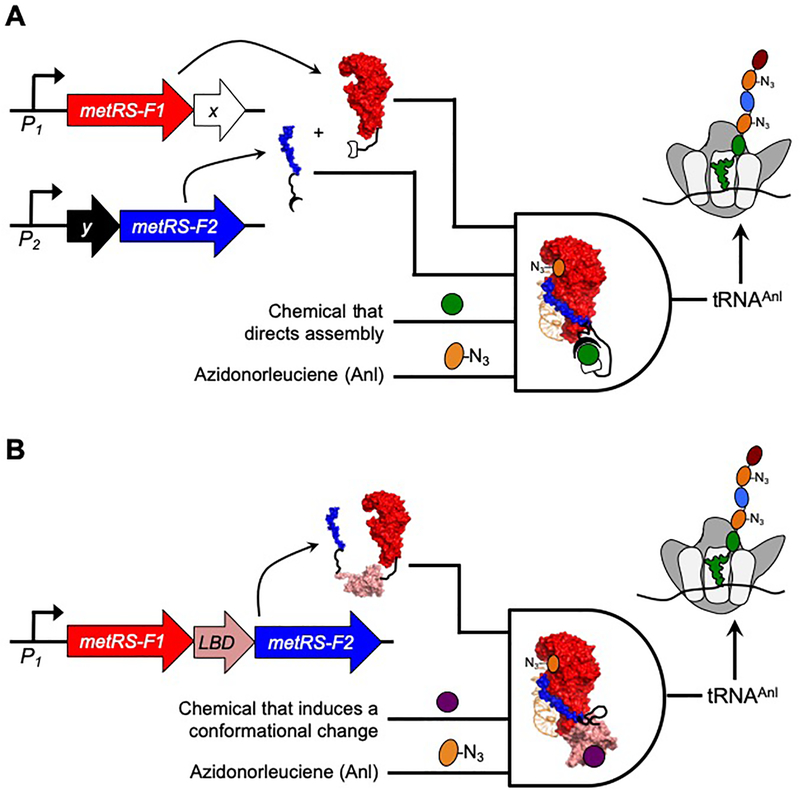
One strategy that can be used to engineer proteins whose activities are regulated by ligand binding is to split proteins into fragments that are inactive unless they are fused to a pair of proteins whose association is stabilized by ligand binding (Figure 1a).22 Alternatively, a pair of protein fragments can be fused to the different termini of a ligand-binding domain to create a single polypeptide whose activity is switched on and off in response to ligand-dependent conformational changes (Figure 1b).23 Neither of these approaches has yet been applied to an aaRS. Herein we describe the identification of split aaRS whose fragments require fusion to ligand-binding proteins for maximal activity. As an initial proof of concept application, we show that a three-hybrid system involving a rapamycin-stabilized protein-protein interaction24,25 can be used to switch on metabolic labeling of newly synthesized proteins with Anl in E. coli. We also demonstrate that the activity of this split aaRS is enhanced when fused to different pairs of interacting proteins, suggesting that it could be coupled to a wide range of protein-protein interactions to regulate non-canonical amino acid incorporation. Finally, we show that MetRS fragments can be fused to the termini of the ligand-binding domain (LBD) from the estrogen receptor (ER), and we demonstrate that the resulting polypeptide displays metabolic labeling activity that is enhanced by 4-hydroxytamoxifen, an estrogen receptor modulator.26
Figure 1. Strategies used to create aaRS that regulate BONCAT using ligand binding.
(A) Assisted NLL-MetRS fragment complementation can be used to switch BONCAT on and off. In this AND gate, two promoters (P1 and P2) are used to control the transcription of NLL-MetRS fragments (F1 and F2) which are expressed as fusions to a pair of proteins (X and Y). In cases where a post-translational binding reaction is needed to stabilize the X-Y complex, metabolic labeling requires four inputs. The transcriptional inputs P1 and P2 need to be on, the chemical that stabilizes the assembly of proteins X and Y must be present, and cells must be provided the non-canonical amino acid Anl. (B) When NLL-MetRS fragments are fused to the termini of a ligand-binding domain (LBD), BONCAT efficiency depends upon ligand-induced conformation changes. In this AND gate, one promoter (P1) controls the transcription of an NLL-MetRS containing an inserted LBD. In cases where ligand binding alters the conformation and activity of NLL-MetRS, metabolic labeling requires three inputs. The transcriptional input P1 must be on, the chemical that binds to the LBD needs to be present, and cells must be provided the non-canonical amino acid Anl.
Results and Discussion
Combining MetRS fission with fusion to peptides.
One way to discover split proteins that require assistance from protein-protein interactions to associate and function is to build all possible split variants of a protein and screen for variants that retain parent-like structure and function when the different fragments are expressed as fusions to interacting proteins.27 To test whether this approach could be used to discover MetRS fragments whose activity depends upon a protein-protein interaction, we constructed a library that expresses randomly fragmented MetRS fused to a pair of peptides (IAAL-E3 and IAAL-K3) that associate strongly, KD = 70 nM.28 This EK library was built using a one step procedure by modifying a previously described library of vectors that express different fragmented variants of truncated MetRS (Figure S1), which were generated by randomly fragmenting a MetRS truncation made up of residues 1–548 (designated MetRS herein).18 This truncation was used to avoid variants that require only a single fragment for activity, since it represents the minimal MetRS fragment that can be generated with parent-like activity and stability.29 In the EK library, the IAAL-E3 and IAAL-K3 peptides were fused to the termini created by protein fission, rather than the original termini of the parental protein. In each variant, the C-terminus of the MetRS fragment that precedes the backbone fission site was fused to the IAAL-E3 peptide, while the N-terminus of the fragment following the fission site was fused to the IAAL-K3 peptide. Both peptides were fused through glycine-rich linkers because the residues in each peptide that become covalently attached to the linkers are separated by 30 Å in the structure of the IAAL-E3/K3 complex.30
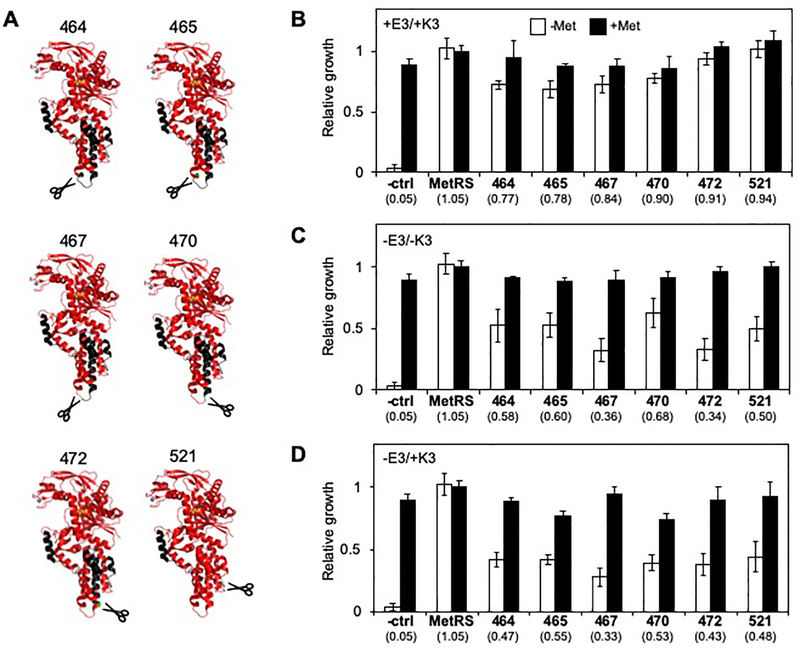
To identify bipartite MetRS that are functional when fused to associating peptides, we transformed our EK library into E. coli CS50, a strain with a chromosomal mutation that decreases MetRS affinity for methionine.31 Because this strain grows poorly on minimal medium lacking methionine (Figure S2), we selected the library for vectors that complemented CS50 growth on minimal medium lacking methionine. In this strain, the Trc and T7 promoters used to control transcription of the MetRS fragments are both repressed by the lac repressor, so their expression can be induced by the addition of isopropyl β-D-1-thiogalactopyranoside (IPTG). However, these promoters were sufficiently leaky to allow for selection of split MetRS that complemented E. coli CS50 in the absence of IPTG. Sequencing colonies obtained from this selection identified six unique split MetRS variants, which arose from backbone fragmentation after residues 464, 465, 467, 470, 472, and 521. The locations of the backbone fission sites are all within the MetRS anti-codon binding domain (Figure 2A),32 and many cluster within the region of the anti-codon binding domain that is most distal from the catalytic site. These backbone cleavage sites all differ from those previously identified in a study that analyzed the functional tolerance of MetRS to backbone fission in the absence of protein fusion.18 This previous study identified a larger number of active bipartite MetRS, which arose from fragmentation within all four MetRS domains (Figure S3), including the Rossmann fold, connective peptide, KMSKS, and anti-codon binding domains.
Figure 2. Effect of peptide fusion on MetRS fragment complementation.
(A) Structural models for fragmented MetRS created using PDB structure 3H9B.32 The models illustrate the N- and C-terminal fragments in red and black, respectively. Split MetRS are named based on the last residue within the N-terminal fragment that precedes the peptide backbone cleavage site, e.g., MetRS-464 represents a split protein that arose from fission after MetRS residue 464. In all cases the backbone cleavage sites (green) were all distal from Anl (orange). (B) The activities of MetRS fragments fused to IAAL-E3 and IAAL-K3 were assessed using bacterial complementation. For each variant, the relative growth was calculated as the area under the growth curve over a 24 hour incubation in minimal medium lacking (open bars) and containing methionine (closed bars). The values below the bars represent the ratio of growth in the absence and presence of methionine. All split MetRS significantly enhanced the growth ratio compared with E. coli CS50 cells alone (p<0.001, two-tailed Welch’s t test). (C) MetRS fragments lacking IAAL-E3 and IAAL-K3 and (D) fused to only the IAAL-K3 presented complementation that was lower than that observed with MetRS fragments fused to both peptides. When fused to only one peptide, all split MetRS presented growth ratios that were significantly lower than those observed when fused to two peptides (p<0.05, two-tailed Welch’s t test). For each split protein, the error bars represent ±1σ from 6 experiments, while the controls represent ±1σ from 24 experiments.
Fragmented MetRS require assistance for activity.
The discovery of novel split MetRS that retain activity when fused to peptides that associate suggested that some of these two-fragment MetRS might require assistance from the IAAL-E3 and IAAL-K3 coils to function. This idea was tested by comparing the complementation of each variant in the presence and absence of fusions to associating peptides. For this complementation analysis, we monitored the extent to which each variant complemented E. coli CS50 grown in liquid cultures. This approach was used to analyze complementation because it provided quantitative information on growth in medium containing and lacking methionine, including growth rate, growth delay, and area under the growth curve. With this analysis, the ratio of each growth parameter +/− methionine was used as a proxy for split MetRS activity. This comparison was chosen because it considered the growth complementation by each variant (-methionine) as well as expression and fitness burdens arising from translation of protein fragments (+methionine).
A comparison of split MetRS complementation of E. coli CS50 ±IAAL-E3/K3 revealed complementation strengths that were dependent on fusion to associating peptides. When fused to both IAAL-E3 and IAAL-K3 (Figure 2B, S4), all six of the variants enhanced E. coli CS50 growth (-methionine) to a level that was significantly higher than cells transformed with an empty vector. Removal of the IAAL-E3 and IAAL-K3 coils significantly decreased complementation in all of the variants (Figure 2C, S5), albeit to varying extents, suggesting that the N-terminal fragments alone are not sufficient for maximal complementation. This observation led us to assess the relative translation rates ±peptide fusions using a thermodynamic model.33 This calculation revealed that peptide removal differentially attenuates the strength of the ribosomal binding site (RBS) used to initiate translation of the second fragment (Figure S6), although it has no effect on the RBS used to initiate translation of the first fragment. To avoid changes in translation initiation when disrupting the IAAL-E3 and IAAL-K3 interaction, we also examined the effect of removing IAAL-E3 (but not IAAL-K3) on complementation by each split MetRS. Removal of the IAAL-E3 peptide also significantly decreased complementation by all six variants (Figure 2D, S7). This finding suggests that all six of the split MetRS require assistance from interacting peptides for maximal activity.
One advantage of our library construction approach is that it generates constructs that can be modified in a single step to express protein fragments fused to different pairs of interacting proteins (Figure S8). We took advantage of this characteristic to create vectors that express all six split MetRS as fusions to Thermotoga maritima chemotaxis proteins (CheY and the CheY binding domain of CheA). CheA and CheY were chosen for this analysis because they display high affinity for one another, KD = 200 nM,34 and the residues in CheA and CheY that become covalently attached to MetRS through peptide linkers are separated by 30 Å in the structure, similar to the IAAL-E3/K3 complex.30,34 When each split MetRS was fused to CheA and CheY, a wider range of complementation strengths were observed compared with the IAAL-E3 and IAAL-K3 fusion proteins (Figure S9). MetRS-521 complemented E. coli CS50 growth strongly across all growth metrics analyzed, and this split MetRS was used for all subsequent measurements.
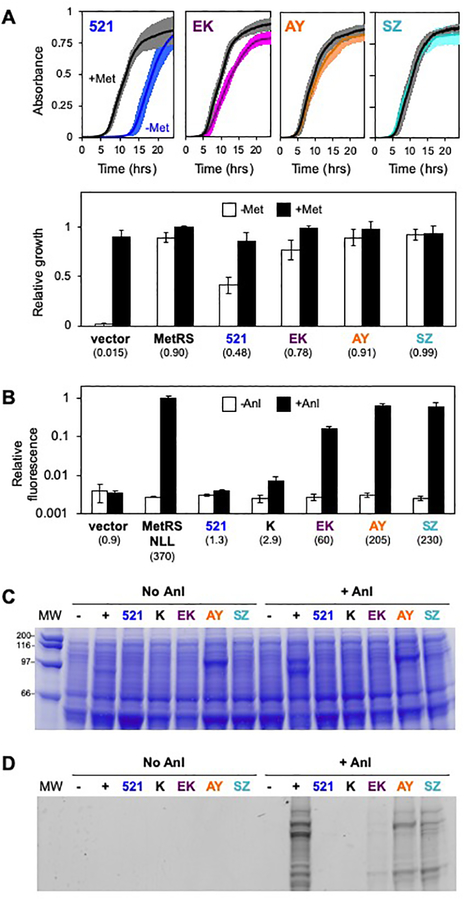
To test whether interacting proteins with a smaller separation between their fused termini also enhance MetRS-521 activity, we fused its fragments to SYNZIP-17 and SYNZIP-18,35,36 which display high affinity (KD <10 nM), and compared its complementation of E. coli CS50 with the complementation observed with other protein fusions. The termini that become fused to MetRS fragments in the SYNZIP-17/18 peptide complex are thought to have a smaller separation (<10 Å) compared with IAAL-E3 and IAAL-K3 because they associate into an antiparallel coiled-coil.35,36 When MetRS-521 fragments were fused to SYNZIP-17 and SYNZIP-18, we observed strong complementation across all growth metrics analyzed (Figure 3A, S10). This finding suggests that the SYNZIP peptides restore MetRS-521 activity more than IAAL-E3/K3 and CheA/Y.
Figure 3. Effect of protein fusion on NLL-MetRS-521 Anl-labeling activity.
(A) Culture absorbance at 600 nm was used to track the growth of E. coli CS50 transformed with vectors that express MetRS-521 fragments alone (521) and as fusion to IAAL-E3 and IAAL-K3 (EK), CheA and CheY (AY) and SYNZIP-17 and SYNZIP-18 (SZ). The relative growth was calculated as the area under growth curve measured in medium containing (closed bars) and lacking methionine (open bars). The values below each label represent the ratio of growth ±methionine. When MetRS-521 was fused to interacting proteins, the growth ratio was significantly higher than that observed with MetRS-521 fragments alone (p<0.0001, two tailed Welch’s t test). Error bars represent ±1σ from 8 experiments. (B) Anl metabolic labeling in cells transformed with vectors that express NLL-MetRS-521 fragments lacking protein fusions (521), as fusion to pairs of interacting proteins (EK, AY, and SZ), and with one fragment fused to the IAAL-K3 peptide (K). Anl labeling of total cellular protein was performed by growing cells in medium containing 2 mM Anl and 4 μM IPTG. Labeling was visualized by reacting cell lysates from each culture with TAMRA-DIBO, measuring fluorescence, analyzing total protein in each sample using the Bio-Rad Protein Assay, calculating the protein-normalized fluorescence to obtain relative fluorescence, and comparing the signals with those obtained in identical experiments performed in the absence of Anl. For each construct, the ratio of TAMRA fluorescence ±Anl is provided below the bar graph. Like full-length NLL-MetRS, NLL-MetRS-521 fragments fused to three different pairs of interacting proteins presented a ratio that is significantly higher than that obtained with cells transformed with an empty vector (p<0.05, two tailed Welch’s t test). In contrast, NLL-MetRS-521 lacking protein fusions showed no significant increase in Anl labeling compared with that observed in cells transformed with an empty vector. Samples were subjected to (C) SDS-PAGE analysis and (D) in-gel imaging to visualize TAMRA-labeled proteins. The error bars represent ±1σ from three experiments.
Effect of active site mutations on MetRS-521 specificity.
A previous study showed that the substrate specificity of a split MetRS can be directed towards the non-canonical amino acid Anl by incorporating L13N, Y260L, and H301L (NLL) mutations.18 To determine if MetRS-521 specificity can also be directed towards Anl, we incorporated these NLL mutations into our different MetRS-521 constructs and examined how Anl metabolic labeling by the resulting protein (NLL-MetRS-521) is influenced by fusion of fragments to IAAL-E3 and IAAL-K3, IAAL-K3 alone, CheA and CheY, and SYNZIP-17 and SYNZIP-18. Labeling experiments were performed by transforming NLL-MetRS-521 constructs into E. coli CS50, growing cells to mid-log phase in minimal medium containing methionine, and initiating metabolic labeling by transferring cells to minimal media containing Anl but lacking methionine (Figure S11). After 18 hours of growth at 37°C, cells were harvested, lysed, and reacted with a dibenzocyclooctyne fluorescent TAMRA dye to visualize labeling. Anl labeling was quantified by measuring the fluorescence of total protein after precipitating, washing each sample to remove unbound dye, and normalizing the fluorescence signal to the total protein in each sample (Figure 3b). To control for TAMRA reactivity with unlabeled protein, we performed identical experiments where we did not add Anl. This analysis revealed NLL-MetRS-521 lacking peptide fusions yields a protein-normalized fluorescence signal that is not significantly greater than that observed with cells that do not express NLL-MetRS. In contrast, NLL-MetRS-521 fused to different pairs of interacting proteins (IAAL-E3/K3, CheA/Y, and SYNZIP-17/18) yielded labeling that was significantly higher than that observed with NLL-MetRS-521 fragments lacking fusions to associating proteins. However, the signal varied across the different NLL-MetRS-521 fragment fusions. Fragments fused to SYNZIP-17 and SYNZIP-18 yielded a fluorescence signal in the presence of Anl that was 230-fold higher than that observed in experiments lacking Anl, a labeling enhancement that corresponds to 62% of the value observed with NLL-MetRS. The signal enhancements with fragments fused to CheA/Y and IAAL-E3/K3 were 200-fold and 60-fold, respectively. Analysis of each sample using SDS-PAGE and in-gel imaging revealed similar trends (Figures 3c–d), with the SYNZIP-17/18 and CheA/Y yielding the highest signals among the different variants tested. Taken together, these findings suggest that the total activity of NLL-MetRS-521 fragments depends upon the affinity of the pair of proteins fused that are fused to their termini, the physicochemical properties of the fused proteins, and the distances between the residues in the interacting proteins that are fused to the NLL-MetRS-521 fragments.
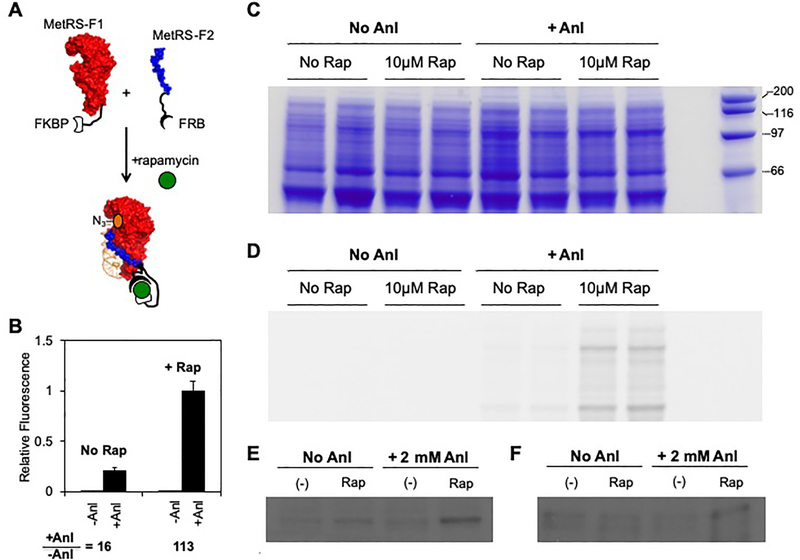
To determine if MetRS-521 activity can be coupled to a conditional protein-protein interaction whose stability depends upon ligand binding (Figure 4a), we analyzed metabolic labeling in cells expressing NLL-MetRS-521 fragments as fusions to the FKBP12 and the FKBP-rapamycin binding domain of mTOR (FRB). FKBP and FRB have strong affinity in the presence of rapamycin (KD = 12 nM) but weak affinity in its absence.25 To control for the effects of rapamycin on metabolic labeling, we first investigated how the addition of rapamycin affects metabolic labeling by NLL-MetRS. Similar metabolic labeling was observed in cells expressing NLL-MetRS in the presence and absence of rapamycin (Figure S12). To investigate whether Anl labeling can be controlled by rapamycin, we evaluated how the addition of rapamycin influences metabolic labeling in cells expressing NLL-MetRS-521 fragments fused to FKBP12 and FRB. We found that labeling experiments performed in the presence of 10 μM rapamycin yielded Anl incorporation that was significantly higher (7-fold) than that observed in the absence of rapamycin (Figure 4b). For these experiments, we induced protein fragment expression using a low level of IPTG (4 μM), which was found to be optimal for a rapamycin-dependent labeling signal (Figure S13). Decreased labeling efficiency was observed when higher levels of IPTG were used to induce MetRS labeling. This trend was interpreted as arising because high levels of IPTG lead to a greater consumption of cell resources for MetRS expression, thus limiting the resources for synthesis of subsequent labeled proteins by MetRS. SDS-PAGE and in-gel fluorescence analysis of the protein lysate from each experiment revealed trends consistent with our solution measurements (Figures 4c–d). In addition, immunoblot analysis of the NLL-MetRS-521 fragments revealed that both fragments were expressed in the presence and absence of rapamycin (Figure 4e–f). This finding suggests that the 7-fold increase in labeling observed in the presence of rapamycin arises because rapamycin stabilizes the NLL-MetRS-521 fragment complex.
Figure 4. Using rapamycin to regulate Anl labeling.
(A) A scheme illustrating how rapamycin was used to regulate BONCAT by stabilizing the NLL-MetRS-521 fragment complex. (B) Effect of rapamycin on Anl labeling in cells transformed with a vector that expresses NLL-MetRS-521 fragments fusions to FKBP and FRB. Protein-normalized TAMRA fluorescence was measured in cell lysates that were derived from cells that had been grown in the presence and absence of 10 μM rapamycin and 2 mM Anl. All experiments contained 4 μM IPTG. The ratio of the signals obtained ±Anl is significantly higher in cell lysates derived from rapamycin treated cells compared with lysates derived from cells that lacked rapamycin (p< 1×10−8 two tailed Welch’s t test). Samples subjected to (C) SDS-PAGE to assess relative protein levels, (D) in-gel imaging to visualize TAMRA-labeled proteins, (E) immunoblot analysis using an anti-Tetra His antibody to determine the relative levels of the N-terminal fragments, and (F) immunoblot analysis using a anti-FRB antibody to establish the relative levels of the C-terminal fragments. The error bars represent ±1σ from 8 experiments.
To understand the response time of our engineered protein, we evaluated how long it takes to observe labeling at different times following the addition of rapamycin. Within ten minutes following rapamycin addition, the labeling in the presence of Anl was 2-fold higher than that observed in the absence of Anl (Figure S14). In contrast, no significant labeling was observed at this time point in the absence of rapamycin.
Using domain insertion to diversify ligand control.
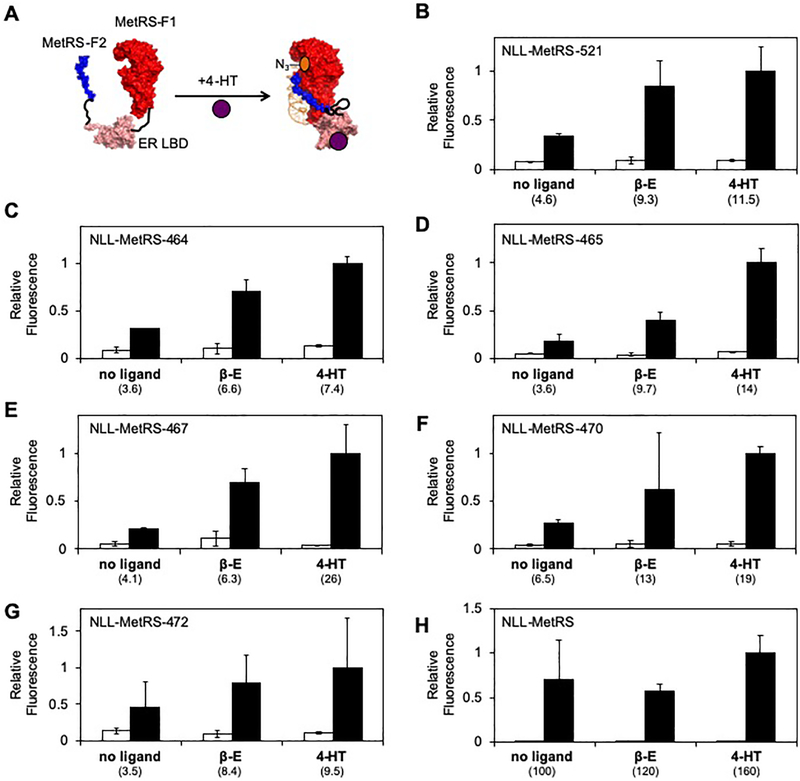
In nature, ligand-dependent protein activity is thought to evolve through domain insertion, i.e., the insertion of a ligand-binding domain into an enzyme. Protein switches can also be engineered in the lab by mimicking this evolutionary process.23 For example, a recent study showed that ligand control can be achieved over Cas9 by inserting the ligand-binding domain (LBD) of human estrogen receptor (ER).37 This LBD was appealing to use for domain insertion with MetRS because it undergoes a large conformational shift upon ligand binding that alters the distance between the N- and C-termini.26,38,39 In the case of β-estradiol (β-E) binding, there is a ~27 Å movement (from 64 to 37 Å), while 4-hydroxytamoxifen (4-HT) binding yields a larger movement (from 64 to 21 Å).37
We hypothesized that the backbone fragmentation sites yielding active split MetRS upon fusion to interacting proteins would also yield proteins that retain detectable activity upon insertion of the ER LBD at the same sites. To test this idea, we created vectors that expressed MetRS with the ER LBD inserted at six sites (after residues 464, 465, 467, 470, 472, and 521) and assessed whether these proteins complemented the growth of E. coli CS50 (Figure S15). This LBD was fused using Gly-rich linkers that were identical in length to those used for fusing the individual MetRS fragments to SYNZIP 17/18. All six MetRS harboring an inserted ER LBD complemented E. coli CS50 growth on minimal medium lacking methionine to ≥30% of the complementation observed with cells expressing MetRS. This finding provides evidence that MetRS retains some enzymatic activity even when the ER LBD is inserted at these six different native sites, although it suggests that these proteins have decreased activities (or expression) compared with that observed when the same split MetRS are fused to interacting proteins. We additionally examined whether β-E and 4-HT affect complementation. Complementation was observed in the presence of these ligands, although the magnitude of this complementation was not significantly higher than that observed without ligands.
To test whether a NLL-MetRS having the ER LBD inserted after residue 521 displays ligand-dependent metabolic labeling, we created a vector that expresses this protein and assayed Anl labeling activity in the presence and absence of 4-HT and β-E (Figure 5a). Anl incorporation was increased in the presence of 4-HT (2.5-fold) and β-E (2.0-fold) compared with that observed in the absence of ligands (Figure 5b). However, these increases were not statistically significant. We next investigated whether insertion of the ER LBD at other locations might yield NLL-MetRS that display ligand-dependent metabolic labeling. For these experiments, we targeted the other five sites where MetRS fission was non-disruptive to activity when the fragments were fused to interacting proteins. With each of these variants, the average metabolic labeling observed in the presence of 4-HT and β-E was also higher than that observed in the absence of ligands (Figures 5c–g). However, 4-HT only increased the Anl labeling activity significantly with the proteins created through insertion of ER LBD after NLL-MetRS residues 464 (2.1-fold), 465 (3.9-fold), and 467 (6.3-fold). In the case of β-E, increased labeling was also observed. However, the enhanced labeling was not significantly higher than that observed in the absence of ligand. To control for the effects of 4-HT and β-E on metabolic labeling, we also measured how these ligands affect metabolic labeling by NLL-MetRS. No significant changes in labeling were observed upon addition of either 4-HT or β-E (Figure 5h).
Figure 5. Using 4-hydroxytamoxifen to regulate Anl labeling.
(A) Scheme showing how 4-HT was used to regulate Anl labeling using NLL-MetRS created by domain insertion. The effect of 4-HT and β-E on Anl labeling in cells expressing NLL-MetRS having the ER LBD inserted after residue (B) 521, (C) 464 (D) 465, (E) 467, (F) 470, and (G) 472. (H) Metabolic labeling in cells expressing NLL-MetRS is not altered in the presence of 4-HT or β-E (p>0.2, two-tailed Welch’s t test). For each experiment, protein-normalized TAMRA fluorescence was measured in cell lysates that were derived from cells that had been grown in the presence and absence of 4-HT (10 μM), β-E (10 μM), and Anl (2 mM). All samples were induced with 4 μM IPTG. The error bars represent the standard deviation from three experiments. NLL-MetRS containing the ER LBD inserted after residues 464, 465, and 467 present a significant increase in labeling (i.e., the ratio of signals obtained ±Anl) upon addition of 4-HT compared to the vehicle control (p<0.05, two-tailed Welch’s t test).
Implications.
Mapping the non-disruptive backbone fragmentation sites identified in our combinatorial experiment onto protein structure suggests that all six split MetRS require both of their fragments for catalytic activity. For example, the backbone fragmentation site that yielded MetRS-521, the variant arising from backbone fission closest to the C-terminus, occurs in an unstructured region of MetRS that connects alpha helices 14 and 15, which are in the anticodon binding and KMSKS domains, respectively.40 This polypeptide is thought to be critical to the activity of MetRS because it contains a residue that has been implicated in hydrogen bonding directly with the tRNA.41 Additionally, this polypeptide makes multiple residue-residue contacts with the anticodon binding domain,32,42 suggesting that residues in this peptide are important for the structure and stability of the anticodon binding domain. Because a previous truncation study showed that this polypeptide cannot be removed without abolishing MetRS activity,29 these findings suggest that the low activity observed when the NLL-MetRS-521 fragments are fused to proteins that do not associate (e.g., K alone) arise because these fragments associate weakly in the absence of assistance.
Like the previously described NLL-MetRS-247, which does not require assistance from a protein interaction for Anl labeling,18 all of the split proteins described herein should be useful as a two-input transcriptional AND gate for controlling BONCAT when their fragments are fused to pairs of proteins that form a stable complex, such as IAAL-E3 and IAAL-K3. Our findings also suggest that NLL-MetRS-521 should be generally useful for controlling BONCAT using post-translational binding that stabilize the MetRS fragment complex. Our results with FKBP and FRB protein fusions provide evidence for this application. When NLL-MetRS-521 fragments were fused to FKBP and FRB, enhanced Anl metabolic labeling was observed in the presence of rapamycin, which stabilizes the FKBP-FRB complex.24,25 Taken together with the finding that NLL-MetRS-521 activity can be enhanced by fusion to different pairs of interacting proteins, this observation suggests that NLL-MetRS-521 activity could be coupled to other pairs of proteins whose association depend upon metabolite binding,43 metal coordination,44 or post-translational modifications.45 Additionally, response regulators from bacterial two-component systems that sense a wide range of chemicals could be used to control NLL-MetRS-521 activity,46 since many of these proteins displayed enhanced dimerization as part of sensing mechanism.47,48 While response regulators could already be used to control the activity of the full-length NLL-MetRS through transcriptional mechanisms, post-translational regulation is expected to allow for control over metabolic labeling on a shorter time scale compared with transcriptional approaches.49 Another potential advantage of using post-translational regulation rather than transcriptional control is that this approach allows for enhanced levels of aaRS in cells that lack non-canonical amino acids, i.e., the NLL-MetRS variant responsible for labeling can accumulate prior to metabolic labeling. With transcriptional control, NLL-MetRS is likely synthesized using non-canonical amino acids as they accumulate in cells, which could impact its ability to label other proteins. Future studies will be needed to better understand the benefits of using post-translational regulation for metabolic labeling and to establish the diversity of protein-protein interactions that can regulate NLL-MetRS-521 activity.
Our results illustrate how a combinatorial library encoding split proteins can be used to rapidly identify bisected proteins that require assistance from interacting proteins for maximal activity to create ligand-stabilized bipartite protein AND gates. Selection of our EK library yielded six variants that all required the peptide fusions for maximal activity. Half of these variants (467, 472, and 521) displayed enhanced activity when fused to globular proteins that interact, CheA and CheY.34 These findings are similar to those from a recent study examining the effects of fission and fusion on the sensory core of a baceriophytochrome,27 where a large fraction of the two-fragment bacteriophytochromes that exhibited enhanced fluorescence when fused to IAAL-E3 and IAAL-K3 also displayed increased fluorescence when fused to CheA and CheY.27 A comparison of our EK library results with studies of MetRS fission in the absence of fusion illustrates the benefits of using peptide assembly to assist with fragment complementation in combinatorial libraries. All of the split MetRS discovered in our EK library arose from fission at locations distinct from split MetRS that retain activity without assistance.18 This observation suggests that IAAL-E3 and IAAL-K3, or their peptide linkers, are disruptive to the function of many split MetRS that are active in the absence of peptide fusions. The underlying cause of this trend is not known. However, the structure of the IAAL-E3/IAAL-K3 complex suggests one possible mechanism.30 The IAAL-E3 and IAAL-K3 termini that become fused to MetRS fragments have a large 30 Å separation,30 which could prevent the fused termini in the MetRS fragments from achieving an active structure even though they bring the MetRS fragments into close proximity. Consistent with this idea, the SYNZIP-17 and SYNZIP-18 peptides, whose termini are expected to have a smaller separation,35,36 enhanced the activity of MetRS-521 to a greater extent. Additional studies will be needed to determine if the SYNZIP-17/SYNZIP-18 peptides are disruptive to the function of some split MetRS that retain activity in the absence of peptide fusions.
Multiple mechanisms could be responsible for the increased azidonorleucine incorporation observed when NLL-MetRS-521 fragments were expressed as fusions to pairs of proteins that associate, i.e., IAAL-E3/IAAL-K3, SYNZIP17/18, and CheA/Y. One or both of the MetRS fragments might display increased solubility when expressed as fusion to other proteins, similar to that observed with split GFP that have been developed as solubility screens.50 Alternatively, the NLL-MetRS-521 fragments could be soluble in the absence of protein fusions, but require assistance to form a stable active complex. A comparison of our metabolic labeling by NLL-MetRS-521 fragments that had been fused to different proteins suggests that both mechanisms might underlie the activity of this split protein. In the case of our FKBP/FRB protein fusions, a low level of metabolic labeling was observed in the absence of rapamycin. The activity observed when NLL-MetRS-521 fragments were fused to these globular proteins was higher than that observed when only one fragment was fused to a peptide (e.g., IAAL-K3) or when both fragments lacked protein fusions. Taken together, these findings suggest that fusing both fragments to globular proteins that do not form a complex enhances their solubility. With the FKBP/FRB protein fusions, we also found that addition of rapamycin enhanced activity. This observation provides evidence that the stabilization of the FKBP/FRB complex correlates with metabolic labeling activity.
We provide evidence that ligand regulation of metabolic labeling by NLL-MetRS can be diversified using domain insertion. When the ER LBD was inserted at the backbone fragmentation sites that yielded our original six functional split proteins, half of the proteins displayed metabolic labeling activity that was significantly enhanced in the presence of 4-HT. This labeling enhancement was statistically significant compared with that observed in the absence of 4-HT. The best protein designed using this approach (NLL-MetRS-467) yielded a 6.3-fold change in labeled protein. This increase in Anl labeling activity is similar to that achieved with our rapamycin-dependent system (7-fold), which was built using a different split variant (NLL-MetRS-521). However, the enhancement of Anl labeling in response to these two different chemicals is smaller than that observed with full-length NLL-MetRS (370-fold) and NLL-MetRS-521 having its fragments fused to the SYNZIP17 and SYNZIP18 peptides (230-fold), which strongly associate.36 This finding indicates that our library design approach can identify MetRS backbone fragmentation sites to target for domain insertion when creating protein switches, and it suggests that additional proteins could be inserted at these NLL-MetRS backbone fragmentation to diversify the ligand-dependence of NLL-MetRS activity, such as the periplasmic binding proteins whose chemical-dependent conformational changes have been used to control the emission from fluorescent protein reporters, such as maltose binding protein.51 However, the protein switches created using this approach will require additional optimization to decrease their background labeling while simultaneously improving the maximal level of labeling activity in the presence of ligand. In cases where optimization is needed, cell-surface display of non-canonical amino acids can be used as a screen to discover NLL-MetRS-521 mutants with optimized regulation.7
The split proteins described herein are expected to be useful for extending control over BONCAT in a range of organisms. The parental NLL-MetRS used to build our split protein can metabolically label newly synthesized proteins with Anl in E. coli,15 the microbial pathogen Yersinia enterocolitica,52 and mammalian cells.53 Post-translational regulation of BONCAT could be extended to other organisms by subjecting other mutant aaRS to random fission and directed fusion using a recently described transposase mutagenesis approach.27 Mutations have been identified that direct the substrate specificity of insect and murine MetRS towards Anl, which allow for genetic control over BONCAT in a range of eukaryotes (fly, hamster, monkey, and human).54,55
Methods
Materials.
All enzymes used for molecular biology were from New England Biolabs and ThermoFisher, synthetic oligonucleotides were from Integrated DNA Technologies, nucleotides for PCR were from Roche, bacterial growth medium components were from Sigma-Aldrich and Amresco, and antibiotics were from Research Products International. DNA clean up and plasmid purification kits were from Zymo Research and Qiagen, respectively.
Library construction.
To create vectors that express different split MetRS as fusions to IAAL-E3 and IAAL-K3, a previously described vector library that expresses different two-fragment MetRS without fusions was digested with NotI,18 the NotI-flanked insert in this library was removed, and the ek-kanR insert coding for IAAL-E3 and IAAL-K3 was ligated in its place.27 The resulting EK vector library was transformed into E. coli using electroporation, spread on LB-agar medium containing 25 μg/mL kanamycin, and incubated at 37°C. After 16 hours, >10,000 colonies were obtained. Assuming sampling with replacement, we estimate that the EK library sampled ~95% of the variants in the original library digested with NotI. All colonies were harvested and pooled, and DNA encoding this EK library was purified using Qiagen miniprep kit.
Bacterial complementation.
Electroporation was used to transform the EK vector library into a DE3 lysogen of E. coli CS50, a strain that displays reduced growth rates on minimal medium lacking methionine because it has a chromosomal mutation that decreases the affinity of MetRS for methionine.56 Cells were spread onto M9-agar plates containing every amino acid except methionine, and plates were incubated at 37°C. Approximately 50 colonies appearing after 18 hours were used to inoculate LB cultures containing 50 μg/mL kanamycin. DNA from each culture was purified after overnight growth and sequenced, and sequenced variants with both fragments in frame were subjected to further analysis. Split proteins were named based on the last MetRS residue in the fragment that precedes the backbone fission site. This residue is also the first residue in the second fragment, because the transposase method used for library construction creates a five base pair duplication at the site of DNA insertion.57
Growth analysis.
E. coli CS50 were transformed with sequence-verified plasmids and spread on LB-agar plates containing 50 μg/mL of kanamycin. After overnight growth at 37°C, colonies from each transformation were used to inoculate M9 liquid cultures (1 mL) containing all twenty amino acids and 50 μg/L ampicillin in deep 96 well plates. Six or more colonies were used for each experimental condition unless noted otherwise. Plates were grown at 37°C while shaking at 250 rpm. After 16 hours, a 96-pin replicator (V&P Scientific, Inc.) was used to transfer 0.2 μL of the overnight cultures to fresh M9 medium (+/-methionine) containing 50 μg/mL ampicillin in shallow flat-bottom 96 well plates. When comparing the effects of different chemicals on growth (±methionine and ligand), experiments represent biological replicates from unique colonies. Culture growth was measured in these plates by monitoring absorbance at 600 nm every five minutes for 24 hours while shaking at 216 rpm using a Tecan M1000 Pro plate reader. Growth data was fit to a simple exponential model using the software R to obtain different growth parameters (rate, delay). In addition, the integrated absorbance over the full 24 hours of growth was used to calculate area under each growth curve.
Vector construction.
Constructs from the EK library that complemented E. coli CS50 were digested with NotI to remove the DNA encoding the IAAL-E3 and IAAL-K3 coiled coils (ek-kanR), and different NotI flanked DNA inserts (f1-kanR, k-kanR, ay-kanR)27,57 were subcloned in place of this DNA to create vectors that express split MetRS fragments without fusions (f1-kanR), with only one fragment fused to the IAAL-K3 coil alone (k-kanR), and with the pair of fragments fused to the chemotaxis proteins CheA and CheY (ay-kanR). NotI-flanked DNA inserts encoding the SYNZIP-17/SYNZIP-18 peptides and FKBP12/FRB were synthesized as gBlocks by IDT, the kanR selectable marker was inserted into these synthetic DNA using standard cloning methods to create sz-kanR and ff-kanR, and the resulting DNA were cloned in the vector expressing MetRS-521 to create vectors that express MetRS fragments fused to these pairs of proteins. To create vectors that express NLL-MetRS-521 fragments as fusions to these different proteins, we cloned the gene encoding NLL-MetRS into pPROEX1 (Life Technologies), the resulting vector was PCR amplified using primers that insert a NotI site after the codon encoding residues 464, 465, 467, 470, 472, and 521. The different NotI-flanked inserts were cloned into this restriction site. A NotI-flanked DNA insert encoding residues 302 to 552 of the human estrogen receptor, corresponding to the ligand binding domain, was synthesized as a gBlock by IDT. The ER-LBD gBlock was PCR-amplified and inserted into pPROEX1 containing MetRS or NLL-MetRS split after the following residues: 464, 465, 467, 470, 472, and 521. All constructs were sequence verified.
Anl preparation.
Boc-azidonorleucine (Sigma-Aldrich) dissolved in dichloromethane was incubated with aq HCl (6M) while stirring in the dark. Reaction progress was monitored by thin layer chromatography (TLC) After 24 h, the reaction appeared complete. Azidonorleucine was isolated as its hydrochloride salt after removal of the volatiles under vacuum. Deprotection of 509 mg of boc-Anl resulted in 448.5 mg of unpurified product. As the tert-butyl cation was not purified from the resulting product, our yield was estimated to be 88%. Data was consistent with previously reported data,58 and the salt was used without further purification.
Anl protein labeling.
The different NLL-MetRS-521 constructs were transformed into E. coli CS50 and plated onto LB-agar medium containing 50 μg/mL kanamycin. Colonies obtained on these plates were used to inoculate liquid cultures (4 mL) containing M9 medium, all twenty amino acids, and 50 μg/mL ampicillin. After overnight growth at 37°C while shaking at 250 rpm, the cultures were diluted 100-fold into M9 media containing all twenty amino acids and 50 μg/mL ampicillin and grown to mid log phase (OD ~0.5). Cultures were then induced with 4 μM IPTG. After 30 minutes, cultures were pelleted and split into two samples, and these samples were resuspended in M9 medium (4 mL) containing 19 amino acids, 4 μM IPTG, 50 μg/mL ampicillin, and either 0 or 2 mM Anl. Cultures starting at an OD of ~0.25 were grown overnight at 37°C while shaking at 250 rpm to allow for Anl incorporation. While the MetRS fragments were constitutively produced at sufficient levels to complement E. coli CS50 growth in the absence of IPTG, protein fragment expression required induction for Anl metabolic labeling. A comparison of labeling using a protocol where samples were resuspended in medium containing 19 (-methionine) versus 20 amino acids revealed no significant difference in the labeling activity of NLL-MetRS (p=0.47, two-tailed Welch’s t test; Figure S16) under these different conditions. For each experiment, a unique colony harboring the indicated protein expression plasmid was used for analysis, i.e., all data represents biological replicates.
Visualizing labeled proteins.
Cultures grown ±Anl were pelleted and resuspended in 50 mM Tris lysis buffer pH 8 containing DNase I (0.04 mg/mL) and cOmplete™ Mini, EDTA-free Protease Inhibitor Cocktail (1 tablet per 20 mL). Each mixture was frozen at −20°C, thawed on ice, and vortexed for five minutes. Total protein was determined in diluted aliquots (4-fold) using the Bio-Rad protein assay, in which the diluted lysate (10 μL) was reacted with 200 μL of Bio-Rad protein assay reagent in triplicate, and absorbance at 595 nm was analyzed using a Tecan M1000 plate reader. The remaining undiluted lysate was treated with sodium dodecyl sulfate (SDS) to a concentration of 1%. The lysate was vortexed for 5 minutes and pelleted, and the supernatant was then treated with 0.5 M iodoacetamide for two hours to alkylate cysteines and prevent non-specific reactions of the dibenzocyclooctyne dye with the native protein. Proteins were precipitated by mixing supernatant (100 μL) and methanol (400 μL), vortexing the mixture, adding chloroform (100 μL), vortexing until well mixed, adding water (300 μL) and vortexing until well mixed. The resulting mixture was centrifuged for two minutes at 14,000×g, the upper aqueous layer was discarded, methanol (400 μL) was added to the lower organic layer containing the precipitated protein, the mixture was vortexed until well mixed, and the mixture was centrifuged at 14,000×g for two minutes. All liquid was removed from above the protein pellet, and the pellet was left to dry. The resulting pellet was resuspended in Tris-Buffered Saline (TBS), which contained 50 mM Tris 150 mM NaCl, and 1% SDS with a pH of 7.6. The solution was vortexed for 15 minutes until the pellet was completely dissolved. Tetramethylrhodamine dibenzocyclooctyne (TAMRA-DIBO) was added to a concentration of 30 μM and left to react for one hour at 23°C in the dark. Proteins were precipitated as described above and resuspended in TBS. This process was repeated two times to remove free TAMRA-DIBO.
Imaging of TAMRA-DIBO Modified Proteins.
Labeled proteins were transferred into a shallow 96 well plate and analyzed for fluorescence (λex = 542 nm; λem = 568 nm) using a Tecan M1000 Pro plate reader. Fluorescence values obtained from this analysis were normalized to the amount of protein in each sample, which was determined using the Bio-Rad Protein Assay. After imaging, proteins were analyzed using a 12% Novex Bis-Tris polyacrylamide gel under denaturing conditions. The gel was excited at 370nm using an ImageQuant LAS400 Imager, and the resulting fluorescence was imaged through an L41 filter, which absorbs light <410 nm. After measuring in gel fluorescence, the gel was then incubated with coomassie blue stain to visualize total protein (labeled and unlabeled). Each measurement analyzed the signal arising from samples derived from different colonies that expressed the indicated proteins.
Chemical-induced Anl labeling.
The vector expressing NLL-MetRS-521 fragments as fusions to FKBP and FRB were transformed into E. coli CS50 and plated onto LB-agar medium containing 50 μg/mL kanamycin. The resulting colonies were grown overnight in M9 medium containing all twenty amino acids and 50 μg/mL ampicillin. After overnight growth, cultures were diluted 100-fold into M9 media containing all twenty amino acids and 50 μg/mL ampicillin, and cultures were grown until they reached mid log phase (OD ~ 0.5). Protein fragment expression was induced by adding IPTG (4, 20, and 100 μM). Rapamycin dissolved in DMSO was added to concentrations of 10 μM. A vehicle control was also used at 0.1% DMSO. After 30 minutes, cultures were pelleted and resuspended into two separate 4mL cultures with M9 medium containing 19 amino acids, 50 μg/mL ampicillin, IPTG (4, 20, and 100 μM), 10 μM rapamycin or 0.1% DMSO, and either 0 or 2 mM azidonorleucine. Cultures were subsequently grown for 0, 10 min, 30 min, 1 hour, 3 hours or 24 hours overnight at 37°C while shaking at 250 rpm to allow for Anl incorporation. The concentration of rapamycin used in this study (10 μM) is higher than the level (1 μM) found to be optimal previously.59 The previous study read out the activity of a rapamycin-stabilized split DHFR through the complementation of E. coli growth. Our requirement for a higher rapamycin concentration could arise from strain differences or because the strategies used to read out rapamycin stabilized split proteins required different levels of protein fragment association.
Vectors expressing the different NLL-MetRS containing inserted ER LBD were transformed into E. coli CS50 and plated onto LB-agar medium containing 100 μg/mL ampicillin. The resulting colonies were grown overnight in M9 medium containing all twenty amino acids and 50 μg/mL ampicillin. After overnight growth, cultures were diluted 100-fold into M9 media containing all 20 amino acids and 50μg/mL ampicillin, and cultures were grown until they reached mid log phase (OD ~ 0.5). Protein expression was induced by adding IPTG (4μM). β-estradiol and 4-hydroxytamoxifen were dissolved in DMSO and added to a concentration of 10 μM. A vehicle control (0.1% DMSO) was used. After 30 minutes, cultures were pelleted and resuspended into two separate 4 mL cultures with M9 medium containing 19 amino acids, 50 μg/mL ampicillin, 4 μM IPTG, 10 μM β-E or 10 μM 4-HT or 0.1% DMSO, and either 0 or 2mM azidonorleucine. Cultures were subsequently grown overnight at 37°C while shaking at 250 rpm to allow for Anl incorporation.
Immunoblot analysis.
Tetra-His (BSA-Free, Qiagen) and mTOR (human FRB Domain, Enzo Life Sciences) antibodies were used to detect the MetRS-521 N- and C-terminal fragments, respectively. Total protein analyzed using SDS-PAGE was transferred to Protran nitrocellulose (Whatman), washed with 20 mL of TBST buffer (100 mM Tris pH 7.5, 150 mM NaCl, and 0.1% Tween 20) for 5 min three times and blocked with 10% dry milk. Membranes containing identical samples were incubated with each antibody (1:1000 and 1:2000 dilutions, respectively) for 1 hour, and the membranes were incubated with a 1:1000 dilution of a secondary goat anti-mouse IgG conjugated to peroxidase (Calbiochem) and a 1:5000 dilution of a secondary goat anti-rabbit IgG conjugated to peroxidase (Calbiochem), respectively. The signals were visualized using the ECL Western blotting substrate (GE Healthcare) and imaged using film (Bio-Excell).
Translation initiation calculations.
The effect of protein fusions on the translation initiation rates of MetRS-521 fragments was determined using a thermodynamic model.33 For these calculations, the rate of translation initiation at the start codon for each fragment was calculated using sequence that encompasses 35 base pairs before and after the start codon.
Supplementary Material
Acknowledgements
We are grateful for financial support from the National Science Foundation (1150138), Robert A. Welch Foundation (C-1614), John S. Dunn Collaborative Research Award, and National Aeronautics and Space Administration (NNX15AL28G). EET was partially funded by a training fellowship from the Keck Center of the Gulf Coast Consortia, on the Houston Area Molecular Biophysics Program, National Institute of General Medical Sciences (NIGMS) T32GM008280.
Footnotes
Supporting Information
This material is available free of charge via the Internet.
Supporting Figures S1–S16 (PDF)
References
- (1).Pandey A, and Mann M (2000) Proteomics to study genes and genomes. Nature 405, 837–846. [DOI] [PubMed] [Google Scholar]
- (2).Aebersold R, and Mann M (2003) Mass spectrometry-based proteomics. Nature 422, 198–207. [DOI] [PubMed] [Google Scholar]
- (3).Yuet KP, and Tirrell DA (2014) Chemical Tools for Temporally and Spatially Resolved Mass Spectrometry-Based Proteomics. Ann. Biomed. Eng 42, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kiick KL, Saxon E, Tirrell DA, and Bertozzi CR (2002) Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc. Natl. Acad. Sci. U.S.A 99, 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Dieterich DC, Link AJ, Graumann J, Tirrell DA, and Schuman EM (2006) Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT). Proc. Natl. Acad. Sci. U.S.A 103, 9482–9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, and Schuman EM (2007) Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat. Protoc 2, 532–540. [DOI] [PubMed] [Google Scholar]
- (7).Link AJ, Vink MKS, Agard NJ, Prescher JA, Bertozzi CR, and Tirrell DA (2006) Discovery of aminoacyl-tRNA synthetase activity through cell-surface display of noncanonical amino acids. Proc. Natl. Acad. Sci. U.S.A 103, 10180–10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Link AJ, and Tirrell DA (2003) Cell surface labeling of Escherichia coli via copper(I)-catalyzed [3+2] cycloaddition. J. Am. Chem. Soc 125, 11164–11165. [DOI] [PubMed] [Google Scholar]
- (9).Dieterich DC, Hodas JJL, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, and Schuman EM (2010) In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat. Neurosci 13, 897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Truong F, Yoo TH, Lampo TJ, and Tirrell DA (2012) Two-strain, cell-selective protein labeling in mixed bacterial cultures. J. Am. Chem. Soc 134, 8551–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ibba M, and Hennecke H (1995) Relaxing the substrate specificity of an aminoacyl-tRNA synthetase allows in vitro and in vivo synthesis of proteins containing unnatural amino acids. FEBS Lett. 364, 272–275. [DOI] [PubMed] [Google Scholar]
- (12).Sharma N, Furter R, Kast P, and Tirrell DA (2000) Efficient introduction of aryl bromide functionality into proteins in vivo. FEBS Lett. 467, 37–40. [DOI] [PubMed] [Google Scholar]
- (13).Datta D, Wang P, Carrico IS, Mayo SL, and Tirrell DA (2002) A designed phenylalanyl-tRNA synthetase variant allows efficient in vivo incorporation of aryl ketone functionality into proteins. J. Am. Chem. Soc 124, 5652–5653. [DOI] [PubMed] [Google Scholar]
- (14).Grammel M, Zhang MM, and Hang HC (2010) Orthogonal alkynyl amino acid reporter for selective labeling of bacterial proteomes during infection. Angew. Chem. Int. Ed. Engl 49, 5970–5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tanrikulu IC, Schmitt E, Mechulam Y, Goddard WA, and Tirrell DA (2009) Discovery of Escherichia coli methionyl-tRNA synthetase mutants for efficient labeling of proteins with azidonorleucine in vivo. Proc. Natl. Acad. Sci. U.S.A 106, 15285–15290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ngo JT, Champion JA, Mahdavi A, Tanrikulu IC, Beatty KE, Connor RE, Yoo TH, Dieterich DC, Schuman EM, and Tirrell DA (2009) Cell-selective metabolic labeling of proteins. Nat. Chem. Biol 5, 715–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Ngo JT, Babin BM, Champion JA, Schuman EM, and Tirrell DA (2012) State-selective metabolic labeling of cellular proteins. ACS Chem. Biol 7, 1326–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Mahdavi A, Segall-Shapiro TH, Kou S, Jindal GA, Hoff KG, Liu S, Chitsaz M, Ismagilov RF, Silberg JJ, and Tirrell DA (2013) A Genetically Encoded AND Gate for Cell-Targeted Metabolic Labeling of Proteins. J. Am. Chem. Soc 135, 2979–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Anderson JC, Voigt CA, and Arkin AP (2007) Environmental signal integration by a modular AND gate. Mol. Syst. Biol 3, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Nielsen AAK, Der BS, Shin J, Vaidyanathan P, Paralanov V, Strychalski EA, Ross D, Densmore D, and Voigt CA (2016) Genetic circuit design automation. Science 352, aac7341. [DOI] [PubMed] [Google Scholar]
- (21).Younger AKD, Dalvie NC, Rottinghaus AG, and Leonard JN (2017) Engineering Modular Biosensors to Confer Metabolite-Responsive Regulation of Transcription. ACS Synth. Biol 6, 311–325. [DOI] [PubMed] [Google Scholar]
- (22).Michnick SW, Ear PH, Manderson EN, Remy I, and Stefan E (2007) Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat. Rev. Drug Discov 6, 569–582. [DOI] [PubMed] [Google Scholar]
- (23).Ostermeier M (2005) Engineering allosteric protein switches by domain insertion. Protein Eng. Des. Sel 18, 359–364. [DOI] [PubMed] [Google Scholar]
- (24).Michnick SW, Rosen MK, Wandless TJ, Karplus M, and Schreiber SL (1991) Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin. Science 252, 836–839. [DOI] [PubMed] [Google Scholar]
- (25).Banaszynski LA, Liu CW, and Wandless TJ (2005) Characterization of the FKBP.rapamycin.FRB ternary complex. J. Am. Chem. Soc 127, 4715–4721. [DOI] [PubMed] [Google Scholar]
- (26).Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, and Greene GL (1998) The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95, 927–937. [DOI] [PubMed] [Google Scholar]
- (27).Pandey N, Nobles CL, Zechiedrich L, Maresso AW, and Silberg JJ (2015) Combining random gene fission and rational gene fusion to discover near-infrared fluorescent protein fragments that report on protein-protein interactions. ACS Synth. Biol 4, 615–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Litowski JR, and Hodges RS (2002) Designing heterodimeric two-stranded alpha-helical coiled-coils. Effects of hydrophobicity and alpha-helical propensity on protein folding, stability, and specificity. J. Biol. Chem 277, 37272–37279. [DOI] [PubMed] [Google Scholar]
- (29).Mellot P, Mechulam Y, Le Corre D, Blanquet S, and Fayat G (1989) Identification of an amino acid region supporting specific methionyl-tRNA synthetase: tRNA recognition. J. Mol. Biol 208, 429–443. [DOI] [PubMed] [Google Scholar]
- (30).Lindhout DA, Litowski JR, Mercier P, Hodges RS, and Sykes BD (2004) NMR solution structure of a highly stable de novo heterodimeric coiled-coil. Biopolymers 75, 367–375. [DOI] [PubMed] [Google Scholar]
- (31).Ahmed A (1973) Mechanism of repression of methionine biosynthesis in Escherichia coli. I. The role of methionine, s-adenosylmethionine, and methionyl-transfer ribonucleic acid in repression. Mol. Gen. Genet 123, 299–324. [DOI] [PubMed] [Google Scholar]
- (32).Schmitt E, Tanrikulu IC, Yoo TH, Panvert M, Tirrell DA, and Mechulam Y (2009) Switching from an induced-fit to a lock-and-key mechanism in an aminoacyl-tRNA synthetase with modified specificity. J. Mol. Biol 394, 843–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Salis HM, Mirsky EA, and Voigt CA (2009) Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol 27, 946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Park S-Y, Beel BD, Simon MI, Bilwes AM, and Crane BR (2004) In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved. Proc. Natl. Acad. Sci. U.S.A 101, 11646–11651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Reinke AW, Grant RA, and Keating AE (2010) A synthetic coiled-coil interactome provides heterospecific modules for molecular engineering. J. Am. Chem. Soc 132, 6025–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Thompson KE, Bashor CJ, Lim WA, and Keating AE (2012) SYNZIP protein interaction toolbox: in vitro and in vivo specifications of heterospecific coiled-coil interaction domains. ACS Synth. Biol 1, 118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Oakes BL, Nadler DC, Flamholz A, Fellmann C, Staahl BT, Doudna JA, and Savage DF (2016) Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nat. Biotechnol 34, 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Tanenbaum DM, Wang Y, Williams SP, and Sigler PB (1998) Crystallographic comparison of the estrogen and progesterone receptor’s ligand binding domains. Proc. Natl. Acad. Sci. U.S.A 95, 5998–6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Wärnmark A, Treuter E, Gustafsson J-A, Hubbard RE, Brzozowski AM, and Pike ACW (2002) Interaction of transcriptional intermediary factor 2 nuclear receptor box peptides with the coactivator binding site of estrogen receptor alpha. J. Biol. Chem 277, 21862–21868. [DOI] [PubMed] [Google Scholar]
- (40).Serre L, Verdon G, Choinowski T, Hervouet N, Risler JL, and Zelwer C (2001) How methionyl-tRNA synthetase creates its amino acid recognition pocket upon L-methionine binding. J. Mol. Biol 306, 863–876. [DOI] [PubMed] [Google Scholar]
- (41).Ghosh A, and Vishveshwara S (2007) A study of communication pathways in methionyl- tRNA synthetase by molecular dynamics simulations and structure network analysis. Proc. Natl. Acad. Sci. U.S.A 104, 15711–15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Larson ET, Kim JE, Zucker FH, Kelley A, Mueller N, Napuli AJ, Verlinde CLMJ, Fan E, Buckner FS, Van Voorhis WC, Merritt EA, and Hol WGJ (2011) Structure of Leishmania major methionyl-tRNA synthetase in complex with intermediate products methionyladenylate and pyrophosphate. Biochimie 93, 570–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Chan ACK, Lelj-Garolla B, I Rosell F, Pedersen KA, Mauk AG, and Murphy MEP (2006) Cofacial heme binding is linked to dimerization by a bacterial heme transport protein. J. Mol. Biol 362, 1108–1119. [DOI] [PubMed] [Google Scholar]
- (44).Hoff KG, Culler SJ, Nguyen PQ, McGuire RM, Silberg JJ, and Smolke CD (2009) In vivo fluorescent detection of Fe-S clusters coordinated by human GRX2. Chem. Biol 16, 1299–1308. [DOI] [PubMed] [Google Scholar]
- (45).Gao R, Tao Y, and Stock AM (2008) System-level mapping of Escherichia coli response regulator dimerization with FRET hybrids. Mol. Microbiol 69, 1358–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Gao R, and Stock AM (2009) Biological insights from structures of two-component proteins. Annu. Rev. Microbiol 63, 133–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Laub MT, and Goulian M (2007) Specificity in two-component signal transduction pathways. Annu. Rev. Genet 41, 121–145. [DOI] [PubMed] [Google Scholar]
- (48).Podgornaia AI, and Laub MT (2013) Determinants of specificity in two-component signal transduction. Curr. Opin. Microbiol 16, 156–162. [DOI] [PubMed] [Google Scholar]
- (49).Olson EJ, and Tabor JJ (2012) Post-translational tools expand the scope of synthetic biology. Curr. Opin. Chem. Biol 16, 300–306. [DOI] [PubMed] [Google Scholar]
- (50).Cabantous S, Terwilliger TC, and Waldo GS (2005) Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol 23, 102–107. [DOI] [PubMed] [Google Scholar]
- (51).Marvin JS, Schreiter ER, Echevarría IM, and Looger LL (2011) A genetically encoded, high-signal-to-noise maltose sensor. Proteins 79, 3025–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Mahdavi A, Szychowski J, Ngo JT, Sweredoski MJ, Graham RLJ, Hess S, Schneewind O, Mazmanian SK, and Tirrell DA (2014) Identification of secreted bacterial proteins by noncanonical amino acid tagging. Proc. Natl. Acad. Sci. U.S.A 111, 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Ngo JT, Schuman EM, and Tirrell DA (2013) Mutant methionyl-tRNA synthetase from bacteria enables site-selective N-terminal labeling of proteins expressed in mammalian cells. Proc. Natl. Acad. Sci. U.S.A 110, 4992–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Erdmann I, Marter K, Kobler O, Niehues S, Abele J, Müller A, Bussmann J, Storkebaum E, Ziv T, Thomas U, and Dieterich DC (2015) Cell-selective labelling of proteomes in Drosophila melanogaster. Nat Commun 6, 7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Mahdavi A, Hamblin GD, Jindal GA, Bagert JD, Dong C, Sweredoski MJ, Hess S, Schuman EM, and Tirrell DA (2016) Engineered Aminoacyl-tRNA Synthetase for Cell-Selective Analysis of Mammalian Protein Synthesis. J. Am. Chem. Soc 138, 4278–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Somerville CR, and Ahmed A (1977) rel-dependent methionine requirement in revertants of a methionyl-transfer RNA synthetase mutant of Escherichia coli. J. Mol. Biol 111, 77–81. [DOI] [PubMed] [Google Scholar]
- (57).Segall-Shapiro TH, Nguyen PQ, Santos, Dos ED, Subedi S, Judd J, Suh J, and Silberg JJ (2011) Mesophilic and hyperthermophilic adenylate kinases differ in their tolerance to random fragmentation. J. Mol. Biol 406, 135–148. [DOI] [PubMed] [Google Scholar]
- (58).Yang R, Bi X, Li F, Cao Y, and Liu C-F (2014) Native chemical ubiquitination using a genetically incorporated azidonorleucine. Chem. Commun. (Camb.) 50, 7971–7974. [DOI] [PubMed] [Google Scholar]
- (59).Pelletier JN, Campbell-Valois FX, and Michnick SW (1998) Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc. Natl. Acad. Sci. U.S.A 95, 12141–12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.